Abstract
Gene expression shows a significant variation (noise) between genetically identical cells. Noise depends on the gene expression process regulated by the chromatin environment. We screened for chromatin factors that modulate noise in S. cerevisiae and analyzed the results using a theoretical model that infers regulatory mechanisms from the noise vs. mean relationship. Distinct activities of the Rpd3(L) and Set3 histone deacetylase complexes were predicted. Both HDACs repressed expression. Yet, Rpd3(L)C decreased the frequency of transcriptional bursts, while Set3C decreased the burst size, as did H2B mono-ubiquitination (ubH2B). We mapped the acetylation of H3 Lysine 9 (H3K9ac) upon deletion of multiple subunits of Set3C and Rpd3(L)C, and of ubH2B effectors. ubH2B and Set3C appear to function in the same pathway to reduce the probability that an elongating PolII produces a functional transcript (PolII processivity), while Rpd3(L)C likely represses gene expression at a step preceding elongation.
Introduction
Cells that are genetically identical may still behave differently under identical conditions (Barkai and Shilo, 2007; Raser and O'Shea, 2005). This non-genetic variability is largely due to noise in gene expression (Bar-Even et al., 2006; Elowitz et al., 2002; Ozbudak et al., 2002). Noise varies between genes. To a first approximation, it decreases with mean abundance. Yet, many genes deviate from this general trend (Bar-Even et al., 2006; Newman et al., 2006). For example, the low noise of essential genes and the high noise of stress-related genes are not explained by differences in mean abundance, but instead may depend on differences in the underlying gene expression mechanisms.
The prevailing model of gene expression noise assumes that proteins are made in “bursts”: short time intervals in which proteins are produced, interspaced by periods of negligible production (Blake et al., 2003; Cai et al., 2006; Tan and van Oudenaarden, 2010; Zenklusen et al., 2008). The main stochastic event is burst initiation. Noise is amplified by the burst-size (number of proteins made per burst), such that for a given level of mean expression, variability increases in proportion to burst size (Paulsson, 2004; Tan and van Oudenaarden, 2010). Burst frequency and burst size can be estimated from the distribution of expression levels: Let μ and η2 denote the respective mean and coefficient of variation (noise) of the expression distribution. The predicted burst size and burst frequency are estimated by η2*μ and η−2, respectively. Burst size is therefore the normalized variance, accounting for the inherent link between the noise and mean expression (Friedman et al., 2006; Raj et al., 2006; Tan and van Oudenaarden, 2010).
A key implication of this model is that gene expression can be regulated in two principally different ways. Regulation of burst frequency will coordinately modify mean expression and noise. By contrast, regulation of burst size will change mean expression but will not alter the coefficient of variation. Therefore, when studying the effect of a regulator of interest on gene expression, it may be beneficial to examine both mean expression and noise. Mean expression will distinguish between activators and repressors while noise may distinguish between regulators of burst frequency vs. regulators of burst size.
Identifying regulators of burst size is of a particular interest, as it provides insight to noise control. We used the model organism S. Cerevisiae to search for regulators of burst size amongst chromatin-associated factors. Chromatin affects gene expression directly, by restricting DNA accessibility, and indirectly, by recruiting other factors (Henikoff and Shilatifard, 2011). Previous reports implicated chromatin in noise regulation. First, genes of high-noise are associated with promoters that lack the typical nucleosome-free region (NFR). These promoters, termed Occupied Proximal Nucleosome (OPN), show increased sensitive to regulation by multiple chromatin factors (Blake et al., 2006; Cairns, 2009; Field et al., 2009; Tirosh and Barkai, 2008). Furthermore, individual deletions of three chromatin factors, the acetyl-transferase GCN5 and the chromatin remodelers SNF6 and ARP8 increased expression noise driven by the inducible PHO5 promoter (Raser and O'Shea, 2004). Recent systematic assays for noise regulators using a specific reporter also pointed to chromatin-associated factors (Rinott et al., 2011).
Motivated by this data, we screened 137 non-essential chromatin factors for modifiers of the normalized noise (the predicted burst size). We initially hypothesized that burst size is regulated primarily at the level of burst duration, likely depending on the promoter or 5’ end of genes. Surprisingly, the modification that had the strongest predicted (repressive) effect on burst size was H2B mono-ubiquitination (ubH2B) which is generally associated with transcription elongation (Fleming et al., 2008; Pavri et al., 2006; Shilatifard, 2006) and is found primarily within the coding region (Schulze et al., 2009). The second process identified was Set3C-dependent deacetylation. Similar to ubH2B, Set3C was also associated with transcription elongation. Further, at least in certain cases, its recruitment depends on H3K4 dimethylation, which is promoted by ubH2B (Kim and Buratowski, 2009; Wang et al., 2002).
PolII processivity is an elongation-related process affecting burst size. This measure defines the probability that an elongating PolII will produce a functional transcript, rather than terminate prematurely, (Mason and Struhl, 2005). When burst events are well separated in time, burst size increases linearly with PolII processivity. Notably, since burst size is the total number of proteins made per burst, other aspects affecting elongation (e.g. elongation velocity) will modulate burst size only through their effect on PolII processivity. We therefore hypothesized that both ubH2B and Set3C-dependent deacetylation repress PolII processivity, and examined this hypothesis using several high-throughput datasets.
Set3C is one of multiple histone deacetylation complexes (HDACs) expressed in S. cerevisiae. HDAC complexes are extensively studied, yet their individual functions are only partially understood (Kurdistani and Grunstein, 2003). The predicted role of Set3C in decreasing burst size was of particular interest to us, as it differed from the predicted function of other HDACs. The well studied Rpd3(L) complex, for example, repressed the predicted burst frequency and not the burst size. Furthermore, while Rpd3(L)C is known to act as a repressor of gene expression (Kurdistani and Grunstein, 2003; Robyr et al., 2002), Set3C is required for the rapid induction of the Gal1 gene (Kim and Buratowski, 2009; Wang et al., 2002). Our data, on the other hand, suggests that Set3C, like Rpd3(L)C, acts primarily as a repressor of gene expression, albeit through different means. We therefore explored further the distinct activities of these two complexes.
HDACs activities can be distinguished by their effect on the genome-wide histone acetylation. Such mapping revealed, for example, a ‘division of labor’ between HDACs acting on different gene promoters (Robyr et al., 2002; Wang et al., 2002). Existing data reporting acetylation profiles in Rpd3(L)C and Set3C mutant background, however, mapped acetylation at promoter only, and is of a low spatial resolution. To examine the role of Set3C in transcription elongation and PolII processivity, and to test whether ubH2B and Set3C function in the same pathway, we wished to examine acetylation profiles in mutants affecting histone Acetylation and ubH2B. We therefore generated a high-resolution map of H3K9 acetylation in five mutants deleted of Set3C and Rpd3(L)C components, and in four mutants deleted of ubH2B effectors. This data was analyzed in combination with existing functional genomic datasets. Based on this, we now provide evidences that ubH2B and Set3-dependent deacetylation function in the same pathway to reduce PolII processivity, while Rpd3(L)C key role in gene repression precedes elongation.
