Abstract
Voltage-gated ion channels are responsible for transmitting electrochemical signals in both excitable and non-excitable cells. Structural studies of voltage-gated potassium and sodium channels by X-ray crystallography have revealed atomic details on their voltage-sensor domains and pore domains, and were put in context of disparate mechanistic views on the voltage-driven conformational changes in these proteins. Functional investigation of voltage-gated channels in membranes, however, showcased a mechanism of lipid-dependent gating for voltage-gated channels, suggesting that the lipids play an indispensible and critical role in the proper gating of many of these channels. Structure determination of membrane-embedded voltage-gated ion channels appears to be the next frontier in fully addressing the mechanism by which the voltage sensor domains control channel opening. Currently electron crystallography is the only structural biology method in which a membrane protein of interest is crystallized within a complete lipid-bilayer mimicking the native environment of a biological membrane. At a sufficiently high resolution, an electron crystallographic structure could reveal lipids, the channel and their mutual interactions at the atomic level. Electron crystallography is therefore a promising avenue toward understanding how lipids modulate channel activation through close association with the voltage sensor domains.
Introduction
The superfamily of voltage-gated ion channels consists of integral membrane proteins that contain four voltage-sensor domains (VSDs) and a central ion-conducting pore domain1–2. Members of this superfamily have been identified in all cells, and play critical roles in a variety of cellular physiology, from muscle contraction to neuronal activity to T cell activation in inflammatory (immune) response. Voltage-gated ion channels are divided into two broad groups: the hyperpolarization-activated and the depolarization-activated channels. Biophysical studies have shown that the VSDs in these two groups work in a similar way3. In both cases, the VSDs undergo significant conformational changes driven by electrical energy. These conformational changes are coupled to the pore domain, to close or open the ion channel in response to electrical stimuli3–6. The hyperpolarization-driven state of the VSD is called the “DOWN” conformation (also resting or closed), and the depolarization-stabilized state is named the “UP” conformation (also activated or open)7•• Understanding the structural basis for the voltage sensor function in membranes not only is fundamentally important for revealing the exquisite electrical control of protein structure, but also will forge the foundation for developing new therapeutical strategy for human diseases caused by the dysfunction of these channels8–9.
All known VSDs are made of four helical transmembrane segments (S1–S4) with highly conserved charged residues on the second (S2) and fourth (S4) helices. During voltage-dependent gating, the charged residues on S4 translocate from one side of the electric field to the other while the VSDs switch their conformations and couple the charge movement to the opening and closing of the channel pore6,10–11. Within each VSD there are water-accessible crevices from either side of the membrane12–13. The transmembrane electric field penetrates into these crevices to establish a certain degree of electric focusing14. In the UP conformation the gating charges (mainly on S4) are in the extracellular crevice and in the DOWN conformation in the intracellular one. Switching between the UP and DOWN conformations requires a significant energy input from the electric field, ~7.5 kcal/mol per VSD15–18.
While a number of different structures of voltage-gated ion channels have been determined it remains unclear how the VSDs couple the charge movement to the pore opening and closing6. Three different groups of mechanistic models have been proposed and experimentally supported: I. the voltage sensor paddle model; II. the transporter-like model and III. the helical-translocation/helical-screw model. The voltage sensor paddle model argues for a 15–20Å motion of the paddle (the helix-loop-helix motif composed of the S3b, the S3S4 linker and the extracellular half of S4) along the membrane normal19•,20. It does not exclude lateral motion or rotation of the S4, nor does it specify how the other parts of the VSD adjust to accommodate the major structural changes in membranes. The transporter-like model stemmed from intramolecular distance measurements, and argues that the toggling of the fixed gating charges from the outward-facing to the inward-facing state needs a small-scale (4–6Å or less) vertical movement of S4, traversing a narrow hydrophobic septum (plug) in the gating pore21••,23•. The transmembrane electric field is thought to be highly focused across such a short distance14,18. The third group of models proposed a vertical displacement of the S4 inside the gating pore with varying distances, and the helical screw model adds a ~180° rotation of S4 in order to reorient the charged residues on S417,24.
