Abstract
Methylmercury (MeHg) is a persistent environmental toxicant that is commonly encountered through dietary fish and seafood. While the fetal nervous system is a well-known primary target for MeHg toxicity, the risks of MeHg exposures that are commonly experienced today through diet and environmental exposure remain uncertain. Despite knowledge of numerous cellular processes that are affected by MeHg, the mechanisms that ultimately influence tolerance or susceptibility to MeHg in the developing fetus are not well understood. Using transcriptomic analyses of developing brains of MeHg tolerant and susceptible strains of Drosophila, we previously identified members of the cytochrome p450 (CYP) family of monooxygenases/oxidoreductases as candidate MeHg tolerance genes. While CYP genes encode Phase I enzymes best known for xenobiotic metabolism in the liver, several classes of CYPs are required for synthesis or degradation of essential endobiotics, such as hormones and fatty acids, that are critical to normal development. We now demonstrate that variation in expression CYP genes can strongly influence MeHg tolerance in the developing fly. Importantly, modulating expression of a single CYP, CYP6g1, specifically in neurons or the fat body (liver equivalent) is sufficient to rescue development in the presence of MeHg. We also demonstrate a conserved function for CYP3A4, a human homolog of CYP6g1, in conferring MeHg tolerance to flies. Finally, we show that pharmacological induction of CYPs with caffeine parallels an increase in tolerance to MeHg in developing flies. These findings establish a previously unidentified role for CYPs in MeHg toxicity and point to a potentially conserved role of CYP genes to influence susceptibility to MeHg toxicity across species.
Keywords: Methylmercury, Drosophila, Cytochrome P450, CYP6g1, CYP3A4
1. Introduction
Methylmercury (MeHg) is a persistent environmental toxin that is most commonly encountered through intake of dietary fish and seafood. It has long been understood that the fetal nervous system is a primary target for MeHg toxicity. Yet, the risks of MeHg exposures that are commonly experienced in the human population today remain uncertain due to an incomplete understanding of factors that influence toxic outcomes during development. Resolving the scientific basis for susceptibility to MeHg is a significant challenge. Most studies to date have focused on identifying cellular targets of MeHg as a way of gaining insight. As a result, the list of proteins and pathways targeted by MeHg is long, consistent with its high affinity for thiols and its ubiquitous reactivity (Castoldi et al., 2001; Eldefrawi et al., 1997; Hughes, 1957; Miura et al., 2000; Sarafian, 1999; Sirois and Atchison, 2000). Despite knowledge of numerous cellular processes that are affected by MeHg, the mechanisms that ultimately influence tolerance or susceptibility to MeHg in the developing fetus are not well understood.
In a previous study we identified candidate MeHg tolerance genes via a transcriptomic analyses of developing brains of tolerant and susceptible strains of Drosophila (Mahapatra et al., 2010). This previous study resolved that the members of the cytochrome p450 (CYP) family of monooxygenases/oxidoreductases are significantly upregulated in MeHg tolerant flies. CYP genes constitute a large family (57 human CYPs and 87 Drosophila CYPs) that share an overall high degree of structural and functional conservation in metazoans and in plants (Graham and Peterson, 1999; Werck-Reichhart and Feyereisen, 2000). CYP genes encode Phase I metabolizing enzymes, which are best known for carrying out covalent modification of xenobiotics in the liver (Smart and Hodgson, 2008). In the context of pharmaceuticals, CYPs can either invoke activation of a favorable drug or convert a compound to a highly toxic metabolite. However, several classes of CYPs are essential for synthesis or degradation of essential endobiotics, such as hormones and fatty acids, and thus critical to normal development (Guittard et al., 2011; Sanderson, 2009). Importantly, CYPs are implicated in neuroprotection, e.g. In the metabolism of MPTP, a toxic entity that provokes onset of Parkinson’s disease (Mann and Tyndale, 2010; Viaggi et al., 2006).
While predominantly expressed in the liver, CYPs are expressed in extra-hepatic tissues, including the nervous system, albeit at lower levels (Hedlund et al., 2001; Pavek and Dvorak, 2008). Yet, a number of CYPs show inducible expression in response physiological and pathophysiological factors, such as stress and inflammation, as well as in response to xenobiotics (Xu et al., 2005). Despite the central role CYPs play in xenobiotic and endobiotic metabolism, a potential role for CYPs in MeHg toxicity has been little explored.
In this study we investigate a role for CYP gene expression in influencing tolerance to MeHg using a Drosophila development assay. We focus on Drosophila CYP6g1, the most highly expressed CYP among the candidate MeHg tolerance genes identified in our previous Drosophila screen. We also investigate a conserved function in CYP3A4, a human homolog of CYP6g1. Using transgenic and pharmacological means of altering CYP expression we show an overall ability for elevated CYP levels to invoke MeHg tolerance in developing flies. Importantly, this apparent MeHg tolerance activity of CYPs is seen when expression is targeted to neurons as well as when targeted to hepatic tissue. These findings reveal a previously unidentified role for CYPs in MeHg toxicity and establish a role for CYPs as genes able to influence susceptibility to MeHg toxicity during development.
2. Methods
2.1. Transcriptome analysis of CYP gene family expression
Relative levels of expression of CYP genes in larval central nervous system of laboratory selected MeHg-tolerant and non-tolerant flies were determined using Affymetrix Drosophila Genome 2 microarrays in the study by Mahapatra et al. (2010), and are presented here in table form. It is of note that a concentration of 15 μM MeHg was employed where MeHg treatments of larva were analyzed in this dataset. This MeHg concentration is where differences in MeHg tolerance can be clearly distinguished, as seen in analyses of transgenic flies and treatments in conjunction with caffeine shown herein.
2.2. Fly strains
The following wild strains and transgenic lines of flies were obtained from the Bloomington Drosophila Stock Center (Indiana University): Canton S (#1), Hikone R (#4267), VAG1 (#3875), Swedish-C (#4271), Reids 3 (#3868), PVM (#3861), Wild 1B (#3879), Wild 5C (#3887), Wild 5A (#3885) and Wild 10E (#3892), Elav-Gal4 (#458), c754-Gal4 (fat body expression Hrdlicka et al., 2002 #6984). The UAS–CYP6g1 (#86 insertion) strain was a gift from Philip Daborn (Daborn et al., 2007 University of Melbourne, Australia). UAS–CYP6g1RNAi strain was obtained from the Vienna Drosophila RNAi Stock Center (Stock #v4615, http://stockcenter.vdrc.at/control/main).
