Abstract
Metal transporters are a central component in the interaction of algae with their environment. They represent the first line of defense to cellular perturbations in metal concentration, and by analyzing algal metal transporter repertoires, we gain insight into a fundamental aspect of algal biology. The ability of individual algae to thrive in environments with unique geochemistry, compared to non-algal species commonly used as reference organisms for metal homeostasis, provides an opportunity to broaden our understanding of biological metal requirements, preferences and trafficking. Chlamydomonas reinhardtii is the best developed reference organism for the study of algal biology, especially with respect to metal metabolism; however, the diversity of algal niches necessitates a comparative genomic analysis of all sequenced algal genomes. A comparison between known and putative proteins in animals, plants, fungi and algae using protein similarity networks has revealed the presence of novel metal metabolism components in Chlamydomonas including new iron and copper transporters. This analysis also supports the concept that, in terms of metal metabolism, algae from similar niches are more related to one another than to algae from the same phylogenetic clade.
1. Introduction
Metal transporters are essential for all organisms. According to the RNA-world hypothesis, at some point in the early history of life, metal-dependent ribozymes were encapsulated by a vesicle wherein these primitive enzymes could work in concert as a functional unit competing with other protocells for nutrients [1]. Since catalysis was now compartmentalized, protocells had to rely on the selective permeability of the membrane to acquire substrates and expel wastes. The concentrations of some nutrients such as transition metal ions, however, need to be tightly controlled to avoid deleterious reactions due to excess, on the one hand, or enzyme inactivity due to deficiency, on the other, necessitating the evolution of homeostatic mechanisms.
The modern cell represents the victor in the competition between these early protocells. Instead of a semi-permeable membrane, contemporary cells have a phospholipid bilayer membrane, which is impermeable to most hydrophilic molecules. The only way these molecules can enter or exit the cell is through protein transporters that span the lipid bilayer, and in this way the exchange of molecules such as metal ions with the environment can be tightly controlled. Metal transporters are also found inside the cell. After engulfing and maintaining another cell (an endosymbiosis event), the first eukaryotes had to provide the necessary nutrients to sustain the symbiont without sacrificing their own fitness. Enzymes and entire pathways can be specific to an organelle, the remnant of a symbiont, and these intracellular membrane-bound compartments contain transporters to mediate the distribution of solutes.
Transporters represent the first line of defense to perturbations in cellular and subcellular metal homeostasis. When metal reserves are depleted, transporters provide the route to specifically supply and distribute the needed cofactor before deficiency symptoms appear, and when the concentration of metal within the cell exceeds the cell's buffering capacity, transporters provide the route to expel excess cofactors before toxicity occurs.
1.1. Metal homeostasis
Metal cofactors provide chemical activities that are not easily achieved with the functional groups found in the side chains of amino acids, but the chemical properties of metal ions can also lead to cytotoxicity. Unchelated redox-active metal ions, such as iron and copper, can generate reactive oxygen species (ROS). Even though zinc is less harmful by comparison, zinc toxicity symptoms have been observed, which could be due to mis-incorporation of zinc into proteins or generation of protein aggregates [2]. Although metal storage proteins and small molecules inside the cell can buffer the concentration of metal ions to a certain degree, the influx and efflux of each metal must be tightly coordinated to ensure the right metal at the correct concentration is present in each compartment.
1.1.1. Transport: where it all begins and ends
Like all proteins, the function of a metal transporter has both a molecular and a physiological dimension. Characterizing the metal selectivity of the transporter, the speciation of the transported metal (such as oxidation state or whether ligand-bound), and its orientation in the membrane (which speaks to direction of transport) describes the molecular dimension. Determining under what conditions the transporter is present, at what abundance, and in which membrane(s) describe the physiological dimension.
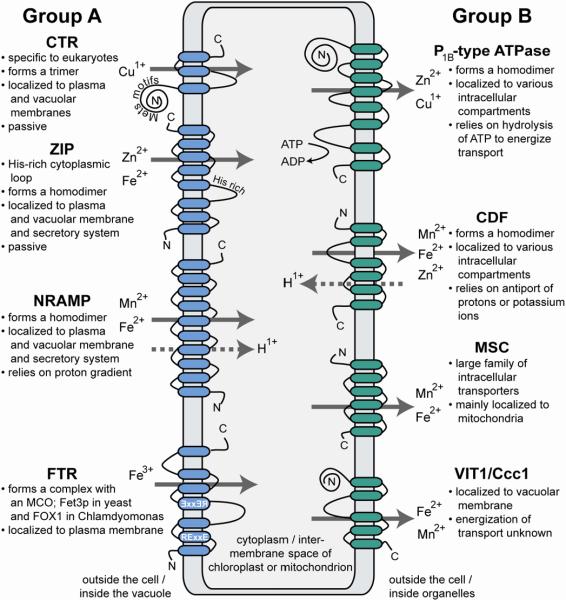
Permeases can be divided into two main functional categories (Figure 1). Group A is responsible for moving metal ions into the cytoplasm. These transporters include members from the NRAMP (Natural Resistance-Associated Macrophage Proteins), ZIP (Zrt-, Irt-like Proteins), FTR (Fe TRansporter) and CTR (Cu TRansporter) families. Within this group are assimilative transporters found in the plasma membrane. These transporters increase the intracellular concentration of metal when the equilibrium between chelating sites and metal ions is perturbed due to deficiency. Group A transporters are also found in the vacuole membrane and have the same role as assimilative transporters except that the source of metal is an intracellular storage compartment versus the external environment. Group B transporters decrease the cytoplasmic concentration of metal. Within this group are distributive transporters, which provide metal for organelle-localized metal-dependent proteins. When present in the membranes of the secretory pathway, group B transporters can mediate the exocytosis of excess metal. This group includes members from the CDF (Cation Diffusion Facilitator), P1B-type ATPases, FPN (FerroPortiN) and Ccc1 (Ca(II)-sensitive Cross-Complementer 1)/ VIT1 (Vacuolar Iron Transporter 1) families. In general, transporter families fall into one group or the other. However, as discussed in sections 3.1.2.2 and 5.1.1. some members of the NRAMP and ZIP families appear to have atypical roles. These deviants highlight our limited understanding of transporter mechanism, topology and structure, and how interaction with other proteins and the cellular environment might determine or modulate transporter properties.
Figure 1. Common transporter families in eukaryotes.
Predicted membrane topologies and common substrates for each transporter family are shown. Arrows represent the direction of metal ion transport, which is either into the cytoplasm for group A transporters or out of the cytoplasm for group B transporters. In the case of transporters localized to the inner membrane of either the chloroplast or mitochondrion, group B transporters would transport metal ions from the inter-membrane space into the stroma or matrix, respectively.
Techniques from various fields advance our knowledge of metal transporter function. Because these proteins can have low substrate specificity in vitro, an understanding of function in the context of the cell is required. Because absence or over-abundance of these proteins can cause pleotrophic effects within the cell, a biochemical approach is required. The first level of a metal transporter study commonly surveys the pattern of gene expression. The presence or absence of the transporter's substrate in the growth medium often influences gene expression. For instance, deficiency induces genes for assimilatory transporters, whereas a metal excess situation induces genes for transporters that pump the metal out of the cell or into the vacuole. While this is a simple concept, these data must be interpreted in the context of metal crosstalk; the reduced concentration of one metal ion can cause a secondary deficiency in another metal ion. In Chlamydomonas, expression of iron transport genes during copper-deficiency is proposed to be a result of secondary iron-deficiency caused by a less active copper-dependent component of the high-affinity iron transporter [3]. The identification of a metal-responsive transcription factor and direct interaction between the regulator and the promoter region of a putative metal transporter gene can reinforce results from a transcriptome study. However, as seen for the distributive copper transporter-encoding gene CCC2 from Saccharomyces cerevisiae [4], expression can be controlled by a transcription factor that responds to a different metal, in this case iron (because copper is required for iron uptake).
To complement gene expression studies, functional complementation studies in S. cerevisiae are typical. Growth defects attributed to the disruption of specific metal transporters in S. cerevisiae have been characterized. The ability of a non-native gene to rescue these growth defects can inform the potential function of the “test” protein. As with gene expression studies, these complementation studies have caveats. They assume that the test transporter localizes to the same compartment in S. cerevisiae as in the native organism and that rescue is because the test transporter and the S. cerevisiae transporter have the same function.
In addition to S. cerevisiae, metal transport assays can be performed using either the Xenopus oocyte assay system or sealed vesicles containing reconstituted test protein. In the Xenopus oocyte assay, cDNA or in vitro-transcribed mRNA encoding the putative transporter is injected into an oocyte, and a change in conductance induced by metal transport across the membrane is measured [5]. Conductance in the presence and absence of the cDNA is compared to determine the contribution of the protein (encoded by the cDNA) to transport. An alternative to the oocyte assay is the use of sealed lipid vesicles reconstituted with the recombinantly expressed transporter protein. By using metal radioisotopes or a metal-selective chromogenic or fluorogenic dye, the transport of the substrate metal ion into the vesicle by the putative transporter can be quantified and subject to kinetic analyses [6, 7]. Other types of complementary evidence for the physiological function include changes in metal uptake rates or intracellular metal content due to knock-down or knock-out of the gene. Transport assays with radioactive substrates have been used, but these are not always straightforward (because of lack of availability of useful substrates or non-specific binding of ions to whole cells). Comparison of metal content of mutant versus wild-type strains by Inductively coupled plasma mass spectrometry (ICP-MS) is another approach.
1.2. The role of algae in advancing our understanding of metal homeostasis
Plastid-containing organisms represent a unique system for studying cellular and intracellular metal homeostasis. The eukaryotic oxygen-evolving cell has two metal-rich, membrane-bound powerhouses to which metal must be differentially and selectively delivered. The chloroplast is the site of photosynthesis, a process that is dependent on the trace metals iron and copper for electron transfer, manganese for the water-splitting reaction of photosystem II, and zinc for carbonic anhydrase and other enzymes. These metals are also needed in other roles such as ROS detoxification and gene expression. The mitochondrion also houses a metal-rich electron transport chain and numerous metal-dependent enzymes. Because of these two organelles, photosynthetic organisms compared to heterotrophic organisms have a high demand for metal and distributive metal transporters, and the individual demands of these compartments can lead to competition in a deficiency situation [8, 9].
Studies that monitor responses to perturbations of homeostasis, predominantly the metal-excess and metal-limitation situations provide the bulk of our understanding of metal metabolism. In the laboratory, the metal-excess state is simple to achieve by just adding metal ions to the culture medium, but achieving metal-limitation is relatively difficult (e.g. [10]). Contaminating metal in glassware or in medium ingredients are often sufficient to sustain the growth of most organisms, because these nutrients are required in trace amounts [11]. Many experimentalists have chosen to employ metal chelators, which are convenient but notoriously lack complete specificity for a single metal ion [12, 13].
Since photosynthetic cells have a higher demand for metal, creating the metal-limitation state can be easier and does not necessarily require the use of chelators. Nevertheless, it is usual to buffer metal ions in trace element solutions with a low concentration of chelator for stability and longer shelf life. Since photosynthetic organisms make their own metabolites, it is not necessary to supplement the medium with amino acids or serum, which also facilitates metal-deficiency studies. Finally, inorganic nutrients are the only nutrients (beside sunlight, water and CO2) provided to plants and algae, and there has accordingly been considerable historical interest in the minimal nutrition of plants [14]. Recently, small flowering plants in the genus Arabidopsis have gained prominence as reference organisms for understanding trace metal homeostasis in land plants and for discovering fundamental metal homeostasis mechanisms [15–18], but in some cases, the discoveries are only relevant to closely related species.
Therefore, it is valuable to compare evolutionarily divergent photosynthetic organisms to reveal core conserved processes and instances of specialization. Algae make such comparisons possible, because they inhabit diverse environments with contrastingly different metal abundance and have complex evolutionary histories involving plastid loss and gain. Novel strategies for acclimating or adapting to iron-, copper- and zinc-deficiency have been revealed by studying algae. These discoveries not only enrich our understanding of metal homeostasis but can also provide new resources such as novel genes for metal biofortification in food crops.
1.2.1. What are algae?
Photosynthetic eukaryotes can be divided into two general groups. Embryophyta, the land plant lineage, is a monophyletic group of complex, multicellular organisms that are capable of forming embryos [19]. All other O2-evolving eukaryotes are referred to as algae and these include diverse organisms from the plant-like seaweeds that form underwater forests to the tiny picoplankton that are no bigger than a bacterial cell. Although cyanobacteria have also been referred to as blue-green algae, the term `algae' is properly reserved for eukaryotes [20].
Algae are polyphyletic, since the group does not share a common algal ancestor, and are paraphyletic, meaning that some taxa also contain non-alga members (Figure 2). The primary plastid is thought to have originated from a single engulfment of an ancient cyanobacterium by a heterotrophic ancestor giving rise to the Archaeplastida (ancient plastid) lineage [21]. Direct descendents of this ancestor include the red, green and glaucophyte algae from Rhodophyta, Chlorophyta and Glaucophyta, respectively. The established plastid was then transferred to other eukaryotic lineages through subsequent endosymbiotic events (Figure 2 and recently reviewed in [22, 23]). Euglenids (in Excavata) and chlorarachniophytes (in Rhizaria) have plastids of green algal origin, while the chromalveolates (such as diatoms and stramenopiles) have plastids of red algal origin [24]. The Archaeplastida lineage is not the only group of algae with a primary plastid; a recent primary endosymbiosis event has occurred in the amoeba Paulinella chromatophora in Rhizaria [25]. Because we do not know the exact history of algal evolution, the number of events leading to the gain of a plastid or the loss of a plastid in the various lineages is continually revisited. For instance, similarity between a subset of genes in sequenced diatom genomes suggests that an ancestor also engulfed a green alga at some point [26] and the plastids in the separate chromalveolate lineages may have arisen from a single or multiple endosymbiotic events [24, 27]. As new algae are characterized and genomes are sequenced, our understanding of algal evolution will inevitably be modified.
Figure 2. Simplified representation of the eukaryotic evolutionary tree.
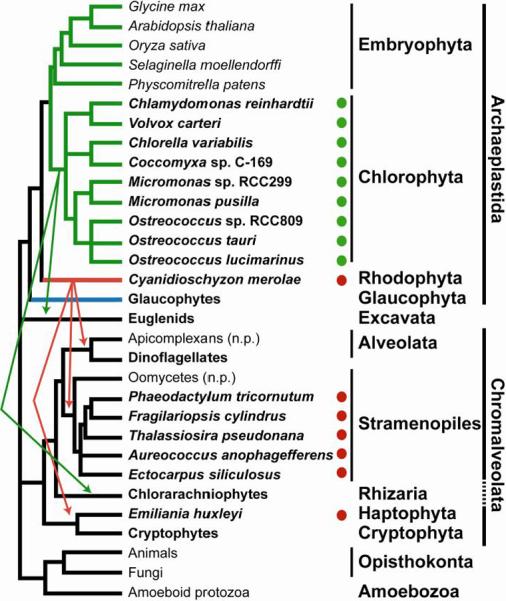
Shown is a tree representing one hypothesis for the relatedness of alga-containing lineages. Individual algae (for which sequenced genomes are available) and groups containing algae are shown in bold. Green and red arrows represent the movement of primary plastids of green and red algal origin to a heterotrophic ancestor of photosynthetic organisms in Excavata, Chromalveolata and Rhizaria. Lineages that have lost the ability to photosynthesize are indicated with “n.p”. Solid circles indicate that the algal genomes that were queried for homologs of known metal transporters; green circles for green algae and red circles for the red alga C. merolae and algae with plastids that originated from the engulfment of a red alga. Branches representing lineages with primary plastids are colored green for Viridiplantae, red for Rhodophyta and blue for Glaucophyta. Branch lengths do not indicate evolutionary distance [215].