Results
Screening for chromatin regulators of gene expression noise
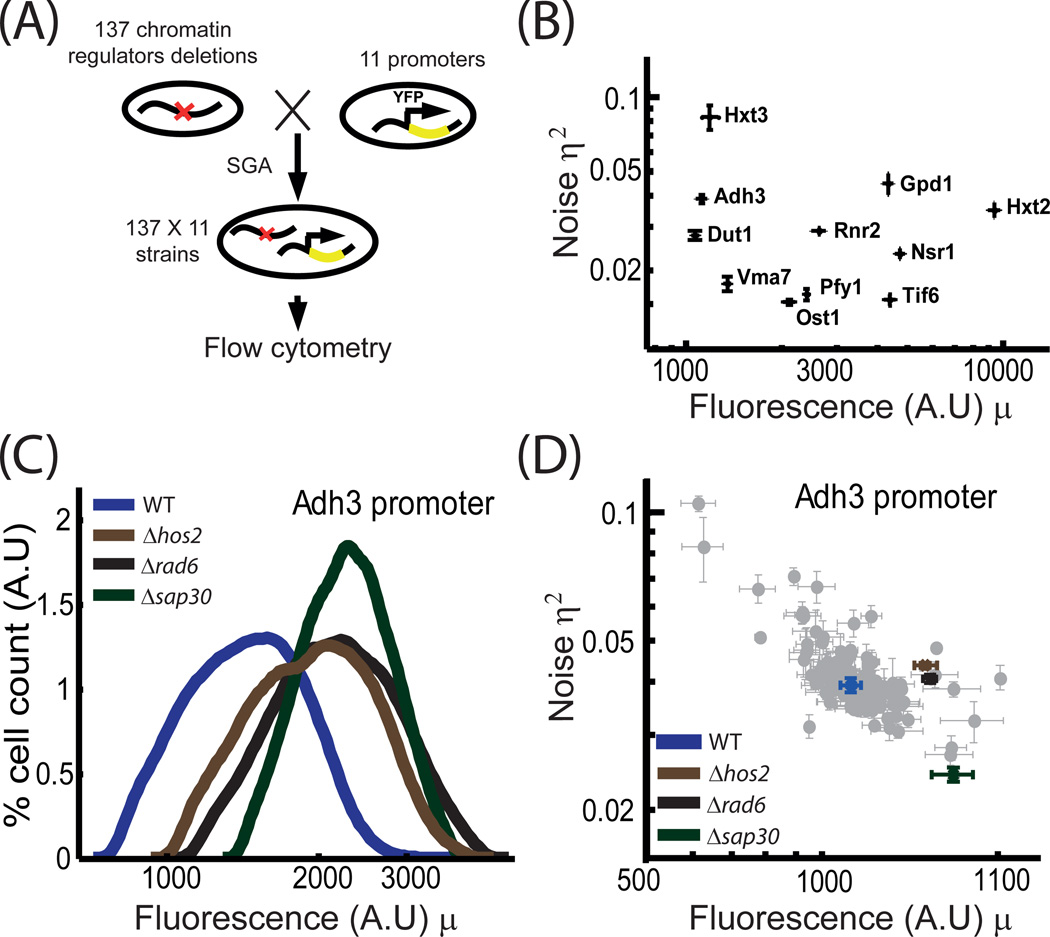
We selected 137 chromatin factors and tested how their individual deletions modulate the expression (mean and variance) of a fluorescence reporter driven by one of eleven representative promoters (figure 1A). Our screen covered most of the non-essential chromatin modifiers, including regulators of histone acetylation, methylation, phosphorylation or ubiquitination, chromatin remodelers, histone variant, exchange factors, elements of the general transcription machinery, and chromatin silencing genes (table S1). The eleven promoters used as reporters spanned a range of intermediate expression levels that are high enough to be detected by flow-cytometry yet not too high to ensure a significant contribution of noise intrinsic to the transcription process (Figure 1B). All promoters were inserted into the HIS3 locus upstream of the reporter, and were combined with the deletion mutants using Synthetic Genetic Array (SGA, (Tong and Boone, 2006)). Altogether, we generated a set of 137X11 strains, each deleted of one chromatin regulator and carrying one YFP-driving promoter.
Figure 1. Screen for chromatin regulators that change gene expression noise.
A. Screen design: 137 strains deleted of individual chromatin-associated factors were obtained from the yeast deletion library. Each mutant was combined with eleven strains carrying YFP under a specific reporter promoter using the SGA method. All together, 137*11 strains were analyzed. For each strain, the single-cell distribution of fluorescence levels was measured using flow cytometry.
B. Noise vs. mean relationship for the reporters used in the screen: The distribution of single-cell YFP expression levels driven by the reporter promoters was measured in a wild-type background. Error bars represent standard error over multiple independent measurements. We define the noise η2 as the coefficient of variation.
C. Distribution of expression levels: histograms of single-cell YFP expression levels driven by ADH3 promoter for the strains indicated. Cells were gated by the Forward-scattering and Side-scattering measures, to ensure a similar cell-cycle phase and cell-size.
D. Noise vs. mean relationship for the ADH3 promoter in all mutant backgrounds: Each point represents the noise and mean of YFP expression driven by the ADH3 promoter in one of the 137 mutant backgrounds. Error bars represent standard error over 3 independent measurements
We used flow cytometry to measure the single-cell reporter expression in each strain. Promoters changed their expression in about ~20–30% of the deletions (figure S1). The expression changes were moderate (~30%) and did not correlate with promoters’ mean abundance or noise. This is consistent with a recent study showing that individual deletions of most chromatin regulators have a minor effect on the transcription profile (Lenstra et al., 2011).
Burst-size is regulated by H2B ubiquitination and Set3-dependent deacetylation
From the distribution of expression levels, we calculated the predicted burst size and burst frequency. For this, we measured the coefficient of variation, while gating for cell population of similar cell cycle phase and size. Burst size was calculated by multiplying the coefficient of variation by the mean expression while burst frequency was calculated as the inverse of the coefficient of variation, as noted above (figure S2A). Significant effects of the deletions on burst size vs. burst frequency were consistent between the different promoters.