UAS–CYP3A4 flies were generated from a construct created in the pUAST-attB transformation vector (Bischof et al., 2007 gift from Johannes Bischof and Konrad Basler, University of Zurich, Switzerland). cDNA encoding the CYP3A4 in the pREP10 vector (Invitrogen) was provided by Erin Schuetz (St. Judes Children’s Research Hospital, Memphis, TN). CYP3A4 sequence was excised from the pREP10 vector and cloned directly into pUASTattB via Xho/Kpn restriction sites. The resulting pUAST–CYP3A4 construct was used to create transgenic flies utilizing the phiC31 integrase system (Bischof et al., 2007) via the embryo injection services of Rainbow Transgenic Flies, Inc. (Camarillo, CA).
Flies were maintained at 25 °C on a standard preparation of cornmeal, molasses and agar medium with yeast. Crosses were performed between virgin females and males of the desired combinations of Gal4 and UAS transgenic parental lines.
2.3. MeHg tolerance assays
MeHg tolerance was assayed using a previously describe Drosophila eclosion assay (Mahapatra et al., 2010). Briefly, on day one first instar larvae were seeded on food vials (20–50 larvae/vial) containing various concentrations of MeHg (0–20 μM). Assays at each MeHg concentration were carried out in triplicate (three vials = 60–150 larvae/MeHg concentration). The number of successfully developed and eclosed adult flies was tallied on day 13. Eclosion was expressed as a percentage of the larvae that successfully develop to adults. Thus, eclosion on higher MeHg concentrations in food indicates tolerance to MeHg, while low eclosion rates indicate MeHg susceptibility. Data are expressed as the mean and standard deviation (s.d.) of eclosion percentages for the three trials. Statistical analyses were done by pairwise comparison of eclosion rates between controls (e.g. EG4 > wt) and transgene expressing combinations (e.g. EG4 > CYP6g1) at each MeHg concentration using the Student’s t-test.
Additional assays investigated effects of caffeine in MeHg toxicity. Caffeine (LKT Laboratories) was prepared in water as a 100 mM stock. Toxicity of Caffeine was assayed in eclosion assays by addition of various concentrations of caffeine (0–20 mM) to food preparations as previously described (Mahapatra et al., 2010). Subsequent eclosion assays were conducted with caffeine-supplemented food (0, 1, 2, 5 and 10 mM) prepared with various MeHg concentrations (0–20 μM) described above.
2.4. Gene expression
CYP gene expression was analyzed by quantitative RT-PCR (qRT-PCR) of RNA isolated from either whole larvae or indicated tissue samples by described methods (Mahapatra et al., 2010). RNA extractions were performed using pooled samples of tissues harvested from 20–35 larvae of the indicated genotype. For CYP6g1 expression under the EG4 driver line RNA extracts were prepared from adult heads, which are more highly enriched for neural tissue as compared to whole larva that were used in other determinations. Total RNA was isolated using Trizol reagent (Invitrogen) and cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen). The cDNA was used to perform qRT-PCR using SYBR®-Green JumpStart Taq ready mix (Sigma) on an ABI PRISM 7500 Fast Sequence Detection System (Applied Biosciences). Gene expression levels were determined by the comparative CT method (Livak and Schmittgen, 2001). Primers used were: CYP6g1, 5′-CATGGCATATCAACCCTTC-3′ and 5′-CATGGCATATCAACCCTTC-3′; CYP12d-1, 5′-GGGTTACCGGGTGCCCAAGG-3′ and 5′-ACGTGAAGGGGCTGACCTGC-3′; CYP3A4, 5′-TGCTCTTCACCGTGACCCAAAGTA-3′ and 5′-AGAGCAAACCTCATGCCAATGCAG-3′.
3. Results
3.1. The CYP gene family is upregulated in expression in MeHg tolerant flies
We previously identified the CYP family of genes as being more highly expressed in MeHg tolerant flies (Mahapatra et al., 2010). It is important to note that, in this previous study, analyses were done on RNA transcripts isolated from CNS tissue at the larval stage, and thus MeHg tolerance is associated with expression levels in the nervous system at an intermediate stage of development. To elaborate on the nature of CYP gene family expression and its potential role in MeHg tolerance these previously determined expression levels were compiled in Table 1. Twenty-one (21) of the 83 CYP genes represented on the Affymetrix Drosophila 2 Genechip array show a change in expression across the various pairwise comparisons of selection and MeHg exposure in the developing brains of MeHg tolerant and non-tolerant flies (Table 1). Importantly, 12 of these genes are seen to be upregulated in the comparison of the tolerance-selected (S20) versus non-selected (S0) flies when both are exposed to MeHg (Table 1, Column 2). Notably, CYP6g1 demonstrates a more than 6-fold higher expression in tolerant flies compared to non-tolerant flies in this comparison. It is also of note that, in the selected tolerant strain (S20), the six most highly upregulated CYPs appear to be induced with exposure to MeHg (Table 1, Column 4). This is in contrast to a ubiquitous down regulation of expression that occurs across the CYPs with MeHg exposure in the non-tolerant strain (S0) (Table 1, Column 8). Thus, the data indicate that MeHg tolerance corresponds with an overall upregulation of CYP activity in the developing brain, with CYP6g1 exhibiting the greatest change in expression in MeHg tolerant flies.
Table 1. Transcriptome analysis of CYP gene family expression.
The relative level of expression of CYP genes in larval central nervous system of laboratory selected MeHg-tolerant and non-tolerant flies, from determinations performed in our previous study employing Affymetrix Drosophila Genome 2 microarrays (Mahapatra et al., 2010), is assembled and presented here in tabular form. Values for the 21 CYP genes (of 83 total CYP genes) that show expression changes are shown. Columns indicate the pairwise comparisons of either MeHg treatment (+ versus −MeHg), selection (S20 versus S0), or the combination of MeHg treatment and selection (S20 versus S0, +/−MeHg) and are expressed as fold change (S20: tolerant strain grouping; S0: non-tolerance strain grouping; nc: no change in expression level).