1.2.2. Chlamydomonas as a reference organism
Chlamydomonas reinhardtii (herein referred to as Chlamydomonas) is a unicellular, green alga from the Chlorophyta lineage and is the best-developed reference organism for studying algal biology. Because the genome has retained genes from the last common ancestor to both the plant and animal lineages, Chlamydomonas is a premiere reference organism for chloroplast function on the one hand and ciliary biology and function on the other [28]. It can be grown in a simple, chemically-defined medium to a high cell density, and both metal-deficiency and metal-limitation situations, which are assessed by monitoring the expression of characterized marker genes, are easily achieved [29]. The single, large chloroplast is biochemically equivalent to land plant chloroplasts but Chlamydomonas uniquely maintains the photosynthetic machinery in the dark and is able to grow heterotrophically (acetate/dark), autotrophically (CO2/light) or mixotrophically (acetate/light), enabling the investigation of metal homeostasis mutants involved in photosynthetic function that would not be viable in higher plants.
1.2.3. Algal metal nutrition
“…the habitat is the mold into which the organism fits.” - Victor E. Shelford [30] Just as this statement was true in the early 1900's for animal behavior, it is true today when attempting to understand the metal requirements of algae. Because all organisms rely on metal cofactors to catalyze many of the reactions essential to life, the abundance and bioavailability (i.e. accessibility) of metal are fundamental characteristics of each niche, and to be successful, an alga must adapt to the geochemistry (i.e. elemental abundance and speciation) of the niche.
Both global and local metal bioavailabilities have shaped the metal-dependency of algae. Of the various biologically active transition metals, iron and copper in particular have had a large impact on the evolution of algae. In the anoxic atmosphere of primitive earth, iron was predominately found in the soluble (bioavailable) +2 oxidation state, and a dependency on the redox activity of iron was established early in protein evolution [31]. In contrast, copper was not readily bioavailable and is predicted to be absent in primitive cells [32]. The appearance of photosynthesis and the advent of dioxygen reversed the solublities of iron and copper [33], but on an evolutionary timescale, copper entered the pool of potential nutrients relatively late, and few proteins have evolved that require a copper cofactor. Instead, most algae invest a large amount of energy overcoming the limited bioavailability of iron.
The gene repertoires of algae and regulation of those genes are reflections of geochemical pressure. A unique environment is the open-ocean where algae are not only limited by the bioavailbility of iron but also its abundance. One way algae have adapted to this niche is reduction of their iron quota by substituting iron proteins with catalytically equivalent copper-dependent proteins. The presence or absence of major iron and copper proteins is therefore indicative of the metal bioavailability and geochemistry of an alga's environment. Photosynthetic eukaryotes can be divided into three groups based on the presence of genes encoding iron-dependent cytochrome c6 and copper-dependent plastocyanin [34] (Table 1). In this classification, cytochrome c6 is distinguished from the non-orthologous cytochrome c6-like proteins of unknown function from plants and green algae (Supplemental figure 1). In one scenario, these cytochrome c6-like proteins may have arisen from a duplication event in the last common ancestor of Viridiplantae prior to the loss of cytochrome c6 in land plants and the prasinophytes.
Table 1.
Algae with sequenced genomes.
| Lineage | Organism | Genome Browser | Unicellular? | Ref. | PCY | CYC6 | Cu/Zn SOD | Fe SOD | Mn SOD | FOX1/Fet3p | FTR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Viridiplantae | |||||||||||
| Chlorophyta | |||||||||||
| Chlorophyceae | Chlamydomonas reinhardtii | Phytozome1 / v4.3 | Y | [28] | Y | Y | N | Y | Y | Y | Y |
| Volvox carteri | Phytozome / v2.0 | N | [216] | Y | N | N | Y | Y | Y | Y | |
| Trebouxiophyceae | Chlorella variabilis NC64A | JGI2 / v1.0 | Y | [217] | Y | Y | N | Y | Y | Y | Y |
| Coccomyxa sp. C-169 | JGI / V2.0 | Y | Y | Y | N | Y | Y | Y | Y | ||
|
| |||||||||||
| Mamiellophyceae | Micromonas sp. RCC299 | JGI / V3.0 | Y | Y | N | Y | N | Y | N | N | |
| Micromonas pusilla CCMP 1545 | JGI / V3.0 | Y | [218] | Y | N | Y | N | Y | N | N | |
| Ostreococcus tauri | JGI / V2.0 | Y | Y | N | Y | N | Y | N | N | ||
| Ostreococcus lucimarinus | JGI / V2.0 | Y | [122] | Y | N | Y | N | Y | N | N | |
| Ostreococcus sp. RCC809 | JGI / V2.0 | Y | Y | N | Y | N | Y | N | N | ||
|
| |||||||||||
| Rhodophyta | |||||||||||
| Bangiophyceae | Cyanidioschyzon merolae | U. of Tokyo3 | Y | [219] | N | Y | N | N | Y | N | N |
|
| |||||||||||
| Stramenopiles | |||||||||||
| Bacillariophyta | |||||||||||
| Bacillariophyceae | Phaeodactylum tricornutum | JGI / V2.0 | Y | [220] | N | Y | Y | N | Y | N | N |
| Fragilahopsis cylindrus | JGI / v1.O | Y | Y | Y | Y | N | Y | Y | Y | ||
| Coscinodiscophyceae | Thalassiosira pseudonana | JGI / V3.0 | Y | [221] | N | Y | N | Y | Y | Y | Y |
| Aureococcus | N | Y | Y | N | Y | Y | Y | ||||
| Pelagophyceae | anophagefferens | JGI / v1.0 | Y | [155] | |||||||
| Ectocarpus siltulosus | BOGAS4 | N | [222] | N | Y | Y | Y | Y | N | N | |
| Haptophyceae | |||||||||||
| Emiliania huxleyi | Y | Y | N | N | Y | N | N | ||||
| Isochrysidales | CCMP1516 | JGI / v1.0 | Y | ||||||||
The presence or absence of the indicated genes is represented with Y (present) or N (absent).
Group 1 algae contain genes for both cytochrome c6 and plastocyanin, group 2 algae only have cytochrome c6-encoding genes, and group 3 algae only have plastocyanin-encoding genes. In the context of iron and copper bioavailabilityover time, cytochrome c6 is proposed to have evolved first but was later replaced in some photosynthetic lineages by plastocyanin [35]. However, inclusion in one group or another is more indicative of iron and copper availabilities in an alga's environment than of its evolutionary history [36].
Characterized members of group 1 are terrestrial green algae. As characterized for Chlamydomonas, the plastocyanin-encoding gene is constitutively expressed, but under copper-deficiency, plastocyanin is degraded and replaced with cytochrome c6 [37, 38]. The cytochrome c6 gene is maintained as a back-up for times when the alga enters a copper-deficient environment.
The red alga Cyanidioschyzon merolae belongs to group 2 and presumably never acquired plastocyanin or Cu/Zn SOD. This primitive alga inhabits sulphur-rich, acidic hot springs where copper may not be as readily available as iron, and the presence of these copper-dependent proteins would not provide a selective advantage.
Group 3 algae are commonly found in the phytoplankton communities of the open-ocean, where iron is a limiting nutrient, and these algae have dispensed cytochrome c6. The presence of a gene encoding Cu/Zn superoxide dismutase (SOD) or Fe-dependent SOD is also contrastingly different between terrestrial and marine green algae; group 1 never acquired a Cu/Zn SOD but has maintained Fe-SOD, whereas group 3 has lost Fe-SOD and gained a Cu/Zn SOD (Table 1). Like marine green algae, land plants belong to group 3; they have acquired plastocyanin and Cu/Zn SOD but have lost the canonical cytochrome c6.
Algae from Chromalveolata are found in all three groups. Groups 1 and 2 inhabit predominately coastal waters where iron is relatively more abundant (Table 1) compared to the open ocean where group 3 algae predominate [36, 39]. Because the plastid was derived from a red alga(e), chromalveolates appear to have acquired plastocyanin and lost cytochrome c6 in a niche-specific manner. The oceanic diatom Thalassiosira oceanica only has plastocyanin [36], and the coastal diatoms Thalassiosira weissflogii [40] and Thalassiosira pseudonana only have cytochrome c6. Most chromalveolates do have a Cu/Zn SOD-encoding gene regardless of the presence of plastocyanin and this gene was likely present in the genome of the ancestral heterotrophic host. Closely related species of algae can inhabit a wide range of environments with different geochemistries, and the assorted occurrence of plastocyanin and cytochrome c6 in algae underlines the difficulty of using lineage-specific generalities.
2. Methods
Advances in DNA sequencing have impacted our understanding of algal metabolism and evolution. Compared to popular plant, animal and bacterial reference organisms little is known regarding the extent to which algae employ mechanisms characterized in these organisms or have novel routes for metal acquisition and distribution. With sixteen algal genomes currently available, comparative genomic approaches can provide some clues. Beyond genome sequencing, the transcriptomes of algae (particularly Chlamydomonas) under different growth conditions, including metal deficiency, are being sequenced. Not only are we gaining a clearer picture of transcriptional responses to environmental stimuli, but we are able to improve gene models and discover genes [3]. Gene models are generally based on sequence similarity and, in the absence of transcript data, can be incomplete. This shortcoming is especially relevant for genes encoding highly regulated proteins such as metal transporters whose transcripts are not present or present at low abundance when the cells are grown under standard laboratory conditions, which means that ESTs derived from these genes are rare.
To reanalyze the metal transporter repertoire of Chlamydomonas (Figure 3), we have taken advantage of gene models [41] (available as of 12/2011), which are based on transcript sequences from a pooled resource that includes transcripts isolated from metal-deficient growth conditions (http://www.phytozome.net/chlamy.php; http://genomes-merchant.mcdb.ucla.edu/Cre454/). These models were also manually curated with Illumina-generated RNA-Seq data from copper– [3], iron– (Urzica and Merchant, unpublished) and zinc– (Malasarn and Merchant, unpublished) deficient cells.
Figure 3. Assimilative and distributive metal transport in Chlamydomonas.
Putative metal transporters, metal-binding proteins and chaperones encoded in the Chlamydomonas genome and their known or predicted intracellular locations are shown. Arrows are used to illustrate the proposed directions of transport. Descriptions of proteins are available in table 2 or the text.
In an attempt to build a complete picture of cellular metal transport for a particular organism, determining the presence of a putative metal transporter-encoding gene is only the first step. The second step is predicting the function of that transporter. Many metal transporter families are multi-functional with members mediating the transport of different metal ions, while some families are a small subgroup of a larger multifunctional superfamily with functions distinct from metal transport. To predict the function of a protein, one approach is to establish orthology (genes that arose by a speciation event) between a characterized protein and a putative protein by iterative BLAST or phylogenetic tree building. However, paralogs (two genes that arose by a duplication event) can cause ambiguity, and because of sampling bias (relatively few algal genomes are available) or inaccurate gene models, algal proteins may share relatively little sequence similarity with characterized proteins and are commonly found branching near the root of phylogenetic trees leading to uncertainty as to which functional subgroup they belong.
To visualize function across metal transport families that contain genes in the Chlamydomonas and other available algal genomes, we have used protein similarity networks [42]. BLAST was used to identify protein transporter homologs from the algae depicted in Figure 2 and listed in Table 1. Protein similarity networks were then created from an all-vs-all BLAST analysis (pairwise alignment between all pairs of proteins) for each transporter family. Protein sequences were accessed through NCBI [43], Uniprot [44], Phytozome [45] or the Joint Genome Institute Genome Portal [46], unless specified otherwise. The network was created in Cytoscape version 2.8 [47] with the BLAST2SimilarityGraph plug-in [48]. Nodes (representing a protein) are connected with an edge (line) if the E-value between two sequences was at least as good as the given value. A custom sequence database was used in each case and the E-values calculated during the all-vs-all BLAST are not necessarily the same as would be reported using large databases such as the non-redundant protein sequences (nr) database commonly used with the blastp tool on the NCBI website.
To visualize the substructure of the network, the yFiles organic layout provided with Cytoscape was used. The organic layout clusters nodes that are relatively more similar to one another, i.e. edges between nodes that have more connections to one another will be shorter than between nodes that have few connections [49]. This layout reveals groups of proteins that are more similar to one another than to the rest of the proteins in the network. Functional information from the literature was then overlaid on the network and the function of some algal proteins could be predicted by inclusion in a cluster with characterized transporters.
3. Iron
Several strategies have evolved to mobilize iron, which is the most common redox-active metal cofactor found in proteins. Different transporters are employed depending on the type of iron chelate and, because the coordination chemistries of ferrous (2+) versus ferric (3+) ions are distinct, sequential redox reactions are central to iron mobilization [50].
3.1. Group A iron transporters
3.1.1. Ferric reductases
In an aerobic environment both on land and in the ocean, iron is predominately insoluble (as Fe(III)-oxides or other chelates). The first step in iron transport is to solubilize available iron through the activity of ferric reductases on the plasma membrane. Characterized ferric reductases transfer one electron to Fe(III) in complexes such as Fe(III)-citrate, and most use cytosolic NAD(P)H as the electron donor [50]. The reduction of ligand-bound Fe(III) causes dissociation of the complex and release of Fe(II) [51].
Putative assimilatory ferric reductases (ferric reductases involved in uptake of iron from the environment) in algae have been identified based on the transcriptional response of their corresponding genes to iron-nutrition. However, it should be noted that various ferric reductases are involved in activities independent of iron mobilization such as signaling [52] and enzyme catalysis [53], and the role `assimilative ferric reduction' should only be assigned to proteins on the plasma membrane. Three types of ferric reductases are responsible for reduction of Fe(III)-chelates in eukaryotes, NADPH oxidases (NOX; cytochrome b558), cytochrome b5 reductases and cytochrome b561. Based on transcript analysis of cells from iron–replete and –deficient growth conditions, proteins from each ferric reductase family are implicated in iron mobilization in algae.
The first NOX enzyme to be described in ferric reduction is Fre1p (Ferric reductase 1) from S. cerevisiae. The corresponding gene is regulated by iron and was isolated by identifying the mutation responsible for ferric reductase deficiency in a mutagenized strain [54, 55]. Fre1p and the subsequently identified Fre2p are responsible for the bulk of cell surface metal reductase activity in S. cerevisiae [56]. Fre1p and Fre2p belong to a seven member gene family that also includes Fre3p, a plasma membrane-localized reductase that facilitates acquisition of siderophore-bound iron [57], and Fre6p, a vacuolar membrane-localized reductase responsible for the reduction of iron before export into the cytoplasm [58]. An iron-responsive NOX gene is also expressed in the roots of Arabidopsis thaliana (herein referred to as Arabidopsis)(FRO2) [59] and is part of an eight member gene family with putative functions in intracellular and intercellular iron and/or copper mobilization [60]. However, not all NOX enzymes appear to be involved in iron transport and homologous proteins such as the respiratory burst oxidase homolog (RBOH) protein family in Arabidopsis are involved in the generation of superoxide as a signaling molecule and for defense [61, 62].
The involvement of NADH-cytochrome b5 reductase in iron homeostasis was first discovered because defects in activity lead to methemoglobinemia, a disease in which hemoglobin accumulates in the oxidized form, methemoglobin [63]. Without NADH-cytochrome b5 reductase, cytochrome b5 remains in the oxidized form and cannot reduce the ferric iron in methemoglobin. Genes from the cytochrome b5 reductase family have been characterized from various organisms and all appear to encode two isoforms, one membrane-bound and one soluble. For instance, one form of Mcr1p from S. cerevisiae is found bound to the outer membrane of the mitochondrion where the bulk of the protein is exposed to the cytosol, while a second form is found as a soluble protein in the intermembrane space of the mitochondrion [64]. The functions of both Mcr1p isoforms remain unknown, but a direct role of cytochrome b5 reductase-like proteins in ferric-chelate reduction was observed for homologous proteins in plants [65–67]. The pumpkin homolog CmPP36 is also present as both membrane-bound and soluble forms, and expression of the corresponding gene is induced by iron-deficiency in the roots [67]. Since CmPP36 can reduce Fe(III)-chelates, it is tempting to speculate that members of the cytochrome b5 reductase family are involved in controlling the oxidation state of iron as it is distributed within the cell and within the organism [67]. By encoding membrane-bound and soluble isoforms, the reduction of iron as it enters a compartment and travels through that compartment can be controlled by the regulation of one gene. However, these ferric-chelate reductases are not expected to contribute to assimilation of iron from the soil. Instead, they may be involved in intracellular and intercellular mobilization of iron within the plant.
