Abstract
The ATP synthase is one of the most important enzymes on earth as it couples the transmembrane electrochemical potential of protons to the synthesis of ATP from ADP and inorganic phosphate, providing the main ATP source of almost all higher life on earth. During ATP synthesis, stepwise protonation of a conserved carboxylate on each protein subunit of an oligomeric ring of 10–15 c-subunits is commonly thought to drive rotation of the rotor moiety (c10–14γε) relative to stator moiety (α3β3δab2). Here we report the isolation and crystallization of the c14-ring of subunit c from the spinach chloroplast enzyme diffracting as far as 2.8 Å. Though ATP synthase was not previously known to contain any pigments, the crystals of the c-subunit possessed a strong yellow color. The pigment analysis revealed that they contain 1 chlorophyll and 2 carotenoids, thereby showing for the first time that the chloroplast ATP synthase contains cofactors, leading to the question of the possible roles of the functions of the pigments in the chloroplast ATP synthase.
Keywords: ATP synthase, crystallization, membrane proteins, chlorophyll, carotenoid
Introduction
The ATP synthase couples the transmembrane proton potential produced by mitochondria, chloroplasts and bacteria to the synthesis of ATP from ADP and inorganic phosphate. ATP synthase is one of the most important enzymes on earth, and is present in all kingdoms of life from bacteria to plants and animals. All ATP produced in the processes of photosynthesis and respiration is synthesized by this enzyme.
Structure of ATP synthase
The ATP synthase is an integral membrane protein that consists of two distinct structural and functional domains: a membrane intrinsic proton translocation system (the F0 part), which is structurally connected by two “stalks” to the membrane extrinsic domain (the F1 part), which in turn harbors the nucleotide binding sites (1). The structure is widely conserved in the bacterial and chloroplast enzymes while the mitochondrial enzyme includes several additional subunits with putative regulatory functions; furthermore, the stator stalk has a different subunit composition (2), (3), (4).
The head of the ATP synthase, F1, which contains the catalytic nucleotide binding sites, has the subunit composition α3β3γδε. It is composed of a hexameric catalytic “head' (α3β3); each β subunit contains a catalytic site while each α subunit contains a regulatory nucleotide-binding site. The symmetry of the hexamer is broken by an asymmetric axle (γε) and differences in nucleotide binding of the β subunits. The membrane intrinsic proton translocation part of the ATP synthase, F0, has the subunit composition ab2c10–15. It contains an ion-driven rotor (c10–15) which is abutted by and shares 2 ion half channels in subunit a. F0 and F1 are connected by an external stator stalk (b2δ) and an internal rotor (c10γε).In chloroplast, the membrane intrinsic subunits are often named by roman numbers, where subunit a corresponds to subunit IV (5) and subunit c corresponds to subunit III. The chloroplast enzyme has two different subunits (I and II) that correspond to the subunit b in E.coli (6) We will use the E.coli nomenclature in this report
ATP synthase Rotary Mechanism
During ATP synthesis, proton flux through the proton channel causes stepwise protonation of a conserved carboxylate on each member of an oligomeric ring of 10–15 c-subunits. This, in turn, drives rotation of the complete rotor moiety (c10–15γε) relative to a stator moiety (α3β3δab2). This rotation of the asymmetric axle causes a change in each of the three catalytic sites of the enzyme, found on the β subunits, which promotes the synthesis of ATP and release of the product via the binding-change mechanism (7). One complete revolution of the rotor, relative to the stator, drives the synthesis of 3 ATP at the cost of x protons (where x is the number of c-subunits in the oligomer). Under conditions where the membrane is de-energized, the enzyme catalyzes the reverse reaction, driving proton flux at the expense of ATP hydrolysis.
The ATP synthase thereby contains two distinct rotary motors, where the rotation of F1 is driven by hydrolysis of ATP and the rotation of F0 is driven by a transmembrane ΔpH/Δψ. The direction of the enzyme depends on the driving force of the two rotors. It rotates in one direction during ATP-hydrolysis when ATP is present and there is no or only a small ΔpH/ Δψ across the membrane; it rotates in the opposite direction during ATP synthesis when driven by a sufficient ΔpH/ Δψ, (8).
Structure of F1
Detailed structural information on the proton translocating ATP synthase is so far only available for subcomplexes of the enzyme. The first structure of the F1 part of the enzyme from bovine heart has been solved in 1994 (9). In the meantime, the resolution was further improved and the bovine enzyme has been co-crystallized with many substrates and inhibitors, which sheds light on the catalytic mechanism, for example (10) (PDB entry 1ohh), (11) (PDB entry 1h8), and (12) (PDB entry 2jdi)].