| Cyp | S20 versus S0 (+MeHg) | P value | S20 (+ versus −MeHg) | P value | S20 versus S0 (−MeHg) | P value | S0 (+ v. −MeHg) | P value |
|---|---|---|---|---|---|---|---|---|
| Cyp6g1 | 6.24 | 0.001 | 3.75 | 0.006 | −2.1 | 0.067 | −3.5 | 0.008 |
| Cyp4e3 | 3.84 | 0.072 | 1.73 | 0.417 | nc | nc | ||
| Cyp309a1 | 3.02 | 0.029 | 3.96 | 0.011 | −1.76 | 0.206 | nc | |
| Cyp4d1 | 2.83 | 0.031 | 5.88 | 0.003 | −4.53 | 0.006 | −2.18 | 0.084 |
| Cyp6d2 | 2.73 | 0.012 | 1.92 | 0.066 | nc | nc | ||
| Cyp6d5 | 2.27 | 0.22 | 3.18 | 0.1 | −3.92 | 0.06 | −2.8 | 0.135 |
| Cyp12c1 | 2.2 | 0.001 | nc | nc | −2.91 | 0 | ||
| Cyp4e2 | 2.01 | 0.017 | nc | nc | nc | |||
| Cyp6a17 | 1.9 | 0.006 | nc | nc | nc | |||
| Cyp304a1 | 1.57 | 0.002 | nc | nc | nc | |||
| Cyp6a23 | 1.57 | 0.012 | nc | nc | nc | |||
| Cyp12d1-p | 1.54 | 0.027 | nc | nc | nc | |||
| Cyp4ac1 | nc | 1.63 | 0.06 | −2.44 | 0.005 | −1.83 | 0.028 | |
| Cyp9b1 | nc | 1.58 | 0 | nc | nc | |||
| Cyp4e1 | nc | −2.34 | 0.001 | nc | −1.9 | 0.006 | ||
| Cyp6g2 | nc | −2.14 | 0.001 | nc | −2.1 | 0.001 | ||
| Cyp18a1 | nc | −1.83 | 0.001 | nc | −1.85 | 0.001 | ||
| Cyp4p3 | nc | nc | −1.74 | 0 | nc | |||
| Cyp310a1 | nc | nc | nc | −2.21 | 0.001 | |||
| Cyp6a13 | nc | nc | nc | −1.78 | 0 | |||
| Cyp6w1 | nc | nc | nc | −1.71 | 0.104 |
3.2. CYP6g1 expression correlates with MeHg tolerance in wild strains
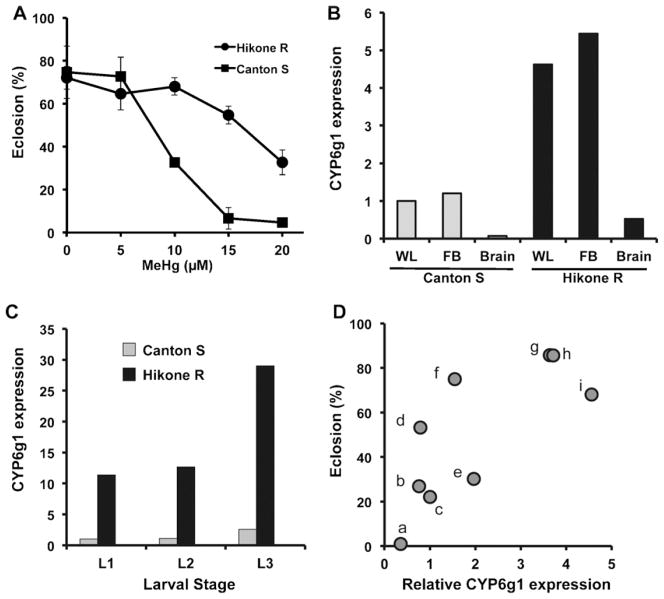
We have focused on CYP6g1 as a candidate to elucidate a potential function for CYPs in conferring MeHg tolerance. To further test the possibility that elevated expression of CYP6g1 plays a role in MeHg tolerance we first examined CYP6g1 expression and MeHg tolerance in the Hikone R wild strain. Hikone R flies have been previously characterized for high expression of CYP6g1 due to an insert known as an Accord element in the enhancer region of the gene (Daborn et al., 2002). Using an eclosion (adult hatching) assay to score development in the presence of MeHg, Hikone R flies are seen to develop and eclose at a much higher rate on high levels of MeHg compared to the standard Canton S strain (Fig. 1A). Levels of CYP6g1 expression measured by qRT-PCR of transcripts isolated from whole larvae shows CYP6g1 is more than 4.5-fold higher in Hikone R flies than in Canton S flies. As CYPs are known to be preferentially expressed in liver we examined CYP6g1 expression in the fat body, which is the organ equivalent of the liver in flies. CYP6g1 showed more than a 5-fold greater expression in the Hikone R fat body compared to the Canton S (Fig. 1B). In addition, overall expression in the whole larva and fat body are roughly equivalent indicating the fat body contributes a large fraction of the CYP6g1 transcripts to the total larval extract. CYP6g1 is expressed at an approximately 20-fold lower level in the isolated central nervous system (brain) compared to the whole larva and fat body (Fig. 1B). Yet, the Hikone R brains show more than 7-fold higher CYP6g1 expression than the Canton S brains (Fig. 1B). Thus, the high MeHg tolerance seen in the Hikone R strain correlates with elevated CYP6g1 in both hepatic and neural tissues.
Fig. 1.
MeHg tolerance and CYP6g1 expression in wild strains of flies. (A) Tolerance to MeHg during development in the Canton S and Hikone R wild strains is determined by eclosion assays. Greater rates of eclosion are seen for Hikone R strain on 10–20 μM MeHg food than for Canton S. (B) Expression levels of CYP6g1 transcripts are determined using total RNA from extracts of whole larvae (WL), fat body (FB) or central nervous system (Brain) of 3rd instar larvae of the Canton S and Hikone R strains. Determinations are done by qRT-PCR and expression levels normalized to Canton S strain WL. (C) Relative expression level of CYP6g1 transcripts across larval developmental stages are determined by qRT-PCR using total RNA from extracts of whole larvae of Canton S and Hikone R strains. Expression levels are normalized to Cantons 1st instar larvae (L1, L2, L3 = 1st, 2nd, 3rd instar larvae, respectively). (D) A comparison of tolerance to MeHg and CYP6g1 expression level is determined in nine wild strains of flies previously reported to have various levels of CYP6g1 (Daborn et al., 2002). Wild strains are analyzed by comparisons of CYP6g1expression (qRT-PCR, normalized to Canton S) and MeHg tolerance determined by eclosion rates on 10 μM MeHg food (Wild strains are: (a) VAG1; (b) Swedish-C; (c) Canton S; (d) PVM; (e) Wild 5A; (f) Reids 3; (g) Hikone R; (h) Wild 1B; (i) Wild 5C).