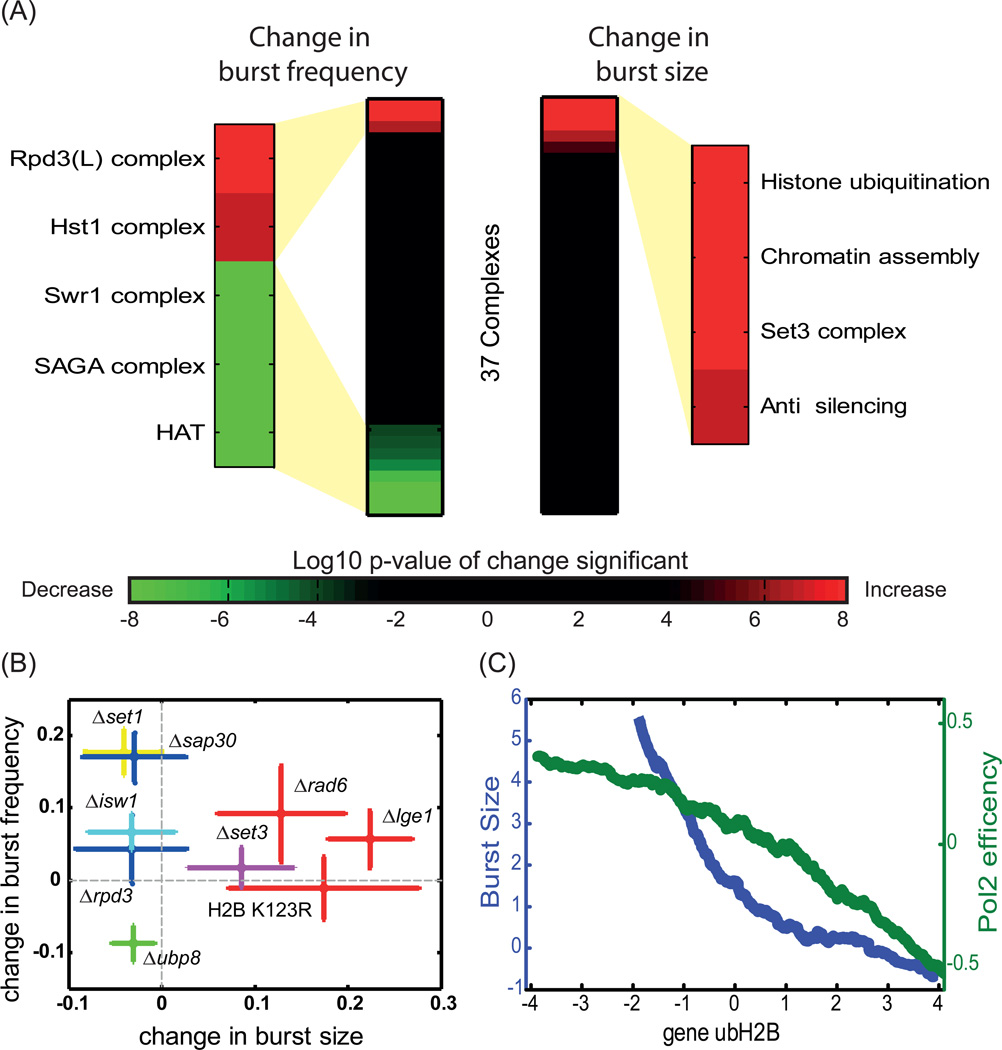
We classified the genes into known complexes and functional groups (table S1) and examined for consistent behavior (Figure 2A). Burst size was increased by deletions of LGE1 and RAD6, genes required for H2B mono-ubiquitination at Lysine residues 123 (ubH2B) (Robzyk et al., 2000). The predicted burst frequency was most strongly affected by histone acetylation: decreasing when acetyltrasferases were deleted (e.g. SAGA complex) and increasing in cells deleted of Rpd3(L)C components (SAP30 and PHO23, although not RPD3 itself) (Keogh et al., 2005). Surprisingly, the deletion of Set3C HDAC components (SET3 and HOS2) increased the predicted burst size, but not burst frequency (figure 1C–D, figure 2A).
Figure 2. Effectors of burst size and burst frequency.
A. Complexes affecting burst-size or burst frequency: The 137 chromatin regulators assayed in the screen were sorted into 37 complexes based on their function and interactions (Table S1). Fisher’s method was used to calculate the affect of each complex on burst size and frequency of all eleven promoters, based on the expression distribution of each of the strains corresponding to its individual constituents. The complexes were sorted and ranked by the strength and the direction of their effect. The color bar corresponds to significance of the changes observed (log10 of p-value).
B. Deletion affecting burst size or burst frequency: The effect of specific deletions on burst size and frequency was verified using additional 200 GFP-fusion reporters. The median change in burst frequency (mutant – wt divided by wt) is plotted as a function of the median change in burst size. The colors correspond to the different complexes/functions: Rpd3(L) histone deacetylation complex (Δsap30, Δrpd3)-blue, SET3 histone deacetylation complex (Δset3) – magenta, histone methylation (Δset1) – yellow, nucleosome remodeling (Δisw1)- cyan, histone deubiquitination (Δubp8) – green, and histone ubiquitination (Δrad6, Δlge1, h2b-K123R – histone mutant unable to go through ubiquitination) – red. Error bars represent standard errors.
C. genomic ubH2B is inversely correlated with burst size and PolI efficiency: Genes were sorted according to the average ubH2B levels throughout the gene. Shown is the normalized noise (predicted burst size) and PolII efficiency, averaged over 400-gene size sliding window.
To further verify these results, we chose nine regulators, including components of the Set3C and Rpd3(L)C, as well as genes involved in ubiquitination, deubiquitination, methylation and histone remodeling, and deleted them individually in ~200 additional reporter strains carrying different GFP-fused proteins (table S2). Consistent with the results of the initial screen, burst frequency increased when deleting the SAP30 component of Rpd3(L)C, while deletion of SET3 or genes required for ubH2B (RAD6, LGE1), increased the predicted burst size. Further, preventing H2B ubiquitination via the H2B-K123R mutation (Robzyk et al., 2000) increased the predicted burst size of most GFP-fused genes (Figure 2B, figure S2B). Taken together, this analysis suggests that ubH2B and Set3C-dependent deacetylation repress burst size in multiple genes.
ubH2B may reduce burst size by limiting PolII processivity
Our analysis assigned ubH2B a role as a repressor of burst size. If the activity of ubH2B in reducing burst size is general, then highly ubiquitinated genes will be of low noise (per mean expression). To examine this prediction, we compared the genome-wide profile of ubH2B (Schulze et al., 2009) with the predicted burst size (normalized noise) of ~2000 yeast GFP-fused proteins (Newman et al., 2006). Indeed, ubH2B levels were inversely correlated with the predicted burst size (c=−0.37, Figure 2C), supporting a general role of ubH2B in reducing burst size. We further noticed elevated levels of ubH2B at highly expressed genes (c=0.44). Together, our results suggest that ubH2B is targeted to genes of high expression where it acts to reduce burst size.
ubH2B can repress burst size by increasing the transition from permissive to non-permissive chromatin state, thereby reducing burst duration. Such regulation would imply a role at gene promoter or 5’ gene end. ubH2B, however, is found primarily within the coding region and is relatively uniform there (Schulze et al., 2009)(Fig S2C). Furthermore, previous studies assigned ubH2B a roles in transcription elongation: it stabilizes nucleosomes, promotes their re-assembly (Chandrasekharan et al., 2009; Fleming et al., 2008) and is required for the elongation-promoting phosphorylation of PolII by Ctk1 (Wyce et al., 2007). Further, limiting H2B ubiquitination increases sensitivity to drugs interfering with transcriptional elongation (Kim and Buratowski, 2009; Wyce et al., 2007). Thus, ubH2B is more likely to affect burst size through its role in transcription elongation.
Burst size depends on PolII processivity, namely the probability that an elongating PolII will produce a functional transcript rather than terminating prematurely. PolII processivity is regulated at the level of transcription elongation and not transcription initiation (Mason and Struhl, 2005; Struhl, 2005; Zhang et al., 2007). We hypothesized that ubH2B reduces burst size by repressing PolII processivity. In support of that, preventing H2B ubiquitination by the htb-K123R mutation led to a PolII processivity defects at the Gal1 gene (Chandrasekharan et al., 2009; Fleming et al., 2008).
A measure that depends on PolII processivity, but is easier to measure, is PolII efficiency, which we define as the number of mRNA transcripts produced per gene-bound PolII (normalized to gene length). We therefore predicted that PolII efficiency will be low at genes that are of high ubH2B. To examine this, we defined PolII efficiency using the genome-wide binding data of the PolII RPB3 subunit or the group of elongation factors (Mayer et al., 2010). mRNA levels were quantified using the data of (Yassour et al., 2009) (supplementary Methods). As predicted, PolII efficiency correlates with burst size (c=0.25; Figure S2F) and is inversely correlated with ubH2B (c= −0.39; Figure 2C, Figure S2F). An independent measure for ubH2B levels is provided by the binding profile of PAF1, an elongation factor facilitating ubH2B (Kim and Roeder, 2009; Warner et al., 2007). PAF1 binding profile was strongly correlated with ubH2B level (c=0.69), and was inversely correlated with PolII efficiency and burst size (Figure S2D).