Besides the uncertainty on the VSD’s mode of action, there is mounting evidence that lipids influence the structural stability and function of the VSDs and therefore the opening and closing of the channel pore. Functional studies of voltage-gated channels in membranes highlight a lipid-dependent gating mechanism. Studies indicate that without any change in transmembrane voltage, manipulating the lipid composition in a membrane switches the VSDs between the DOWN and UP conformations7••. This and other studies suggest that the lipids exert strong gating effects on the voltage-gated channels7••,25••,26••,27••.
In this review we highlight some of the key structural features of voltage-gated ion channels and discuss how lipids were shown to influence channel structure and function. We then highlight electron crystallography as a structural biology technique that could provide information about how the lipids interact with the VSDs to affect channel gating.
An overview of voltage-gated ion channel structures
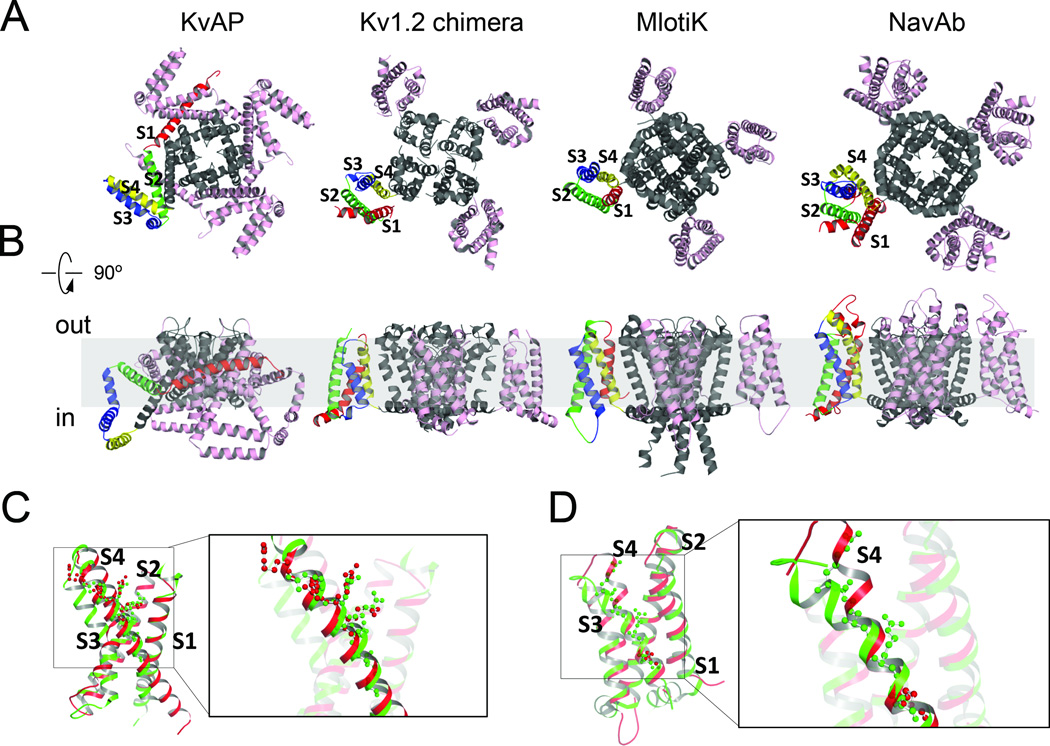
Structures of four channels that contain VSDs or VSD-like domains have been determined to date: KvAP, Kv1.2 (and its chimera), MlotiK and NavAb 19•,28••,29••,30••. MlotiK is a ligand-gated channel with VSD-like domains, but has not been found to be functional yet. KvAP, Kv1.2 and NavAb are functional voltage-gated channels. The four structures confirm the common topology that was previously proposed for the superfamily of voltage-gated ion channel. The channels are tetrameric assemblies (Fig 1 A and B). The first four helices in each monomer constitute the VSD, and the sequence between helices 5 (S5) and 6 (S6) forms the pore domain. The loop connecting S5 and S6 forms the ion selectivity filter. Four pore domains (S5S6 from each monomer) assemble together around the 4-fold axis to create an ion-conducting pore.