The prototypical cytochrome b561 ferric reductase is DCYTB (duodenal cytochrome b) from mammals. Dcytb was obtained by identifying mRNAs present in a mouse that has high rates of iron absorption compared to a sibling that has relatively normal rates [68]. Characterization of Dcytb revealed that expression is induced by iron-deficiency, and DCYTB localizes to the intestinal lining and has ferric reductase activity. Proteins with a cytochrome b561 domain are ubiquitous among eukaryotes and are thought to use cytoplasmic ascorbate as the electron donor [69].
Of the four putative ferric reductase-encoding genes in Chlamydomonas, expression of CBR1 (encoding a cytochrome b5 reductase), RBOL1 and RBOL2 (encoding NOX enzymes) is unresponsive to iron-deficiency, while expression of FRE1 (also a NOX enzyme) is highly induced [70]. In support of a role in iron assimilation, FRE1 localizes to the plasma membrane [71]. The other putative ferric reductases may be involved in intracellular mobilization of iron, providing electrons for various reactions such as desaturation of fatty acid [72] or signaling. CBR1 shares sequence similarity with Mcr1p and Cbr1p from S. cerevisiae, which are localized to the mitochondria [73, 74]. RBOL1 and RBOL2 share sequence similarity with the Arabidopsis RBOH proteins, which are linked to the generation of superoxide as a signaling molecule [52].
Iron-responsive reductase genes are also present in the diatoms T. pseudonana (TpFRE1 and TpFRE2 [75]) and Phaeodactylum tricornutum (PtFRE1 – PtFRE4 [75, 76]). TpFRE1 is similar to cytochrome b5 reductase, and PtFRE3 and PtFRE4 are similar to cytochrome b561. TpFRE2, PtFRE1 and PtFRE2 are similar to NOX proteins, but TpFRE2 and PtFRE1 are more similar to the RBOH protein family of Arabidopsis than to the FRO proteins. Ferric reduction appears to be necessary for iron transport into the chloroplast [77] and out of the vacuole [58]. Therefore, ferric reduction may be a common characteristic of all intracellular membrane-localized iron transport systems. Some of these putative diatom ferric reductases may be involved in assimilation or distribution of iron within the cell, but they might just as well be involved in a different role. Oxidative stress accompanies iron-deficiency in Chlamydomonas, and reductases that are specifically present during iron-deficiency could be involved in responding to ROS [78, 79].
3.1.2. Ferrous transporters
3.1.2.1. ZIP
The ZIP family is so named because the first members to be characterized were the zinc transporters Zrt1p [80] and Zrt2p [81] from S. cerevisiae and IRT1, the major iron uptake protein in Arabidopsis roots [82]. Fourteen homologs have been identified in the human genome and are commonly referred to as either the SLC39 or ZIP family. ZIP members are composed of eight predicted transmembrane regions (TMs), and most have a histidine-rich loop between TMs III and IV that protrudes into the cytoplasm (Figure 1). The exact mechanism of how ZIP members transport metal is currently unknown, but recent work suggests that they are passive transporters driven by a metal concentration gradient [7].
The ZIP family has been divided into four groups based on phylogenetic relationships [83]: Subfamily I, II, GufA and LZT. Subfamily I contains mainly fungal and plant proteins, subfamily II contains plant and animal proteins, GufA from Myxococcus xanthus is the founding member of the GufA subgroup, which contains other prokaryotic members as well as eukaryotic ones, and the LZT subgroup (LIV-1 subfamily of ZIP zinc Transporters [84]) of which LIV-1, a human estrogen-responsive zinc transporter, is the founding member. A protein similarity network produces two additional clusters (Figure 4); one cluster includes the protein of unknown function ZTP50 from Arabidopsis and the other cluster includes Atx2p from S. cerevisiae and ZIP9 from human, which are putative manganese and zinc transporters, respectively, in the Golgi membrane. ZIP family transporters are often able to transport multiple divalent metal cations, such as iron, zinc, manganese, cobalt, copper or cadmium, but inclusion in a particular subgroup does not necessarily equate to shared substrate specificity.
Figure 4. Protein similarity network of the ZIP family.
Six distinct clusters are shown, four of which have been previously distinguished from one another using phylogeny [83]. Proteins from Arabidopsis, human, Chlamydomonas and S. cerevisiae in each cluster are listed under the subgroup name. Additionally, the nodes representing those proteins are labeled; the nodes representing Chlamydomonas ZIPs are light green (for Chlorophyta) and labeled with a name (i.e. ZIP1), human proteins are yellow and labeled, Arabidopsis proteins are green and labeled and S. cerevisiae proteins are light blue and labeled.
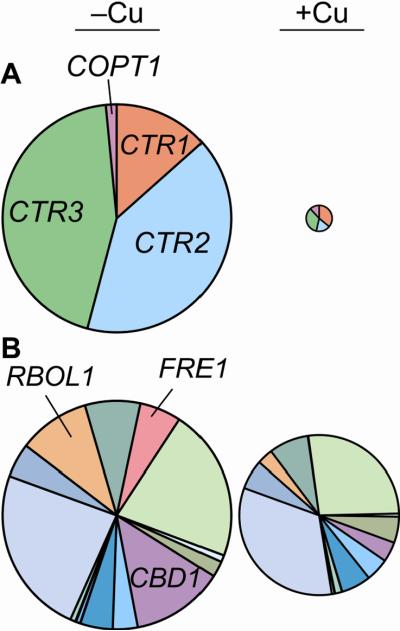
Of the 13 ZIP family members in Chlamydomonas, the abundance of IRT1 and IRT2 transcripts is affected by the iron status of the cell [70]. While ZIP6 is the dominant ZIP transcript in the cell during the metal-replete situation (20 μM Fe), ZIP6 and IRT2 are equally abundant during iron-deficiency (1 μM Fe) (Figure 5A). During the iron limitation state (0.2 μM Fe), which is characterized by reduced growth and chlorosis, IRT1 transcript dominates (Urzica and Merchant, unpublished) (Figure 5A). In addition to distinct patterns of expression in response to iron status, IRT1 and IRT2 are differentially present in cells grown under copper- and zinc-deficiency. IRT1 but not IRT2 is induced by zinc-deficiency (Malasarn and Merchant, unpublished), while IRT2 but not IRT1 is induced by copper-deficiency and is predicted to be a direct target of the copper-responsive transcription factor CRR1 [3]. Because IRT2 is a potential target of CRR1, IRT2 could be an intracellular ferrous transporter responsible for mobilizing iron for cytochrome c6. Alternatively, it may be a back-up route for iron assimilation when the copper-dependent iron transport route (see section 3.1.3.) is impaired due to copper-deficiency.
Figure 5. Abundance of Chlamydomonas ZIP transcripts.
A) Pie charts show the contribution of each ZIP transcript during growth in iron-limited (0.2 μM Fe), iron-deficient (1μM Fe) and iron-replete (20 μM Fe) medium. B) Pie charts show the contribution of each ZIP transcript during growth in zinc-limited (0μM Zn) and zinc-replete (2.5 μM Zn) medium. In (A) and (B), the size of each pie chart represents the sum of all ZIP transcripts during that condition normalized to the iron- or zinc-limited condition. Consequently, the size of the metal-replete chart is different in each case.
IRT1 and IRT2 belong to the GufA subfamily, which also includes the iron-responsive ZIP homolog from the diatom P. tricornutum [76]. The GufA group also contains the vacuole-localized Zrt3p from S. cerevisiae and the ER-localized ZTP29 from Arabidopsis. Zrt3p is responsible for mobilizing zinc from the vacuole in response to zinc-deficiency [85] and ZTP29 is a putative zinc transporter involved in acclimating to salt stress [86]. Since Chlamydomonas has a high-affinity iron transporter (see section 3.1.3.) and IRT1 and IRT2 are members of the GufA subgroup, they may be localized to intracellular membranes.
In contrast to IRT1 and IRT2 from Chlamydomonas, the iron-responsive ZIP genes AtIRT1 and AtIRT2 from Arabidopsis encode proteins from ZIP subfamily I (Figure 4). AtIRT1 is the main iron uptake route for ferrous iron across the root epidermis [87]. AtIRT2 is also induced by Fe-deficiency in the root but, while AtIRT1 localizes to the plasma membrane [87], AtIRT2 is found in intracellular vesicles and is proposed to be involved in compartmentalization once iron is assimilated by AtIRT1 [88].
3.1.2.2. NRAMP
The NRAMP family (also SLC11) is so named because the first member to be studied, natural-resistance-associated macrophage protein 1, is involved in macrophage digestion of intracellular parasites [89]. The role of NRAMP1 in this process remains controversial, and there is still debate as to whether it functions in metal transport into or out of the phagosome [90, 91]. Most NRAMP permeases contain ten or eleven TMs and mediate transport of divalent cations, mainly iron and manganese, into the cytoplasm (Figure 1). Exceptions include rice and sea squirt NRAMPs that transport trivalent metal cations [92, 93] and a putative soybean iron transporter [94] that appears to transport metal out of the cytoplasm. Shortly after the involvement of NRAMP1 in metal transport was reported, NRAMP2 (also referred to as DMT1 and DCT1) was described as an iron-deficiency induced gene in the intestines [5]. But while NRAMP1 appears to be an metal/hydrogen antiporter or symporter [95], NRAMP2 is a symporter [5].
Phylogenetically, the NRAMP family is divided into four main subfamilies: prokaryotic MntH group A, group B and group C and the eukaryotic group [96]. The protein similarity network splits the eukaryotic subgroup into three clusters (Figure 6). Eukaryotic subgroup I contains NRAMPs from the major eukaryotic lineages, while eukaryotic subgroup II is predominately composed on proteins from plants, prasinophytes and C. merolae. The third eukaryotic cluster contains fungal proteins including Smf1p, Smf2p and Smf3p from S. cerevisiae.
Figure 6. Protein similarity network of the NRAMP family.

Six distinct clusters are shown, four of which have been previously distinguished from one another using phylogeny [96]. The E-value giving the best separation of the eukaryotic subgroup from the MntH group C, produced a network without connections to the MntH group B or the fungal NRAMPs. When the threshold is lowered, the fungal proteins cluster with the eukaryotic groups, and MntH group B clusters with the other MntH groups. Proteins from Arabidopsis, human, Chlamydomonas in each cluster are listed under the subgroup name. For S. cerevisiae, the protein names are listed next to the fungal cluster (light blue nodes). The nodes themselves are labeled following the convention described in the legend to figure 4, except the “NRAMP” portion of each name is excluded (i.e. `NRAMP1' is shown as `1').
Most algal genomes contain only one NRAMP, while an NRAMP homolog is missing from the P. tricornutum genome [75]. The Chlamydomonas genome encodes four members, NRAMP1, NRAMP2, RET1 and NRAMP4 (NRAMP4 is missing from earlier releases of the Chlamydomonas genome). NRAMP1 and NRAMP2 are bacterial-like, while NRAMP4 clusters with eukaryotic homologs, AtNRAMP3 and AtNRAMP4 from Arabidopsis (Figure 6). AtNRAMP3 and AtNRAMP4 appear to have different functions in the plant depending on both the developmental stage and tissue type. During germination, these permeases transport vacuolar iron [97], but in adult leaves they mobilize vacuolar manganese [98]. The mechanism underlining the physiological impact on AtNRAMP3 and AtNRAMP4 function is unknown. An ortholog of NRAMP4 from T. pseudonana was found to be highly induced by iron-limitation [75, 99], however the cellular location of this permease is needed to determine whether it functions in assimilation or iron mobilization. RET1 (related to EIN2 [100]; previously referred to as NRAMP3 [101]) contains an NRAMP domain near the N-terminus but resembles the EIN2 proteins of plants and shares conserved residues across the entire length of the protein with EIN2-like proteins from plants. EIN2 (Ethylene Insensitive 2; ORE3) was first identified in Arabidopsis and found to play a central role in ethylene signaling [102, 103]. The NRAMP domain of EIN2 does not appear to have metal transport activity, suggesting that the EIN2 is not involved in metal transport [102]. However, the NRAMP domain may be involved in sensing metal, which could be relevant with respect to the copper requirement of the ethylene receptor [104]. RET1 is expected to function in a different signal transduction pathway in algae, since they are not known to have ethylene receptors.
3.1.3. Ferroxidase dependent
The ability of a transporter to transport more than one metal ion can have detrimental effects on metal homeostasis. Unwanted metal ions can enter the cell or can compete with entry of the desired nutrient [105–107]. In the extreme case, transport of these unneeded metals can lead to a toxicity situation. Many organisms have therefore exploited redox chemistry for selective iron transport [108]. Specifically, iron occurs in two oxidation states under physiological conditions, and a +3 oxidation state is unique to iron compared to other biologically-abundant transition metal ions such as copper and zinc. By coupling two proteins, one that recognizes and oxidizes the divalent form (Fet3p from S. cerevisiae) and one that transports the trivalent form (Ftr1p from S. cerevisiae), an iron-selective assimilation pathway is created.
Translocation of iron by Fet3p/Ftr1p across the membrane is proposed to occur by an iron-channeling mechanism [109]. Fet3p, a multicopper oxidase (MCO), binds ferrous iron and after oxidation hands the ferric iron to Ftr1p, which transports ferric iron into the cytoplasm [110]. Without Fet3p, Ftr1p is unable to transport iron, and the maturation of these two proteins is coupled in the secretory pathway so that only the complex containing both proteins is present in the plasma membrane [111]. High-affinity, copper-dependent iron assimilation is not unique to fungi. Inducible copper-dependent iron uptake was characterized for Chlamydomonas [112] and the diatoms T. oceanica and T. pseudonana [113, 114]. Mammals do not have a protein equivalent to Ftr1p, but as iron is exported into circulation by ferroportin, it is oxidized by the multi-copper ferroxidase ceruloplasmin producing Fe(III) that then binds to transferrrin, the iron-delivery protein [115].
Fet3p contains four copper ions arranged in three different ligand fields, two mononuclear sites and one binuclear site. The sites differ by how the amino acid side groups are arranged around the copper ion(s) and the electronic structure of each site [116]. Additionally, one mononuclear site and the binuclear site are close enough to one another to form a trinuclear site. Multicopper ferroxidases are divided into two types; the Fet3p-type has three domains, while ceruloplasmin has six domains and two extra mononuclear copper sites. The multicopper ferroxidase from Chlamydomonas, FOX1, represents a third type that contains 6 domains like ceruloplasmin, but the domains are shuffled [117] (Figure 7). As observed for Fet3p, iron-deficiency leads to an increase in FOX1 protein (also referred to as FLP) [118] and mRNA abundance [112], while a knock-down of FOX1 gene expression specifically impairs the growth of Chlamydomonas in iron-deficient medium [119]. Further supporting a role in iron assimilation, FOX1 is localized to the plasma membrane [120]. There is a second FOX1-type MCO in Chlamydomonas, but this paralog, FOX2, is encoded by a low-abundant transcript, representing less than 1 mRNA per cell under iron-deficiency (Urzica and Merchant, unpublished).
Figure 7. Multi-copper ferroxidase domains.

The domain architecture of Fet3p from S. cerevisiae, ceruloplasmin (hCp) from human and FOX1 from Chlamydomonas according to [114] is shown. Each circle represents a cupredoxin domain. Residues from the red and blue domains form the trinuclear copper cluster. The blue domains contain a T1 copper cysteine, while the red domains have lost this cysteine and contain only T2 and T3 copper ligands. Domains that contain T1 copper sites and are involved in iron-binding have a dashed border.
FTR permeases contain seven potential TMs. REXXE motifs in TMs I and IV are proposed to be involved in iron translocation and the extracellular loop between TMs six and seven is required for trafficking of iron between the oxidase and the permease [121] (Figure 1). FTR1 homologs are commonly found in algal genomes except the prasinophytes and several chromalveolates (Table 1). In the absence of FOX1 and FTR1, prasinophyte green algae are proposed to use siderophore-dependent iron uptake [122]. The putative FTR1 genes in T. pseudonana are induced by iron-deficiency [99] as is the homolog in Chlamydomonas [112]. Algae with FTR-like permeases should also have partner MCOs. The green algae, Chlorella variabilis, Coccomyxa sp. C-169 and Volvox carteri have at least one FOX1 homolog and T. pseudonana, and Fragilariopsis cylindrus have MCOs more similar to Fet3p. Based on sequence similarity the putative MCO from Aureococcus anophagefferens is distinct from both FOX1 and Fet3p. As observed previously [101], an MCO was not found in the C. merolae genome, suggesting either that transport by FTR in this organism is not dependent on a ferroxidase or C. merolae has an unidentified, unrelated protein functioning as a ferroxidase.
