Abstract
Circulating tumour cells (CTCs) shed into blood from primary cancers include putative precursors that initiate distal metastases1. While these cells are extraordinarily rare, they may identify cellular pathways contributing to the blood-borne dissemination of cancer. Here, we adapted a microfluidic device2 for efficient capture of CTCs from an endogenous mouse pancreatic cancer model3 and subjected CTCs to single molecule RNA sequencing4, identifying Wnt2 as a candidate gene enriched in CTCs. Expression of Wnt2 in pancreatic cancer cells suppresses anoikis, enhances anchorage-independent sphere formation, and increases metastatic propensity in vivo. This effect is correlated with fibronectin upregulation and suppressed by inhibition of Map3k7 (Tak1) kinase. In humans, formation of non-adherent tumour spheres by pancreatic cancer cells is associated with upregulation of multiple Wnt genes, and pancreatic CTCs revealed enrichment for Wnt signaling in 5 of 11 cases. Thus, molecular analysis of CTCs may identify candidate therapeutic targets to prevent the distal spread of cancer.
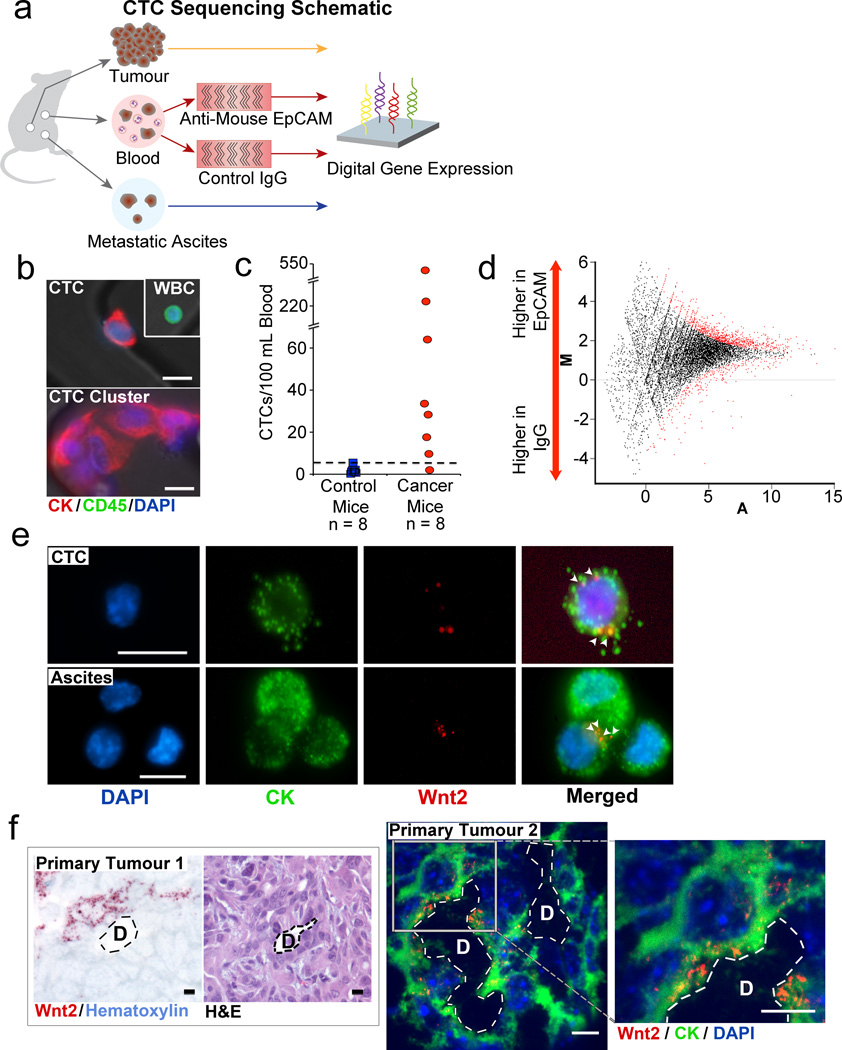
We established the HbCTC-Chip microfluidic capture platform1, to isolate CTCs from blood samples of a genetically engineered mouse pancreatic cancer model2, comparing their expression profile to that of simultaneously harvested primary tumour and metastatic ascites cells (Fig. 1a and Supplementary Fig. 1). Captured cells were stained for epithelial cytokeratin (CK) and the leukocyte marker CD45, with CTCs scored as CK+/ CD45− (Fig. 1b). Eight control tumour-free mice had a median of 1.3 CK+/CD45− cells /100 µL of blood (range, 0.2–5.6; mean, 1.8 ± 0.6) under the selected imaging parameters. Applying a threshold of ≥ 6 cells/100 µL, 7/8 tumour-bearing mice were positive for CTCs (median, 31 cells/100 µL; range, 2–547; mean, 115 ± 67) (Fig. 1b, c). Most CTCs were captured as single cells, although clusters of 5 to 10 cells were seen in ~50% of mouse blood samples. The on-chip purity of captured CTCs ranged from 0.1–6%, due to non-specific binding (NSB) by leukocytes (Supplementary Table 1).
Figure 1. Analysis of mouse pancreatic CTCs identifies Wnt2 as a candidate CTC gene.
a, Schematic representation of strategy for DGE of RNA isolated from primary mouse pancreatic tumour, metastatic ascites, and CTC-enriched blood. b, Immunofluorescence staining of mouse pancreatic CTC, leukocyte (WBC), and CTC cluster captured on HbCTC-Chip (DAPI, blue; Cytokeratin (CK), red; CD45, green). c, Quantitation of CK+ cells captured from control mice and pancreatic cancer bearing mice (dashed line - threshold of ≥ 6 CK+ cells/100 µL). d, DEGseq M-A plot of RNA sequence reads from mouse (MPANC-9) CTCs, comparing anti-EpCAM-captured versus IgG-coated HbCTC-Chips. (M - log2 fold change, A - averaged log2 reads, red dots – differentially expressed genes) e, RNA-ISH of CTCs and metastatic ascites cells co-expressing CK8+18 (green) and Wnt2 (red) transcripts. White arrowheads highlight Wnt2 signals. f, RNA-ISH of primary pancreatic tumours showing a small cluster of Wnt2 mRNA expressing cells near a pancreatic duct (D) (Wnt2, red dots; hematoxylin, light blue). Hematoxylin and eosin (H&E) stained serial section is shown (right). A second primary tumour imaged under fluorescence demonstrating Wnt2 expression in a subpopulation of tumour cells; high magnification shown (right, Wnt2, red; CK, green; DAPI, blue). (Scale bars=10 µm).