Most of the structural work has been done with the enzyme from bovine heart, but there are now also structures available from other organisms like Bacillus PS3 (13), (PDB entry code 1sky) and yeast mitochondria (14) (PDB entry 2hld). In addition, several structures of parts of the stator stalks from E. coli enzyme (15) (PDB entry 1l2p), (16) (PDB entry 1b9u) have been published. A structure has also been published from bovine heart mitochondrial enzyme that shows major parts of the peripheral stalk (2). There are only two structures published of a subcomplex of the chloroplast enzyme, which contains the α3β3 core of F1 (17), (18)
Structural information of F0
No structures have been determined for the complete proton translocation channel of F0 so far. In the consensus model, the oligomeric ring of c-subunits is abutted by the 5–6 transmembrane helices of subunit a (19) as well as two b subunits (b and b' in the chloroplast enzyme) (1). The first subcomplex of the ATP synthase which has been crystallized that contained a subunit of the proton conduction channel was the F1-c10 complex from yeast. This complex was crystallized and the structure was solved to a resolution of 3.9 Å (20). There is also a structure of the transmembrane helix of subunit b (16). No crystal structures of subunit a have yet been determined, but biochemical evidence suggests that this subunit may contain five transmembrane helices (19).
Structural information on subunit c was first provided by solution NMR (21), which showed that the c-subunit consists of 2 helices connected by a hairpin loop. In 2005, the structure of the c11 ring of the Na+ ATP synthase of the thermophilic bacterium I. tartaricus was determined by X-ray structure analysis at 2.4 Å resolution (22). One of the very astonishing facts of the c-ring is that the number of subunits in the ring differs between different species. The individual c-subunits assemble into oligomeric rings of 10–15 subunits. The ring from S. cerevisiae contains 10 subunits (20), the ring from I. tartaricus contains 11 subunits (22), (23), the ring from spinach S. oleracea contains 14 subunits (24), and the ring from S. platensis contains 15 subunits (25). The stoichiometry is important because it is directly related to the proton-to-ATP ratio and the proton-motive-force (p.m.f.) required for each proton to drive the enzyme. Assuming an efficiency nearing 100%, each full rotation of the c-subunit ring drives the synthesis of 3 ATP; the more subunits are present in each ring, the more protons and less p.m.f. per proton are required for the rotation.
We report here the first crystallization of the c-ring of a proton-driven ATP synthase. Although there are no published crystal structures of ATP synthase showing the presence of any non-nucleotide cofactors, the chloroplast enzyme from spinach (S. oleracea) co-purifies with pigments. The chlorophyll and carotenoids are present in the thylakoid membrane extract and follow the protein through a sucrose density gradient, the exposure of the c-ring to strong detergent and heating, and even through to the crystallization of the c-subunit oligomer. The amount of pigment present in a sample is roughly proportional to the ATP synthase content of a sample, which may indicate an important functional or structural role of the pigments.
Materials/Methods
Isolation of chloroplast membranes and thylakoids
Intact and active chloroplast ATP synthase was extracted from spinach with slight modifications as described in (26), (27). Blemish-free spinach leaves weighing a total of 1.5–2.5 kg were stored overnight at 4°C in the dark to reduce the starch content. The leaves were then homogenized in a 4L Waring blender with a minimal volume of homogenization buffer (0.4 M sucrose, 100 mM Tricine-NaOH pH 8.0, 2 mM MgCl2). The homogenate was filtered through 6 layers of cheesecloth before being centrifuged at 12,000 rpm in a SLA1500 rotor for 25 minutes at 4°C. The supernatant was discarded and the pellet was re-suspended in 1L of a solution containing (10 mM Tris-HCl pH 8.0, 0.5 mM MgCl2). The suspension was stirred for 15 minutes at 4°C before being centrifuged at 12,000 rpm in a SLA1500 rotor for 25 minutes at 4°C. The supernatant was discarded and the pellet was re-suspended in 1L of a buffer containing (0.4 M sucrose, 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 mM MgCl2). The solution was then centrifuged at 12,000 rpm in a SLA1500 rotor for 25 minutes at 4°C. The supernatant was carefully discarded and the upper part of the pellet, containing thylakoids was re-suspended in 8–16 mL of a re-suspension buffer (0.4 M sucrose, 50 mM Tricine-NaOH pH 8.0, 2 mM MgCl2). The chlorophyll concentration of the solution was determined using the method of Wellburn and Lichtenthaler (28). The chlorophyll concentration of the solution was adjusted to 5 mg/mL by dilution with re-suspension buffer.
Extraction of the ATP synthase
Solid DTT was added to a final concentration of 50 mM and the suspension was stirred for 15 minutes on ice. An equal volume of extraction buffer was added (20 mM Tricine-NaOH pH 8.0, 200 mM sucrose, 5 mM MgCl2, 400 mM (NH4)2SO4, 2 mM Na2-ATP, 25 mM = 1.08% (w/v)] Na cholate, 60 mM β-D-octylglucoside, 50 mM DTT). The solution was stirred for 30 minutes at 4°C. The extracted proteins were separated from the remaining membranes by centrifugation at 45, 000 RPM for 60 min at 4°C in a Beckman Ti-70 rotor.