MeHg has effects on the rate of larval development as well as the penultimate eclosion event. It can therefore be anticipated that expression levels of CYP6g1 measured at one point of development (e.g. 3rd instar larvae in Fig. 1B) might not accurately reflect the relative expression of CYP6g1 in Canton S and Hikone R flies over the course of larval development. We therefore measured CYP6g1 levels in 1st, 2nd and 3rd instar larvae of the Canton S and Hikone R strains reared on normal food media and observed an overall 10-fold higher expression of CYP6g1 in Hikone R larva at all stages (Fig. 1C). These data indicate that elevated CYP6g1 expression persists in Hikone R larva over the feeding period where MeHg exposure occurs in our assay.
As seen in Fig. 1A eclosion rates at 10–15 μM MeHg are useful to discriminate differences in tolerance to MeHg during development. We proceeded to analyze seven additional wild strains of flies previously characterized for their varied expression of CYP6g1 (Daborn et al., 2002). Evaluating eclosion rates on 10 μM MeHg food, we see an association of elevated expression of CYP6g1 with an increased rate of eclosion across the nine wild strains (Fig. 1D). Together, these data demonstrate that, among a limited sampling of wild derived strains of Drosophila, an enhanced developmental tolerance to MeHg correlates with elevated expression of CYP6g1.
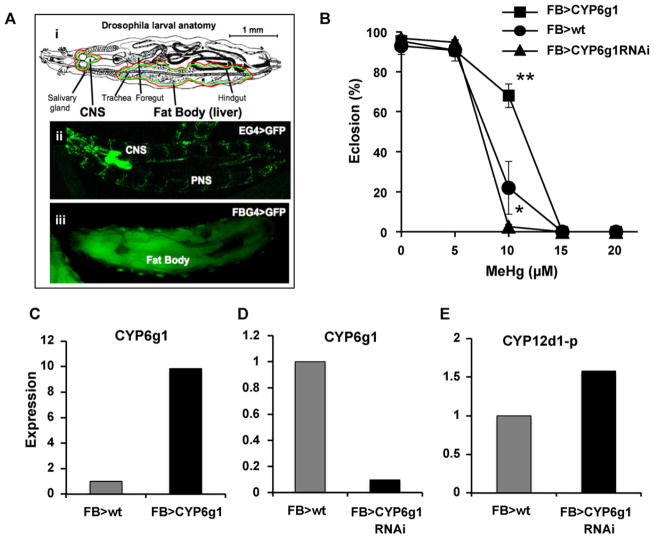
3.3. Transgenic CYP6g1 expression alters MeHg tolerance during development
Our initial identification of CYP6g1 in lab-selected MeHg-tolerant flies was determined with transcripts derived from the larval brain. Yet, expression in the Hikone R strain suggest MeHg tolerance correlates with elevated CYP6g1 expression in both the brain and the fat body. In addition, expression levels in the fat body are more than 10-fold greater than in the brain indicating the fat body is a predominant organ site of action for CYPs. We therefore sought to alter CYP6g1 expression in a targeted manner to evaluate its tissue specific contribution to MeHg tolerance. We used the GAL4 > UAS expression system to direct CYP expression specifically to the fat body or to the nervous system. Two GAL4 “driver” lines were used; the c754-GAL4 line that expresses predominantly in the fat body (designated FBG4), and the elav-GAL4 (EG4) line that drives expression specifically in all central and peripheral neurons, as demonstrated by expression of the UAS-GFP reporter seen in Fig. 2A. We first examined the effects overexpressing CYP6g1 in the fat body. FBG4 > CYP6g1 larvae show a significantly greater rate of eclosion on 10 μM MeHg food (Fig. 2B). This was accompanied by an approximately 10-fold higher expression of CYP6g1 in FBG4 > CYP6g1 larvae compared to control FBG4 > wt larvae (Fig. 2C), consistent with the idea that CYP6g1 activity induces MeHg tolerance. We then investigated the possibility that endogenous levels of CYP6g1 are important for MeHg tolerance by knocking down CYP6g1 expression using RNAi. Larvae carrying the FBG4 > UASCYP6g1RNAi combination demonstrate substantially reduced eclosion on 10 μM MeHg food compared to FBG4 > wt control larvae (Fig. 2B). Correspondingly, CYP6g1 expression level is seen to be reduced approximately 10-fold in FBG4 > UASCYP6g1RNAi larvae compared to control larvae (Fig. 2D). To probe the possibility that CYP6g1 RNAi could have an off-target effect on other CYPs we examined the expression of CYP12d1-p in parallel. CYP12d1-p was chosen since it was moderately upregulated in our previous selection experiment (see Table 1) and it is known to be highly responsive to other stressors in flies (Willoughby et al., 2006). CYP12d1-p expression was seen to be slightly up-regulated with CYP6g1RNAi expression (Fig. 2E) indicating that the CYP6g1 RNAi effect does not reduce CYP expression ubiquitously. While it remains a possibility that other CYPs are affected by CYP6g1 RNAi, these data demonstrate the strong correlation of CYP6g1 expression in hepatic tissue and MeHg tolerance in developing flies.
Fig. 2.
Transgenic modulation of CYP6g1 expression in the fat body influences MeHg tolerance. (A) (i) Larval anatomy highlighting the central nervous system (CNS) and fat body tissues that are targeted for Gal4 > UAS expression below. (ii and iii) Tissue specific transgene expression is demonstrated for the neuron specific driver (EG4, ii) and for the fat body specific driver (FBG4, iii) in combination with a UAS-GFP reporter construct. (B) MeHg tolerance is determined by eclosion assays of flies of the indicated Gal4 > UAS genotypes (n = 150 flies/data point, expressed as mean and s.d. *: p = <0.05; **: p = <0.005 by pairwise Student’s t-test with the FB > wt control at each concentration). (C and D) Expression level of CYP6g1 determined by qRT-PCR of RNA from whole larvae extracts. Levels for over-expression (FB > CYP6g1) or knockdown (FB > CYP6g1 RNAi) are expressed relative to the FB > wt driver line alone. (E) Expression levels of CYP12d1-p in response to CYP6g1 RNA are expressed relative to the driver line alone (FB > wt). Determinations are done on RNA extracted from pooled samples of 20 larvae.