Given the minute amounts and variable purity of CTCs, we applied a sub-microgram RNA based sequencing method using a single molecule “next generation” platform3, to derive a Digital Gene Expression (DGE) profile. We processed each blood sample through paired EpCAM- and mock IgG-functionalized HbCTC-Chips, allowing digital subtraction of matched NSB leukocyte reads from each CTC-enriched DGE profile. Using the DEGseq statistical model5 applied to the MA-plot with a false discovery rate (FDR) of 0.10, we identified 361 transcripts in mouse MPANC-9 that were increased >2-fold in anti-EpCAM- versus IgG-chips, and absent in blood from non-tumour bearing mice (Fig. 1d). This CTC candidate gene set was then compared with DGE profiles of 12 mouse and 15 human primary pancreatic tumours, versus normal mouse and human pancreata6, yielding a set of 9 tumour-associated genes whose expression was markedly increased in CTCs (Supplementary Table 2). One of these, Wnt2, was found to be consistently enriched in pancreatic cancers in the Oncomine™, leading us to pursue this gene as a prototype CTC-enriched transcript. RNA sequencing of metastatic cells in the mouse ascites demonstrated enrichment for Wnt2. A second mouse, MPANC-10, also demonstrated Wnt2 expression in both CTCs and metastatic ascites cells. The lower number of Wnt2 mRNA reads in MPANC-10 was correlated with the fewer cytokeratin transcript reads, suggesting that both transcripts track with the number of CTCs in the blood specimen (Supplementary Table 3).
Given the absence of antibodies for cellular imaging of Wnt2, we developed fluorescent RNA in-situ hybridization (RNA-ISH) to verify Wnt2 expression in CTCs (Supplementary Figs. 2 and 3). RNA-ISH analysis of CTCs using dual staining for Wnt2 and cytokeratin mRNAs identified Wnt2 transcripts in 64% of cytokeratin-expressing cells in 3/4 mice. A comparable Wnt2 mRNA signal was observed in metastatic cells from ascites of mice bearing pancreatic tumours (Fig. 1e). In contrast, within primary tumour specimens, Wnt2 mRNA expression was only detectable in very small localized clusters of cells (1–5% of all cells) in 8/14 primary tumour specimens (Fig. 1f). Histological analysis of rare Wnt2 positive cells within PDACs did not reveal any obvious distinction from other tumour cells. The small number of Wnt2-positive cells in primary tumour specimens was consistent with DGE analysis, which showed rare Wnt2 RNA reads in both mouse (8/12) and human (9/15) PDACs. Thus, Wnt2 positive cells are present within both CTC and metastatic ascites, and represent a rare subset of the primary tumour population.
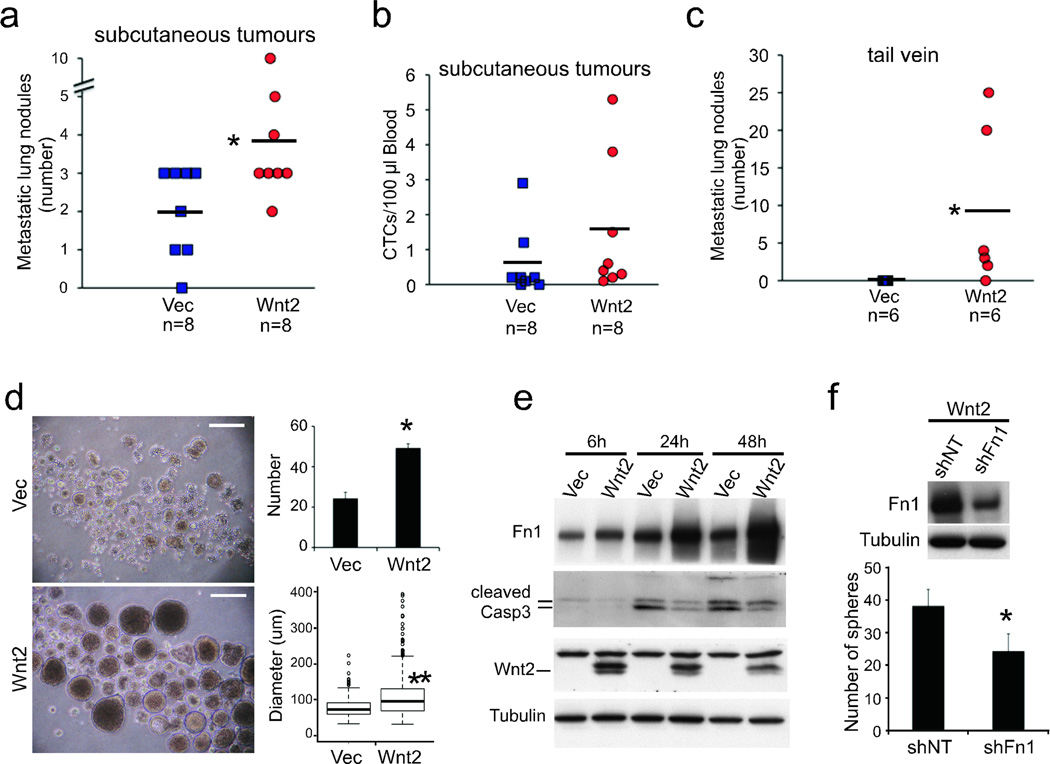
To test the functional consequences of Wnt2 expression in vivo, lentiviral constructs were introduced into NB508 PDAC cells, which lack detectable endogenous Wnt2 expression (Supplementary Fig. 4). Subcutaneous engraftment of GFP-tagged Wnt2-NB508 cells produced significantly more lung metastases in nude mice, compared with vector-transduced cells, despite the larger size of vector-expressing primary tumours (n=8, p<0.05) (Fig. 2a and Supplementary Fig. 4). EpCAM-captured CTCs showed a modest increase in numbers in mice bearing Wnt2-expressing tumours, but this did not reach statistical significance (p=0.25, Fig. 2b), even after normalization for the smaller size of Wnt2-expressing tumours (p=0.17, Supplementary Fig. 4). Thus, Wnt2 expression may increase the metastatic potential of circulating cancer cells, without markedly increasing their generation from primary tumours. Consistent with this hypothesis, direct intravascular inoculation of cells into the tail vein of syngeneic mice, which bypasses the vascular invasion step, showed a striking increase in lung metastases for Wnt2-NB508 cells (5/6 Wnt2-transduced versus 0/6 vector, p<0.05) (Fig. 2c and Supplementary Fig. 4).