Figure 1. The X-ray structures of ion channels containing VSDs.
A) Top views of the structures of KvAP (PDB code 1ORS), Kv1.2/2.1 chimera (PDB code 2R9R), MlotiK (PDB code 3BEH) and NavAb (PDB code 3RW0). One voltage sensor domain in each channel is colored as S1 (red), S2 (green), S3 (blue) and S4 (yellow).
B) Side views of the four channels in a putative lipid bilayer (hashed grey).
C) Comparison of the voltage-sensors of KvAP (red) and Kv1.2 (green) with the side chains of the arginine residues presented as colored balls.
D) Comparison of the voltage-sensors of MlotiK (red) and NavAb (green) in the same view as in C. Figures were produced using PYMOL.
The conformation of the VSD from the full-length KvAP is significantly different than those in the Kv1.2 and its chimera, MlotiK and NavAb (Fig 1A and B). It is fully splayed with helices S1 and S2 wrapped along the side of the pore (Fig 1B, red and green helices). In other three cases the VSDs are folded into a compact 4-helix bundle neatly tucked to the lateral side of the pore (Fig. 1A). The structure of the isolated KvAP VSD resembles closely that of the VSDs from Kv1.2 (as well as the Kv1.2/2.1 chimera; Fig. 1C, overlay). S1, S2 and the top part of S4 overlay very well, but the position of S3b is different, displaying approx a 90° rotation between the two VSDs. Moreover, the positions of S4 arginines differ between KvAP and Kv1.2 as if they are shifted down by one register in the latter (Fig. 1C), and the intracellular half of the S4 in the Kv1.2 chimera structure shows a short 310 helix, which is absent in the same location of the other three VSDs.
The VSD structures of MlotiK and NavAb exhibit good overall fit among all four helices (Fig 1D). MlotiK has only one conserved charge in its S4. Its VSDs appear to be in a permissive “UP” state, leaving the control of the channel pore to the intracellular nucleotide binding domains. In both channels, the N-terminal halves of their S4 segments harbor a short 310 helix. Charged residues along the 310 helix face the same side, which has implications for sliding the S4 across a newly-named charge transfer center without much rotation29••,30••,31•.
Although the conformational change that ensues in the VSDs in response to voltage is not clear, what is agreed upon is that the movement in the VSD helices is tightly coupled to the pore opening/closing. Exactly how the VSD and pore are coupled is not entirely clear. Two different coupling schemes were proposed for Kv1.2 and NavAb28••,29••. The first is based on the observed interaction between the S4–S5 linker and the intracellular half of S6. It was suggested that the sliding motion of S4 pushes the S4S5 linker intracellularly as well as the intracellular end of S6, leading to pore closure at a conserved PVP motif28••. This coupling scheme gained support from both structural and functional studies32. The second coupling scheme is based entirely on structural comparison between Kv1.2/2.1 chimera and NavAb, whose pore domains are in the open and closed states, respectively. It was suggested that wobbling the VSD could lead to a lateral rotation of the S4S5 linker, which in turn exerts a torque on the S5 and S6 to gate the pore with only a limited vertical movement of the S4. In previous biophysical analysis, the first closing step was found to bear weak voltage-dependence (0.5–1.0 e0), which seemingly agrees with the small adjustment of the VSD to close the pore even though it is unclear what contributes to the small charge displacement33–34.
While the available structures helped tremendously in understanding voltage gated ion channels, there are still many unanswered questions. The problem is further complicated by various unexplained effects observed on channels (pore and VSD) in the presence of various detergents and/or lipid molecules. For example, the structure of the VSD from KvAP varied in the presence of different detergents such as β-octyl-glucoside (β-OG) or diheptyl-phosphotidyl-choline (DHPC) and the presence or absence of long acyl chain detergents and lipid molecules used in various studies seem to affect the structural stability and function of the channels28••,35••,36•. It is also unclear how well the voltage-sensor domains are coupled to the pore in the Kv1.2 and NavAb structures, even though the assignment of the conformations appears coherent with biophysics analysis28••,29••.