We next investigated a potential role for CYP6g1 activity in the nervous system in MeHg tolerance. EG4 > UASCYP6g1 larvae are seen to eclose at a significantly greater rate on both 10 μM and 15 μM MeHg food compared to control EG4 > wt larvae (Fig. 3A). This increase is accompanied by a more than 100-fold higher level of CYP6g1 expression, in this instance measured by RNA transcripts isolated from adult heads which are rich in brain tissue (Fig. 3B). In parallel, CYP12d1-p is seen to be down-regulated significantly in the CYP6g1 over expressing flies, indicating a global upregulation of CYPs is not occurring in this transgenic line. It is also notable that EG4 > UASCYP6g1 larvae appear to be more tolerant to 10 μM MeHg than FBG4 > UASCYP6g1 larvae whereas the FB > wt and EG4 > wt controls are nearly identical in their tolerance between these two independent determinations (compare Fig. 2B and Fig. 3A). Together, these data indicate that elevated CYP6g1 expression can confer MeHg tolerance when targeted to either neural or hepatic tissue.
Fig. 3.
Expression of CYP6g1 in neurons confers MeHg tolerance during development. (A) Eclosion assays of larvae carrying indicated Gal4 > UAS combination to target neuron specific expression of CYP6g1 (n = 150 flies/data point, expressed as mean and s.d. *: p = <0.05; **: p = <0.005 by Student’s t-test at each concentration). (B) CYP6g1 and CYP12d1-p levels are determined by qRT-PCR of adult head total RNA (pooled samples of 35 adult heads for each genotype) and expressed relative to the EG4 > wt control.
3.4. CYP6g1 is homologous to human CYP3A enzymes
The apparent MeHg tolerance activity of CYPs in flies prompted investigation of a potentially conserved function for mammalian CYPs. The CYP gene family is noted for its large numbers and diversity across the Phyla (e.g. 57 human CYPs and 83 Drosophila CYPs Nelson, 2009; Tijet et al., 2001). A relationship of the Drosophila CYP6 family and human CYP3 family genes has been noted previously (Daborn et al., 2002). To examine this further we performed BLAST (NCBI) searches to identify the human CYP(s) most homologous to CYP6g1. Results show the highest identity and similarity scores between CYP6g1 and human CYP3A4, CYP3A5 and CYP3A7 (Table 2). This result is consistent with a homology modeling study of CYP6g1 and human CYP3A4, which demonstrates that the quality of the modeling is extremely high with approximately 97% of the residues of CYP6g1 being within the generously allowed regions of the CYP3A4 structure (Jones et al., 2010). Together, these data indicate human CYP3A genes are the homologs of Drosophila CYP6g1.
Table 2. Homology determination for CYP6g1 using BLAST.
BLAST scores for comparison of Drosophila CYP6g1 versus all human CYPs are shown (performed at http://blast.uthsc.edu/index.html). Higher bits score indicates higher degree of identity and similarity across the pairwise comparison of sequences. Higher significance of alignments is reflected in lower E value. Results show the highest identity and similarity scores between CYP6g1 and CYP3A family members, CYP3A4, CYP3A5 and CYP3A7.
| CYP name | Score (bits) | E value |
|---|---|---|
| 3A4 | 267 | 6.0E-74 |
| 3A7 | 262 | 1.0E-72 |
| 3A5 | 255 | 2.0E-70 |
| 4F2 | 113 | 1.0E-27 |
| 11A1 | 99 | 2.0E-23 |
| 2C9 | 87 | 8.0E-20 |
| 2A6 | 85 | 1.0E-18 |
| 27A1 | 84 | 1.0E-18 |
| 2J2 | 83 | 2.0E-18 |
| 1A2 | 78 | 5.0E-17 |
| 2C8 | 75 | 6.0E-16 |
| 2E1 | 72 | 5.0E-15 |
| 2D6 | 71 | 8.0E-15 |
| 7A1 | 42 | 3.0E-06 |
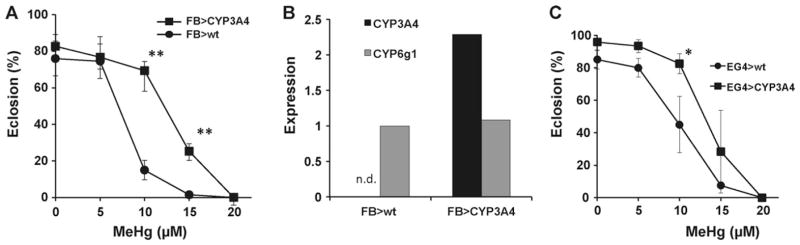
3.5. Expression of CYP3A4 induces MeHg tolerance in flies
Since the CYP3A genes are highly similar in sequence and activity, showing differences predominantly in regulation of expression, we chose to analyze CYP3A4 to investigate a functional relationship of CYP6g1 and CYP3A genes our eclosion assays. We cloned the coding region of human CYP3A4 into the pUAST expression vector and used this to create new lines of transgenic flies carrying this construct (UAS–CYP3A4). We then evaluated the effect of CYP3A4 expression in combination with the FBG4 driver. To gauge the expression of CYP3A4 in FB > UASCYP3A4 larvae we arbitrarily compared the level of qRT-PCR amplification of CYP3A4 transcripts to that of CYP6g1 in the same flies using the delta–delta Ct method (see Section 2). Quantitative amplification of CYP3A4 transcripts shows a 2-fold higher expression level as compared to CYP6g1 in the FBG4 > UASCYP3A4 flies, whereas CYP6g1 levels are seen to be nearly equivalent between the control and FBG4 > UASCYP3A4 larvae (Fig. 4B). CYP3A4 expressing larvae show significantly higher rates of eclosion on 10 μM and 15 μM MeHg food (Fig. 4A) indicating that expression of CYP3A4 is very effective in inducing a MeHg tolerance phenotyope. While FBG4 > UASCYP3A4 flies appear to show more MeHg tolerance than FB > UASCYP6g1 flies (compare Fig. 2B and Fig. 4A), further analyses are needed to validate comparable expression levels of these two transgenes before their relative activities can be compared.
Fig. 4.
Human CYP3A4 confers MeHg tolerance when expressed in Drosophila. (A and C) Eclosion assays of larvae carrying indicated Gal4 > UAS combination to target expression of CYP3A4 to the fat body or to neurons (n = 150 flies/data point for FBG4 > CYP3A4 and 60 flies/data point for EG4 > CYP3A4. Expressed as mean and s.d. *: p = <0.05; **: p = <0.005 by Student’s t-test at each concentration). (B) CYP3A4 expression level, expressed relative to CYP6g1 level (see Section 2), measured by qRT-PCR of RNA from whole larva extracts of indicated genotype (pooled samples of 25 larvae for each genotype).