In general, algal high-affinity iron transporters are structurally divergent from the characterized transporter from yeast, and not only have FOX1-like MCOs experienced unique domain evolution, but algal ferric transport may involve the novel FEA1-like proteins. The absence of a detectable MCO-encoding gene in the genome of the red alga C. merolae also provides another example of how algal iron transport is divergent from common reference organisms.
3.1.4. Algal-specific
In Chlamydomonas, the induced expression of FTR1 and FOX1 in response to iron-deficiency is accompanied by high expression of two algal-specific genes named FEA1 and FEA2 for Fe-assimilation 1 and 2, respectively [70]. Not only are the FEA1 and FEA2 genes highly inducible, but FEA1 and FEA2 are the major secreted proteins during iron-deficiency. Similar proteins have only been found in the genomes of other green algae (C. variabilis, Chlorococcum littorale (HCR1 [123]), Micromonas pusilla CCMP 1545, Ostreococcus lucimarinus, and V. carteri) and chromalveolates (A. anophagefferens, Ectocarpus siliculosus, Heterocapsa triquetra, P. tricornutum, T. pseudonana, F. cylindrus, and Emiliania huxleyi). Although the plastids of chromalveolates are proposed to be of red algal origin and FEA-like proteins are missing from C. merolae, a cryptic endosymbiont event is proposed to have occurred resulting in a substantial “green” contribution to the genomes of these organisms [26], which may explain the presence of FEA-like genes in chromaveolates.
Several lines of evidence have led to the conclusion that the FEA proteins are involved in iron assimilation, although the exact biochemical and molecular role of these proteins remains to be determined. Strains of Chlamydomonas that lack a cell wall secrete FEA1 and FEA2 into the medium, and these strains are more sensitive to iron deficiency compared to strains that have a cell wall and retain FEA1 and FEA2 in the periplasm [70]. Heterologous expression of FEA1 in Arabidopsis and yeast iron-uptake mutants can rescue growth on iron-deficient medium [124]. Uniquely, the FTR1-like protein from A. anophagefferens is a fusion between the FEA1 domain and a partial FTR domain. This protein has seven potential TMs; six TMs are in the FTR domain and contain the conserved REXXE motifs in TMs I and IV. The FEA1 domain is between TMs VI and VII, the typical location of the residues involved in iron trafficking between FTR1 and the oxidase, suggesting that the FEA1 domain could deliver iron to the FTR domain. However, the gene model should be verified to confirm whether the FTR1/FEA1 fusion protein is a genuine Rosetta stone protein. FEA1-like genes are present in the genomes of algae that do not have recognizable FTR1 homologs such as M. pusilla and O. lucimarinus. In these organisms, FEA may be able to facilitate iron transport by otherwise non-specific metal transporters such as permeases from the ZIP or NRAMP families.
3.2. Group B transporters
Once iron enters the cytoplasm, it must be trafficked to its sites of incorporation and, when in excess, transported to sites of storage. The major sinks for iron in algae are the bioenergetic membranes of the chloroplast and mitochondria, where iron is concentrated in the proteins of the electron transport chains. Compared to transport of iron across the plasma membrane, relatively little is known about how iron is trafficked to these compartments and even less is known in algae.
3.2.1. Chloroplast
The mechanism by which iron is transported into the chloroplast remains elusive. Active ferrous transport through the plastid inner membrane has been measured [125], and in Arabidopsis transport is dependent on the ferric reductase FRO7 [77], which implies transport of Fe(II). Arabidopsis PIC1 is proposed to be involved in plastid iron transport. A pic1 mutant exhibits defective chloroplast development, and both PIC1 and an ortholog, sll1656 from the cyanobacterium Synechocystis sp PCC 6803, rescue a yeast mutant defective in iron uptake [126, 127], but how PIC1 affects iron concentration in the chloroplast is not known. Not all algal genomes encode a PIC1 homolog, and PIC1 may actually be involved in translocation of proteins into the plastid [128], a function that could lead to a downstream effect on iron transport.
3.2.2. Mitochondria
The mitochondrial solute carrier (MSC) family is a large family of inner membrane transporters involved in the transport of multiple substrates between several compartments. In humans, this transporter family is referred to as SLC25 and is the largest SLC family (at least 43 members) [129]. Some members of the MSC family transport iron across the inner membrane of mitochondria. These include Mrs3p and Mrs4p in yeast [130, 131], mitoferrin-1 and -2 from zebrafish and mouse [132] and MIT from rice [133]. Deletion of rim2 also a member of the MSC family from yeast leads to mitochondrial iron defects, but this phenotype is attributed to defective pyrimidine transport rather than direct loss of iron transport [134].
A protein similarity network of the MSC family leads to the identification of a cluster of proteins that includes Mrs3/4, mitoferrins and MIT plus predicted iron transporters from plants and algae (Figure 8). The neighboring cluster (in grey) is composed of known S-adenosylmethionine transporters such as SAMC1 and SAMC2 from Arabidopsis and Pet8p from S. cerevisiae. The MTM1 cluster, also shown, is composed of proteins required for the activity of Mn-SOD in the mitochondrion (see section 6.2). The mitoferrin cluster includes three uncharacterized proteins from Arabidopsis At1g07025, At1g07030 and At2g30160 and at least one protein from each alga, including a Chlamydomonas protein which we have renamed MFL1 for MitoFerrin-Like 1. This MSC subgroup appears to be a fundamental characteristic of eukaryote genomes and could represent a ubiquitous mitochondrial iron transporter.
Figure 8. Protein similarity network of the MTM1, Mitoferrin and S-adenosylmethionine transporter subgroups of the MSC family.

Nodes representing proteins that belong to the S-adenosylmethionine transporter cluster are colored grey. Proteins from Arabidopsis, human, Chlamydomonas and S. cerevisiae in the MTM1 or Mitoferrin clusters are listed under the subgroup name and the corresponding nodes are labeled.
Additional mitochondrial iron transporters include members of the CDF family (described in section 5.2.1.). Mmt1p and Mmt2p from S. cerevisiae are closely related to FieF/YiiP from Escherichia coli, a plasma-membrane-localized metal-efflux protein. In vivo studies with deletion and over-expression strains have linked FieF/YiiP to iron efflux, while in vitro studies with membrane vesicles have linked FieF/YiiP to zinc efflux [135]. In S. cerevisiae, over-expression of MMT1 or MMT2 (referred to as MFT1 and MFT2) increases mitochondrial iron content, while deletion of both genes causes sensitivity to the iron-chelator bathophenanthroline sulfonate [136]. While these CDF proteins are found in yeast, land plants (MTP6 from Arabidopsis), and photosynthetic and non-photosynthetic stramenopiles, they are missing from humans and most algae. An interesting exception is Micromonas sp. RCC299, the only sequenced chlorophyte with a FieF-like CDF protein. Unfortunately, the sequenced Micromonas genomes are surprisingly divergent [137], and the significance of Micromonas sp. RCC299 having a FieF-like CDF is currently unknown.
3.2.3. Vacuole
In Arabidopsis, iron is loaded into the vacuole by VIT1 [138], a homologue of the yeast iron/manganese vacuole transporter Ccc1p [139]. A second Arabidopsis protein, FPN2/IREG2, also mediates transport of iron into the vacuole. Unlike VIT1, FPN2/IREG2 expression is regulated by iron status, and the protein pumps iron into the vacuole during iron-deficiency. This counterintuitive activity is proposed to buffer the uptake of iron by AtIRT1 in the root [140]. FPN2/IREG2 is also linked to cobalt and nickel homeostasis[105, 140], which points to a general role in metal sequestration. Additional proteins in Arabidopsis referred to as Nodulin-21-like proteins share sequence similarity with Ccc1p and VIT1 and are regulated by iron status, but their functions remain unknown [141]. The Nodulin-21-like proteins from Arabidopsis and orthologs from other plants form a distinct cluster from Ccc1 and VIT1 in the protein similarity network (Figure 9). All algal genomes analyzed encode at least one Ccc1p/VIT1-like protein and the VIT1 homolog from C. merolae was identified in purified acidic vacuoles (acidocalcisomes) [142]. But while the C. merolae protein clusters with Ccc1p and VIT1, the Chlamydomonas proteins (renamed CVL1 and CVL2 for Ccc1/Vit1-Like) cluster with Pcl1 from Schizosaccharomyces pombe (Figure 9). Pcl1 is also linked to iron homeostasis since the corresponding gene is down-regulated during iron-limitation [143], and Δpcl1 accumulates less iron that WT [144]. Whether Pcl1 and the Chlamydomonas homologs localize to the vacuole like Ccc1p and VIT1 do is yet to be determined.
Figure 9. Protein similarity network of the Ccc1p and VIT1 family.
The similarity network shows three main clusters. Each cluster contains proteins linked to iron homeostasis. Nodes representing Ccc1p from S. cerevisiae, VIT1, Nodulin-like1 (NL1), Nodulin-like2 (NL2), Nodulin-like3 (NL3), Nodulin-like4 (NL4) and Nodulin-like21 (NL21) from Arabidopsis, Pcl1 from S. pompe and CVL1 and CVL2 from Chlamydomonas are labeled. Animal sequences belonging to this family were not found.
4. Copper
4.1. Group A transport family CTR
In eukaryotes, high-affinity copper assimilation is mediated by members of the CTR family (also known as SLC31). These permeases contain three putative TMs and an extracellular N-terminus with organism-specific ratios of methionine, histidine and cysteine that are involved in Cu(I) binding [145] (Figure 1). Transport by CTR is dependent on reduction of copper in the periplasm by a cupric reductase. Upon entering the cytoplasm, small molecules and proteins sequester the copper ions, and the resulting concentration gradient drives transport by CTR. The unique +1 oxidation state of copper lends substrate specificity to CTR [146, 147], but CTR proteins can also mediate the transport of Ag(I) [147] and are thought to be a route of silver toxicity [148, 149]. In addition to uptake, low-affinity CTRs mediate the transport of copper from intracellular stores [150–153].
As is the case for many eukaryotic metal transporters, CTR was discovered in S. cerevisiae (Ctr1p) but unusually, the gene was found with a screen designed to identify mutants with defective iron transport [154]. The reason why disruption of CTR1 led to an iron uptake defect was explained by the concurrent discovery of Fet3p the multicopper ferroxidase required for iron transport [108] (discussed in section 3.1.3.): loading of copper into Fet3p is dependent on copper transport by Ctr1p, therefore copper assimilation is a prerequisite for iron transport in yeast.
All algal genomes analyzed contain at least one CTR gene. An exception is A. anophagefferens, even though this organism is predicted to have more copper-dependent proteins than other closely-related sequenced algae [155]. However, A. anophagefferens does not have a plastocyanin gene and the abundance of copper-dependent proteins in the proteome is not known; perhaps, uptake of copper by low-affinity transporters is sufficient. The size of algal CTRs compared to homologs from plants, fungi and animals suggests an enrichment of assimilative transporters; CTRs on the plasma membrane tend to have an extended N-terminus with more metal binding motifs presumably increasing chelating capacity and therefore copper affinity.
The Chlamydomonas genome contains four genes encoding CTRs: CTR1, CTR2, CTR3, and COPT1. COPT1 is relatively small and more similar to plant and animal proteins than to other algal sequences. COPT1-like proteins are also found in the green algae Coccomyxa sp. C-169 and Micromonas spp. and may be involved in intracellular trafficking of copper. COPT1 transcript is also the least abundant mRNA among the CTR family genes both during copper-replete and copper-deficient growth conditions [3] (Figure10A). Chlamydomonas CTR1, CTR2 and CTR3 are 3 times the size of COPT1 and contain multiple putative copper binding sites indicative of copper assimilation. Unlike COPT1, CTR1 – CTR3 are induced by copper-deficiency, but only CTR1 and CTR2 were localized to the plasma membrane by biochemical fractionation, indicating that CTR1 and CTR2 are the major copper assimilation permeases in Chlamydomonas [146]. CTR3 may have arisen from a partial duplication event of CTR2 and is found in the soluble proteome [3, 146]. The genomes of the green algae V. carteri and C. variabilis also contain CTR3-like genes that encode a putatively soluble protein. CTR3 may act as a periplasmic copper chaperone delivering copper to CTR1 and CTR2. The similarity between CTR3 and CTR1/2 is reminiscent of the sequence similarity between characterized Atx1-like copper chaperones and the N-termini of P1B-type ATPases.
Figure 10. Abundance of Chlamydomonas CTR family and putative ferric reductase transcripts.
A) Pie charts show the contribution of CTR1, CTR2, CTR3 and COPT1 transcript abundance during copper-deficient and copper-replete growth conditions. The sizes of each chart represent the relative total transcript abundance. B) Pie charts show the contribution of putative ferric reductase transcript abundance during copper-deficient and copper-replete growth conditions. The sizes of each chart represent the relative total transcript abundance.
Since CTR permeases transport copper in the +1 oxidation state, copper must be reduced prior to transport in aerobic environments. Ferric reductases on the plasma membrane can also reduce cupric copper. Iron- and copper-deficiency independently induce FRE1 expression, a gene encoding a cytochrome b558 NADPH oxidase from S. cerevisiae (see section 3.1.1.). The CTR gene from the red alga C. merolae uniquely encodes a cytochrome b561 domain that may function as the cupric reductase. In Chlamydomonas, cupric reductase activity is induced in copper-deficient Chlamydomonas cells by two fold [156]. While the abundance of the putative metal reductase-encoding transcripts RBOL2 (respiratory burst oxidase like 2) and CBD1 (Cre14.g609900; Cytochrome b561 DOMON 1) are increased about five fold in copper-deficient versus copper-replete growth conditions (Figure 10B), they are represented by only about one mRNA per cell during copper-deficiency [3]. The presence of a copper chaperone, CTR3, may increase the Cu(I) chelating capacity of the periplasmic space and reduce dependency on cupric reduction.
4.2. Intracellular distribution of copper
In addition to catalyzing potentially harmful reactions, copper has a high affinity for the side chains of metal-binding amino acids, and toxicity can arise through mis-incorporation of copper into proteins. Therefore, copper is delivered by metallochaperones [157] from its site of uptake on the plasma membrane to copper-dependent enzymes or additional transporters. In vitro metal transfer between S. cerevisiae Ctr1p and the metallochaperone Atx1p [158]supports a model where trafficking of copper within the cell is controlled through protein-protein interactions.
4.2.1. Group B transport family P1B-type ATPases
If the final destination of copper is an organelle, then metallochaperones will hand off their cargo to P1B-type ATPases in the organellar membrane [159]. An exception is cytochrome oxidase assembly, which requires the cytosolic copper chaperone COX17 to interact with additional chaperones in the mitochondrion forgoing contact with a distributive transporter [160, 161]. PIB-type ATPases contain six or eight putative TMs and use the phosphorylation of an aspartic acid residue in the ATPase domain to cause structural changes that drive metal transport. The P1B-type ATPases were first recognized as copper transporters because ATP7A and ATP7B in humans lead to inherited copper metabolism disorders (reviewed in [162]). These proteins are located in the trans-Golgi compartment [163], where they are responsible for providing copper to cuproproteins in the secretory pathway, but during copper excess, they move to the plasma membrane and mediate copper efflux [164].The protein similarity network for this family contains three distinct clusters, and clusters 1 and 3 contain members implicated in intracellular copper transport (Figure 11).
Figure 11. Protein similarity network of the P1B-type ATPase (HMA) family.