The influence of lipids on voltage-gated ion channels
Voltage-gated ion channels function in membranes in which lipids associate closely with both the VSDs and pore domains. It is therefore not surprising that lipids can influence the structure and function of these channels. Recent studies suggest that changing lipid composition alone, without a change in transmembrane voltage, caused the VSDs to switch conformations7••. In oocytes expressing Kv2.1 and other Shaker-like channels, sphingomyelinase (SMase) D treatment was able to increase the total number of active channels25••. The action of SMase C instead suppressed the total channel activity by decreasing the number of active channels26•• (Fig. 2A). In other experiments, phospholipids were found to be required for KvAP to reach the open state, and lipids lacking phosphate headgroups (called nonphospholipids hereafter) were found to stabilize the VSDs in the “DOWN” conformation7••,27•• (Fig. 2B). Comparison of the chemical nature of the products from SMase D/C treatment with the lipids that gate the KvAP channel (Fig. 2A and B) suggests that ceramide molecules produced by SMase C catalysis exert lipid-dependent gating on the Shaker-like channels and favor the “DOWN” conformation of the VSDs in these channels. SMase D treatment instead liberated ceramide-1-phosphates which favor the “UP” conformation.
Figure 2. Lipid-dependent gating of voltage-gated ion channels.
A) Sphingomyelinase (SMase) C or D, respectively, immobilizes or mobilizes the VDSs of Kv channels. It was modified from references 25•• and 26••. SMase C produces ceramide and leads to the decrease of total gating charge (Q), indicating a decrease of active channels with mobile voltage sensor domains. SMase D treatment generates ceramide-1-phosphate, and liberates the VSDs in the channels as showed by the increased total gating charge.
B) KvAP voltage sensor domain switches from DOWN (resting) to UP (activated) conformation when it is changed from a nonphospholipid bilayer to a phospholipids membrane. The model at the bottom schematically demonstrates the lipid-dependent gating.
It was proposed that phospholipids directly interact with the arginines on the S427••. The requirement of phospholipids is therefore to stabilize the lipid-facing arginines in the “UP” conformation, as implicated by electron microscopy and single particle reconstruction of detergent solubilized KvAP, as well as in the EPR study of KvAP embedded in vesicles37•,38. Multiple calculations by molecular dynamics simulation proposed that the phospholipids around the voltage sensor may interact with the hydrophobic cation in the guanidium side chains of the arginine residues on S4 that are partitioned to the middle of the membrane39–41. Such phosphate-arginine interaction could lead to a local distortion of the bilayer structure, decrease the energetic cost for the membrane insertion of arginines42–43, and thus stabilize the “UP” conformation in phospholipid membranes.
With nonphospholipid membranes, it is more difficult to stabilize the lipid-facing arginines through charge interactions since the phosphate groups on the lipids are absent. Instead, the interactions between S4 and nonphospholipids are dominated by hydrogen bonding and hydrophobic interactions. In this case the voltage sensors are likely in the “DOWN” conformation where their S4 arginine residues experience less lipid-exposure. This observation is certainly oversimplified because the hydrophobic residues in the voltage sensors, especially those interposed between the arginines in S4, do contribute to the stability of the “DOWN” conformations7••,44. Nevertheless, there are clear stabilizing interactions between the VSD and lipids in both the UP and DOWN conformations. It is thus imperative to study the structures of voltage-gated ion channels while they are embedded in a lipid bilayer.
Lipid molecules have been seen and described in a number of structures of membrane proteins. The lipids often co-purify with the membrane proteins of interest but sometimes the lipids are added to help stabilize the proteins for biochemical and structural analysis. The latter was the case with Kv1.2 and its chimera structure28••. The channel was difficult to handle as it often crashed out of solution making both biochemical and structural analysis difficult. However, addition of phospholipids during the purification stage markedly increased the stability of the channel making structural studies possible. In fact when the structure of Kv1.2 chimera was determined a number of lipid molecules were observed in the density maps. Lipids closely associated with both the voltage sensor domains as well as the pore domain of the channel (Fig. 3A and B, lipids in yellow).
Figure 3. Lipids arranged around the Kv1.2 chimera structure.