We further assessed the activity of CYP3A4 by targeting expression to the nervous system. Flies carrying the EG4 > UASCYP3A4 combination showed significantly enhanced rates of eclosion on 10 μM MeHg food compared to EG4 > wt controls flies (Fig. 4C). In sum, these data show that human CYP3A4, similar to CYP6g1, can induce MeHg tolerance in developing flies when expressed either in hepatic or nervous tissue.
3.6. Caffeine induces CYP expression and MeHg tolerance
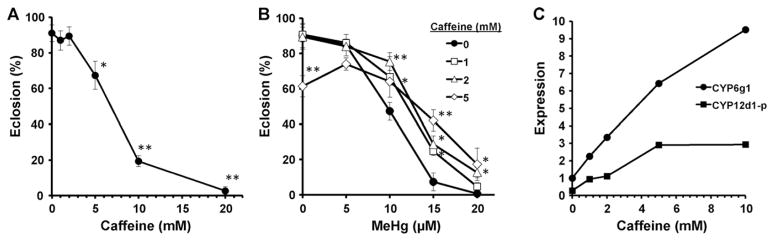
Many CYP genes exhibit induced expression upon exposure to xenobiotics. Previous studies have shown that CYP6g1 and several other Drosophila CYPs are highly responsive to exposure to drugs such as caffeine and phenobarbital, as well as several pesticides, resulting in large increases in CYP expression (Morra et al., 2010; Willoughby et al., 2006). We therefore tested the ability of caffeine to induce MeHg tolerance by enhancing CYP expression. First, we examined the toxicity of caffeine in eclosion assays and determined that doses as high as 2 mM in the food medium showed negligible effects on eclosion rates, whereas an approximately 35% and 80% reduction in eclosion rate was seen with 5 mM and 10 mM caffeine in the medium, respectively (Fig. 5A). Twenty millimolar caffeine elicts almost complete lethality (Fig. 5A). We then examined eclosion rates on MeHg food supplemented with 1, 2 or 5 mM caffeine. An increase in eclosion rate on 10–20 μM MeHg food was seen at all concentrations caffeine, as compared to no caffeine control (Fig. 5B). Notably, on 15 μM MeHg food, a 25%, 27% and 42% eclosion rate was seen with 1, 2, and 5 mM caffeine, respectively, compared to only 7% eclosion in the absence of caffeine (Fig. 5B). It is of note that while 5 mM caffeine alone elicits a significantly reduced eclosion rate, this level of caffeine proves to induce MeHg tolerance.
Fig. 5.
Caffeine induces MeHg tolerance and elevated CYP expression. (A) Developmental toxicity of caffeine is determined by eclosion assays with Canton S larva on food containing indicated concentrations of caffeine. No significant decrease in eclosion rate is seen up to 2 mM caffeine. Significant decreases in eclosion rate are seen for 5, 10 and 20 mM caffeine. (B) Eclosion assays of Canton S larvae reared on food with indicated concentrations of caffeine and MeHg (n = 150 flies/data point for all assays in (A) and (B). Eclosion is expressed as mean and s.d. *: p = <0.05; **: p = <0.005 determined by Student’s t-test at each MeHg concentration relative to no caffeine control). (C) Expression level CYP6g1 and CYP12d1-p levels are determined by qRT-PCR with RNA from extracts of Canton S whole larvae reared on food with the indicated concentration of caffeine (pooled samples of 25 larvae for each caffeine concentration). Expression levels for both genes are normalized to CYP6g1 expression with no caffeine.
We next examined CYP expression in response to caffeine in RNA extracts of whole larvae exposed to 0, 1, 2, 5 and 10 mM caffeine. For these assays we chose to analyze larva at the 2nd instar stage, versus the 3rd instar stage, for the reason that, unlike MeHg which is highly stable, caffeine is rapidly metabolized. Second instar larvae are constantly feeding, and thus experiencing a steady state of caffeine exposure, whereas 3rd instar wandering larvae have ceased feeding and have likely metabolized caffeine consumed at earlier stages. CYP6g1 showed a strong dose dependent increase in expression showing approximately 2.3, 3.4, 6.4 and 9.5-fold higher expression on 1, 2, 5 and 10 mM caffeine, respectively, compared to no caffeine control (Fig. 5C). For comparison we analyzed CYP12d1-p expression. CYP12d1-p showed a similar pattern of induced expression with caffeine as CYP6g1 (Fig. 5C). While CYP12d1-p showed an overall lower expression level relative to CYP6g1, these results raise the possibility that the MeHg tolerance effect of dietary administration of caffeine stems a more widespread induction of CYPs.
4. Discussion
In this study, we characterize MeHg tolerance effects stemming from elevated expression of CYPs in a Drosophila model. We demonstrate that MeHg tolerance correlates with elevated CYP6g1 expression in several wild strains of flies. Transgenic overexpression of CYP6g1, when restricted to neural tissue or to hepatic tissue (fat body) in the developing fly, invokes developmental tolerance to MeHg. Alternatively, diminishing CYP6g1 expression with RNAi results in susceptibility to MeHg. We also identify the human CYP3A family of genes (CYP3A4, CYP3A5, CYP3A7) as the homologs of CYP6g1 and furthermore, that transgenic expression of CYP3A4 in either nervous or hepatic tissue invokes MeHg tolerance in developing flies. Finally, we show that supplementation of food with caffeine, a known inducer of Drosophila CYP expression, results in greater rates of eclosion on MeHg food accompanied by higher levels of CYP6g1 and CYP12d1-p expression in developing flies. Together, these data identify a functional role for CYPs in invoking tolerance to MeHg during development.
A striking finding is that elevated expression of either CYP6g1 or CYP3A4 exclusively in neurons is able to rescue overall development of the fly in the presence of MeHg. This result is consistent with the notion that the developing nervous system is a preferred and sensitive MeHg target in insects, as it is in humans. These data also suggest that CYPs perform a cytoprotective function in neurons. Recent evidence for specific patterns of CYP expression in the mammalian brain point to a central role for CYPs in normal and pathological conditions in the nervous system (Dutheil et al., 2008; Ferguson and Tyndale, 2011). Several possible mechanisms could explain the MeHg tolerance conferred by elevated neuronal CYP expression. One possible mechanism may stem from the ability of MeHg to compromise CYP activity, either via repression of transcription of CYP genes or inhibition of catalytic activity of CYP enzymes (Lucier et al., 1973; Robbins et al., 1978). Consistent with this, we observe an overall reduction of CYP transcripts in the brains of non-tolerant strains of flies exposed to MeHg (see Table 1, Column 8). Thus, the elevated expression of CYPs seen in tolerant flies could act to compensate for MeHg inhibitory action, and sustain the necessary level of CYP activity required for normal neural development.