Figure 2. Wnt2 promotes anchorage-independent cell survival and pancreatic cancer cell metastasis.
a, Number of GFP-positive metastatic nodules in the lungs of mice bearing subcutaneous tumours established with Vec- or Wnt2-expressing NB508 cells. b, Number of GFP-positive CTCs captured from the blood of mice described above. c, Number of GFP-positive lung metastatic nodules in mice, following tail vein injection with Vec- or Wnt2-NB508 cells tagged with both GFP and luciferase. d, Representative images of tumour spheres formed by Vec- or Wnt2-NB508 cells plated at 1,000 cells/well (scale bar = 250 µm). Quantitation of both tumour sphere number and diameter is shown plated at 100 cells/well (n=3). e, Immunoblotting analysis of Vec- and Wnt2-NB508 cells at time intervals following plating under non-adherent conditions. f, Suppression of tumour spheres formed by Wnt2-NB508 cells following infection with lentivirus encoding shRNA targeting Fn1 compared with non-target (NT) shRNA (n=3). Effectiveness of knockdown is shown by western blot above. (mean ± s.d.; * p < 0.01,** p < 0.001).
Modeling the effects of ectopic Wnt2 expression on NB508 PDAC cells cultured in vitro showed marked enhancement of anchorage independent growth, without affecting cellular proliferation, migration or invasion assayed under standard conditions (Supplementary Fig. 5, 6). Tumour sphere formation under non-adherent conditions in serum-free media supplemented with growth factors7,8 was markedly increased by Wnt2 expression, with respect to sphere numbers (p<0.05) and size (p<0.005) (Fig. 2d, Supplementary Fig. 7). Anoikis, the induction of apoptosis in epithelial cells resulting from loss of basement membrane attachment, was attenuated following Wnt2 expression in NB508 cells (Fig. 2e). Resistance to anoikis is thought to be essential for epithelial cell survival in the bloodstream, and epithelial to mesenchymal transition (EMT) has been postulated to mediate this effect. We therefore tested for induction of characteristic mesenchymal markers by Wnt2 at both RNA and protein levels, by DGE analysis and Western blotting respectively. Among classical EMT markers, we only detected induction of fibronectin (Fn1), an extracellular protein implicated in cell-matrix interactions and cellular survival signals (Fig. 2e, Supplementary Figs. 6, 8). shRNA-mediated knockdown of Fn1 suppressed the ability of ectopic Wnt2 to enhance tumour sphere formation (n=6, p<0.05, Fig. 2f), suggesting that Fn1 contributes to Wnt2-mediated anoikis resistance. Two transcription factors, Mycn and the known Fn1 regulator Etv4, were induced by Wnt2, with their knockdown suppressing Fn1 expression, suggesting that they contribute to mediating the Wnt2 effect (Supplementary Fig. 9).
Wnt2 may signal through either canonical (β-catenin/TCF dependent) or non-canonical pathways. While canonical Wnt signaling expression signatures have been developed, there are no such signatures for non-canonical Wnt pathways. We therefore generated a customized non-canonical Wnt signaling gene set (Supplementary Table 4)9 and used this to interrogate the CTC DGE data. Significant enrichment was observed for components of the non-canonical Wnt signaling pathway in mouse pancreatic CTCs, metastatic ascites cells, and Wnt2-NB508 cells grown under non-adherent sphere-forming conditions (Supplementary Figs. 10, 11 and Supplementary Table 5). Similar enrichment was noted for a Wnt and TGF-β driven signature10 (Supplementary Table 6). No such enrichment was evident for signatures of canonical Wnt signaling in CTCs and metastatic ascites cells, although Wnt2-NB508 cells showed a mild increase in canonical Wnt signaling (Supplementary Table 6, Fig. 12).
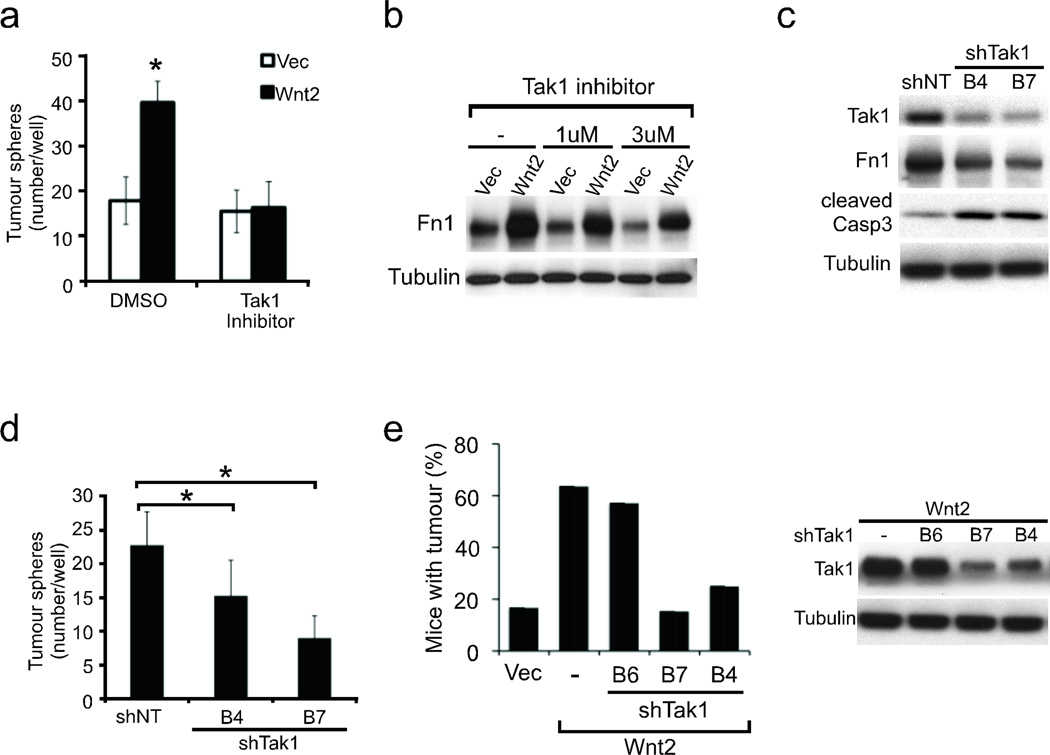
We tested a panel of inhibitors of Wnt-related pathways to identify small molecule inhibitors capable of suppressing the Wnt2 effect on anoikis. Among these, 5Z-7-Oxozeaenol, an inhibitor of Map3k7, also known as Tak1 (TGF-β activated kinase 1)11, was remarkable in completely abrogating Wnt2-induced tumour spheres, without suppressing baseline sphere formation (Fig. 3a and Supplementary Figs. 12, 13). 5Z-7-Oxozeaenol also suppressed Fn1 expression in Wnt2-NB508 cells (Fig. 3b). To validate the effect of the Tak1 inhibitor in reversing the prosurvival phenotype conferred by Wnt2, shRNA constructs targeting Tak1 were introduced into Wnt2-NB508 cells. Tak1 shRNAs suppressed Fn1 expression in Wnt2-NB508 cells with an associated increase in cleaved caspase-3 (Fig. 3c and Supplementary Fig. 9d). Effective suppression of Wnt2-induced tumour sphere formation was observed and correlated with the degree of Tak1 knockdown by the shRNA constructs (Fig. 3d). Consistent with these in vitro findings, the generation of metastases following tail vein inoculation of Wnt2-expressing cells was suppressed as a function of shRNA knockdown of Tak1 (Fig. 3e and Supplementary Fig. 14). Together, these results support the role of Wnt2 in mediating metastasis-associated survival signals, which depend, at least in part, on a Tak1-Fn1 signaling axis.