A) The Kv1.2/2.1 chimera structure is viewed laterally from within the membrane. The protein density (PDB code 2R9R) is presented as a white surface map, and the partial (the majority) and full lipid molecules are showed as balls with yellow carbon atoms and red glycerol backbones. A putative lipid bilayer is marked grey. B) The same model viewed from the extracellular side.
Electron crystallography of membrane proteins
Electron crystallography is the only structural biology technique in which the membrane protein of interest is crystallized within a complete lipid bilayer that mimics biological membranes45–46. Moreover, electron crystallography is the only electron cryomicroscopy (cryoEM) technique capable of delivering atomic information about membrane proteins. It has been used to provide important insights into the structure and function of several membrane proteins belonging to different protein families47••. Together with recent advance in hardware and methodology this approach shows a lot of promise. As discussed before, lipids are clearly important in the structural stability and function of voltage-gated ion channels. It is imperative therefore to begin to study the high-resolution three-dimensional structures of such channels by electron crystallography.
While many reviews have been written about the crystallization process for electron crystallographic studies this step remains challenging48–52. The most common way to achieve reconstitution and therefore crystallization of the membrane protein of interest is by slow dialysis. Here the detergent-solubilized purified membrane protein is mixed with detergent-solubilized lipids. The detergent is then slowly removed by dialysis against a crystallization buffer lacking the detergent. As the detergent is removed, lipids begin to form membranes and the protein of interest is integrated into this membrane. Under certain conditions (which have to be determined empirically) the protein molecules will pack tightly into two-dimensional (2D) arrays or crystals. Obviously the choices of detergents, lipids, lipid amount (measured in lipid-to-protein ratio), buffer composition and temperature all play a role in the 2D crystallization process. And 2D crystallization is no less complex than that for 3D crystals used in X-ray crystallography. Once large and well-ordered 2D crystals are obtained, data are collected under cryogenic conditions with low electron dose.
A recent study highlighted the potential of using electron diffraction for rapid structural analysis of membrane proteins47••. Here the authors relied on images of 2D crystals to supply initial low-resolution (but accurate) phase information. Electron diffraction to atomic resolution was then collected to provide accurate amplitude data. Polyalanine α-helical fragments were then placed into the low-resolution map and new phases were calculated and extended to the resolution limit of the electron diffraction data. The structures of three different membrane proteins were determined rapidly by this method following several cycles of phase combination, density modification, model building and refinement. When the resolution of the electron crystallographic study is sufficiently high (better than ~3Å), the structures of both protein and lipids can be determined46 (Fig. 4A). In both bacteriorhodopsin and the water channel aquaporin-0 (AQP0), complete lipid bilayers were observed surrounding the protein and detailed lipid-protein interactions were described (Fig. 4B). A few key features were observed, for example, charge complementation between lipid and protein as well as hydrophobic matching principles.
Figure 4. Phase extension in electron crystallography.
A) Phase extension for bacteriorhodopsin and aquaporin-0 (top and bottom, respectively). The method was recently developed for rapid structure determination by electron crystallography to atomic resolution47••.
B) Complete lipid bilayers were seen and modeled surrounding both bacteriorhodopsin and aquaporin-0 (top and bottom, respectively). Given sufficiently high resolution electron crystallography can reveal details on lipid-protein interactions and possibly how lipids influence the structure and function of membrane proteins.
Membrane proteins are dynamic as they undergo various conformational changes to carry out their biological function. This is beautifully illustrated by the work on VSDs and how they alter their conformations in the membrane to open or close the coupled ion-conducting pore. Perhaps one of the most important applications of electron crystallography in studying voltage-gated ion channels is in fact the presence of a complete lipid bilayer around the protein in the 2D crystals. Indeed a theme that is emerging in electron crystallography is that crystal contacts in membranes are mediated by lipids with very little or no direct protein-protein contacts45. Lipids in 2D crystals therefore buffer the protein from its neighbors and as such conformational changes of the protein in response to activators or inhibitors may not affect crystalline order. Structures of the protein of interest can then be determined under a variety of physiologically important functional states simply by incubating the crystalline membranes with activators or inhibitors, changing pH, or adding a variety of substrates. Functional as well as structural analysis of the protein of interest can therefore be carried out from the very same preparations. Moreover, electron crystallography can be used to visualize the atomic charged states of amino acids45, a characteristic of this technique that has not been exploited yet but holds a great promise for the study of various voltage-gated ion channels to fully characterize their activation/deactivation and therefore function.