Alternatively, CYPs could serve a more direct neuroprotective role by facilitating clearance of toxic metabolites arising from MeHg insult. A related activity has been demonstrated for CYP3A4 and CYP2D6 in mammalian neural derived cell lines. 1-Methyl-4-phenylpyridinium (MPP+) is a neurotoxic derivative of MPTP and an agent used to induce dopaminergic neuron death in models of Parkinson’s disease. Inhibition of CYP3A4 or CYP2D6 activity in SH-SY5Y human neuroblastoma cells enhances the neurotoxicity of MPP+ (Mann and Tyndale, 2010), whereas overexpression of CYP2D6 in PC12 cells can reduce the toxicity of MPP+ (Matoh et al., 2003). It remains to be resolved what the relevant CYP substrates are in the context of MeHg insult. Nonetheless, these data indicate that neurotoxic outcomes of environmental agents are likely to be influenced by levels of CYP expression and activity in the brain.
The observation that CYP6g1 and CYP3A4 expressed in the fat body increases eclosion of developing flies on MeHg food points to a role for liver CYP activity in MeHg tolerance. One interpretation of this effect is that the liver is essential for clearance of toxic metabolites generated by MeHg exposure. MeHg is known to invoke oxidative stress, yielding reactive oxygen species (ROS) that compromise structure and function of lipids and proteins. It is likely that CYP enzyme activity aids in clearance of these potentially toxic by-products via covalent modifications that enhance their excretion. CYP activity in the fat body is also essential for endobiotic metabolism of key signaling molecules. Notable is the CYP-mediated hydroxylation of ecdysteroid hormones that are essential for molting in insects (Guittard et al., 2011). Perturbing this essential CYP function in the fat body may be an outcome of MeHg exposure. In which case, enhanced CYP levels could overcome this MeHg toxic effect.
An important consideration is the potential for MeHg tolerance to stem from a combinatorial activity of several CYPs. Here, we have focused on the activity of just one CYP gene family, CYP6g1 and its homolog CYP3A4. Our data demonstrate that MeHg tolerance can be influenced by altered expression of just one CYP. Yet, several CYPs are upregulated in MeHg tolerant flies (Table 1), indicating MeHg tolerance could arise out of the activity of more than one CYP gene family. Thus, going forward it will be important to characterize the ability of other CYP families to influence the MeHg tolerance trait, both independently and in combinations.
Our findings predict that CYPs could influence MeHg toxic outcomes in mammals and humans. It is plausible that the lessons learned in the pharmacogenomics of drug metabolism could apply to understanding CYP influence on MeHg toxicity in the fetus and young children. Polymorphic variants of a number of human CYPs give phenotypes of either extensive or poor metabolism of common drugs, and thus dictate the toxicity profile of these drugs on an individual basis (Smart and Hodgson, 2008). It follows that a particular genotype of CYP polymorphisms could influence the outcome of MeHg exposure in people. Particularly relevant to our findings here are polymorphic variants in CYP3A family genes that predict higher or lower expression and/or activity (e.g. CYP3A4*1B and CYP3A5*3B, CYP3A7*2). It is also interesting to note that the frequency of these CYP3A polymorphisms vary widely among people of Caucasian versus Asian and African decent (Lamba et al., 2002; Rodríguez-Antona et al., 2005). Thus, as in pharmacogenomics, CYP genotypes may prove useful for determining disposition to MeHg toxicity in a population, or even on an individualized basis.
We observe a significant caffeine-induced MeHg tolerance in flies. Numerous studies have demonstrated neuroprotective effects of caffeine, notably in models of chemical-induced dopaminergic neurodegeneration that model Parkinsons disease. Our finding that caffeine gives a robust induction of both CYP6g1 and CYP12d1-p are consistent with the notion that CYP activity contributes to the MeHg tolerance phenotype. Previous studies have shown caffeine treatment of Drosophila larvae results in upregulation of a number of CYPs, including CYP6g1 and CYP12d1-p (Willoughby et al., 2006). However, caffeine also induces expression of other detoxification genes (e.g. Gst, Willoughby et al., 2006) and can enhance glutathione production in neurons (Aoyama et al., 2011). Thus, it is likely that a number of genes contribute to MeHg tolerance stemming from caffeine. Nonetheless, our observations highlight the potential for CYPs to contribute to MeHg tolerance and suggest that dietary or pharmacological inducers of CYP expression could have beneficial effects in averting MeHg toxicity.
In summary, we demonstrate an activity for elevated CYP gene expression in conferring tolerance to MeHg toxicity during development. This CYP activity in flies shows to be conserved between Drosophila CYP6g1 and its human homolog CYP3A4. Importantly, targeted CYP expression in neurons has profound effects on overall tolerance to MeHg during development. These findings predict that further analyses of human CYP3A genes, and other CYP family members, will reveal fundamental mechanisms that influence the susceptibility to MeHg in people.
Acknowledgments
Funding
This work was supported by NIEHS R01-ES015550 awarded to M.D.R.
We wish to thank Ben Moody for technical assistance. We are grateful to Philip Daborn for the UAS–CYP6g1 flies and to Erin Schuetz for CYP3A4 cDNA. We thank Johannes Bischof and Konrad Basler for providing the pUAST-attB plasmid and advice on cloning.
Footnotes
Conflicts of interest
All the authors have no conflicts to declare.