Figure 3. Association of Wnt2-prosurvival phenotype with non-canonical Wnt signaling and inhibition by suppression of Map3K7 (Tak1).
a, Suppression of Wnt2-induced, but not baseline, tumour sphere formation by NB508 cells, following treatment with 5Z-7-Oxozeaenol, a small molecule inhibitor of Tak1 (n=6). b, Immunoblot showing suppression of Fn1 expression following treatment of non-adherent cultures of Vec- and Wnt2-NB508 cells with increasing concentrations of 5Z-7-Oxozeaenol. c, Immunoblot analysis of Tak1, Fn1 and cleaved caspase 3 expression in Wnt2-NB508 cells following lentiviral infection with two different shRNAs targeting Tak1 or non-target (NT). d, Enumeration of tumour spheres formed by cells in c (n=6). e, Percentage of mice with detectable metastases by bioluminescence imaging for luciferase-producing tumor cells in mice 6 weeks following tail vein injection of Vec-NB508, Wnt2-NB508, and Wnt2-NB508 cells infected with three different shRNAs targeting Tak1 (n=12, 11, 14, 13, 12 respectively). Immunoblot of Tak1 knockdown by shRNAs B6, B7, and B4 is shown (right). (mean ± s.d., * p < 0.05)
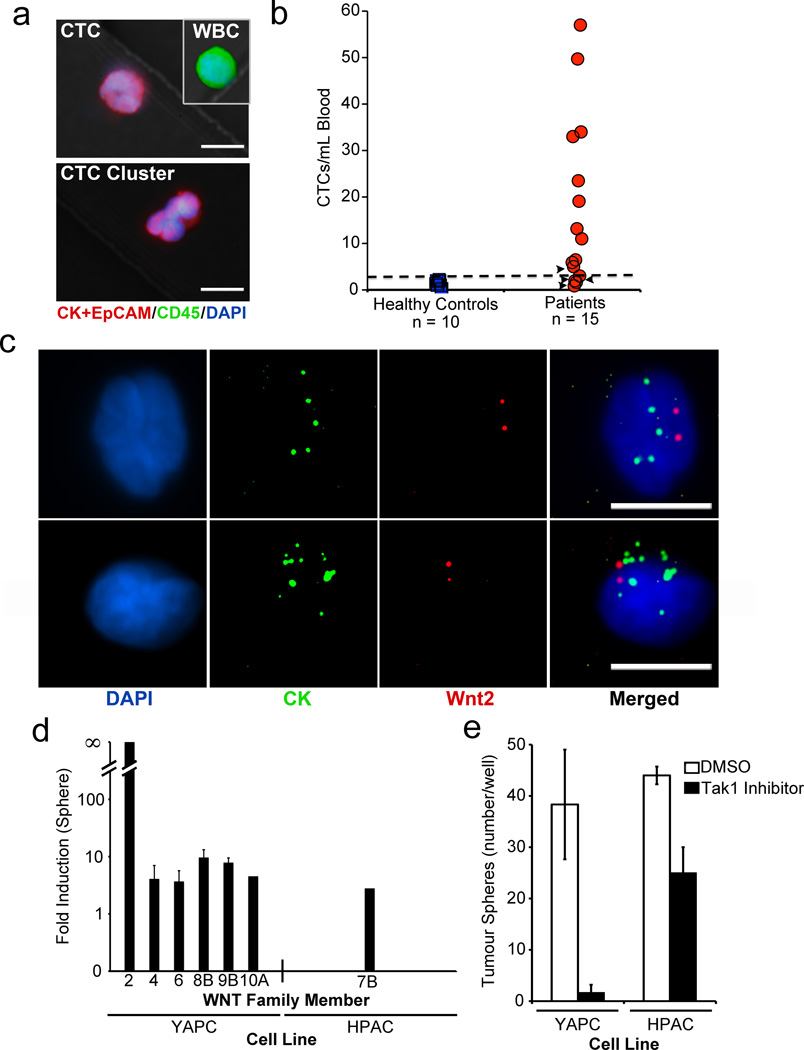
To extend our analyses to human pancreatic cancer, we captured CTCs from blood specimens of patients with metastatic pancreatic cancer using the HbCTC-Chip and stained the cells with a combination of antibodies against cytokeratin and intracellular EpCAM residues (Fig. 4a). Under conditions optimized for human pancreatic CTC analysis, 10 healthy controls had a median 1.2 CK+EpCAM+/CD45− cells (range, 0 to 2.2; mean, 1.3 ± 0.2). Of 15 patients with metastatic pancreatic cancer, 11 had CTC counts above threshold (≥ 3 CK+EpCAM+/CK45− cells/mL), with a median 11 cells/mL (range, 1 to 57; mean, 18 ± 5; on-chip median purity, 0.5%) (Fig. 4b and Supplementary Table 7). We tested for Wnt2 expression in human pancreatic CTCs using RNA-ISH identifying Wnt2 transcripts in 23 of 66 (35%) CK-positive CTCs from 2 of 8 metastatic PDAC patients (Fig. 4c). To derive DGE profiles for human pancreatic CTCs, we followed the strategy defined for mouse CTC analyses, analyzing blood specimens from 12 patients and 4 healthy controls using parallel anti-EpCAM versus mock IgG-functionalized HbCTC-Chips, followed by low quantity RNA-based single molecule sequencing analysis. No individual Wnt transcript was consistently enriched across all CTC-enriched populations (FDR 0.10), but among 11 patients with CTC counts above threshold, 5 (45%) had significant enrichment of non-canonical Wnt signaling pathway components and 8 (73%) showed enrichment for the Wnt and TGF-β-driven profile (Supplementary Table 8). No such enrichment was present in 4 healthy controls or in one patient without detectable CTCs (Supplementary Tables 9, 10, 11).