Concluding Remarks
As the field of structural membrane biology expands and more and more structures of membrane proteins are determined our understanding of membrane biology grows. Yet at the same time our understanding is limited because the vast majority of structures are determined without the stabilizing effects of a surrounding lipid bilayer. Lipids and protein are an integral part of biological membranes: they are coevolved and function together to support life. Both the functions and structures of many membrane proteins are dependent on the presence of a lipid bilayer and it is imperative to study the structures of such proteins within the context of real membranes by electron crystallography. This is especially true in the case of voltage-gated ion channels for a complete elucidation of the mechanism underlying their lipid-dependent gating.
Highlights.
Current structures of voltage gated ion channels reviewed
Voltage sensor domains highlighted
The influence of lipids on voltage gated ion channels discussed
Electron crystallography described as a tool for studying lipid-protein interactions
Acknowledgements
Due to limited space we were not able to cite the large body of published work by many of our colleagues on the topics discussed here. The authors want to thank Dr. Liang Shi for his help in preparing the figures. The cryoEM work in Dr. Jiang’s lab on the Kv channels is funded by NIH (R01GM088745 and R01GM093271). Dr. Gonen’s group is supported by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol Rev. 2005;57:387–395. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- 2.Hille B. Ion Channels of Excitable Membranes. 3rd edn. Sinauer Associates, Inc.; 2001. [Google Scholar]
- 3.Mannikko R, Elinder F, Larsson HP. Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature. 2002;419:837–841. doi: 10.1038/nature01038. [DOI] [PubMed] [Google Scholar]
- 4.Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 5.Vemana S, Pandey S, Larsson HP. S4 movement in a mammalian HCN channel. J Gen Physiol. 2004;123:21–32. doi: 10.1085/jgp.200308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezanilla F. Voltage-gated ion channels. IEEE Trans Nanobioscience. 2005;4:34–48. doi: 10.1109/tnb.2004.842463. [DOI] [PubMed] [Google Scholar]
- 7. Zheng H, Liu W, Anderson LY, Jiang QX. Lipid-dependent gating of a voltage-gated potassium channel. Nat Commun. 2011;2:250. doi: 10.1038/ncomms1254. •• This paper first demonstrated that the lipid-dependent gating in a Kv channel.
- 8.McGivern JG. Advantages of voltage-gated ion channels as drug targets. Expert Opin Ther Targets. 2007;11:265–271. doi: 10.1517/14728222.11.3.265. [DOI] [PubMed] [Google Scholar]
- 9.Wickenden AD, McNaughton-Smith G. Kv7 channels as targets for the treatment of pain. Curr Pharm Des. 2009;15:1773–1798. doi: 10.2174/138161209788186326. [DOI] [PubMed] [Google Scholar]
- 10.Schoppa NE, McCormack K, Tanouye MA, Sigworth FJ. The size of gating charge in wild-type and mutant Shaker potassium channels. Science. 1992;255:1712–1715. doi: 10.1126/science.1553560. [DOI] [PubMed] [Google Scholar]
- 11.Islas LD, Sigworth FJ. Voltage sensitivity and gating charge in Shaker and Shab family potassium channels. J Gen Physiol. 1999;114:723–742. doi: 10.1085/jgp.114.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 13.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 14.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Yang N, George AL, Jr, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 16.Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 17.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16(2):387–397. 1610. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 18.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol. 2001;117:469–490. doi: 10.1085/jgp.117.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. • This paper proposed the voltage-sensor paddle model for channel gating.
- 20.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 21. Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. • The transporter-like model was presented.
- 22. Yang N, George AL, Jr, Horn R. Probing the outer vestibule of a sodium channel voltage sensor. Biophys J. 1997;73:2260–2268. doi: 10.1016/S0006-3495(97)78258-4. • The movement of S4 in the gating pore.