PolII efficiency depends on PolII processivity but may also be regulated by additional processes. For example, slowing elongation will decrease processivity (and burst size) only if increasing the probability of premature PolII termination, but will reduce efficiency even if not effecting processivity. As an additional, more direct measure of PolII processivity, we examined the decrease in PolII density along the gene, using the high-resolution data recently published (Churchman and Weissman, 2011). Indeed, consistent with ubH2B repressing PolII processivity, the decrease in PolII density along the gene was inversely correlated with ubH2B levels (c=−0.28) and also with PAF1 binding (c=−0.39) (Figure S2G–H).
Genome-wide functions of Set3C and Rpd3(L)C
Our screen for noise regulators indicated that Set3C, like ubH2B effectors, reduces burst size. In contrast, Rpd3(L)C was predicted to reduce burst frequency, suggesting fundamentally different modes of action of these two HDACs. To more generally characterize the differences between Set3C and Rpd3(L)C we applied Chip-Seq to map H3K9 acetylation (H3K9ac) in five mutants, individually deleted for components of Set3C (SET3 and HOS2) or Rpd3(L)C (SAP30, PHO23 and RPD3). Note that while the SAP30 and PHO23 are specific components of Rpd3(L)C, RPD3 itself participates also in Rpd3(S) HDAC complex (Keogh et al., 2005). We profiled the H3K9ac mark since it correlates with gene expression (Liu et al., 2005; Robyr et al., 2002) and was previously used to analyze the activity of Set3C at individual genes (Kim and Buratowski, 2009; Liu et al., 2005).
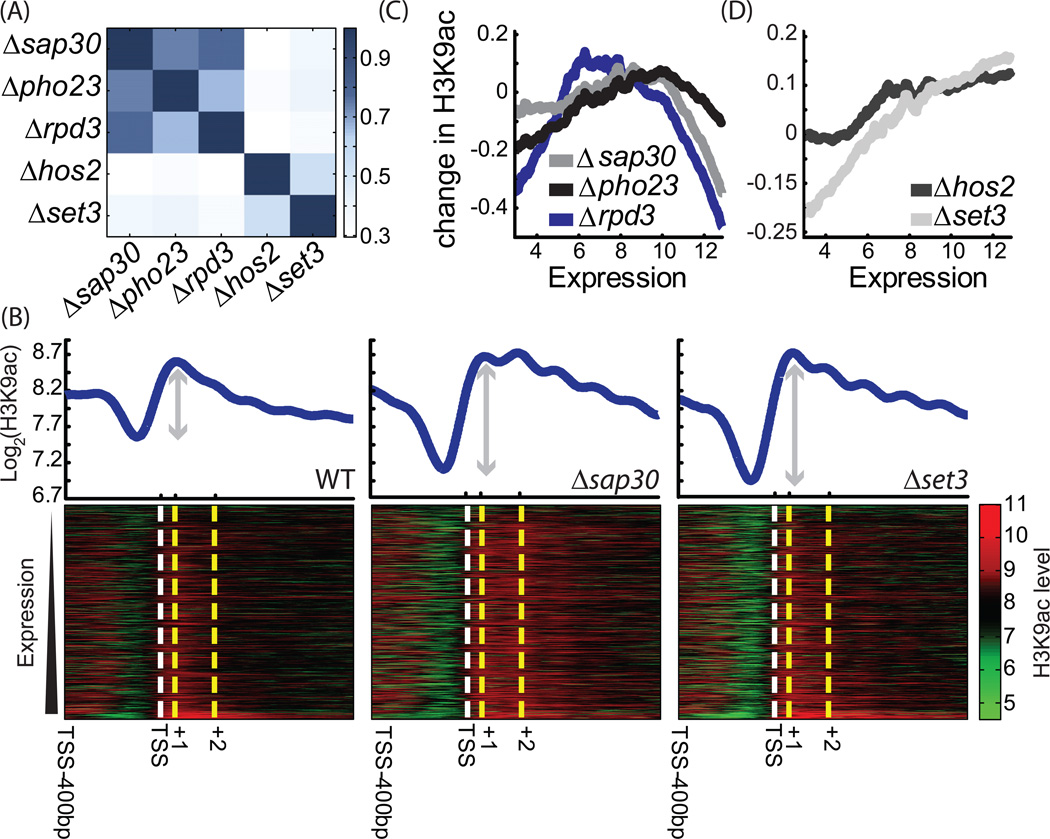
Changes in acetylation were highly correlated between same-complex deletions (Figure 3A, Figure S3A). For Rpd3(L)C, the two specific subunits (SAP30 and PHO23) correlated significantly better than the non-specific subunit RPD3, although all three were closer to each other than to the two Set3C components.
Figure 3. H3K9ac profiles in SET3C and RPD3(L)C HDAC deletions.
A. Consistent profiles of Rpd3(L) and Set3C components: correlation matrix (Pearson) between the log-ratio change in the acetylation profiles in the mutants relative to wild-type. Acetylation levels were averaged over the coding region, for up to 800 bp from the transcription start site (Xu et al., 2009).
B. The pattern of H3K9ac: H3K9ac profile in wild-type and two mutants, as indicated. Genes were sorted by their wild-type expression (Yassour et al., 2009). The average acetylation along each genes is plotted at the top panel.