Figure 4. Detection of Wnt2 mRNA expression and non-canonical Wnt signature in human pancreatic CTCs.
a, Immunofluorescence staining of human pancreatic CTC, leukocyte (WBC), and CTC cluster captured on anti-human EpCAM HbCTC-Chip (DAPI nuclear stain, blue; CK and EpCAM cocktail, red; CD45, green). b, Enumeration of CK+EpCAM positive human CTCs (CTCs/mL) captured from patients with metastatic pancreatic cancer. Arrowheads indicate lower CTC numbers in patients responding to therapy. Blood samples from healthy donors were used to establish the threshold of ≥ 3 CK+EpCAM+/CK45− cells/mL (dashed line). c, RNA-ISH analysis of human pancreatic CTCs, showing co-expression of mRNAs for cytokeratins (CK) 8, 18, 19 and 23 (pooled probes in green) and Wnt2 (red). (DAPI, blue). (Scale bars = 10 µm). d, Induction of Wnt transcripts in pancreatic cancer cells grown as non-adherent tumour spheres compared to standard conditions (fold increase). e, Quantification of tumour spheres with or without Tak1 inhibitor (3µM 5-Z-7-Oxozeanol). (n=3; mean ± s.d.)
To test whether multiple Wnt genes contribute to anoikis resistance in human pancreatic cancer, we measured expression of all known family members in six cell lines grown under non-adherent tumour sphere conditions. Five of six cell lines showed upregulation of multiple Wnt genes following shift from 2D to 3D culture, an effect that was correlated with sensitivity to the Tak1 inhibitor and to Tak1 shRNA (Fig. 4d, e and Supplementary Figs. 15–17, Table 12). Thus, in human pancreatic cancer cells, the induction of multiple Wnt family members appears to circumvent anoikis following loss of adherence, an effect suppressed by inhibition of Tak1.
In summary, we have applied microfluidic CTC isolation from a genetically engineered mouse model to generate an unbiased RNA sequencing profile for CTC-specific expression patterns. The cellular pathways activated during the haematogenous spread of human pancreatic cancer are likely to be complex, and our analysis is limited by technological hurdles inherent in the molecular characterization of partially purified CTC populations. Nonetheless, our results suggest that non-canonical Wnt signaling pathways may contribute to metastasis in human pancreatic cancer. The effectiveness of Tak1 inhibition in suppressing this effect identifies a novel potential drug target for metastasis suppression.
Methods
Mice, tumour tissues, and cell lines
Mice with pancreatic cancer used in these experiments express Cre driven by Pdx1 or P48, LSL-KrasG12D, and Tp53lox/+ or Tp53lox/lox as previously described 2. Normal B6/129, FVB, and Nu/Nu mice were purchased from Jackson Laboratory. For cardiocentesis, animals were sedated with isofluorane, the chest wall was sterilized with ethanol and a skin incision was made above the rib cage. A 23-gauge needle was used to draw approximately 1 mL of blood into a syringe primed with 100 µL of PBS-10mM EDTA pH 7.4 (Gibco). Animals were then euthanized per animal protocol guidelines. Pancreatic tumours and normal tissues were extracted and separated using flash freezing with liquid nitrogen for RNA extraction and fixation with 4% PFA followed by paraffin embedding (FFPE) for RNA-ISH and histochemistry. Ascites if present was sterilely removed with a 23G needle syringe and cells were pelleted by centrifugation. Pelleted cells were flash frozen in liquid nitrogen for RNA extraction. Cell lines generated from mouse tumours and human pancreatic cancer cell lines were cultured in RPMI-1640 + 10% FBS + 1% Pen/Strep (Gibco/Invitrogen).
HbCTC-Chip Production, Preparation and Blood Processing
HbCTC-Chips were manufactured on site at the MGH Cancer Center/ BioMEMS Resource Facility, and chips were functionalized using biotinylated antibodies as previously described 1. Mouse HbCTC-Chips were functionalized with biotinylated anti-mouse EpCAM (CD326) antibody (Biolegend 118204) or isotype control biotinylated Rat IgG2a (Biolegend 400504). Human HbCTC-Chips were functionalized with biotinylated polyclonal goat anti-human EpCAM (R&D Systems BAF960) or control goat IgG antibody (R&D Systems BAF108).
For mouse samples, approximately 1 mL of mouse blood was diluted 1:1 with PBS-10 mM EDTA pH 7.4 and a total volume of 1 mL of diluted blood was processed on each HbCTC-Chip, using a Harvard Apparatus volumetric syringe pump set at 1.5 mL/hr. After processing the blood, devices were washed with PBS followed by analyses as described below.
Human blood specimens obtained for CTC analysis was obtained after informed consent, per IRB protocol (05–300) at the Massachusetts General Hospital (MGH). A maximum of 20 mL of blood were obtained at any given blood draw, using EDTA vacutainers. Blood specimens were processed through the HbCTC-Chip within 4 hours of blood draw. Approximately 3 mL of blood was processed through the HbCTC-Chip, using the pressure controlled apparatus, at a target flow rate of 1.5 mL/hr as previously described 2.