References
- Aoyama K, Matsumura N, Watabe M, Wang F, Kikuchi-Utsumi K, Nakaki T. Caffeine and uric acid mediate glutathione synthesis for neuroprotection. Neuroscience. 2011;181:206–215. doi: 10.1016/j.neuroscience.2011.02.047. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized trans-genesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi AF, Coccini T, Ceccatelli S, Manzo L. Neurotoxicity and molecular effects of methylmercury. Brain Res Bull. 2001;55:197–203. doi: 10.1016/s0361-9230(01)00458-0. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Lumb C, Boey A, Wong W, Ffrench-Constant RH, Batterham P. Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome p450 genes by transgenic over-expression. Insect Biochem Mol Biol. 2007;37:512–519. doi: 10.1016/j.ibmb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, Tijet N, Perry T, Heckel D, Batterham P, Feyereisen R, Wilson TG, ffrench-Constant RH. A single p450 allele associated with insecticide resistance in Drosophila. Science (New York, NY) 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Dutheil F, Beaune P, Loriot MA. Xenobiotic metabolizing enzymes in the central nervous system: contribution of cytochrome p450 enzymes in normal and pathological human brain. Biochimie. 2008;90:426–436. doi: 10.1016/j.biochi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Eldefrawi MENAMATE. Interaction of Acetylcholine Receptors with Organic Mercury Compounds. Plenum Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- Ferguson CS, Tyndale RF. Cytochrome p450 enzymes in the brain: emerging evidence of biological significance. Trends Pharmacol Sci. 2011;32:708–714. doi: 10.1016/j.tips.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SE, Peterson JA. How similar are P450s and what can their differences teach us? Arch Biochem Biophys. 1999;369:24–29. doi: 10.1006/abbi.1999.1350. [DOI] [PubMed] [Google Scholar]
- Guittard E, Blais C, Maria A, Parvy JP, Pasricha S, Lumb C, Lafont R, Daborn PJ, Dauphin-Villemant C. CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev Biol. 2011;349:35–45. doi: 10.1016/j.ydbio.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Hedlund E, Gustafsson JA, Warner M. Cytochrome p450 in the brain; a review. Curr Drug Metab. 2001;2:245–263. doi: 10.2174/1389200013338513. [DOI] [PubMed] [Google Scholar]
- Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, Perrimon N. Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis. 2002;34:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- Hughes WL. A physicochemical rationale for the biological activity of mercury and its compounds. Ann N Y Acad Sci. 1957;65:454–460. doi: 10.1111/j.1749-6632.1956.tb36650.x. [DOI] [PubMed] [Google Scholar]
- Jones RT, Bakker SE, Stone D, Shuttleworth SN, Boundy S, McCart C, Daborn PJ, ffrench-Constant RH, van den Elsen JM. Homology modelling of Drosophila cytochrome p450 enzymes associated with insecticide resistance. Pest Manag Sci. 2010;66:1106–1115. doi: 10.1002/ps.1986. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lucier GW, Matthews HB, Brubaker PE, Klein R, McDaniel OS. Effects of methylmercury on microsomal mixed-function oxidase components of rodents. Mol Pharmacol. 1973;9:237–246. [PubMed] [Google Scholar]
- Mahapatra CT, Bond J, Rand DM, Rand MD. Identification of methylmercury tolerance gene candidates in Drosophila. Toxicol Sci. 2010;116:225–238. doi: 10.1093/toxsci/kfq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A, Tyndale RF. Cytochrome p450 2D6 enzyme neuroprotects against 1-methyl-4-phenylpyridinium toxicity in SH-SY5Y neuronal cells. Eur J Neurosci. 2010;31:1185–1193. doi: 10.1111/j.1460-9568.2010.07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoh N, Tanaka S, Takehashi M, Banasik M, Stedeford T, Masliah E, Suzuki S, Nishimura Y, Ueda K. Overexpression of CYP2D6 attenuates the toxicity of MPP+ in actively dividing and differentiated PC12 cells. Gene Expr. 2003;11:117–124. doi: 10.3727/000000003108749017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Himeno S, Koide N, Imura N. Effects of methylmercury and inorganic mercury on the growth of nerve fibers in cultured chick dorsal root ganglia. Tohoku J Exp Med. 2000;192:195–210. doi: 10.1620/tjem.192.195. [DOI] [PubMed] [Google Scholar]
- Morra R, Kuruganti S, Lam V, Lucchesi JC, Ganguly R. Functional analysis of the cis-acting elements responsible for the induction of the Cyp6a8 and Cyp6g1 genes of Drosophila melanogaster by DDT, phenobarbital and caffeine. Insect Mol Biol. 2010;19:121–130. doi: 10.1111/j.1365-2583.2009.00954.x. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The Cytochrome p450 homepage. Human Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavek P, Dvorak Z. Xenobiotic-induced transcriptional regulation of xenobiotic metabolizing enzymes of the cytochrome p450 superfamily in human extrahepatic tissues. Curr Drug Metab. 2008;9:129–143. doi: 10.2174/138920008783571774. [DOI] [PubMed] [Google Scholar]
- Robbins MS, Hughes JA, Sparber SB, Mannering GJ. Delayed teratogenic effect of methylmercury on hepatic cytochrome p-450-dependent monooxygenase systems of rats. Life Sci. 1978;22:287–294. doi: 10.1016/0024-3205(78)90135-2. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Antona C, Jande M, Rane A, Ingelman-Sundberg M. Identification and phenotype characterization of two CYP3A haplotypes causing different enzymatic capacity in fetal livers. Clin Pharmacol Ther. 2005;77:259–270. doi: 10.1016/j.clpt.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Sanderson JT. Placental and fetal steroidogenesis. Methods Mol Biol. 2009;550:127–136. doi: 10.1007/978-1-60327-009-0_7. [DOI] [PubMed] [Google Scholar]
- Sarafian TA. Methylmercury-induced generation of free radicals: biological implications. Met Ions Biol Syst. 1999;36:415–444. [PubMed] [Google Scholar]
- Sirois JE, Atchison WD. Methylmercury affects multiple subtypes of calcium channels in rat cerebellar granule cells. Toxicol Appl Pharmacol. 2000;167:1–11. doi: 10.1006/taap.2000.8967. [DOI] [PubMed] [Google Scholar]
- Smart R, Hodgson E. Molecular and Biochemical Toxicology. Chapter 9 John Wiley and Sons; Hoboken, NJ: 2008. pp. 147–172. [Google Scholar]
- Tijet N, Helvig C, Feyereisen R. The cytochrome p450 gene superfamily in Drosophila melanogaster: annotation, intronexon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/s0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- Viaggi C, Pardini C, Vaglini F, Corsini GU. Cytochrome p450 and Parkinson’s disease: protective role of neuronal CYP 2E1 from MPTP toxicity. J Neural Transm (Suppl) 2006:173–176. doi: 10.1007/978-3-211-45295-0_27. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochromes p450: a success story. Genome Biol. 2000;1:REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby L, Chung H, Lumb C, Robin C, Batterham P, Daborn PJ. A comparison of Drosophila melanogaster detoxification gene induction responses for six insecticides, caffeine and phenobarbital. Insect Biochem Mol Biol. 2006;36:934–942. doi: 10.1016/j.ibmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]