C. Rpd3(L)C preferentially targets genes of mid expression: Genes were sorted by their wild-type expression. The (log2) change in gene acetylation is plotted for the indicated mutants, averaged over a sliding window of 400 genes. The decrease in Rpd3(L)C effect at highly expressed genes remains also when ribosomal proteins are excluded(Figure S4A)
D. Set3C preferentially targets genes of high expression: Same as C for deletion of Set3C components
See also figure S3
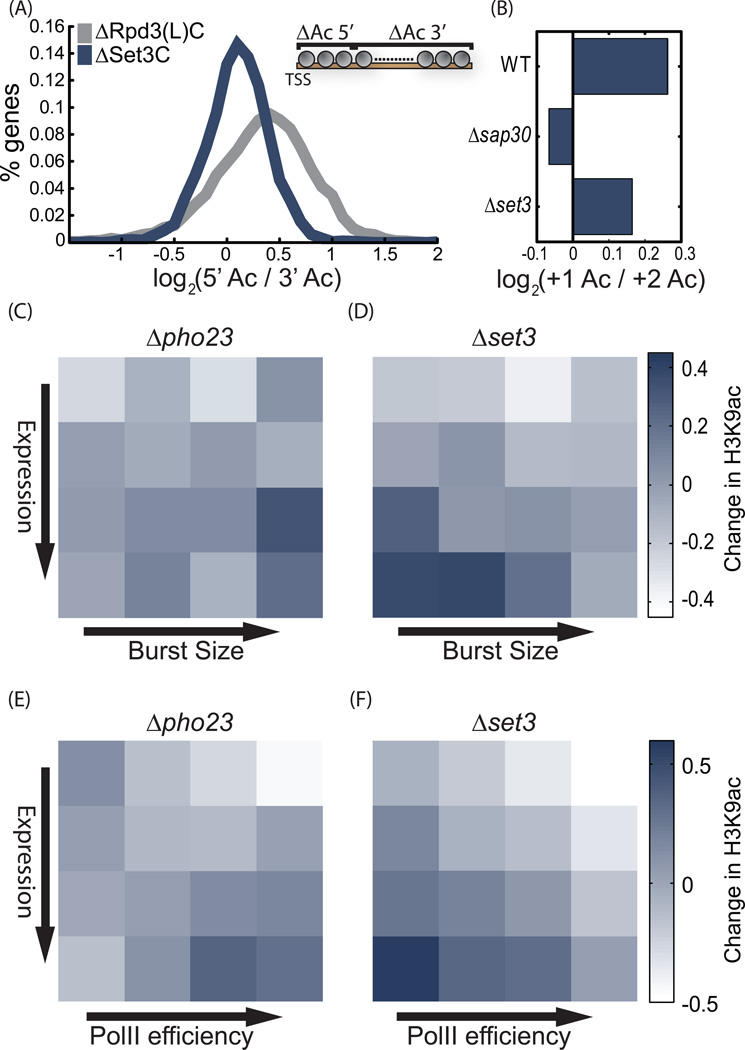
The data we obtained was of high enough resolution to define acetylation of single nucleosomes. H3K9ac is strongest at the +1 nucleosome, directly downstream of the transcription start site, and decreases gradually within the coding region (Figure 3B, Figure S3C). A notable drop between the +1 and +2 nucleosomes suggests a +1 specific acetylation or +2 nucleosome specific deacetylation. All deletions increased acetylation primarily within the coding regions, as reflected by the sharper increase in acetylation between promoter and coding region in the mutants relative to wild-type (Figure 3B, Figure S3C). This is in contrast with the common view that Rpd3(L)C acts preferentially in promoters, but is consistent with the recent work showing Rpd3(L) binding also to coding regions (Drouin et al., 2010).
Gene expression correlates with H3 acetylation only at the 5’ end of genes
Both Set3C and Rpd3(L)C were predicted to act as repressors of gene expression in our screen. Yet, we find them to preferentially deacetylate genes of high (Set3C) or medium (Rpd3(L)C) expression (Figure 3C–D, Figure S4A). Furthermore, while the role of Rpd3(L)C as a repressor is well established, previous studies have shown that Set3C is required for the rapid activation of the GAL1 gene (Kim and Buratowski, 2009; Wang et al., 2002). To verify their role as repressors of gene expression, we examined the relationship between H3K9ac, HDAC activity and gene expression.
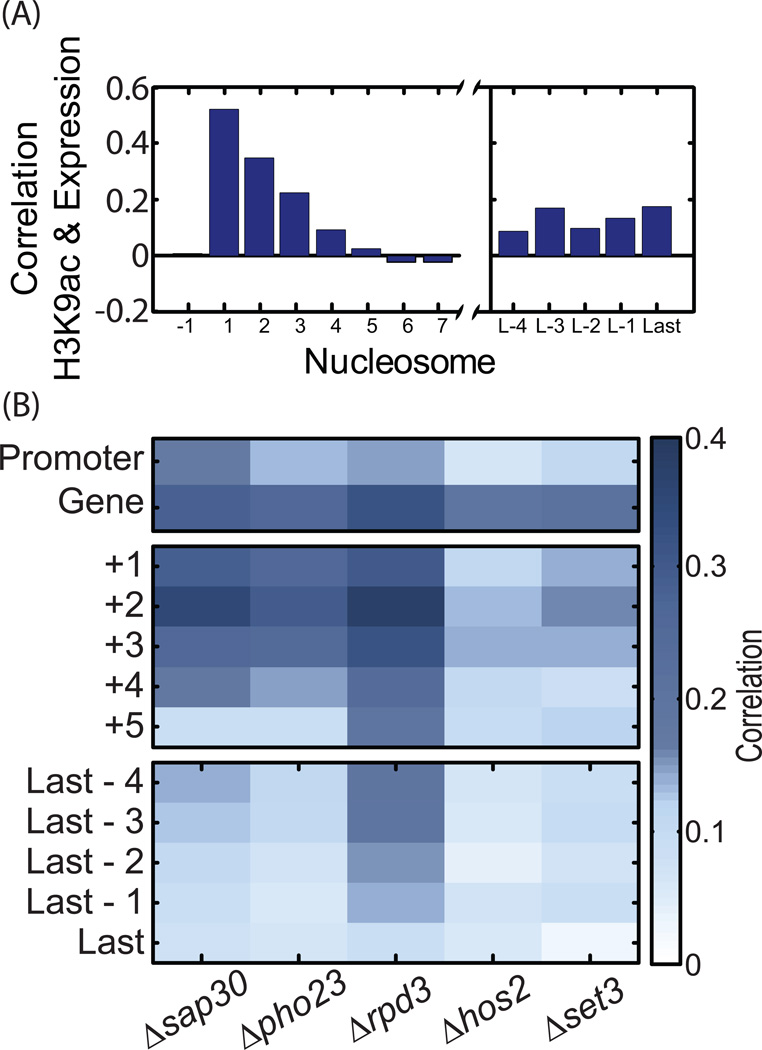
The total H3K9ac over a gene increases with gene expression (c=0.4), consistent with previous reports (Liu et al., 2005; Pokholok et al., 2005). Our high-resolution data demonstrates, however, that this correlation varies along gene positions, with high correlation at the 5' end but considerably lower correlations at downstream nucleosomes (Figure 4A). In fact, gene expression does not correlated with H3K9ac downstream of the +4 nucleosome. Thus, H3K9ac peaks at the +1 nucleosome where it is highly correlated with gene expression, and gradually decreases at subsequent nucleosomes while losing the correlation with gene expression.
Figure 4. Correlations between gene expression, H3K9ac and HDACs activity.
A. Gene expression correlates with H3K9ac only at the 5’ gene end: Pearson correlations between wild-type expression and H3K9ac levels at each nucleosome. Note that H3K9ac signal strength at the +5,+6 and +7 is the same as at the 3’ gene end where H3K9ac and gene expression are correlated.
B. Changes in expression are correlated with changes in H3K9ac: gene expression data for the different were taken from published dataset (Lenstra et al., 2011), and were correlated with the change in H3K9ac. Promoter acetylation is approximated by the H3K9ac levels at the -1 nucleosome. Changes in acetylation were averaged over all genes at the relevant regions.
We next used published gene expression profiles in these mutants (Lenstra et al., 2011) and compared the expression changes with the changes in H3K9ac. In all mutants, changes in expression and acetylation were correlated. The correlations are rather small, in particular for the Set3C deletions that had a moderate effect on gene expression (Figure 4B). In all mutants, changes of +2 nucleosome acetylation best predicted expression changes. Together, these results support the notion that both Rpd3(L)C and, to a lesser extent, Set3C act as repressors of gene expression.
Evidences that Set3C reduce burst size by repressing PolII processivity
We predicted that genes targeted by Set3C are of low normalized noise, reflecting Set3C role in reducing burst size. Indeed, comparing our acetylation profiles with the available large-scale noise measurements (Newman et al., 2006) we find that Set3C preferentially targets genes of low predicted burst size. In contrast, Rpd3(L)C preferentially targets genes of high predicted burst size (Figure 5C–D, Figure S4B).