CTC staining and enumeration
Following blood processing, captured cells on the HbCTC-Chip were fixed with 4% paraformaldehyde and washed with PBS. The fixed cells were then permeabilized with 1% NP40 in PBS, blocked with either 5% donkey serum (mouse samples) or 3% goat serum/2% BSA (human samples), and then immunostained with the relevant primary antibodies. Primary antibodies for the mouse CTC analyses were rabbit anti-wide spectrum cytokeratin (1:50, Abcam ab9377), chicken anti-GFP (1:1000, Abcam ab13970) and goat anti-mouse CD45 (1:500, R&D systems AF114). Primary antibodies for the human CTC analyses were rabbit anti-wide spectrum cytokeratin (1:50, Abcam ab9377), rabbit anti-EpCAM (1:500, Abcam ab71916), and mouse IgG1 anti-CD45 (1:1000, BD 55480). Secondary immunofluorescent-tagged antibodies were used for signal amplification. For the mouse, these were donkey anti-rabbit Alexa Fluor 594 (1:500, Invitrogen A-21207), donkey anti-chicken Dylight™ 488 (1:500, Jackson ImmunoResearch 703-486-155) and donkey anti-goat Alexa Fluor 488 (1:500, Invitrogen A-11055). For human, the secondary antibodies were goat anti-rabbit Alexa Fluor 594 (1:500, Invitrogen A-11012) and goat anti-mouse IgG1 Alexa Fluor 488 (1:500, Invitrogen A-21121). Nuclei were then stained with DAPI and the devices were washed with PBS and stored at 4°C. The devices were imaged under 10× magnification using the BioView Ltd. automated imaging system (Billerica, MA) as well as an automated upright fluorescence microscope (Eclipse 90i, Nikon, Melville, NY). Positive staining for CK or EpCAM, without CD45 staining, was required for scoring potential CTCs, which were then manually reviewed. Threshold and baseline signals were established using specimens from non-tumour bearing mice or healthy human controls.
RNA Extraction and Purification from the HbCTC-Chip and from tissues
RNA from CTC-enriched cell populations was isolated from the devices using a modified protocol for the Qiagen RNeasy MinElute kit (Qiagen). Briefly, 375 µl of the RLT buffer was pipetted into the HbCTC-Chip with mixing between the inlet and outlet of the device. The resulting RLT lysate was then loaded onto the Qiagen RNeasy MinElute column and RNA was purified per protocol. RNA was eluted in a total of ~ 10 µL RNase free water.
To isolate RNA from fresh frozen tissues, the material was pulverized with a sterile pestle in a microfuge tube on dry ice. RNA from cell lines and fresh frozen tumour and normal tissues were all processed in the same manner. RNA was extracted using the TRIzol® Reagent (Invitrogen) per manufacturer’s specifications.
Single Molecule Sequencing
Purified RNA was subjected to Digital Gene Expression (DGE) sample prepping and analyzed on the HeliScope™ Single Molecule Sequencer from Helicos BioSciences.
For mouse samples, a low quantity on surface RNA capture followed by cDNA synthesis and sequencing method was developed and used on RNA from EpCAM HbCTC-Chip, IgG HbCTC-Chip, primary tumour, and metastatic ascites. This method has been previously described 3. Briefly, the purified RNA in RNAse free water was hybridized in 10µl volume to Helicos poly(dT)-coated sequencing flow cell channels in 1X SSC, 0.05% SDS at 37°C for 30 minutes. First-strand cDNA was synthesized with the SuperScript III first-strand cDNA synthesis kit (Invitrogen) using manufacturer’s recommendations, except that no additional primers were added, and the incubation steps were modified as follows: 37°C 15 minutes, and 55°C 45 minutes. Subsequent to cDNA synthesis, hot water was passed through the channels to degrade and melt away the RNA strands. Guanine tailing was performed using terminal transferase, by adding 500µM guanine in 20µl volume in 1X TdT buffer, 2.5mM CoCl2 and 20 units terminal transferase per channel. The reaction took place at 37°C for 30 minutes, followed by 3’ blocking with 100µM ddGTP and ddATP under the same reaction conditions. The 18 nt poly-C primers were hybridized at 50nM in 1X SSC, 0.05% SDS at 55°C for 30 minutes, followed by step-wise “fill” steps with 500µM cytosine and adenine nucleotides with 5 units Klenow fragment (NEB) in 1X NEB2 buffer and 20µl reaction volume per channel. The lock step was then performed with virtual terminator guanine and thymidine nucleotide analogs. Single molecule sequencing by synthesis was then initiated using standard procedures 12.
For human CTC samples, initial low quantity on surface methods did not have sufficient yield, so application of a low quantity RNA-seq method was utilized as previously described 12. Briefly, the purified RNA in RNAse free water was converted to first-strand cDNA with the SuperScript III first-strand cDNA synthesis kit (Invitrogen) using random hexamers according to the manufacturer’s recommendations. RNA was digested and single stranded cDNA was purified using a combination of QIAquick nucleotide removal kit (Invitrogen) and ethanol precipitation with ammonium acetate and glycogen. Single stranded cDNA was denatured and then a poly-A tail was added to the 3’ end using terminal transferase (New England Biolabs). Tailed cDNAs were then hybridized to the sequencing flow cell followed by “Fill and Lock” and subjected to single molecule sequencing by synthesis.
Bioinformatics
DGE
To compute digital gene expression (DGE) from the Helicos sequence data, we used the DGE pipeline of the HeliSphere 1.1.498.63 software (open.helicosbio.com) using the Human.Txome and Mouse.Txome (ftp://ftp.helicosbio.com/pub/distribution) reference files 13. We ignored DGE output corresponding to mitochondrial and ribosomal RNA and Helicos control spike-ins.
DEGSeq
DEGseq comparisons of DGE profiles were run using the DEGexp function of version 1.0.5 of the Bioconductor (www.bioconductor.org) DEGseq package. The “method” argument was set to “MARS” for the MA-plot-based method with Random Sampling 5 and the threshold for statistical significance was a Benjamini-Hochberg q-value (i.e., FDR estimate) of 10%.
hGSEA
We performed hypergeometric gene set enrichment analysis (hGSEA) as follows. To determine whether gene set A was enriched in gene set B, we used the hypergeometric distribution to test whether the overlap of A and B was larger than would be expected by chance had A and B been drawn randomly from all the genes quantitated by the DGE. To account for multiple hypothesis testing, we applied the method of Benjamini and Hochberg to the p-values so generated for each analysis, yielding false discover rate (FDR) estimates. The odds ratio (OR) is the odds of a gene being in A given it is in B divided by the odds of that gene being in A given it is not in B. Gene sets evaluated were obtained from MSigDB (Broad Institute).