A) The similarity network shows three main clusters. Proteins from Arabidopsis, human, Chlamydomonas, and S. cerevisiae in each cluster are listed under the subgroup name. The nodes themselves are labeled following the convention described in the legend to Figure 4. In addition, the PIB-type ATPases from E. coli, ZntA and CopA, are labeled. B) A separate network was built with the known and putative Cu-transporters from (A). The E-value used in part A as a similarity cutoff was lowered, leading to the loss of connections between less similar proteins. Because of the higher stringency, Cu transporters localized to the chloroplast and those found in cyanobacteria separate from Cu transporters localized to the secretory system and those from noncyanobacterial bacteria, forming two clusters.
Cluster 1 can be further divided into two subgroups: Cu-A contains proteins localized to the chloroplast such as PAA1 and PAA2 from Arabidopsis (Figure 11B), and Cu-B contains proteins localized to the secretory pathway such as ATP7A and ATP8A. The chloroplast subgroup is shared among land plants, green algae and cyanobacteria but missing from C. merolae and most of the sequenced chromalveolates. Plastocyanin is a soluble protein in the thylakoid lumen, and copper must be pumped across two plastid membranes. In Arabidopsis, PAA1 localizes to the inner membrane of the chloroplast and pumps copper into the stroma [165], while PAA2 localizes to the thylakoid membrane and pumps copper into the lumen [166]. The function of these transporters in copper transport is supported in part by the observation that a paa1 mutant is defective in plastocyanin maturation, which occurs in the thylakoid lumen, and stroma-localized Cu/Zn SOD activity, while a paa2 mutant is only defective in plastocyanin maturation [166]. C. merolae does not have a gene encoding plastocyanin or a gene for Cu/Zn SOD, and orthologs of either PAA1 or PAA2 are not found. Since most chromalveolates do have a Cu/Zn SOD, the absence of a PAA1 ortholog would suggest that this SOD is not localized to the chloroplast. Only E. siliculosus has both a Cu/Zn SOD and a single transporter from the Cu-A subgroup. Two chromalveolates, E. huxleyi and F. cylindrus have plastocyanin (top BLAST hit for both proteins is the plastocyanin from the diatom T. oceanic), but genes encoding Cu-A subgroup proteins were not found in their genomes. Therefore, if plastocyanin from E. huxleyi and F. cylindrus localizes to the chloroplast, then the pathway for copper transport to plastocyanin may have evolved independently in these algae.
Cluster 3 contains proteins from organisms in Archaeplastida. The one characterized member, HMA1 from Arabidopsis, localizes to the chloroplast envelope, but the function of this protein remains controversial. An initial study of HMA1 suggested that it transports copper into the stroma [166], but a subsequent study proposed that HMA1 transports zinc out of the stroma into the cytoplasm [167].
Three proteins, multi-copper ferroxidase (FOX1, on the plasma membrane), cytochrome oxidase (COX2A/B, in the inner mitochondrial membrane) and plastocyanin (PCY1, in the thylakoid lumen) make up the major portion of the copper quota in Chlamydomonas under copper-replete growth conditions [3, 168]. ATX1 is predicted to deliver copper to the P1B-type ATPase CTP1 in the trans-Golgi compartment where FOX1 is matured, as has been shown for orthologous proteins from S. cerevisiae and mammals [169] (Figure 3). The function of Chlamydomonas ATX1 is supported by functional complementation studies; expression of ATX1 can rescue growth of an atx1Δ strain of S. cerevisiae on iron-depleted medium [112]. CTP1 belongs to the Cu-B subgroup in the similarity network and is similar to Ccc2p, which is responsible for providing copper to Fet3p, the FOX1 equivalent from S. cerevisea. The C. merolae genome does not encode a FOX1 homolog and a gene encoding ATX1 was not found, but it does have a P1B-type ATPase similar to ATP7B, which could be involved in copper efflux. Based on similarity to PAA1 and PAA2, the Chlamydomonas protein CTP2 is predicted to pump copper across the inner membrane and the fourth identified P1B-type ATPase, renamed CTP4, is predicted to pump copper across the thylakoid membrane (Figure 3). Since the Chlamydomonas genome lacks a Cu/Zn SOD-encoding gene, the main functions of CTP2 and CTP4 are predicted to be loading of plastocyanin with copper. A fourth P1B-type ATPase is encoded in the Chlamdyomonas genome. This protein, CTP3, is similar to HMA5 from Arabidopsis (Figure 11) and therefore may be involved in detoxifying copper through the secretory pathway (Figure 3).
A chaperone for delivery of copper to plastocyanin has not been characterized but a transcriptomic analysis of Cu-deficient Chlamydomonas cultures identified PCC1 as a likely candidate [3]. PCC1 shares sequence similarity with ATX1 including a conserved copper-binding motif, and PCC1 orthologs are found in all currently sequenced green algal genomes but not the genomes of C. merolae (lacks plastocyanin), chromalveolates (most lack plastocyanin) or land plants. By expressing two copper chaperones, Chlamydomonas may be able to differentially direct copper to plastocyanin versus FOX1 [3].
5. Zinc
Zinc is one of the most abundant trace metal nutrients in biology. This metal ion is prevalent in hydrolytic enzymes, where it provides a chemical functionality (electrophile) that is not easily provided with amino acid side chains, and because zinc lacks redox chemistry, it is commonly used to stabilize protein structures. Eukaryotes in particular have taken advantage of the latter feature leading to an expansion of zinc proteins, particularly zinc-finger transcription factors, in their proteomes [170].
5.1. Group A
5.1.1. ZIP
In eukaryotes, members of the ZIP family mediate zinc transport across the plasma membrane. As expected from the fundamental role in zinc assimilation, all sequenced algal genomes contain ZIP genes, but the green algae C. variabilis and Coccomyxa sp. C-169 are the only algae that contain ZIP genes from all subfamilies (see section 3.1.2.1.). ZIP transporters from algae are expected to have similar roles as have been found for other organisms, such as zinc assimilation and intracellular distribution. There are 14 ZIP family proteins encoded in the Chlamydomonas genome; one of which (ZIP14) only shows remote similarity to the family [101]. Expression of the genes encoding the low- and high-affinity zinc transporters Zrt1p and Zrt2p from S. cerevisiae is inducible by zinc-deficiency, and assimilative zinc transporters in mammals are also responsive to zinc nutrition. In contrast, the iron transporter-encoding gene IRT1 from Arabidopsis is regulated by iron nutrition. Therefore, metal responsiveness to iron-deficiency and zinc-deficiency at the transcription level was assessed for the 13 ZIP genes in Chlamydomonas leading to the present nomenclature [70] (Malasarn and Merchant, unpublished) (Urzica and Merchant, unpublished) (Table 2). Of the ZIP genes with increased mRNA abundance during zinc-deficiency, ZRT1, ZRT2, ZRT3 and ZRT5 cluster with subfamily I proteins and are similar to the assimilative zinc transporters Zrt1p and Zrt2p from S. cerevisiae and OsZIP4 [171] and OsZIP5 [172] from rice (Figure 4). ZRT4 is a member of subfamily II and is similar to ZIP1 – ZIP3 from human. Based on transcript abundance, ZRT1 appears to be the main assimilative zinc transporter during zinc-deficiency (Figure 5B).
Table 2.
Proteins involved in metal transport in Chlamydomonas.
| Protein Family | Putative Role | Subgroup | Name& | Gene Model | Other Names | Ref.# |
|---|---|---|---|---|---|---|
|
Group A
| ||||||
| Zn transport | Subfamily I | ZRT1 | Cre07.g351950 | CrZIP1 | [100],a | |
| Subfamily I | ZRT2 | Cre01.g000150 | CrZIP2 | [100],a | ||
| Subfamily I | ZRT3 | Cre13.g573950 | CrZIP3 | [100],a | ||
| Subfamily II | ZRT4 | Cre06.g299600 | CrZIP6 | a | ||
| Subfamily I | ZRT5 | Cre07.g355150 | CrZIP4 | [100],a | ||
|
|
||||||
| ZIP | unknown | Subfamily I | ZIP1 | Cre07.g355100 | CrZIP5 | [100],a |
| Subfamily II | ZIP2 | Cre13.g576050 | CrZIP13 | [100],a | ||
| GufA | ZIP3 | CreO3.g189550 | CrZIP8 | [100],a | ||
| GufA | ZIP4 | CreO2.g127650 | CrZIP12 | [100],a | ||
| GufA | ZIP6 | Cre23.g766350 | CrZIP9 | a | ||
| ZTP50 | ZIP7 | Cre06.g281900 | CrZIP7 | a | ||
|
|
||||||
| Fe transport | Gufa | IRT1 | Cre12.g530400 | CrZIP11 | [70] | |
| IRT2 | Cre12.g530350 | CrZIP10 | [70] | |||
|
| ||||||
| NRAMP | Fe/Mn transport | MntH - C | NRAMP1 | Cre17.g707700 | DMT1 | [201,204] |
| MntH - C | NRAMP2 | Cre07.g321950* | [201] | |||
| Eukaryotic I | NRAMP4 | Cre05.g248300* | ||||
|
| ||||||
| FTR | Fe transport | FTR1 | CreO3.g192050 | [117] | ||
|
| ||||||
| MCO | Fe transport | FOX1 | Cre09.g393150* | [117,119] | ||
|
| ||||||
| CTR | Cu assimilation | CTR1 | Cre13.g570600 | [146] | ||
| CTR2 | Cre10.g434350 | [146] | ||||
|
| ||||||
| Putative Cu chaperone | CTR3 | Cre10.g434650 | [146] | |||
|
| ||||||
| Cu distribution | COPT1 | Cre05.g247050 | [146] | |||
|
| ||||||
|
Group B
| ||||||
| MSC | Fe transport | Mitoferrin | MFL1 | Cre16.g695950 | ||
|
| ||||||
| Mn transport | MTM1 | MML1 | Cre06.g267800 | |||
|
| ||||||
| Ccc1/VIT1 | Fe transport | Pcl1 | CVL1 | Cre02.g099500 | ||
| CVL2 | CreO2.g107550 | |||||
|
| ||||||
| P1B-type ATPase | Cu transport | Cu-B | CTP1 | Cre01.g064350* | CrHMA2 | |
| Cu-B | CTP3 | Cre09.g406400* | ||||
| Cu-A | CTP2 | Cre10.g424800 | CrHMA3 | |||
| Cu-A | CTP4 | Cre10.g422200 | ||||
|
|
||||||
| Unknown | HMA1 | HMA1 | Cre17.g720400 | CrHMA1 | ||
|
| ||||||
| ATX1 | Cu chaperone | ATX1 | CreO2.g128200 | [112] | ||
| PCC1 | Cre05.g248600 | [3] | ||||
|
| ||||||
| CDF | Zn transport | Zn | MTP1 | Cre01.g058300 | [201] | |
|
| ||||||
| Mn transport | Mn | MTP2 | CreO3.g160800 | CrMTP8 | [201] | |
| MTP3 | CreO3.g160750 | CrMTP8.1 | [201] | |||
| MTP4 | CreO3.g160550 | CrMTP8.2 | [201] | |||
|
| ||||||
| unknown | ZnT9/AtMTP7 | MTP5 | Cre06.g289150 | CrMTP7 | [201] | |
Protein names in bold are new annotations.
References are given for proteins associated with experimental data.
Davin Malasarn and SM, unpublished.
Because of gaps in the genome assembly, these gene models are incorrect and were manually curated using ESTs before inclusion in the protein similarity network.
In addition to the plasma membrane, ZIP permeases are also found in intracellular membranes. ZIP-mediated efflux of zinc across the membranes of the Golgi, endoplasmic reticulum and vacuole has been characterized [85, 129, 173]. Although the consequence of ZIP activity is usually zinc transport into the cytoplasm, Yke4p, a yeast transporter, is bi-directional and mediates zinc transport into or out of the endoplasmic reticulum depending on the zinc status of the cell [173], and GmZIP1 from soybean can transport zinc out of the cytoplasm into a specialized nitrogen-fixing compartment called a symbiosome [174]. As passive transporters, ZIP activity would be highly dependent on metal speciation within the cell and structural determinants are only part of the equation.
5.2. Group B
5.2.1. CDF
Most CDF proteins are composed of six putative TMs and transport metal out of the cytoplasm and into intracellular compartments (Figure 1). The term “cation diffusion facilitator” was coined to refer to four zinc, cobalt and cadmium transporters in bacteria and S. cerevisiae with shared sequence similarity and roles in metal tolerance [175]. Although called facilitators, these transporters are actually antiporters and rely on a proton gradient across the membrane to mediate efflux of metal. Characterization of bacterial CDF proteins, specifically YiiP/FieF from E. coli, has provided novel insight into the transport mechanism and how the cytoplasmic concentration of metal can regulate transport activity [176, 177].The first CDF gene from mammals, ZnT-1, was discovered in a complementation assay of a cell line that displayed increased sensitivity to zinc [178]. Ten ZnT-like proteins (also referred to as SLC30) have now been described in mammals, most of which have roles in zinc efflux [179]. ZAT1 (Zn transporter of Arabidopsis thaliana; also referred to as CDF1) was the first member of the CDF family found in a plant [180] and an ensuing analysis of ZAT-like genes in plants resulted in the renaming of ZAT to AtMTP1 and creation of the MTP (Metal Tolerance Protein) family of plant CDF proteins [181].
Like the ZIP family, CDF proteins can transport multiple divalent metal cations, but the family can be divided phylogenetically into the zinc, manganese and zinc/iron permease groups [182]. Characterized plant and fungal members of the zinc subgroup are involved in sequestering zinc in the vacuole [85, 183, 184], while mammalian ZnT proteins are found in the plasma membrane plus various intracellular compartments such as the vacuole and secretory vesicles [179]. Although most CDF proteins are involved in tolerance to excess metal by mediating efflux across the plasma membrane or into the vacuole, some of these transporters are more abundant during zinc deficiency. These proteins include Zrc1p, Msc2p and Zrg17p from S. cerevisia e[185, 186], ZnT-5 and ZnT-7 from humans [187, 188] and AtMTP2 from Arabidopsis [189]. Zrc1p localizes to the vacuole and protects against zinc shock, which can occur when the cell transitions from a zinc-deficient environment to a zinc-replete environment [190]. Msc2p, Zrg17p, ZnT-5 and ZnT-7 transport zinc into the ER or Golgi during zinc-deficiency and participate in the unfolded protein response (UPR). By analogy, AtMTP2 may also sequester zinc within an intracellular compartment.
In comparison to plants and mammals, algae have few genes encoding CDF proteins. MTP1 is the only putative zinc-specific CDF protein in Chlamydomonas [101] Since MTP1is induced by Zn-deficiency (Malasarn and Merchant, unpublished), this putative zinc transporter could be involved in pumping zinc into either the vacuole or endomembrane system. The red alga C. merolae has two MTP1-like CDF proteins, and one, CMF058C, was identified in purified acidocalcisomes [142]. Acidocalcisomes are calcium- and inorganic phosphate-rich acidic vacuoles that can contain various metal ions including zinc [191]. Therefore, CMF058C is likely involved in zinc detoxification by pumping zinc into acidocalcisomes. The group 3 green algae M. pusilla CCMP1545, O. lucimarinus and Ostreococcus tauri are missing a zinc-specific CDF, indicating loss in only a subset of prasinophytes (marine green alga). This gene loss may relate to the metal content of their natural environment. The fact that zinc-sparing mechanisms, such as replacement of zinc-dependent carbonic anhydrases with cobalt- or cadmium-containing enzymes [192, 193], occur in nature indicates that portions of the ocean may be zinc-limited, just as portions are iron-limited [194, 195]. Algae in such zinc-limited environments may not need CDF proteins in zinc homeostasis.
5.2.2. P1B-type ATPases
Besides their role in copper efflux (see section 4.2.1), P1B-type ATPases can efflux zinc out of the cytoplasm, either into the vacuole as described for Arabidopsis HMA3 [196] or across the plasma membrane as described for Arabidopsis HMA2 and HMA4 [197]. Both HMA3, HMA4 and HMA2 belong to cluster 2 in the protein similarity network (Figure 11). Like the green algae C. variabilis and V. carteri and the red alga C. merolae, Chlamydomonas does not have an HMA3- or HMA2-like P1B-type ATPase, while homologs are present in the prasinophytes and most chromalveolates. A second cluster similar to the HMA2/HMA3 cluster includes the Arabidopsis protein HMA1, which is also involved in tolerance to excess zinc but is localized to the chloroplast membrane [167]. The green algae and red alga that are missing a HMA2/HMA3-like protein contain a HMA1-like protein instead, while prasinophytes have both. The HMA1 group, however, is specific to the Archaeplastida lineage (see section 4.2.1).