Fractionated Ammonium Sulfate Precipitation
The extracted thylakoid membrane proteins were enriched in ATP synthase by ammonium sulfate precipitation. A saturated ammonium sulfate solution (pH 8.0) was added drop-wise to the protein on ice until it reached a concentration of 1.2 M. The protein solution was centrifuged at 10,000 × g in an SS-34 rotor at 4 °C for 15 minutes and the precipitate was discarded. Additional ammonium sulfate was added to the protein solution until it reached a concentration of 1.8 M. The protein solution was again centrifuged for 15 minutes at 10,000 × g in an SS-34 rotor at 4 °C. The pellet was re-suspended in 4–5 mL of freezing buffer (30 mM NaH2PO4-NaOH pH 7.2, 200 mM sucrose, 2 mM MgCl2, 0.5 mM Na2 EDTA, 4 mM dodecylmaltoside) before being flash-frozen in liquid nitrogen and stored at −80 °C.
Sucrose Density Gradient centrifugation
Protein fractions from the ammonium sulfate precipitation were thawed and mixed with an equal volume of density gradient buffer (30mM NaH2PO4-NaOH pH 7.2, 2 mM MgCl2, 0.5 mM Na2-EDTA, 4 mM dodecylmaltoside). Dodecylmaltoside was added as a concentrated stock solution until the solution clarified (~0.2 %).
Sucrose gradients with steps of 12, 15, 18, 21, 24, 27, and 30% (w/v) sucrose in the density gradient buffer were prepared and ~3.5 mL diluted protein solution was added to the top (for a total volume of ~38.5 mL). These samples were centrifuged in a Beckman SW-32 rotor at 32,000 rpm for 22 hours at 4 °C. The yellow band containing the ATP synthase was collected in a solution containing ~24% (w/v) sucrose, 30 mM NaH2PO4-NaOH pH 7.2, 2 mM MgCl2, 0.5 mM Na2-EDTA, 4 mM dodecylmaltoside. The ATP synthase solution was flash frozen and stored in liquid nitrogen.
Functional characterization of ATP synthase
The activity of the enzyme was measured using the assay described in (26). Liposomes were prepared according to (29) by first preparing a mixture of phosphatidylcholine (PC) and phosphatidic acid (PA) in a 19:1 ratio. The lipids were then dissolved at 18 mg/ml in a detergent-containing solution (7.2 mg/ml cholic acid, 3.6 mg/ml sodium deoxycholate, 500 μM DTT, 100 μM EDTA and 10 mM Tricine-NaOH, pH 8.0) and sonicated in an ice bath (3 times at 30 seconds) under a stream of nitrogen gas. This solution was dialyzed against a 5000-fold volume of buffer (2.5 mM MgCl2, 250 μM DTT, 200 μM EDTA acid and 10 mM Tricine-NaOH, pH 8.0) at room temperature for 5 hours using a Spectrapor membrane (12kDa MWCO) and then frozen at −20° C in 125 μL aliquots.
ATP synthase was inserted into the liposomal membranes using a method adapted from (30). This method uses the detergent Triton X-100 to destabilize the liposomal membranes, allowing the protein-detergent micelles to incorporate, before Biobeads are added to remove the detergent and re-stabilize the membranes. The method was carried out as follows: The liposome aliquots were thawed at room temperature before being combined with 5–20 μL enzyme-micelle solution (2–5 mg/mL), 125 μL buffer (20 mM Tricine, 20 mM succinate, 0.6 mM KOH, 80 mM NaCl, 5 mM MgCl2, pH 8.0 with NaOH), and 20 μL of Triton X-100 (10% w/v). Biobeads (80 mg) were added and the solution was stirred at room temperature for 1 hour before the solution was decanted off.
The proteo-liposomes (10 μL) were incubated with 50 μL acidic buffer (20 mM succinate 0.6 mM KOH, 2.5 mM MgCl2, 20 μM valinomycin, 10 mM NaH2PO4, titrated to pH 4.5) for 2 minutes. ATP synthesis was triggered in a ΔpH/Δψ jump by the addition of 200 μL of basic buffer (200 mM Tricine, 130 mM KCl, 10 mM NaH2PO4, 2.5 mM MgCl2, 0.1 mM ADP, titrated to pH 8.8) and halted by the addition of 250 μL tricholoacetic acid (6% w/v) at discrete time intervals (0.5, 1.0, 2.0, 3.0, 5.0, 10.0 sec). Basic buffer without ADP (500 μL) was added to adjust the pH of the solution towards neutral for the ATP-determination assay.
The ATP content of each sample was determined by the luciferin-luciferase assay. The activity of the enzymes was determined by dividing the rate of ATP synthesis, [ATP/s] from each sample by the protein content of each sample. Typical peak activities achieved for the chloroplast enzyme were ~200 ATP per enzyme per second.