QuantiGene ViewRNA in-situ hybridization on HbCTC-Chip
Following blood processing, HbCTC-Chips were fixed with 4% paraformaldehyde, washed with PBS and dehydrated with ethanol at increasing concentrations (50%, 70%, and 100%). HbCTC-Chips were submerged in 100% ethanol at −20° C for storage. Of note, mouse metastatic ascites fluid was also run on HbCTC-Chips for RNA-ISH analysis. Before further analysis, cells were rehydrated using decreasing concentrations of ethanol (70% and 50%) and washed with PBS. ISH was performed using QuantiGene ViewRNA protocols. HbCTC-Chip were permeabilized with Working Detergent Solution (Affymetrix, Santa Clara, CA), and digested with Protease (Affymetrix, Santa Clara, CA) at 1:2000 dilution in PBS. The HbCTC-Chips were hybridized for 3 hours at 40° C with custom-designed QuantiGene ViewRNA probes against Wnt2 and Cytokeratins 8 and 18. Unbound probes were flushed out with Wash Buffer (Affymetrix, Santa Clara, CA) and HbCTC-Chips were stored overnight at 4° C in Storage Buffer (Affymetrix, Santa Clara, CA). The bound probes were amplified the following day through PreAmp (Affymetrix, Santa Clara, CA) hybridization for 1 hour at 40° C, followed by Amp (Affymetrix, Santa Clara, CA) hybridization for 1 hour at 40° C. Label Probes (Affymetrix, Santa Clara, CA) targeting the individual probe types were added for 1 hour at 40° C. HbCTC-Chips were stained with DAPI, and images were taken using a Nikon 90i microscope.
QuantiGene ViewRNA in-situ hybridization on FFPE tissue
Mouse tissue for FFPE was prepared per standard protocol by the MGH Clinical and Research Pathology Cores. ISH was performed using QuantiGene ViewRNA protocols. 5 micron sections were cut, fixed in 10% formaldehyde (Fisher Scientific, Pittsburgh, PA), deparaffinized, boiled in pre-treatment solution (Affymetrix, Santa Clara, CA) and digested with proteinase K (Affymetrix, Santa Clara, CA). Sections were hybridized for 3 hours at 40° with a custom designed QuantiGene ViewRNA probes against Wnt2 and the control gene Ubc (Affymetrix, Santa Clara, CA). Bound probes were then amplified per protocol from Affymetrix using PreAmp and Amp molecules. Multiple Label Probe oligonucleotides conjugated to alkaline phosphatase (LP-AP Type 1) are then added and Fast Red Substrate is used to produce signal (red dots, Cy3 fluorescence). For two color assays, an LP-AP type 6 probe is used with Fast Blue substrate (blue dots, Cy5 fluorescence) followed by LP-AP type 1 probe with Fast Red Substrate (red dots, Cy3 fluorescence) to produce a dual colorimetric and fluorescent signal. Wnt2 probes were used in type 1/Fast Red and pooled CK 8 and 18 were used in type 6/Fast Blue. Slides are then counterstained with Hematoxylin. Serial sections were also subjected to Hematoxylin & Eosin staining per standard histology protocol to confirm the identity of cells in the region of RNA-ISH. Images were taken by a Nikon 90i scope with color camera.
Constructs and Viral Infection
The mouse Wnt2 open reading frame was cloned into pENTRI plasmid as a BamHI-XhoI fragment. pWPI plasmid carrying Wnt2 open reading frame was generated by gateway cloning following the manufacturer’s protocol (Invitrogen). Luciferase gene was expressed in pMSCV plasmid containing hygromycin selection marker. 7TFP construct (7xTcf-FFluc) used in TopFlash assay was from addgene. pLKO shRNAs targeting the mouse FN1 and Tak1 were from the RNAi consortium at the Broad Institute. Control shRNA constructs for Luciferase (Catalog# SHC007) and non-target (Catalog# SHC016) were purchased from Sigma. Target sequences for Fn1: 5’-GCCTAGAAATACCTTTCTCTT-3’; for Tak1: B4 5’-AGGCAAAGCAACAGAGTGAAT-3’; B7 5’-TCTGAGAGGAAGGCTTTCATT-3’; B6 5’-CAGCCCTAGTGTCAGAATGAT -3’. Vesicular stomatitis virus glycoprotein-pseudotyped retroviruses or lentiviruses were generated using 293T cells as packaging cell lines following protocols described in the RNAi Consortium (Broad Institute). NB508 cells were infected with pWPI virus followed by GFP-positive cell sorting. pMSCV-luciferase was introduced into the cells and followed by 200 µg/ml hygromycin B selection. Cells transduced with pLKO shRNA virus were selected with 2 µg/ml puromycin. On-target plus siRNA constructs were purchased from Dharmacon (MYCN: L-058793-01; ETV4: L-048237-01; non-targeting pool: D-001810-10-20) and transfected into cells with Lipofectamine RNAiMax (Invitrogen).
Tumourigenicity and Experimental Metastasis Assays
The animal protocol was approved by the MGH Subcommittee on Research Animal Care. Six-week old Nu/Nu mice were anesthetized by isofluorane, 1 × 106 NB508 cells expressing vector, or Wnt2 in 100 µl of PBS were injected subcutaneously on the right flank of the mice. Primary tumours, CTCs, and lungs were sampled after 2 weeks. Six to 8-week old FVB mice were used for tail vein injection. NB508 cells (5×104) expressing luciferase and vector, or Wnt2 in 100 µl of PBS were injected into lateral tail vein of the mice. Tumour formation in the lung was monitored weekly by bioluminescence using IVIS Lumina II (Caliper Life Science). Mice were euthanized after 6 weeks and lungs were sampled. Comparison between groups was performed using t-test.
Tumour Sphere Assay
Cells were plated as single cell suspension in ultralow attachment 6-well or 96-well plates (Corning) and grown in RPMI medium (serum free) supplemented with 20 µl/ml B27 (Invitrogen), 20 ng/ml EGF and 20 ng/ml bFGF. Fresh media (1 ml or 30 µl) was added every 3 days. Tumour spheres were counted and photographed at day 10. Inhibitors were purchased from Sigma and used at 1 µM (for mouse cell lines) and 0.3, 1, and 3 µM (for human cell lines) (5Z-7-Oxozeaenol), 1.8 µM (IWR1), and 1.4 µM (Y-27632). For immuofluorescent staining, tumour spheres were deposited onto a microscope slide by cytospin (Shandon), fixed and stained. Comparison between groups was performed using t-test analysis (number differences) or Wilcoxon analysis (size differences).
Anoikis Assay
Plates were coated with 10 ml of 20 mg/ml poly-HEMA (Sigma) and air-dried. Cells (2×106) were plated on the poly-HEMA coated plates. Cells were collected at different time points and proteins extracted.