6. Manganese
Manganese is essential for phototrophic growth. The two main sites of manganese utilization in algae are the polynuclear cluster of photosystem II and manganese-dependent superoxide dismutases (Mn-SOD). As shown in Table 1, while the presence of genes encoding Cu/Zn-SOD and Fe-SOD can vary from one alga to another, all sequenced algae have at least one Mn-SOD and one of the five Mn-SODs (MSD1 – MSD5) in Chlamydomonas is in the mitochondria [198] as is the case for most eukaryotes. In the diatom T. pseudonana, Mn-SOD is the dominant SOD in the cell and is localized to the chloroplast [199]. Reliance on Mn-SOD instead of the Fe-SOD, also encoded in the genome, is proposed to be an adaptation to low iron in T. pseudonana. In addition to the five Mn-SODs, Chlamydomonas has a single Fe-SOD, but unlike in T. pseudonana, it is the dominant SOD in the chloroplast under standard laboratory conditions [198]. Under iron deficiency, however, MSD3 is induced ~103 fold compared to iron-replete growth conditions, suggesting that expression of MSD3 is part of a low-iron-acclimation strategy [200].
6.1. Group A manganese transporters
In most organisms, the predominant route for manganese uptake is proposed to involve members of the NRAMP family. AtNRAMP1 from Arabidopsis is localized to the plasma membrane and expression is induced by manganese deficiency [201]. The S. cerevisiae NRAMP, Smf1p, is also localized to the plasma membrane, and the presence of manganese (and to a lesser extent iron) causes targeting to and subsequent degradation of the transporter in the vacuole [202]. The expression of two NRAMP genes (encoding NRAMP1 and NRAMP2) is induced by manganese limitation in Chlamydomonas, about 8- and 3-fold, respectively [200]. NRAMP1 was able to rescue a yeast strain deficient in manganese transport and partially rescue a strain deficient in iron uptake [203], but the locations of these transporters and their functions within the algal cell remain unknown. As mentioned in section 3.1.2.2., the developmental stage determines the physiological role of AtNRAMP3 and AtNRAMP4; they are involved in iron homeostasis in seeds, but manganese homeostasis in adult leaves. Whether analogous dual-function NRAMPs exist in algae is not known.
In contrast to the putative iron transporter NRAMP4 which clusters with eukaryotic NRAMP proteins, NRAMP1 and NRAMP2 cluster with bacterial transporters (Figure 6). The bacterial NRAMP, often referred to as MntH, was first described in E. coli, and although it can transport iron, it has a preference for manganese [204]. The MntH subgroup C is predominately composed of prokaryotic NRAMPs but contains several eukaryotic proteins in addition to Chlamydomonas NRAMP1 and NRAMP2: two homologs from Physcomitrella patens, one from V. carteri, one from Coccomyxa sp. C-169 and one from C. merolae. NRAMPs from other green algae belong solely to the eukaryotic groups, and most are more similar to animal NRAMPs than to plant or algal proteins.
As in yeast [205], Mn(II) may also enter the algal cell as MnHPO4 via a phosphate transporter. Manganese deficiency results in a lower phosphate quota in Chlamydomonas, suggesting that manganese may be the preferred counter ion for a phosphate transporter, and expression of PTA3 and PTA4, encoding putative phosphate transporters, are induced by manganese-deficiency [200].
6.2. Group B manganese transporters
In S. cerevisiae, the Golgi plays an important role in manganese trafficking to Mn-SOD in the mitochondrion. The NRAMP protein Smf2p pumps manganese out of Golgi-like vesicles [206] and is essential for activation of Mn-SOD in the mitochondria [207, 208]. Mn-SOD metallation and activity in the mitochondria also require the presence of a mitochondrial carrier protein, Mtm1p, which may serve as a manganese transporter in the mitochondrial membrane [208, 209]. However, deletion of MTM1 does not affect the concentration of manganese in the mitochondria, suggesting that if it is a manganese transporter then it is specific for maturation of Mn-SOD. A distinct cluster containing Mtm1p and the functional ortholog AtMTM1 from Arabidopsis is present in the protein similarity network of the mitochondrial solute carrier family (Figure 8). All sequenced algae have a Mn-SOD and an Mtm1p-like protein, except A. anophagefferens, which does not have a recognizable Mtm1p-like protein as determined by the cluster analysis. SLC25A39 and SLC25A40 from human and a second Arabidopsis protein At2g46320 also clustered with Mtm1p and AtMTM1. We have renamed the Chlamydomonas protein MML1 for MTM1-Like 1. The presence of genes encoding members this transport family subgroup suggests that, like in yeast, Mn-SOD localizes to the mitochondria in most algae.
Members of the CDF family are also predicted to play a role in manganese transport. Based on phylogenetic relationships, the Chlamydomonas CDF proteins MTP2, MTP3 and MTP4 are predicted to transport manganese [182, 210] (see section 5.2.1.), while MTP2 and MTP5 transcripts are 3 and 5 fold more abundant in manganese-limited medium than manganese-replete [200]. MTP5 is part of the ZnT9/AtMTP7 cluster in the CDF protein network (Figure 12) and proteins from this subgroup are uncharacterized. ZnT9 (also referred to as HUEL; Human Embryonic Lung) was localized to the cytoplasm and nucleus depending on the cell-cycle and is proposed to be involved in DNA synthesis or transcriptional regulation because of the presence of a DNA repair motif [211]. While ZnT9-like proteins are found in animals, plants and algae, the partial DNA repair domain is only conserved in animals and the alveolate Perkinsus marinus. Like the EIN2 proteins from plants, that have an NRAMP domain but function in a signaling cascade, ZnT9-like proteins may have adopted the metal transport domain for a function other than metal trafficking.
Figure 12. Protein similarity network of the CDF family.
The similarity network shows four main clusters. Proteins from Arabidopsis, human, Chlamydomonas, and S. cerevisiae in each cluster are listed under the subgroup name. The nodes themselves are labeled following the convention described in the legend to figure 4. In addition, the CDF proteins from E. coli, FieF and ZitB, are labeled.
7. Nickel and cobalt
Nickel and cobalt-dependent enzymes are relatively rare. For several organisms, enzymes that are dependent on these metals have become redundant by replacement with analogous enzymes, such as substitution of B12 (cobalt)-dependent methionine synthase with a B12-independent enzyme. B12-dependent enzymes appear to have been lost by all land plants, most unicellular eukaryotes and insects [212]. Even eukaryotes that use B12-dependent enzymes do not need to transport Co(II) since these organisms obtain the porphyrin-bound form (B12) from their diet. Nickel does need to be transported by organisms that contain urease, which includes plants and many algae except the non-chromeolovate algae Chlamydomonas, V. carteri, Coccomyxa sp. C-169, C. variabilis, C. merolae and M. pusilla CCMP 1545. Although nickel-enzymes were not found to be encoded in these algal genomes (as of 2/2012), proteins with similarity to the assimilative nickel transporter NicO were found. In prokaryotes, the NicO family is composed of a high-affinity nickel/cobalt transporter, which supplies metal for cobalamin biosynthesis, Ni-Fe hydrogenase, Ni-dependent superoxide dismutase and/or Ni-urease. The presence of NicO in the genomes of algae that lack Ni-dependent enzymes suggests that either there is an unidentified nickel enzyme or NicO transports a different metal.
In the absence of a nutritional requirement for nickel and cobalt, NicO homologs in algae could be iron transporters in the thylakoid membrane. A NicO homolog (named ZN) is responsible for the zebra-necrosis mutant phenotype in plants, which is a green/yellow striped pattern on the leaf caused by defective chloroplast biogenesis [213]. ZN localizes to the thylakoid membrane in rice and in Arabidopsis it may interact with PIC1, which has been implicated in plastid iron homeostasis [127]. In Chlamydomonas, transcript abundance of NicO increases with decreasing iron concentration but not in zinc- or copper-deficiency (Urzica, Malasarn and Merchant, unpublished). Therefore, NicO is an appealing candidate for plastid iron transport.
8. Concluding remarks
Algae represent a relatively unexplored group of photosynthetic eukaryotes with respect to molecular biology and biochemistry. They are ideal for comparative studies of trace metal homeostasis, and the genome is an accessible tool for initiating such an analysis. As with other genomes, however, most genes are uncharacterized, and the first challenge is to predict a function for each gene. For protein families with multiple functions, protein similarity networks can be used in a first-level functional assessment in which an uncharacterized protein is grouped with characterized or partially characterized proteins. In some cases, the function of one or more characterized proteins can be extrapolated to uncharacterized proteins in the same cluster. Using this approach, we have identified six new putative metal transporters in Chlamydomonas. We predict NRAMP4 and MFL1 are involved in iron homeostasis, while CVL1 and CVL2 may be involved in iron distribution either into the vacuole or into another intracellular compartment. MML1 could be a mitochondrion-localized transporter, and from the similarity network of the MSC family, it is apparent that the MTM1 cluster (to which MML1 belongs) is similar to the mitoferrin cluster (to which MFL1 belongs) (Figure 8). Therefore, MML1 could also be a metal transporter, except unlike mitoferrin, MML1 may transport manganese. We predict CTP4 localizes to the thylakoid membrane and provides copper for plastocyanin, while the previously identified CTP3 is predicted to localize to the secretory system.
The presence and absence of proteins involved in metal homeostasis can provide the initial clues as to how algae are able to thrive in diverse environments with unique geochemistries. In terms of metal metabolism, algae have taken divergent paths, which is explicit from the presence of iron- or copper-containing proteins and types of metal transporters. For instance, the red alga C. merolae and some chromalveolates do not have plastocyanin and accordingly do not have recognizable orthologs PAA1 or PAA2. We hypothesize that chromalveolates that have plastocyanin but not PAA1 or PAA2 orthologs may have a novel route for plastocyanin metallation.
By examining the presence of metal transporters, we can also gain insight into other aspects of metal metabolism, such as the subcellular location of the different types of SODs. The presence of MTM1 orthologs and at least one Mn-SOD suggests that all algae have a Mn-SOD localized to the mitochondria. The absence of PAA1 orthologs but the presence of Cu/Zn SODs supports the notion that, in contrast to land plants, the Cu/Zn SOD in most chromalveolates is not localized to the chloroplast.
Two putative transport proteins with novel domains were also identified. The fusion of a cytochrome b561 domain to the CTR protein of C. merolae suggests that a cytochrome b561 protein may be responsible for copper reduction in other algae. The fusion of FEA1 and FTR domains in an A. anophagefferens protein supports the role of FEA1 in iron assimilation, and the FEA1 domain appears to be on the periplasmic side of the membrane and could, therefore, bind ferrous iron and transfer it to the permease. Assuming the FEA1-FTR1 fusion is genuine and not due to an incorrect gene model, the combination of both domains in one protein indicates that FEA1 performs a supportive role in iron transport. In algae where it is a separate protein, FEA1 may function to deliver iron to a permease.
Supplementary Material
Protein similarity network of cytochrome c6 and cytochrome c6-like proteins encoded in the genomes of selected photosynthetic organisms.
Highlights
Comparative genomic approach reveals the diversity of metal metabolism among algae.
Algal genomes encode transporters related to plants, animals and bacteria.
Similarity networks are used to predict functions for putative metal transporters.