Isolation of c14 Subcomplex
Several methods were initially applied for the disassembly of the ATP synthase complex and isolation of the c-ring, including exposure to NaBr, EDTA, low pH, high pH, urea, and SDS. The ATP synthase was exposed to the different treatments and the c-ring was separated from the remaining protein subunits by gel fitration chromatography. Combinations of several methods had been successfully used for the isolation; however the yields were rather low. After the structure of the c11-ring of I. tartaricus was published, (23) we also tried this method and determined that the chloroplast c-ring can be isolated with good yields with slight modifications of the published procedure. Briefly, 0.8–1% (w/v) lauroylsarcosine was added to the protein (typically 3–4 mg/mL) and the solution was heated to 50–65 °C for 10 minutes. The protein was cooled to room temperature and then ammonium sulfate was added to a final concentration of 2.5 M. The solution was then centrifuged in a SH-3000 rotor at 3,5000 rpm for 15 minutes at 20 °C. The supernatant was collected and filtered through a 0.45 μm filter. The filtrate was dialyzed against a solution of 50 mM NaCl, 20 mM HEPES, pH 7.0, and 0.03% dodecylmaltoside.
The dialyzed protein was concentrated to ~5 mg/mL using a centrifugal concentrator (Vivaspin 20, 50 kDa MWCO) before being applied to a Superdex 200 gel filtration size-exclusion column (GE Healthcare), equilibrated with buffer containing 50 mM NaCl, 20 mM HEPES, pH 7.0, 0.03% dodecylmaltoside (volume of column: 24 mL, flow rate: 0.4 mL/min). The major peak, eluting at μ28 minutes, which contained the c-ring, was collected and then concentrated with a centrifugal concentrator (Vivaspin 500, Sartorius 100 kDa MWCO). An absorption spectrum (280–800 nm) was collected and the protein preparations were further analyzed by SDS-PAGE and MALDI.
Analysis of the pigment content by reverse-phase HPLC
For the analysis of the pigments, the protein was denatured and the pigments extracted by 80% acetone/20% water treatment for 10 minutes at RT. The pigment content of the crystals and protein batches was determined by HPLC in principle as described in (31), (32) on a HP-1100 Chemstation using a Waters Spherisorb S5ODS2 (250× 4 mm) column filled with C-18 reverse-phase silica gel.
Crystallization of the c-ring
Initial screens of crystallization conditions were conducted on the intact and active ATP synthase using the vapor diffusion method. The systematic screens included the variation of pH, precipitant, ionic strength, temperature, protein concentration, detergents and additives (Details of the crystallization procedure will be published elsewhere. Varco-Merth et al, In Preparation). Several crystallization experiments contained thin 20μ50 μm crystals growing out of a “skin”-like phase composed of detergent, PEG, and denatured protein. These conditions were further varied to optimize the crystallization, producing larger and thicker crystals (~0.3–0.4 mm), which were suitable for analysis by SDS-PAGE. We also observed that the crystals had a strong yellow color.
SDS gel electrophoresis showed that the crystals were determined to be composed of the c-subunit oligomer. Therefore, further efforts were undertaken to further purify the subcomplex (see above) in order to improve the quality of the crystals and enable the use of re-crystallization as a final purification step. Crystals prepared from the purified c-ring grew larger in drops with less “skin” observed in the drops. They showed improved X-ray diffraction quality as they diffracted to higher resolution and showed less anisotropy than the crystals grown from the intact ATP synthase.
Characterization of the c-ring crystals by X-ray diffraction
The first X-ray data on crystals were measured using the home X-ray source (Bruker) that contains a Cu-anode as X-ray source. These thin hexagonal plates (50–100 μm in diameter, 10–20 μm in height) diffracted as far as 5 Å in the direction of one crystal axis. However they showed strong anisotropy, diffracting to less than 20 Å in other dimensions. Optimization of the growth conditions, further purification of the subunit c oligomer, and the use of synchrotron radiation led to diffraction data with higher resolution and less anisotropy. The best diffracting crystals showed X-ray diffraction as far as 2.8 Å in one direction and 3.4 Å in the 90 degree rotation, reflecting the inner anisotropy of the crystals. Therefore, the evaluation of the data is currently limited to 3.45 Å.
X-ray diffraction data on the crystals were collected using synchrotron X-ray radiation at beamlines 8.3.1, 5.0.3, 8.2.1, and 8.2.2 at the Advanced Light Source at Berkeley National Lab and Beamlines 19-ID and 22-ID at the Advanced Photon Source at Argonne National Laboratory.
The primary data evaluation was done using the software package HKL 2000 version 1.93 (for indexing, integration and merging of the data). (33, 34). The scaled data were further evaluated with programs from the CCP4i family (35).