Figure 5. Gene targets of Rpd3(L)C and Set3C.
A. Activities of Set3C and Rpd3(L)C along the coding region: For each mutant, the ratio between the (log2) change in H3K9ac at gene beginning (TSS to TSS+400bp) and at gene end (TSS+400 to gene end) was measured. Shown is the histogram of this ratio, averaged for two mutants of each complex (SAP30, PHO23 for Rpd3(L)C and HOS2, SET3 for Set3C).
B. The decrease in H3K9ac at the +2 vs. +1 nucleosomes depends on Rpd3(L)C: the (Log2) ratio between H3K9ac levels at the +1 vs. the +2 nucleosomes in the indicated mutants.
C–F. Burst size and PolII efficiency of target genes: Genes were ordered into 16 groups based on gene expression and burst size (or PolII efficiency, as indicated). Shown is the average change in H3K9ac for each group and indicated mutant. For burst size, the analysis was restricted to the ~2000 genes where data is available, while PolII efficiency was defined for all genes. In each mutant, changes in H3K9ac were normalized to a mean of 0 and standard deviation of 1, to allow easier comparisons.
See also figure S4
We hypothesize that, similarly to ubH2B, Set3C represses burst size of highly expressed genes by reducing PolII processivity. Consistent with a role in elongation, Set3C acts preferentially within the coding region and affects nucleosomes relatively uniformly (Figure 5A). In contrast, Rpd3(L)C acts preferentially at the 5’ end of genes, and specifically at the +2 nucleosomes. In fact, the decrease in H3K9ac between the +1 and +2 nucleosomes is completely lost in all three strains deleted of Rpd3(L)C components (Figure 5B, Figure S3B). The activity of Rpd3(L)C at the +2 nucleosome suggests a post-recruitment step in transcription initiation. Notably, it was shown that genes of medium expression accumulate PolII at the 5’ gene end, consistent with these genes being the preferred targets of Rpd3(L)C (Venters and Pugh, 2009; Wade and Struhl, 2008).
Furthermore, we find that Set3C preferentially targets genes of low PolII efficiency. In contrast, Rpd3(L)C targets are of mid-to-high PolII efficiency (Figure 5E–F, Figure S4C-D). As explained above, PolII efficiency is a surrogate for PolII processivity, supporting the idea that Set3C reduces burst size by increasing the chance that the polymerase will abort transcription. Consistent with that, the spatial decay of PolII throughout genes was faster in Set3C targets (figure S2H).
ubH2B promotes Set3-dependent H3K9ac
Previous studies linked Set3C recruitment to ubH2B. First, at least in some cases, Set3C was recruited by H3K4 dimethylation, a modification that requires SET1 recruitment by ubH2B (Kim and Buratowski, 2009). Second, ubH2B may also promote Set3C function through PAF1, in a SET1-indepenent manner (Lenstra et al., 2011). We therefore hypothesized that ubH2B and Set3C repress burst size through the same pathway, and asked whether our data supports such a link.
As a first indication that ubH2B promoters Set3C function, we observed that Set3C (but not Rpd3(L)C) preferentially deacetylates genes of high ubH2B and high PAF1 occupancy (Figure 6D). We further expected an inverse correlation between acetylation and ubH2B, but reasoned that such correlation may be hindered by the mutual dependence of both H3K9ac and ubH2B on gene expression (Henikoff and Shilatifard, 2011; Kim and Buratowski, 2009; Lickwar et al., 2009). Following our observations that expression correlates with H3K9ac at gene beginning only, we examined the correlation between H3K9ac and ubH2B at individual nucleosomes, expecting to observe an (inverse) correlation between ubH2B and H3K9ac at downstream nucleosomes.
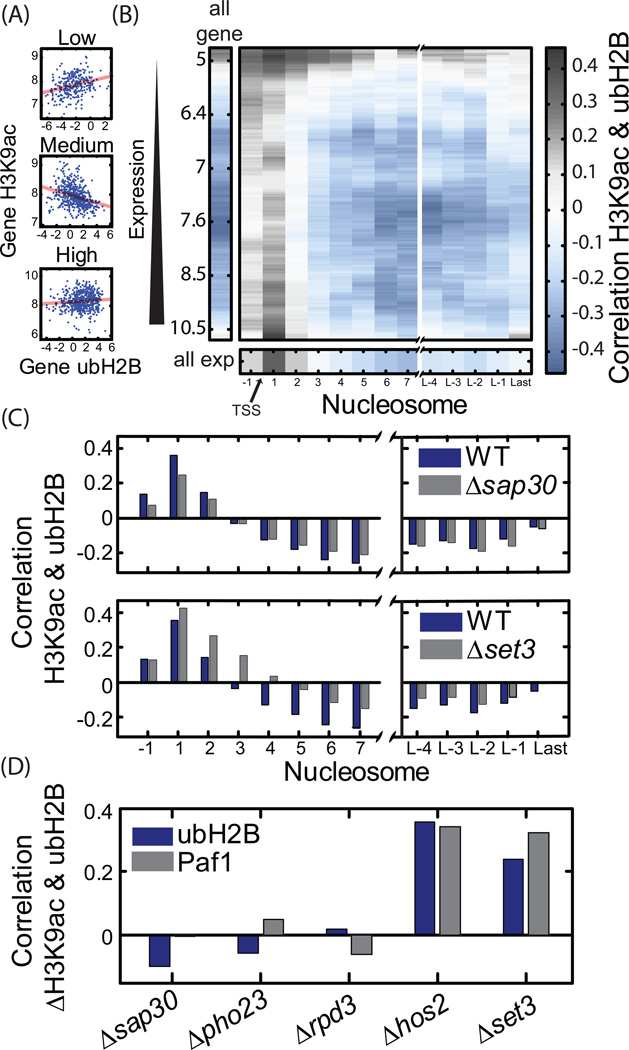
Figure 6. Correlation between ubH2B and H3K9ac.
A–B. The correlation between H3K9ac and ubH2B depends on gene expression and nucleosome position: (A) H3K9ac profile as a function of gene ubH2B for the 10% lowest expressed genes, 10% of medium expressed genes and 10% highest expressed genes, as indicated. Linear fit is shown in red. (B) In the center plot, the correlation between H3K9ac at particular nucleosome and the gene average ubH2B for gene groups of different expression. Each correlation value was measured for a window of 500 genes, centered at the indicated expression. The correlation of ubH2B with gene average H3K9ac is shown at the left, whereas the correlation between ubH2B and the nucleosome-specific H3K9ac for all expression level is shown on the bottom (See also figure S5A for a similar plot describing correlation between H3K9ac and PAF1 binding).
C. Deletion of Set3C eliminates the negative correlation between H3K9ac and ubH2B: Correlation between gene average ubH2B and H3K9ac at different nucleosomes is plotted for WT (same as in B bottom), as well as strains deleted of SAP30 or SET3.
D. Set3C preferentially targets genes of high ubH2B: The average Rank correlation between gene average ubH2B (or Paf1 binding) and changes in gene H3K9ac is shown for the different mutants.
Also see figure S5
As predicted, we find that gene ubH2B levels correlate with H3K9ac at the +1 nucleosome but this correlation vanished rapidly at downstream nucleosomes (Figure 6B, Figure S5A). In fact, an inverse correlation between the gene average ubH2B and H3K9ac was apparent for all positions downstream of + nucleosomes. Thus, in most regions inside genes, higher average ubH2B is associated with lower acetylation, suggesting that ubH2B promotes deacetylation, as predicted.