Some transporters are fused to other domains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- [2].Maret W, Li Y. Coordination dynamics of zinc in proteins. Chem. Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- [3].Castruita M, Casero D, Karpowicz SJ, Kropat J, Vieler A, Hsieh SI, Yan W, Cokus S, Loo JA, Benning C, Pellegrini M, Merchant SS. Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell. 2011;23:1273–1292. doi: 10.1105/tpc.111.084400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
- [5].Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- [6].Raimunda D, González-Guerrero M, Leeber BW, Argüello JM. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals. 2011;24:467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lin W, Chai J, Love J, Fu D. Selective electrodiffusion of zinc ions in a Zrt-, Irt-like protein, ZIPB. J. Biol. Chem. 2010;285:39013–39020. doi: 10.1074/jbc.M110.180620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Terauchi AM, Peers G, Kobayashi MC, Niyogi KK, Merchant SS. Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 2010;105:39–49. doi: 10.1007/s11120-010-9562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Naumann B, Busch A, Allmer J, Ostendorf E, Zeller M, Kirchhoff H, Hippler M. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics. 2007;7:3964–3979. doi: 10.1002/pmic.200700407. [DOI] [PubMed] [Google Scholar]
- [10].Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J. Biol. Chem. 2009;284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cox CD. Deferration of laboratory media and assays for ferric and ferrous ions. In: Virginia PMB, Clark L, editors. Methods in Enzymology. Academic Press; 1994. pp. 315–329. [DOI] [PubMed] [Google Scholar]
- [12].Sigdel T, Easton J, Crowder M. Transcriptional response of Escherichia coli to TPEN. J. Bacteriol. 2006;188:6709–6713. doi: 10.1128/JB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Haas KL, Franz KJ. Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 2009;109:4921–4960. doi: 10.1021/cr900134a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marschner H. Mineral nutrition of higher plants. Academic Press; 1995. [Google Scholar]
- [15].Atkinson A, Guerinot M, Murphy AS, Schulz B, Peer W. The Plant Plasma Membrane. Springer Berlin / Heidelberg; 2011. Metal Transport; pp. 303–330. [Google Scholar]
- [16].Hassinen VH, Tervahauta AI, Schat H, Kärenlampi SO. Plant metallothioneins--metal chelators with ROS scavenging activity? Plant Biol. (Stuttg) 2011;13:225–232. doi: 10.1111/j.1438-8677.2010.00398.x. [DOI] [PubMed] [Google Scholar]
- [17].Hanikenne M, Nouet C. Metal hyperaccumulation and hypertolerance: a model for plant evolutionary genomics. Curr. Opin. Plant Biol. 2011;14:252–259. doi: 10.1016/j.pbi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- [18].Krämer U. Metal hyperaccumulation in plants. Annu. Rev. Plant Biol. 2010;61:517–534. doi: 10.1146/annurev-arplant-042809-112156. [DOI] [PubMed] [Google Scholar]
- [19].Kenrick P. Embryophyta (Land Plants), eLS. John Wiley & Sons, Ltd; 2001. [Google Scholar]
- [20].Stanier RY, Cohenbarzire G. Phototrophic prokaryotes - cyanobacteria. Ann. Rev. Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- [21].Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- [22].Chan CX, Gross J, Yoon HS, Bhattacharya D. Plastid origin and evolution: new models provide insights into old problems. Plant Physiol. 2011;155:1552–1560. doi: 10.1104/pp.111.173500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu. Rev. Plant. Biol. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- [24].Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- [25].Bodył A, Mackiewicz P, Stiller JW. The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles? Trends Microbiol. 2007;15:295–296. doi: 10.1016/j.tim.2007.05.002. [DOI] [PubMed] [Google Scholar]
- [26].Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- [27].Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- [28].Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kropat J, Hong-Hermesdorf A, Casero D, Ent P, Castruita M, Pellegrini M, Merchant SS, Malasarn D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shelford VE. Animal communities in temperate America. University of Chicago Press; Chicago, IL: 1913. [Google Scholar]
- [31].da Silva JJRF, Williams RJP. The biological chemistry of the elements: The inorganic chemistry of life. 2nd ed. Oxford University Press; Oxford: 2001. [Google Scholar]
- [32].Crichton RR, Pierre JL. Old iron, young copper: from Mars to Venus. Biometals. 2001;14:99–112. doi: 10.1023/a:1016710810701. [DOI] [PubMed] [Google Scholar]
- [33].Bekker A, Holland HD, Wang PL, Rumble D, Stein HJ, Hannah JL, Coetzee LL, Beukes NJ. Dating the rise of atmospheric oxygen. Nature. 2004;427:117–120. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- [34].Sandmann G, Rsck H, Kessler E, Boger P. Distribution of plastocyanin and soluble plastidic cytochrome-c in various classes of algae. Arch. Microbiol. 1983;134:23–27. [Google Scholar]
- [35].De la Rosa MA, Navarro JA, Díaz-Quintana A, De la Cerda B, Molina-Heredia FP, Balme A, Murdoch P.e.S., Díaz-Moreno I, Durán RV, Hervás M. An evolutionary analysis of the reaction mechanisms of photosystem I reduction by cytochrome c(6) and plastocyanin. Bioelectrochemistry. 2002;55:41–45. doi: 10.1016/s1567-5394(01)00136-0. [DOI] [PubMed] [Google Scholar]
- [36].Peers G, Price NM. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature. 2006;441:341–344. doi: 10.1038/nature04630. [DOI] [PubMed] [Google Scholar]
- [37].Merchant S, Bogorad L. Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J. Biol. Chem. 1986;261:15850–15853. [PubMed] [Google Scholar]
- [38].Wood PM. Interchangeable copper and iron proteins in algal photosynthesis. Studies on plastocyanin and cytochrome c-552 in Chlamydomonas. Eur. J. Biochem. 1978;87:9–19. doi: 10.1111/j.1432-1033.1978.tb12346.x. [DOI] [PubMed] [Google Scholar]
- [39].Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, Morales R, Allen AE, Armbrust EV. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U. S. A. 2012 doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Inda L, Erdner D, Peleato M, Anderson D. Cytochrome c(6) isolated from the marine diatom Thalassiosira weissflogii. Phytochemistry. 1999;51:1–4. [Google Scholar]
- [41].Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- [42].Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Consortium U. Ongoing and future developments at the Universal Protein Resource. Nuc. Acids Res. 2011;39:D214–219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nuc. Acids Res. 2011 doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. The Genome Portal of the Department of Energy Joint Genome Institute. Nuc. Acids Res. 2011 doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wittkop T, Emig D, Truss A, Albrecht M, Böcker S, Baumbach J. Comprehensive cluster analysis with Transitivity Clustering. Nat. Protoc. 2011;6:285–295. doi: 10.1038/nprot.2010.197. [DOI] [PubMed] [Google Scholar]
- [49].Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kosman DJ. Redox cycling in iron uptake, efflux, and trafficking. J. Biol. Chem. 2010;285:26729–26735. doi: 10.1074/jbc.R110.113217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schröder I, Johnson E, de Vries S. Microbial ferric iron reductases. FEMS Microbiol. Rev. 2003;27:427–447. doi: 10.1016/S0168-6445(03)00043-3. [DOI] [PubMed] [Google Scholar]
- [52].Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R. Respiratory burst oxidases: the engines of ROS signaling. Curr. Opin. Plant Biol. 2011;14:691–699. doi: 10.1016/j.pbi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- [53].Castignetti D, Smarrelli J. Siderophores, the iron nutrition of plants, and nitrate reductase. FEBS Lett. 1986;209:147–151. [Google Scholar]
- [54].Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dancis A, Roman DG, Anderson GJ, Hinnebusch AG, Klausner RD. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. U. S. A. 1992;89:3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Georgatsou E, Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell Biol. 1994;14:3065–3073. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yun CW, Bauler M, Moore RE, Klebba PE, Philpott CC. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:10218–10223. doi: 10.1074/jbc.M010065200. [DOI] [PubMed] [Google Scholar]
- [58].Singh A, Kaur N, Kosman DJ. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 2007;282:28619–28626. doi: 10.1074/jbc.M703398200. [DOI] [PubMed] [Google Scholar]
- [59].Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- [60].Jeong J, Connolly EL. Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci. 2009;176:709–714. [Google Scholar]
- [61].Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- [62].Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- [63].Scott EM, McGraw JC. Purification and properties of diphosphopyridine nucleotide diaphorase of human erythrocytes. J. Biol. Chem. 1962;237:249–252. [PubMed] [Google Scholar]
- [64].Hahne K, Haucke V, Ramage L, Schatz G. Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell. 1994;79:829–839. doi: 10.1016/0092-8674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- [65].Bagnaresi P, Basso B, Pupillo P. The NADH-dependent Fe(3+)-chelate reductases of tomato roots. Planta. 1997;202:427–434. doi: 10.1007/s004250050146. [DOI] [PubMed] [Google Scholar]
- [66].Sparla F, Bagnaresi P, Scagliarini S, Trost P. NADH:Fe(III)-chelate reductase of maize roots is an active cytochrome b5 reductase. FEBS Lett. 1997;414:571–575. doi: 10.1016/s0014-5793(97)01073-9. [DOI] [PubMed] [Google Scholar]
- [67].Xoconostle-Cázares B, Ruiz-Medrano R, Lucas WJ. Proteolytic processing of CmPP36, a protein from the cytochrome b(5) reductase family, is required for entry into the phloem translocation pathway. Plant J. 2000;24:735–747. doi: 10.1046/j.1365-313x.2000.00916.x. [DOI] [PubMed] [Google Scholar]
- [68].McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- [69].Su D, Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273:3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- [70].Allen MD, del Campo JA, Kropat J, Merchant SS. FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell. 2007;6:1841–1852. doi: 10.1128/EC.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Reinhardt I, Haebel S, Herbik A, Buckhout T. Proteomic studies under iron stress: iron deficiency-induced regulation of protein synthesis in the green alga Chlamydomonas reinhardtii. :371–393. [Google Scholar]
- [72].Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Haucke V, Ocana CS, Hönlinger A, Tokatlidis K, Pfanner N, Schatz G. Analysis of the sorting signals directing NADH-cytochrome b5 reductase to two locations within yeast mitochondria. Mol. Cell Biol. 1997;17:4024–4032. doi: 10.1128/mcb.17.7.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kustka A, Allen A, Morel F. Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J. Phycol. 2007;43:715–729. [Google Scholar]
- [76].Allen AE, Laroche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10438–10443. doi: 10.1073/pnas.0711370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jeong J, Cohu C, Kerkeb L, Pilon M, Connolly EL, Guerinot ML. Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10619–10624. doi: 10.1073/pnas.0708367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Busch A, Rimbauld B, Naumann B, Rensch S, Hippler M. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 2008;55:201–211. doi: 10.1111/j.1365-313X.2008.03490.x. [DOI] [PubMed] [Google Scholar]
- [79].Horemans N, Foyer CH, Asard H. Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 2000;5:263–267. doi: 10.1016/s1360-1385(00)01649-6. [DOI] [PubMed] [Google Scholar]
- [80].Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- [82].Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14:251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- [84].Taylor KM, Nicholson RI. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim. Biophys. Acta. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- [85].MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wang M, Xu Q, Yu J, Yuan M. The putative Arabidopsis zinc transporter ZTP29 is involved in the response to salt stress. Plant Mol. Biol. 2010;73:467–479. doi: 10.1007/s11103-010-9633-4. [DOI] [PubMed] [Google Scholar]
- [87].Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Vert G, Barberon M, Zelazny E, Séguéla M, Briat JF, Curie C. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta. 2009;229:1171–1179. doi: 10.1007/s00425-009-0904-8. [DOI] [PubMed] [Google Scholar]
- [89].Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- [90].Forbes JR, Gros P. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- [91].Kuhn DE, Lafuse WP, Zwilling BS. Iron transport into mycobacterium avium-containin phagosomes from an Nramp1(Gly169)-transfected RAW264.7 macrophage cell line. J. Leukoc. Biol. 2001;69:43–49. [PubMed] [Google Scholar]
- [92].Xia J, Yamaji N, Kasai T, Ma JF. Plasma membrane-localized transporter for aluminum in rice. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18381–18385. doi: 10.1073/pnas.1004949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ueki T, Furuno N, Michibata H. A novel vanadium transporter of the Nramp family expressed at the vacuole of vanadium-accumulating cells of the ascidian Ascidia sydneiensis samea. Biochim. Biophys. Acta. 2011;1810:457–464. doi: 10.1016/j.bbagen.2010.12.006. [DOI] [PubMed] [Google Scholar]
- [94].Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J. 2003;35:295–304. doi: 10.1046/j.1365-313x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- [95].Goswami T, Bhattacharjee A, Babal P, Searle S, Moore E, Li M, Blackwell JM. Natural-resistance-associated macrophage protein 1 is an H+/bivalent cation antiporter. Biochem. J. 2001;354:511–519. doi: 10.1042/0264-6021:3540511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cellier MF, Bergevin I, Boyer E, Richer E. Polyphyletic origins of bacterial Nramp transporters. Trends Genet. 2001;17:365–370. doi: 10.1016/s0168-9525(01)02364-2. [DOI] [PubMed] [Google Scholar]
- [97].Lanquar V, Lelièvre F, Bolte S, Hamès C, Alcon C, Neumann D, Vansuyt G, Curie C, Schröder A, Krämer U, Barbier-Brygoo H, Thomine S. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lanquar V, Ramos MS, Lelièvre F, Barbier-Brygoo H, Krieger-Liszkay A, Krämer U, Thomine S. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010;152:1986–1999. doi: 10.1104/pp.109.150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Thamatrakoln K, Korenovska O, Niheu AK, Bidle KD. Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ. Microbiol. 2011 doi: 10.1111/j.1462-2920.2011.02468.x. [DOI] [PubMed] [Google Scholar]
- [100].Merchant S, Allen M, Kropat J, Moseley J, Long J, Tottey S, Terauchi A. Between a rock and a hard place: Trace element nutrition in Chlamydomonas, BBA-Mol. Cell Res. 2006;1763:578–594. doi: 10.1016/j.bbamcr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- [101].Hanikenne M, Krämer U, Demoulin V, Baurain D. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 2005;137:428–446. doi: 10.1104/pp.104.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- [103].Benavente LM, Alonso JM. Molecular mechanisms of ethylene signaling in Arabidopsis. Mol. Biosyst. 2006;2:165–173. doi: 10.1039/b513874d. [DOI] [PubMed] [Google Scholar]
- [104].Hirayama T, Alonso JM. Ethylene captures a metal! Metal ions are involved in ethylene perception and signal transduction. Plant Cell Physiol. 2000;41:548–555. doi: 10.1093/pcp/41.5.548. [DOI] [PubMed] [Google Scholar]
- [105].Schaaf G, Honsbein A, Meda AR, Kirchner S, Wipf D, von Wirén N. AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 2006;281:25532–25540. doi: 10.1074/jbc.M601062200. [DOI] [PubMed] [Google Scholar]
- [106].Nishida S, Tsuzuki C, Kato A, Aisu A, Yoshida J, Mizuno T. AtIRT1, the primary iron uptake transporter in the root, mediates excess nickel accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2011;52:1433–1442. doi: 10.1093/pcp/pcr089. [DOI] [PubMed] [Google Scholar]
- [107].Hart BA, Bertram PE, Scaife BD. Cadmium transport by Chlorella pyrenoidosa. Environ Res. 1979;18:327–335. doi: 10.1016/0013-9351(79)90109-9. [DOI] [PubMed] [Google Scholar]
- [108].Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell. 1994;76:403–410. doi: 10.1016/0092-8674(94)90346-8. [DOI] [PubMed] [Google Scholar]
- [109].Kwok EY, Severance S, Kosman DJ. Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry. 2006;45:6317–6327. doi: 10.1021/bi052173c. [DOI] [PubMed] [Google Scholar]
- [110].Van Ho A, Ward DM, Kaplan J. Transition metal transport in yeast. Annu. Rev. Microbiol. 2002;56:237–261. doi: 10.1146/annurev.micro.56.012302.160847. [DOI] [PubMed] [Google Scholar]
- [111].Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- [112].La Fontaine S, Quinn JM, Nakamoto SS, Page MD, Göhre V, Moseley JL, Kropat J, Merchant S. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell. 2002;1:736–757. doi: 10.1128/EC.1.5.736-757.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Peers G, Quesnel S, Price N. Copper requirements for iron acquisition and growth of coastal and oceanic diatoms, Limnol. Oceanogr. 2005;50:1149–1158. [Google Scholar]
- [114].Maldonado MT, Allen AE, Chong JS, Lin K, Leus D, Karpenko N, Shannon LH. Copper-dependent iron transport in coastal and oceanic diatoms, Limnol. Oceanogr. 2006;51:1729–1743. [Google Scholar]
- [115].Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem. Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- [117].Terzulli AJ, Kosman DJ. The Fox1 ferroxidase of Chlamydomonas reinhardtii: a new multicopper oxidase structural paradigm. J. Biol. Inorg. Chem. 2009;14:315–325. doi: 10.1007/s00775-008-0450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Herbik A, Bölling C, Buckhout TJ. The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 2002;130:2039–2048. doi: 10.1104/pp.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chen J, Hsieh S, Kropat J, Merchant S. A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell. 2008;7:541–545. doi: 10.1128/EC.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Terzulli A, Kosman DJ. Analysis of the high-affinity iron uptake system at the Chlamydomonas reinhardtii plasma membrane. Eukaryot Cell. 2010;9:815–826. doi: 10.1128/EC.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Severance S, Chakraborty S, Kosman DJ. The Ftr1p iron permease in the yeast plasma membrane: orientation, topology and structure-function relationships. Biochem. J. 2004;380:487–496. doi: 10.1042/BJ20031921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, Zhou K, Otillar R, Merchant SS, Podell S, Gaasterland T, Napoli C, Gendler K, Manuell A, Tai V, Vallon O, Piganeau G, Jancek S, Heijde M, Jabbari K, Bowler C, Lohr M, Robbens S, Werner G, Dubchak I, Pazour GJ, Ren Q, Paulsen I, Delwiche C, Schmutz J, Rokhsar D, Van de Peer Y, Moreau H, Grigoriev IV. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sasaki T, Kurano N, Miyachi S. Cloning and characterization of high-CO2-specific cDNAs from a marine microalga, Chlorococcum littorale, and effect of CO2 concentration and iron deficiency on the gene expression. Plant Cell Physiol. 1998;39:131–138. doi: 10.1093/oxfordjournals.pcp.a029349. [DOI] [PubMed] [Google Scholar]