Results
Isolation and functional characterization of the ATP synthase
The first step in the structure determination of the c-subunit rotor of the ATP synthase was the isolation of the functional intact enzyme from spinach. The isolation was performed as described in materials and methods using a protocol which was modified after published procedure (5), (26). The protein was solubilized from the membrane in the presence of DTT by a combination of 2 detergents (octylglycoside and cholate). The ATP synthase was enriched by use of step-wise ammonium sulfate precipitation. The detergent was changed to the milder detergent dodecylmaltoside upon density gradient centrifugation as the protein showed higher stability and activity in the β-dodecylmaltoside preparations compared to octylglucoside preparations. We have also tried to isolate the ATP synthase exclusively in dodecylmaltoside by replacing the cholate-octylglucoside extraction procedure. However, the results showed that the yields of ATP synthase by extraction with dodecylmaltoside were extremely low and contained mainly Photosystem I and II, leading to the conclusion that these preparations were not useful for structural and functional investigations.
Using the standard density gradient used by (26), the ATP synthase appeared in yellowish band in the lower third of the gradient. It was noted more than 20 years ago that the ATP synthase band in the density gradient is yellow (27), but the fact did not draw any further attention as the pigmentation was generally regarded as a small contamination from the chlorophyll-containing proteins. Attempts to prepare a functional intact “colorless” enzyme failed. While they were still able to hydrolyze ATP all colorless preparations of the ATP synthase were inactive in direction of ATP synthesis (Fromme unpublished results). ATP hydrolysis does not require a tight coupling of the F1 and F0 parts and can therefore not serve as an assay for the determination of an intact enzyme.
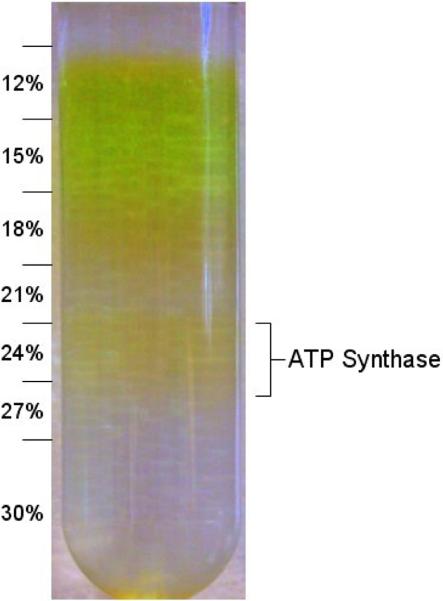
The density gradient centrifugation was optimized testing different linear and step gradients to establish a better separation of the ATP synthase from the major pigment-containing contaminants (Photosystem I and II). The optimized step gradient contained steps of 12,15, 18, 21, 24, 27 and 30% sucrose (for more details see materials and methods). The result is shown in Figure 2. The ATP synthase is contained in the yellow band at 24% sucrose.
Figure 2. Sucrose density gradient centrifugation of the ATP synthase.
Intact ATP synthase is purified using a sucrose density gradient centrifugation. The lower yellow band at ~24% contains the ATP synthase. It has a strong yellow color that is proportional to the amount of protein present. The numbers indicated represent the concentration of the sucrose (w/v) in each step.
The activity of the isolated ATP synthase was measured by investigation of the rate of ATP synthesis in reconstituted liposomes. The protein was reconstituted into liposomes by removal of detergents in the presence of liposomes using Bio-Beads (30). The enzyme preparations showed high rates of ATP synthesis of 200± 30 ATP per enzyme per second.
Crystallization experiments
The isolated intact ATP synthase in the dodecylmaltoside micelle was used for crystallization experiments using a large comprehensive screening procedure. The systematic screen included a broad screen of temperature, ionic strength, pH, different cations and anions, precipitants and additives. The aim was to screen for crystallization of the intact enzyme as well as subcomplexes of the protein, with special interest of crystallization conditions that could lead to the crystallization of subcomplexes containing subunits of the membrane intrinsic part of the ATP synthase.
Very thin hexagonal crystals were first observed as shown in Figure 3A. SDS gel electrophoresis showed that the crystals exclusively contain the c-subunit of the ATP synthase as shown in Figure 4. The crystallization was further optimized to achieve larger crystals, which are suitable for X-ray structure analysis (Details will be published elsewhere. Varco-Merth et al, In Preparation.).