To further support the idea that the spatially-varying correlation pattern we observe reflects the mutual dependence of ubH2B and H3K9ac on gene transcription, we grouped genes based on expression levels, and analyzed the correlation between ubH2B and H3K9ac within each group (figure 6A–B). We found negative correlation between ubH2B and average gene H3K9ac in genes of mid expression, while highly expressed genes showed no correlation and lowly expressed genes showed positive correlation. To further understand the difference between the expression levels, we looked at the correlations at individual nucleosome. While high expressed genes showed negative correlation downstream inside the gene, low expressed genes didn’t. We hypothesized that the two phenomena are due to the coupling of both modifications to transcription process. While in the low express genes histone ubiquitination is mostly not present, in high express genes the high level of histone acetylase activity dominates the H3K9ac pattern over the effect of deacetylation.
Together, our analysis supports the idea that ubH2B promotes deacetylation. To examine whether ubH2B functions through Set3C, we examined the correlation between ubH2B and H3K9ac in the different deletion mutants. Deletion of Rpd3(L)C-specific subunits (SAP30 or PHO23) had no effect on the correlation pattern, while deletion of the Set3C subunits reduced the negative correlation between ubH2B and H3K9ac in all nucleosomes and at all expression levels (Figure 6C, Figure S5B-D). Notably, even in Set3C mutants, a negative correlation was still observed in nucleosomes closer to the 3’gene end, suggesting that ubH2B recruits additional HDACs.
H3K9ac profiles of mutants deleted of ubH2B effectors support Set3C recruitment by ubH2B
Our analysis supports a genome-wide link between ubH2B and Set3C. To more directly examine the prediction that ubH2B promotes Set3-dependent deacetylation, we mapped the H3K9ac profile in mutants deleted of two H2B ubiquitinating enzymes (BRE1 and RAD6) and two H2B deubquitinating enzymes (UBP8 and UBP10). Due to technical reasons, the resolution this ChIP-seq data was lower than that of the deacetylation mutants analyzed above, and we therefore consider only the gene-averaged profile (rather than single-nucleosomes).
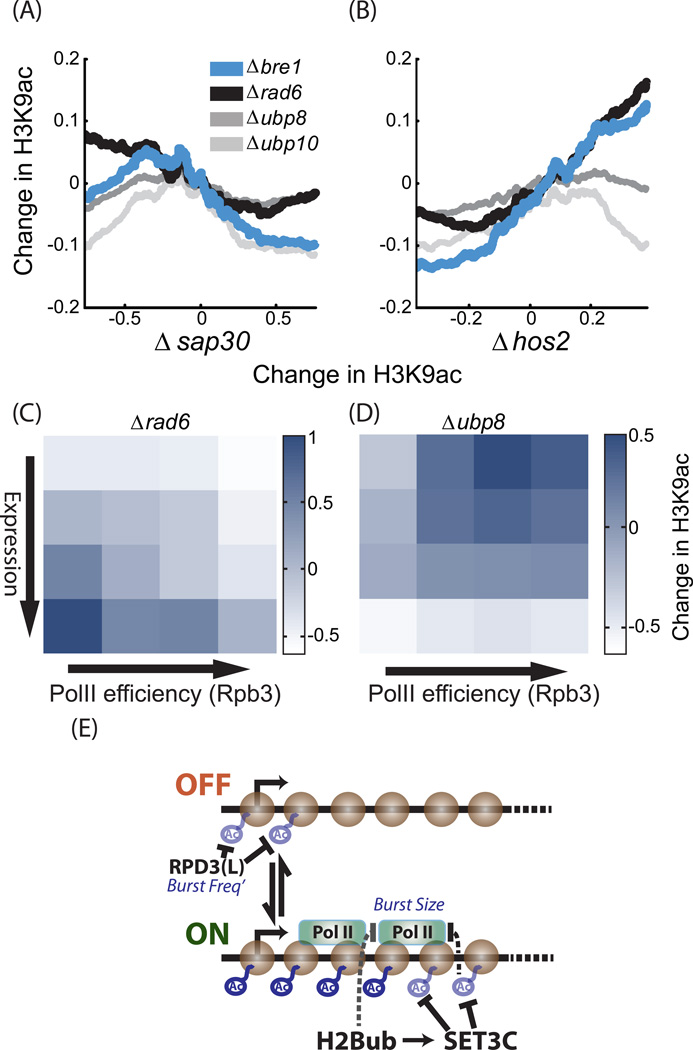
As predicted, deletion of BRE1 or RAD6 increased acetylation preferentially at Set3-dependent genes and genes of high ubH2B (Figure 7A–B, Figure S6B–F). Further, consistent with a function of ubH2B in reducing PolII efficiency, both mutants affected preferentially genes of low PolII efficiency (Figure 7C, Figure S6G–I). Those results directly support the recruitment of Set3C by ubH2B to genes of high expression and low noise.
Figure 7. H3K9ac profiles in mutants deleted of ubiquitination and de-ubiquitination enzymes.
A–B. Deletion of BRE1 or RAD6 increase acetylation of Set3-dependent genes: Genes were sorted by average change in H3K9ac in strains deleted of SAP30 (A) or HOS2(B). The (log2) change in acetylation in the indicated mutants is shown. H3K9ac values were averaged over 400 genes sliding window.
C–D. BRE1 targets genes of high Expression and low PolII efficiency: genes were ordered into 16 groups based on gene expression and PolII efficiency. Shown is the average change in H3K9ac at the indicated mutant for each group. In each mutant, changes in H3K9ac were normalized to a mean of 0 and standard deviation of 1, to allow an easier comparison (See also figure S6G–J for other mutants).
E. Graphic model summarizing the results: In the prevailing model of gene expression, genes are made in bursts, namely period of extensive expression interspaced by time intervals in which expression is negligible. This is captured by assuming that gene can be at two states: a state permissive for expression (ON) and a state that is not permissive (OFF). The rate of transition from the OFF to ON state defines the burst frequency, while the number of proteins made per burst even defines the burst size. Gene expression can be regulated through changes in either burst frequency or burst size. Our results indicated that Rpd3(L) represses burst frequency, probably by deacetylation of nucleosomes at the beginning of a gene (+2 in particular). In contrast, Set3C and ubH2B are predicted to repress burst size by reducing PolII processivity. ubH2B and Set3C likely function within the same pathway, to modulate nucleosomes within the gene.
See also figure S6
Notably, the H3K9ac profiles in the strains deleted of UBP8 or UBP10 did not correlate with the profiles of Set3C deletions (Figure 7B, Figure S6D). This is consistent with the limited impact of Ubp8 on burst size, predicted by our screen, and also with a recent report showing that deleting UBP8 or UBP10 does not change the profiles of PolII binding or histone methylation (Schulze et al., 2011).
Discussion
Our study began with the theoretical observation that gene expression variance (noise) can distinguish mechanisms of gene expression regulation (Friedman et al., 2006; Raj et al., 2006; Tan and van Oudenaarden, 2010). Applying this approach to 137 chromatin-associated factors suggested distinct roles of the Set3C and Rpd3(L)C HDACs. In our assay, both complexes repressed the expression of multiple reporter genes, yet appeared to function through distinct mechanisms: Rpd3(L)C modulated the predicted burst frequency while Set3C modulated the predicted burst size, as did ubH2B. Using genome-wide profiling of H3K9ac, we further characterized the HDAC activities of Set3C and Rpd3(L)C.