- [124].Narayanan NN, Ihemere U, Chiu WT, Siritunga D, Rajamani S, Singh S, Oda S, Sayre RT. The iron assimilatory protein, FEA1, from Chlamydomonas reinhardtii facilitates iron-specific metal uptake in yeast and plants, Frontiers in Plant. Science. 2011;2 doi: 10.3389/fpls.2011.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shingles R, North M, McCarty RE. Ferrous ion transport across chloroplast inner envelope membranes. Plant Physiol. 2002;128:1022–1030. doi: 10.1104/pp.010858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Duy D, Wanner G, Meda AR, von Wirén N, Soll J, Philippar K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell. 2007;19:986–1006. doi: 10.1105/tpc.106.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Duy D, Stübe R, Wanner G, Philippar K. The chloroplast permease PIC1 regulates plant growth and development by directing homeostasis and transport of iron. Plant Physiol. 2011;155:1709–1722. doi: 10.1104/pp.110.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kikuchi S, Oishi M, Hirabayashi Y, Lee DW, Hwang I, Nakai M. A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell. 2009;21:1781–1797. doi: 10.1105/tpc.108.063552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Huang L, Kirschke CP, Zhang Y, Yu YY. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J. Biol. Chem. 2005;280:15456–15463. doi: 10.1074/jbc.M412188200. [DOI] [PubMed] [Google Scholar]
- [130].Mühlenhoff U, Stadler JA, Richhardt N, Seubert A, Eickhorst T, Schweyen RJ, Lill R, Wiesenberger G. A specific role of the yeast mitochondrial carriers MRS3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 2003;278:40612–40620. doi: 10.1074/jbc.M307847200. [DOI] [PubMed] [Google Scholar]
- [131].Froschauer EM, Schweyen RJ, Wiesenberger G. The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane. Biochim Biophys Acta. 2009;1788:1044–1050. doi: 10.1016/j.bbamem.2009.03.004. [DOI] [PubMed] [Google Scholar]
- [132].Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH. Mitoferrin is essential for erythroid iron assimilation. Nature. 2006;440:96–100. doi: 10.1038/nature04512. [DOI] [PubMed] [Google Scholar]
- [133].Bashir K, Ishimaru Y, Shimo H, Nagasaka S, Fujimoto M, Takanashi H, Tsutsumi N, An G, Nakanishi H, Nishizawa NK. The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2011;2:322. doi: 10.1038/ncomms1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yoon H, Zhang Y, Pain J, Lyver ER, Lesuisse E, Pain D, Dancis A. Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria. Biochem. J. 2011;440:137–146. doi: 10.1042/BJ20111036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch. Microbiol. 2005;183:9–18. doi: 10.1007/s00203-004-0739-4. [DOI] [PubMed] [Google Scholar]
- [136].Li L, Kaplan J. Characterization of two homologous yeast genes that encode mitochondrial iron transporters. J. Biol. Chem. 1997;272:28485–28493. doi: 10.1074/jbc.272.45.28485. [DOI] [PubMed] [Google Scholar]
- [137].Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KF, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- [138].Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- [139].Li L, Chen OS, McVey Ward D, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- [140].Morrissey J, Baxter IR, Lee J, Li L, Lahner B, Grotz N, Kaplan J, Salt DE, Guerinot ML. The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell. 2009;21:3326–3338. doi: 10.1105/tpc.109.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Gollhofer J, Schläwicke C, Jungnick N, Schmidt W, Buckhout TJ. Members of a small family of nodulin-like genes are regulated under iron deficiency in roots of Arabidopsis thaliana. Plant Physiol. Biochem. 2011;49:557–564. doi: 10.1016/j.plaphy.2011.02.011. [DOI] [PubMed] [Google Scholar]
- [142].Yagisawa F, Nishida K, Yoshida M, Ohnuma M, Shimada T, Fujiwara T, Yoshida Y, Misumi O, Kuroiwa H, Kuroiwa T. Identification of novel proteins in isolated polyphosphate vacuoles in the primitive red alga Cyanidioschyzon merolae. Plant J. 2009;60:882–893. doi: 10.1111/j.1365-313X.2009.04008.x. [DOI] [PubMed] [Google Scholar]
- [143].Mercier A, Pelletier B, Labbé S. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell. 2006;5:1866–1881. doi: 10.1128/EC.00199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Pouliot B, Jbel M, Mercier A, Labbé S. abc3+ encodes an iron-regulated vacuolar ABC-type transporter in Schizosaccharomyces pombe., Eukaryot. Cell. 2010;9:59–73. doi: 10.1128/EC.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Rubino JT, Chenkin MP, Keller M, Riggs-Gelasco P, Franz KJ. A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics. 2011;3:61–73. [PubMed] [Google Scholar]
- [146].Page MD, Kropat J, Hamel PP, Merchant SS. Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell. 2009;21:928–943. doi: 10.1105/tpc.108.064907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Lee J, Peña MM, Nose Y, Thiele DJ. Biochemical characterization of the human copper transporter Ctr1. J. Biol. Chem. 2002;277:4380–4387. doi: 10.1074/jbc.M104728200. [DOI] [PubMed] [Google Scholar]
- [148].Howe G, Merchant S. Heavy metal-activated synthesis of peptides in Chlamydomonas reinhardtii. Plant Physiol. 1992;98:127–136. doi: 10.1104/pp.98.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Morgan TP, Grosell M, Gilmour KM, Playle RC, Wood CM. Time course analysis of the mechanism by which silver inhibits active Na+ and Cl− uptake in gills of rainbow trout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R234–242. doi: 10.1152/ajpregu.00448.2003. [DOI] [PubMed] [Google Scholar]
- [150].Garcia-Molina A, Andrés-Colás N, Perea-García A, Del Valle-Tascón S, Peñarrubia L, Puig S. The intracellular Arabidopsis COPT5 transport protein is required for photosynthetic electron transport under severe copper deficiency. Plant J. 2011;65:848–860. doi: 10.1111/j.1365-313X.2010.04472.x. [DOI] [PubMed] [Google Scholar]
- [151].Klaumann S, Nickolaus SD, Fürst SH, Starck S, Schneider S, Ekkehard Neuhaus H, Trentmann O. The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 2011;192:393–404. doi: 10.1111/j.1469-8137.2011.03798.x. [DOI] [PubMed] [Google Scholar]
- [152].Bellemare DR, Shaner L, Morano KA, Beaudoin J, Langlois R, Labbe S. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:46676–46686. doi: 10.1074/jbc.M206444200. [DOI] [PubMed] [Google Scholar]
- [153].Rees EM, Lee J, Thiele DJ. Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J Biol Chem. 2004;279:54221–54229. doi: 10.1074/jbc.M411669200. [DOI] [PubMed] [Google Scholar]
- [154].Dancis A, Yuan DS, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner RD. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- [155].Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, Zhang Y, Collier JL, Wurch LL, Kustka AB, Dill BD, Shah M, VerBerkmoes NC, Kuo A, Terry A, Pangilinan J, Lindquist EA, Lucas S, Paulsen IT, Hattenrath-Lehmann TK, Talmage SC, Walker EA, Koch F, Burson AM, Marcoval MA, Tang YZ, Lecleir GR, Coyne KJ, Berg GM, Bertrand EM, Saito MA, Gladyshev VN, Grigoriev IV. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Hill KL, Hassett R, Kosman D, Merchant S. Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 1996;112:697–704. doi: 10.1104/pp.112.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].O'Halloran T, Culotta V. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- [158].Xiao Z, Wedd AG. A C-terminal domain of the membrane copper pump Ctr1 exchanges copper(I) with the copper chaperone Atx1. Chem. Commun. (Camb) 2002:588–589. doi: 10.1039/b111180a. [DOI] [PubMed] [Google Scholar]
- [159].González-Guerrero M, Argüello JM. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- [161].Remacle C, Coosemans N, Jans F, Hanikenne M, Motte P, Cardol P. Knock-down of the COX3 and COX17 gene expression of cytochrome c oxidase in the unicellular green alga Chlamydomonas reinhardtii. Plant Mol. Biol. 2010;74:223–233. doi: 10.1007/s11103-010-9668-6. [DOI] [PubMed] [Google Scholar]
- [162].La Fontaine S, Ackland ML, Mercer JF. Mammalian copper-transporting P-type ATPases, ATP7A and ATP7B: emerging roles. Int. J. Biochem. Cell Biol. 2010;42:206–209. doi: 10.1016/j.biocel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Suzuki M, Gitlin JD. Intracellular localization of the Menkes and Wilson's disease proteins and their role in intracellular copper transport. Pediatr. Int. 1999;41:436–442. doi: 10.1046/j.1442-200x.1999.01090.x. [DOI] [PubMed] [Google Scholar]
- [164].La Fontaine S, Mercer JF. Trafficking of the copper-ATPases, ATP7A and ATP7B: role in copper homeostasis. Arch. Biochem. Biophys. 2007;463:149–167. doi: 10.1016/j.abb.2007.04.021. [DOI] [PubMed] [Google Scholar]
- [165].Shikanai T, Müller-Moulé P, Munekage Y, Niyogi KK, Pilon M. PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell. 2003;15:1333–1346. doi: 10.1105/tpc.011817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Abdel-Ghany SE, Müller-Moulé P, Niyogi KK, Pilon M, Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, Finazzi G, Grunwald D, Rappaport F, Vavasseur A, Joyard J, Richaud P, Rolland N. HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 2006;281:2882–2892. doi: 10.1074/jbc.M508333200. [DOI] [PubMed] [Google Scholar]
- [168].Kim YY, Choi H, Segami S, Cho HT, Martinoia E, Maeshima M, Lee Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009;58:737–753. doi: 10.1111/j.1365-313X.2009.03818.x. [DOI] [PubMed] [Google Scholar]
- [169].Merchant S, Bogorad L. Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell Biol. 1986;6:462–469. doi: 10.1128/mcb.6.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Singleton C, Le Brun NE. Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals. 2007;20:275–289. doi: 10.1007/s10534-006-9068-1. [DOI] [PubMed] [Google Scholar]
- [171].Dupont CL, Butcher A, Valas RE, Bourne PE, Caetano-Anollés G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10567–10572. doi: 10.1073/pnas.0912491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 2005;56:3207–3214. doi: 10.1093/jxb/eri317. [DOI] [PubMed] [Google Scholar]
- [173].Lee S, Jeong HJ, Kim SA, Lee J, Guerinot ML, An G. OsZIP5 is a plasma membrane zinc transporter in rice. Plant. Mol. Biol. 2010;73:507–517. doi: 10.1007/s11103-010-9637-0. [DOI] [PubMed] [Google Scholar]
- [174].Kumánovics A, Poruk KE, Osborn KA, Ward DM, Kaplan J. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:22566–22574. doi: 10.1074/jbc.M604730200. [DOI] [PubMed] [Google Scholar]
- [175].Moreau S, Thomson RM, Kaiser BN, Trevaskis B, Guerinot ML, Udvardi MK, Puppo A, Day DA. GmZIP1 encodes a symbiosis-specific zinc transporter in soybean. J. Biol. Chem. 2002;277:4738–4746. doi: 10.1074/jbc.M106754200. [DOI] [PubMed] [Google Scholar]
- [176].Nies DH, Silver S. Ion efflux systems involved in bacterial metal resistances. J. Ind. Microbiol. 1995;14:186–199. doi: 10.1007/BF01569902. [DOI] [PubMed] [Google Scholar]
- [177].Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- [178].Lu M, Chai J, Fu D. Structural basis for autoregulation of the zinc transporter YiiP. Nat. Struct. Mol. Biol. 2009;16:1063–1067. doi: 10.1038/nsmb.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Palmiter RD, Findley SD. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annu. Rev. Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- [181].van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, Verkleij JA, Hooykaas PJ. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999;119:1047–1055. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Gustin JL, Zanis MJ, Salt DE. Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol Biol. 2011;11:76. doi: 10.1186/1471-2148-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Kobae Y, Uemura T, Sato MH, Ohnishi M, Mimura T, Nakagawa T, Maeshima M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004;45:1749–1758. doi: 10.1093/pcp/pci015. [DOI] [PubMed] [Google Scholar]
- [185].Arrivault S, Senger T, Krämer U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006;46:861–879. doi: 10.1111/j.1365-313X.2006.02746.x. [DOI] [PubMed] [Google Scholar]
- [186].Wu YH, Frey AG, Eide DJ. Transcriptional regulation of the Zrg17 zinc transporter of the yeast secretory pathway. Biochem J. 2011;435:259–266. doi: 10.1042/BJ20102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Ellis CD, Wang F, MacDiarmid CW, Clark S, Lyons T, Eide DJ. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Biol. Chem. 2004;166:325–335. doi: 10.1083/jcb.200401157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Kirschke CP, Huang L. ZnT7, a novel mammalian zinc transporter, accumulates zinc in the Golgi apparatus. J. Biol. Chem. 2003;278:4096–4102. doi: 10.1074/jbc.M207644200. [DOI] [PubMed] [Google Scholar]
- [189].Ishihara K, Yamazaki T, Ishida Y, Suzuki T, Oda K, Nagao M, Yamaguchi-Iwai Y, Kambe T. Zinc transport complexes contribute to the homeostatic maintenance of secretory pathway function in vertebrate cells. J. Biol. Chem. 2006;281:17743–17750. doi: 10.1074/jbc.M602470200. [DOI] [PubMed] [Google Scholar]
- [190].van de Mortel JE, Almar Villanueva L, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MG. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006;142:1127–1147. doi: 10.1104/pp.106.082073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].MacDiarmid CW, Milanick MA, Eide DJ. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 2003;278:15065–15072. doi: 10.1074/jbc.M300568200. [DOI] [PubMed] [Google Scholar]
- [192].Scott DA, Docampo R, Dvorak JA, Shi S, Leapman RD. In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J. Biol. Chem. 1997;272:28020–28029. doi: 10.1074/jbc.272.44.28020. [DOI] [PubMed] [Google Scholar]
- [193].Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, Morel FM. Biochemistry: a cadmium enzyme from a marine diatom. Nature. 2005;435:42. doi: 10.1038/435042a. [DOI] [PubMed] [Google Scholar]
- [194].Yee D, Morel FMM. In vivo substitution of zinc by cobalt in carbonic anhydrase of a marine diatom, Limnol. Oceanogr. 1996;41:573–577. [Google Scholar]
- [195].Bruland K. Complexation of zinc by natural organic-ligands in the Central North Pacific, Limnol. Oceanogr. 1989;34:269–285. [Google Scholar]
- [196].Schulz K, Zondervan I, Gerringa L, Timmermans K, Veldhuis M, Riebesell U. Effect of trace metal availability on coccolithophorid calcification. Nature. 2004;430:673–676. doi: 10.1038/nature02631. [DOI] [PubMed] [Google Scholar]
- [197].Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, Vavasseur A, Richaud P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009;149:894–904. doi: 10.1104/pp.108.130294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 2004;16:1327–1339. doi: 10.1105/tpc.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Kitayama K, Kitayama M, Osafune T, Togasaki RK. Subcellular localization of iron and manganese superoxide dismutase in Chlmaydomonas reinhardtii (Chlorophyceae) J. Phycol. 1999;35:136–142. [Google Scholar]
- [200].Wolfe-Simon F, Starovoytov V, Reinfelder JR, Schofield O, Falkowski PG. Localization and role of manganese superoxide dismutase in a marine diatom. Plant Physiol. 2006;142:1701–1709. doi: 10.1104/pp.106.088963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Allen MD, Kropat J, Tottey S, Del Campo JA, Merchant SS. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 2007;143:263–277. doi: 10.1104/pp.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Liu XF, Culotta VC. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J. Biol. Chem. 1999;274:4863–4868. doi: 10.1074/jbc.274.8.4863. [DOI] [PubMed] [Google Scholar]
- [204].Rosakis A, Köster W. Divalent metal transport in the green microalga Chlamydomonas reinhardtii is mediated by a protein similar to prokaryotic Nramp homologues. Biometals. 2005;18:107–120. doi: 10.1007/s10534-004-2481-4. [DOI] [PubMed] [Google Scholar]
- [205].Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol. Microbiol. 2000;35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- [206].Jensen LT, Ajua-Alemanji M, Culotta VC. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 2003;278:42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- [207].Culotta VC, Yang M, Hall MD. Manganese transport and trafficking: lessons learned from Saccharomyces cerevisiae., Eukaryot. Cell. 2005;4:1159–1165. doi: 10.1128/EC.4.7.1159-1165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Dürr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol Biol Cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Luk E, Carroll M, Baker M, Culotta VC. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10353–10357. doi: 10.1073/pnas.1632471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Su Z, Chai MF, Lu PL, An R, Chen J, Wang XC. AtMTM1, a novel mitochondrial protein, may be involved in activation of the manganese-containing superoxide dismutase in Arabidopsis. Planta. 2007;226:1031–1039. doi: 10.1007/s00425-007-0547-6. [DOI] [PubMed] [Google Scholar]
- [211].Montanini B, Blaudez D, Jeandroz S, Sanders D, Chalot M. Phylogenetic and functional analysis of the Cation Diffusion Facilitator (CDF) family: improved signature and prediction of substrate specificity. BMC Genomics. 2007;8:107. doi: 10.1186/1471-2164-8-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Sim DL, Chow VT. The novel human HUEL (C4orf1) gene maps to chromosome 4p12-p13 and encodes a nuclear protein containing the nuclear receptor interaction motif. Genomics. 1999;59:224–233. doi: 10.1006/geno.1999.5856. [DOI] [PubMed] [Google Scholar]
- [213].Zhang Y, Rodionov DA, Gelfand MS, Gladyshev VN. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics. 2009;10:78. doi: 10.1186/1471-2164-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Li J, Pandeya D, Nath K, Zulfugarov IS, Yoo SC, Zhang H, Yoo JH, Cho SH, Koh HJ, Kim DS, Seo HS, Kang BC, Lee CH, Paek NC. ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J. 2010;62:713–725. doi: 10.1111/j.1365-313X.2010.04183.x. [DOI] [PubMed] [Google Scholar]
- [215].Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc. Biol. Sci. 2012 doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KF, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- [219].Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- [220].Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Oudot-Le Secq MP, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, Van de Peer Y, Grigoriev IV. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- [221].Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- [222].Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Le Bail A, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collén P, Peters AF, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder DC, Ségurens B, Strittmatter M, Tonon T, Tregear JW, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein similarity network of cytochrome c6 and cytochrome c6-like proteins encoded in the genomes of selected photosynthetic organisms.