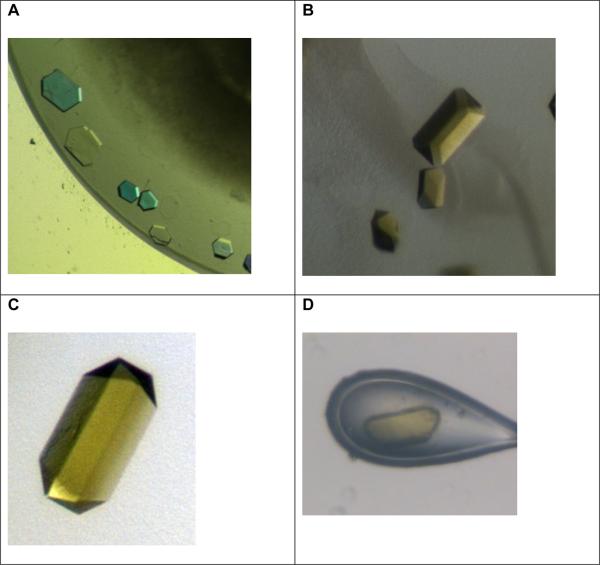
Figure 3. Gallery of crystals of the c-ring of the ATP synthase.
A: Initial small crystals of the c14-ring. The crystals are very thin plates (diameter 50 μM, height less than 10 μM). They were grown from a starting solution that contained the intact ATP synthase. Please note that the picture is taken under polarized light and therefore appear with false colors.
B, C and D: These pictures were taken under white light without polarization filter to show the true colors of the crystals.
B: Improved crystals of the c-ring (diameter 100 μM, height 50 μM), showing a strong yellow color. The crystals were grown from a starting solution that contains the intact ATP synthase.
C and D: Crystals of the isolated c-ring of the ATP synthase. Please note that the crystals show the same strong yellow color as the crystals grown from the intact enzyme.
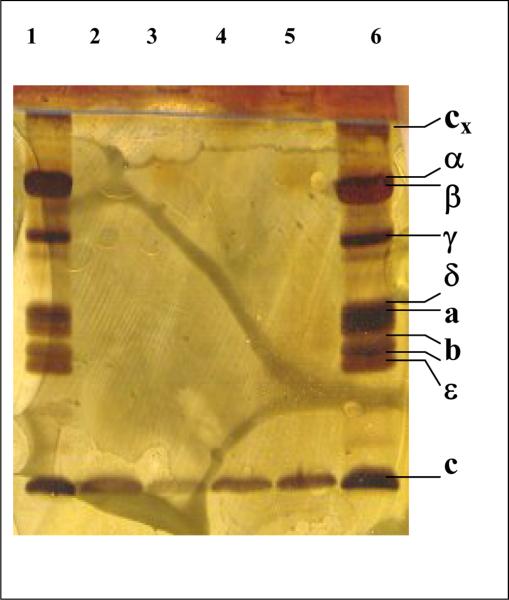
Figure 4. SDS gel electrophoresis of the intact ATP synthase and crystals of the c-ring.
Lanes 1 and 6 contain intact ATP synthase, lanes 2 to 5 contain dissolved crystals. The samples were run on a Phastgel system HD gel and stained with silver.
When we observed the first larger crystals of the chloroplast c-ring, we were very surprised to see that they were yellow. A picture of the crystals is shown in Figure 3B. SDS gel electrophoresis showed the same pattern as the small crystals, i.e. the crystals contained exclusively the c-subunit of the chloroplast ATP synthase. Spectroscopy and HPLC analysis revealed that they contained chlorophyll and carotenoids, however we could not exclude at this time that the pigment were non-specifically bound to the detergent.
An initial screening for X-ray diffraction at the home X-ray source showed X-ray diffraction to about 5.5 A. A complete native data set had been collected at synchrotron sources and could be evaluated to 4.5 Å resolution (data not shown), however, the crystals showed large anisotropy. During the optimization of the crystallization conditions we observed that the crystals grow in hanging drops in a skin, consisting of PEG, detergent, and precipitated protein. No crystals could be grown by batch or dialysis methods, which made it difficult to use crystallization as the last purification step, a method which has been successfully used in the past to grow and improve crystals of Photosystem I and II (5), (36), (37).
Isolation of the c-ring of the ATP synthase
The next step in the experimental scheme was the further purification of the c-ring. There were two major aims in the establishment of an isolation procedure for the c-ring. We wanted to investigate if the pigments are an intrinsic structural part of the c-ring that would sustain a harsh isolation procedure of the c-ring. Furthermore, we wanted to improve the crystal quality by performing the crystallization experiments with a pure preparation of the c-ring. The first step in the isolation process was the disassembly of the intact ATP synthase. The experiments showed that the enzyme could be disassembled in different ways either by use of NaBr, low pH or high pH, EDTA, or strong ionic detergents. The intact c14-ring protein could also be isolated in SDS as first described by Fromme (5), in a method which was used for the AFM studies by N. Dencher and his colleagues (24) that revealed for the first time the 14-fold symmetry of the chloroplast c-ring.