Previous studies associated both ubH2B and Set3C with transcription elongation (Fleming et al., 2008; Kim and Buratowski, 2009). This is also evident from the high correlation between expression level and ubH2B inside the gene. We suggest that this reflects the coupling of ubH2B to the transcription machinery, rather than a role for ubH2B in promoting transcription. Consistent with that, we find that Set3C functions relatively uniformly in coding region. Based on our data, we further refined the role of ubH2B and set3C in elongation, suggesting that they act to decrease PolII processivity, namely the probability that an elongating PolII will produce a functional transcript. Other elongation factors may not influence burst size, or could even have an opposite effect. For example, TFIIS (DST1 in yeast) increases PolII processivity (Mason and Struhl, 2005). Notably, recent work in human have shown that ubH2B inhibits the ability of TFIIS to relieve stalled PolII (Shema et al., 2011). Thus, both the increase of PolII processivity by TFIIS, and its inhibition by ubH2B appear to be conserved between yeast and human.
How could ubH2B and Set3C reduce PolII processivity? Both ubH2B and deacetylation can stabilize nucleosomes (Chandrasekharan et al., 2009; Henikoff and Shilatifard, 2011; Kurdistani and Grunstein, 2003). Higher nucleosome stability may interfere with PolII progression and increase the probability of pausing or premature termination. Alternatively, factors that actively promote premature PolII termination might be recruited in ubH2B or Set3C-depdent manner. There are examples for such active recruitment of the Nrd1-Nab3-Sen1 termination complex (Kim and Levin, 2011; Terzi et al., 2011). Thus, the Paf1 complex, which is required for ubH2B, also recruits Sen1 to repress the FKS2 gene. ubH2B and H3K4 dimethylation were also associated with this recruitment (Kim and Levin, 2011). This pathway, however, functions mostly in the regulation of snoRNA and acts at 5’ end of genes (Sheldon et al., 2005; Steinmetz et al., 2006). In contrast, most of the effects we observe occur within coding regions.
Our data supports a general role of ubH2B in promoting Set3C-dependent deacetylation, suggesting that they function through the same pathway. ubH2B may promote Set3C function indirectly, by recruiting Set1 thereby promoting H3K4 dimethylation. Alternatively, ubH2B promotes Set3C function primarily through a process independent of Set1, as was suggested recently (Lenstra et al., 2011). We favor this latter hypothesis since Set3C did not appear to prefer genes of high H3K4 di- or trimethylation (Pokholok et al., 2005) (Figure S6A). Further, our screen, deletion of Set1 did not increase burst size, although this result is hard to interpret due to the severe growth limitation of this strain. Notably, the effect of ubH2B on burst size is consistently stronger than that of Set3C, suggesting that ubH2B effects burst size both by facilitating Set3C function and by additional means.
More generally, the high resolution of our data enabled characterizing acetylation at the level of single nucleosomes. We found that transcription-dependent acetylation is localized to the 5’ end of genes but is largely absent from mid-gene and downstream. Analysis of these nucleosomes can therefore be used to identify direct interactions between different chromatin marks, as we demonstrated in the context of ubH2B-dependent recruitment of Set3C. We anticipated that, as sequencing technologies become more efficient, similar high-resolution spatial analysis can be used to distinguish direct from indirect effects of chromatin modifications.
Our study suggests that gene expression noise depends on specific regulatory factors that function at only a subset of genes. Noise may therefore be subject to selection independent of mean expression. Tuning noise levels may be beneficial: some genes are required at precise amount, while noise in others could provide diversity to an otherwise genetically identical population. It will be interesting to examine whether noise levels diversified during evolution, and whether this diversity was driven, at least in part, by changing the function or specificity of chromatin factors such as the ubiquitination or deacetylation enzymes.
Materials and methods
Strains and media
Construction of strains for noise screen was done by SGA method (Tong et al., 2001) with minor extensions. GFP-fusion strains were taken from the yeast GFP clone collection. The ChIP-Seq experiments were done on BY4741 strains. Full list on strains can be found in the supplementary tables 1–2.
Flow cytometry
GFP fluorescence was measured for stains in logarithmic growth stage, using LSRII flow cytometer fitted with HTS sampler (Becton Dickinson). Analysis and gating of FACS measurements is described in Supplementary methods.
ChIP-Seq
Chromatin immuno-precipitation was done as described previously (Liu et al., 2005), with some modifications. Samples were sonicated with bioruptor sonicator for 60 minutes (30 sec intervals), and were precipitated with H3K9-acetyl antibody (Abcam ab12179) and conjugated to protein A beads. High throughput sequencing was done using Illumina GAII system for mots samples and HiSeq2000 for Δbre1, Δrad6, Δubp8, Δubp10 and a second WT. Analysis of the high throughput sequencing, definition of polII efficiency and definition of nucleosome and gene regions are described at Supplementary methods.
Accesion numbers
Chip-seq data files described in this study are published in the SRA database, accession number SRA051855.1
Supplementary Material
Highlights.
Gene expression noise can distinguish the role of transcription regulators
Noise-based predictions are verified by genome-wide profiling of H3K9 acetylation.
Evidence are provided that Set3C and ubH2B repress polII processivity
Acknowledgments
We thank Sagi Levy and Ilya Soifer for remarks on the manuscript, Yaron Mosseson and Dalia Rosin-Grunewald for experimental help, Dima Lukatsky for starting the project, Shirley Horn-Saban and members of our group for discussions. This work was supported by NIH (P50GM068763), by the ERC, The Israel Science Foundation, and by the Hellen and Martin Kimmel award for innovative investigations.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Barkai N, Shilo BZ. Variability and robustness in biomolecular systems. Mol Cell. 2007;28:755–760. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS Genet. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011 doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–756. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One. 2009;4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Noise in gene expression: origins, consequences, control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinott R, Jaimovich A, Friedman N. Exploring transcription regulation through cell-to-cell variability. Proc Natl Acad Sci U S A. 2011;108:6329–6334. doi: 10.1073/pnas.1013148108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Hentrich T, Nakanishi S, Gupta A, Emberly E, Shilatifard A, Kobor MS. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Genes Dev. 2011;25:2242–2247. doi: 10.1101/gad.177220.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3' end formation. Mol Cell. 2005;20:225–236. doi: 10.1016/j.molcel.2005.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell. 2011;42:477–488. doi: 10.1016/j.molcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SB, Cloute JP, Brow DA. cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol. 2006;26:2688–2696. doi: 10.1128/MCB.26.7.2688-2696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional activation: mediator can act after preinitiation complex formation. Mol Cell. 2005;17:752–754. doi: 10.1016/j.molcel.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Tan RZ, van Oudenaarden A. Transcript counting in single cells reveals dynamics of rDNA transcription. Mol Syst Biol. 2010;6:358. doi: 10.1038/msb.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 Trimethylation by Set1 Promotes Efficient Termination by the Nrd1-Nab3-Sen1 Pathway. Mol Cell Biol. 2011;31:3569–3583. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci U S A. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol. 2008;15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Klatt A, Henderson AJ, Gilmour DS. Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev. 2007;21:1609–1614. doi: 10.1101/gad.1542707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.