- 23.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 24.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K(+) channels. Proc Natl Acad Sci U S A. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. •• This paper first showed that SMase D treatment increased channel activity.
- 26. Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. •• This paper first compared the SMase D and SMase C effects, suggesting a relation to the phosphate in the products.
- 27. Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. •• The paper first demonstrated a requirement of the phosphate groups for a Kv channel to function.
- 28. Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. •• This paper presented the structure of the Kv1.2/2.1 chimera structure with surrounding partial and full lipid molecules.
- 29. Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. •• This paper showed the structure of a voltage-gated Na channel.
- 30. Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc Natl Acad Sci U S A. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. •• The MlotiK structure showed a bundled structure with the channel pore closed.
- 31. Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. • This paper advanced the concept of a charge transfer center.
- 32.Lu Z, Klem AM, Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J Gen Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoppa NE, Sigworth FJ. Activation of shaker potassium channels. I. Characterization of voltage-dependent transitions. J Gen Physiol. 1998;111:271–294. doi: 10.1085/jgp.111.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezanilla F, Perozo E, Stefani E. Gating of Shaker K+ channels: II. The components of gating currents and a model of channel activation. Biophys J. 1994;66:1011–1021. doi: 10.1016/S0006-3495(94)80882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. •• This paper presented the first structure of a voltage-gated channel.
- 36. Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci U S A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. • This paper showed that short-chain detergents could not support the 4-helix bundle conformation of the KvAP voltage sensor domain.
- 37. Jiang QX, Wang DN, MacKinnon R. Electron microscopic analysis of KvAP voltage-dependent K+ channels in an open conformation. Nature. 2004;430:806–810. doi: 10.1038/nature02735. • This paper first demonstrated that the S4 arginines may face the lipids.
- 38.Cuello LG CD, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 39.Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys J. 2006;91:L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freites JA, Tobias DJ, von Heijne G, White SH. Interface connections of a transmembrane voltage sensor. Proc Natl Acad Sci U S A. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sands ZA, Sansom MS. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 43.Hessa T, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Ramu Y, Lu Z. A shaker K+ channel with a miniature engineered voltage sensor. Cell. 2010;142:580–589. doi: 10.1016/j.cell.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wisedchaisri G, Reichow SL, Gonen T. Advances in structural and functional analysis of membrane proteins by electron crystallography. Structure. 2011;19:1381–1393. doi: 10.1016/j.str.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichow SL, Gonen T. Lipid-protein interactions probed by electron crystallography. Curr Opin Struct Biol. 2009;19:560–565. doi: 10.1016/j.sbi.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wisedchaisri G, Gonen T. Fragment-based phase extension for three-dimensional structure determination of membrane proteins by electron crystallography. Structure. 2011;19:976–987. doi: 10.1016/j.str.2011.04.008. •• This manuscript describes a new method for rapid structure determination for electron crystallography. The basis of this study is phase extension taking advantage of relatively straight forward imaging of 2D crystals to provide high quality phase information and extending this information to atomic resolution against diffraction data.
- 48.Fujiyoshi Y, Unwin N. Electron crystallography of proteins in membranes. Curr Opin Struct Biol. 2008;18:587–592. doi: 10.1016/j.sbi.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unger VM, Kumar NM, Gilula NB, Yeager M. Expression, two-dimensional crystallization, and electron cryo-crystallography of recombinant gap junction membrane channels. J Struct Biol. 1999;128(1):98–105. doi: 10.1006/jsbi.1999.4184. 2788. [DOI] [PubMed] [Google Scholar]
- 50.Jap BK, et al. 2D crystallization: from art to science. Ultra microscopy. 1992;46:45–84. doi: 10.1016/0304-3991(92)90007-7. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlbrandt W. Two-dimensional crystallization of membrane proteins. Q Rev Biophys. 1992;25:1–49. doi: 10.1017/s0033583500004716. [DOI] [PubMed] [Google Scholar]
- 52.Fujiyoshi Y. The structural study of membrane proteins by electron crystallography. Adv Biophys. 1998;35:25–80. 755. doi: 10.1016/s0065-227x(98)90004-1. [DOI] [PubMed] [Google Scholar]