However, the protein in SDS micelles was not suitable for crystallization experiments. The best detergent for crystallization of the isolated c-ring was the very mild detergent dodecylmaltoside, which had also been used to grow the first c-ring crystals starting with the intact ATP synthase. While we were optimizing the isolation procedure for the chloroplast c-ring, the crystal structure for a c11-ring of the Na+ ATP synthase from I. tartaricus was published (22). We also tested the isolation procedure described for the Na+-driven ATP synthase c11-ring for the chloroplast enzyme and could achieve a disassembly of the intact enzyme and crude purification of the c14-ring of the chloroplast ATP synthase following their procedure with minor modifications. This was astonishing, as spinach grows in cool climates while I. tartaricus is a thermophilic prokaryote. The protein was further purified by gel chromatography following dialysis. The harsh detergent lauroylsarcosine was replaced by the mild anionic detergent dodecylmaltoside during the dialysis and gel chromatography steps. The chromatogram showed two peaks (data not shown). The major peak, eluting at ~28 min, contained the c-ring. We were very excited to see that the pigments largely remained in the c-ring during the harsh detergent and heat treatment. This gives a clear indication that the pigments are not non-specifically attached to detergent micelle, but must be located inside the c-ring, being an intrinsic cofactor of the ATP synthase.
Crystallization of the isolated c-ring
The crystals grown from the isolated c-ring are shown in Figures 3C and 3D. They crystallize in the same space group and show the same strong yellow color as the crystals grown form the intact ATP synthase. While the crystallization drops of the intact enzyme always contained a lot of denatured protein in form of amorphous precipitation, the crystallization experiments of the isolated c14 ring were free of amorphous precipitate.
Pigment analysis of the crystals of the c14-ring
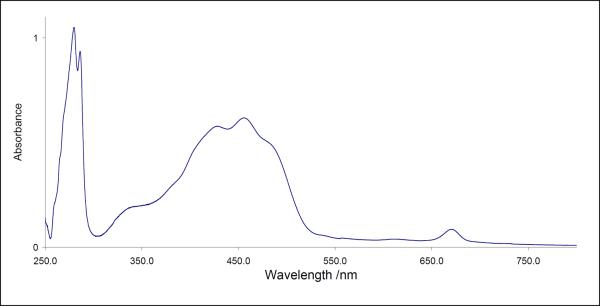
The crystals still grow out of a skin of detergent and PEG, but this skin was diminished and they could be easily separated from it. This allowed us to used seeding techniques to further improve the size and quality of the crystals and analyze the pigment content in more detail. The spectrum of the dissolved crystals is shown in Figure 5. The absorption spectra indicated a pigment content of 1 ± 0.3 chlorophyll per 2 ± 0.5 carotenoids. As the determination of the pigment content from the spectrum alone is not very precise due to the overlap of the chlorophyll and carotenoid bands, we have analyzed the pigment content by reverse-phase HPLC chromatography. The results are shown in Figure 6. The HPLC data confirmed the pigment content, but also showed that the sample contains pheophytin. The pheophytinization is probably caused by loss of the central Mg2+ during the extraction procedure of the pigments for the HPLC analysis. The protein/ chlorophyll ratio was calculated on the basis of the ratio between the protein absorption at 280nm, using a molar extinction coefficient of 20860 for the c ring and an absorption coefficient of the chlorophyll of 64 000 at 665 nm. The determination of the ratio of Chl/protein in the crystals was difficult as the crystals had to be completely separated from the PEG-skin and were also difficult to re-solubilize. Furthermore, more than 200 crystals had to be dissolved for one absorption spectrum, so this method did not allow the determination of the Chl/protein ratio of individual crystals. Therefore, we can currently only give a rough estimate of the Chl/protein ratio of different crystal batches. The chlorophyll/protein was in most preparations less than stochiometric, caused by the harsh treatment of the c-ring during disassembly and isolation process. The crystals with the best diffraction quality (< 3 Å) had the highest chlorophyll/protein content Chl/c14 ring, while crystal-batches with a low pigment content (< 0.1 Chl/c14 ring) showed generally only very weak X-ray diffraction quality (resolution limit 8–12 Å).
Figure 5. Absorption spectrum of dissolved crystals of the c14 ring of the ATP synthase.
The evaluation of the spectrum revealed a pigment content of ~2 carotenoids per 1 chlorophyll. The stoichiometric ratio of pigments to protein oligomers was in some preparations less than 1 to 1. It could be caused by removal of pigments by the strong methods required to dissociate the subunit c oligomer from the rest of the ATP synthase complex.
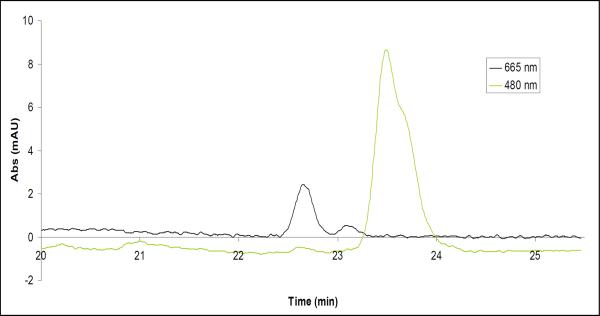
Figure 6. Reverse-phase HPLC analysis of the pigment content of the c-ring crystals.