Antibodies and Immunoblot Analysis
Antibodies used were β-catenin (BD), activated β-catenin (Millipore), Fibronectin, Wnt2 (Sigma), β-tubulin (Santa Cruz), and cleaved Caspase-3, p-Tak1 (Cell Signaling), and Tak1 (Cell Signaling). Cells were lysed in RIPA lysis buffer [20mM Tris, pH8/150mM NaCl/10mM NaF/0.1% SDS/1% Nonidet P-40/1x protease inhibitor mixture (Roche)]. For conditioned media, Vec-NB508 or Wnt2-NB508 cells were seeded at 1×106 in regular growth media (10% FBS) for 2 days in culture, and culture medium was replaced with low-serum media (0.2% FBS). After 24 hours, conditioned medium was collected and cell debris was removed by centrifugation. Proteins were concentrated using Amico Ultracel 30k column (Millipore) and resuspended in RIPA buffer to detect the Wnt2 protein by immunoblot. Lysates were run on an SDS/4–15% polyacrylamide gel (Bio-Rad) and transferred onto PVDF membranes (Invitrogen), and immunoblots were visualized with a Western Lightning Plus chemiluminescence kit (PerkinElmer). For phospho-Tak1 level, 1ug of protein lysate was immunoprecipitated with Tak1 antibody (Bethyl Laboratories) and blotted with p-Tak1 antibody (Cell Signaling).
SYTO60 Assay
Cells were seeded in 96-well plates in standard growth media at 1,000 cells /well or serum-free media at 5,000 cells /well. Cells were fixed every day after plating and incubated with Syto60 red fluorescence nucleic acid staining (Invitrogen) at 1:8000 dilution in 1xPBS for 30 minutes in the dark. Red fluorescence nucleic acid staining was measured using a plate reader (Molecular Probes).
Transwell Migration and Invasion Assays
Cells (5×104) were seeded on 8-µm pore size Transwell filters (Corning) in regular growth media or low-serum media (0.2% FBS). Invasion chambers coated with Matrigel (Corning) were used for invasion assays with cells plated in regular growth media. Two days after seeding, cells on top of the chamber were stripped off with cotton tips and cells on the bottom of the chambers were fixed in 4% PFA and stained with crystal violet.
Real-time qRT-PCR
Total RNA (2 µg) was reverse transcribed using a cDNA synthesis kit (GE Healthcare). Real-time quantitative RT-PCR was performed using SYBR green in an ABI PRISM 7500 sequence detection system with 96-block module and automation accessory (Applied Biosystems). Gapdh was used as an internal control gene. The primers used are shown below: Gapdh: 5′- TGGTGAAGCAGGCATCTGAG -3′ and 5′-TGCTGTTGAAGTCGCAGGAG -3′. Etv4: 5′- CGACTCAGATGTCCCTGGAT-3′ and 5′- GGAATGGTCGAAGGGATTTT -3′. Mycn: 5′- GATGAGGATGACGAGGAGGA-3′ and 5′- ACGCACAGTGATCGTGAAAG -3′. Fn1: 5′-GATAAATCAGCAGTGGGAACGG -3′ and 5′-CAAAGCAAGTCTCTTCAGGCTCAG -3′.
TOPflash Assay
Vec or Wnt2-NB508 cells were infected with lentivirus carrying 7TFP construct and selected with puromycin. Cells were plated in 96-well plate at 5×103 cells per well. Small molecule reagents were added into media 1 day after plating and cells were imaged in the IVIS Lumina II (Caliper Life Science) the following day.
Low quantity high throughput qRT-PCR
RNA was isolated from adherent monolayer cells and tumour spheres cultured in non-adherent plate using Qiagen RNeasy kit (Qiagen). Reverse transcription was carried out using 200ng of RNA with SuperScriptIII (Invitrogen) per manufacturer’s protocol. Pre-amplification of cDNA was done with 1.25 µl of cDNA using pooled 48 TaqMan Assays at a final concentration of 0.2X for each assay. The pre-amplification PCR was performed at one cycle 95°C for 10 min, 14 cycles at 95°C for 15 sec and then 60°C for 4 min. After pre-amplification PCR, the product was diluted 1:5 with dH2O and stored at −80°C until needed. qPCR was carried out using the 48.48 dynamic array (Fluidigm Corporation, CA, USA) following the manufacturer's protocol.
Supplementary Material
Acknowledgements
We thank C. Koris and the MGH clinical research coordinators for help with clinical studies, J. Gentry for computer informatics support, L. Libby for mouse studies, J. Walsh for microscopy expertise, Z. Nakamura for technical support, D. Jones for sequencing, and M. Rivera and S. Akhavanfard for RNA-ISH. This work was supported by Stand Up To Cancer (D.A.H., M.T., S.M.), Howard Hughes Medical Institute (D.A.H), NIBIB Quantum Grant 5R01EB008047 (M.T.), NIH CA129933 (D.A.H.), Pancreatic Cancer Action Network – AACR Fellowship (D.T.T.), the Warshaw Institute for Pancreatic Cancer Research (D.T.T.), and the K12 Paul Calabresi Award for Clinical Oncology Clinical Research Career Development Program NIH 5K12CA87723-09 (D.T.T.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.Y., D.T.T., S.M. and D.A.H. designed and conducted the study, analyzed data, and prepared the manuscript. S.L.S. and M.T. contributed to the microfluidic system. D.T.T., B.S.W., F.O., B.W.B. and P.M.M. performed Helicos digital gene expression analysis. B.S.W. and S.R. performed bioinformatic and statistical analysis. S.P., J.C.C., M.E.S., D.W., A.J.G., M.J.U. and K.X. provided technical assistant for microfluidic experiments. M.Y., D.T.T., B.W.B. and J.C.C. performed RNA-ISH analysis. D.T.T., D.P.R. and L.V.S. acquired clinical samples. G.C., B.A. and N.B. provided the mouse model and cell line. S.L.S., B.S.W., P.M.M., S.R. and M.T. commented on the manuscript.
The authors declare no competing financial interests.
References
- 1.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stott SL, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozsolak F, et al. Amplification-free digital gene expression profiling from minute cell quantities. Nat Methods. 2010;7:619–621. doi: 10.1038/nmeth.1480. 10.1038/nmeth.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 6.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 8.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. 17/10/1253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 10.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ninomiya-Tsuji J, et al. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. M207453200 [pii] [DOI] [PubMed] [Google Scholar]
Additional references in methods
- 12.Bowers J, et al. Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods. 2009;6:593–595. doi: 10.1038/nmeth.1354. 10.1038/nmeth.1354 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozsolak F, et al. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20:519–525. doi: 10.1101/gr.102129.109. 10.1101/gr.102129.109 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipson D, et al. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27:652–658. doi: 10.1038/nbt.1551. 10.1038/nbt.1551 [doi] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.