Protein crystals were briefly subjected to a solution of 80% acetone/20% water to denature the protein and extract the pigments. The solution was then separated using a reverse-phase HPLC (as described in Methods) and two major peaks were observed. The peak observed by the 665 nm detector was identified as chlorophyll and pheophytin by spectral analysis. The peak observed by the 480 nm detector was identified as beta-carotene by spectral analysis.
Preliminary characterization of X-ray diffraction
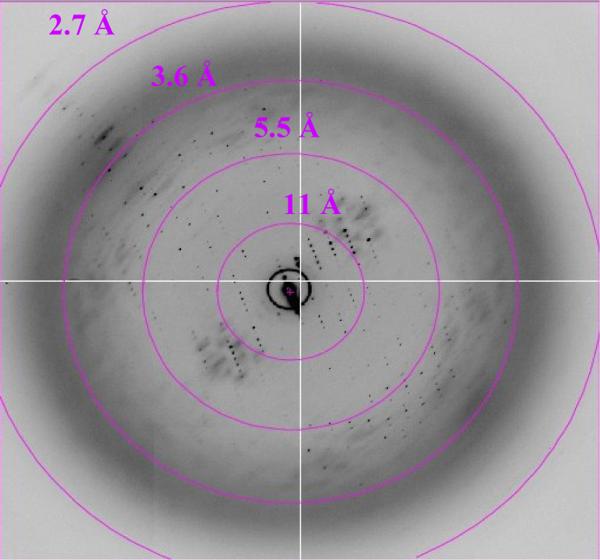
Data on the improved crystals have recently been collected at synchrotron X-ray sources. The resolution of the crystals has been dramatically improved and diffraction along the c-axis is observed to 2.8 Å resolution (see Figure 7). The diffraction is still anisotropic with diffraction in the a and b planes to 3.5 Å. A native data set has been collected and evaluated to 3.45 Å resolution. The native Patterson map of the data set revealed a 14-fold symmetry, which is in agreement with the stochiometry of 14 c-subunits per ring as determined by AFM studies of the chloroplast enzyme (24). We are currently in the process of determining the phases by a combination of molecular modeling, molecular replacement and experimental phases using Nobel gases and other heavy atoms.
Figure 7. X-ray diffraction pattern of the crystals of the c-ring of the ATP synthase.
This data was collected at Beamline 8.3.1 at the Advanced Light Source at Lawrence Berkeley National Laboratory (see Acknowledgements).
Discussion of potential functions of the pigments in the chloroplast ATP synthase
The spectral analysis of the pigments in dissolved crystals of the re-crystallized protein revealed the presence of chlorophylls and carotenoids. This is, to our knowledge, the first time that results have been presented that the chloroplast ATP synthase may contain pigments as integral parts of the enzyme. This situation is somewhat similar to the case of the cytochrome b6f complex, where crystallization and structure determination revealed that one chlorophyll and one carotenoid are essential pigments deeply buried inside the protein complex (38), (39), The function of the chlorophyll and carotenoid in the cytochrome b6f complex is still under discussion (40).
In the case of the chloroplast c14 ring, the pigments are tightly bound to the c14 ring and may be located in the inside of the ring, as they are resistant to removal by the harsh detergent treatment during the isolation of the c14 ring. We want to speculate now on the potential functions of the pigments in the c14 ring of the chloroplast ATP synthase. The possibility, which we want to discuss first, is that the chlorophyll and carotenoids may function as “lipid” filling (together with lipids) the inside of the c-ring. In this case they may stabilize the hydrophobic “inner” side of the N-terminal helix of the ring and the major function would be the stabilization of the complex. It might also be considered that the presence of the pigments (and especially the large chlorophyll head group) may play a role in the determination of the size of the c-ring. We are currently working on the reconstitution of the c-ring with different stoichiometry of pigments to test this hypothesis. We are also currently undertaking a study to determine whether the stochiometry of the ring depends on the photosynthetic pigments or special assembly factors by attempting heterologous over-expression of the chloroplast c-subunit in E. coli. Another possibility is that the pigments (and especially the carotenoids) may play a photo-protective role preventing the photo damage of the enzyme under high light intensity. The pigments may also interact with the bottom part of the γε subunits, which form the central stalk of the ATP synthase (41), (20) thereby providing stabilization at the coupling site between the c-ring rotor in F0 and the γε rotor in F1. If the pigments play a role in the interaction of the central stalk with the c-ring the question may be raised if the pigments could play a role in the light regulation of the ATP synthase (42), (43). However, it is clear that many more experiments have to be done to rule out or confirm some of these hypotheses, which leaves a large field for further investigations and the structure determination, which is in progress, may finally show the location of the pigments.
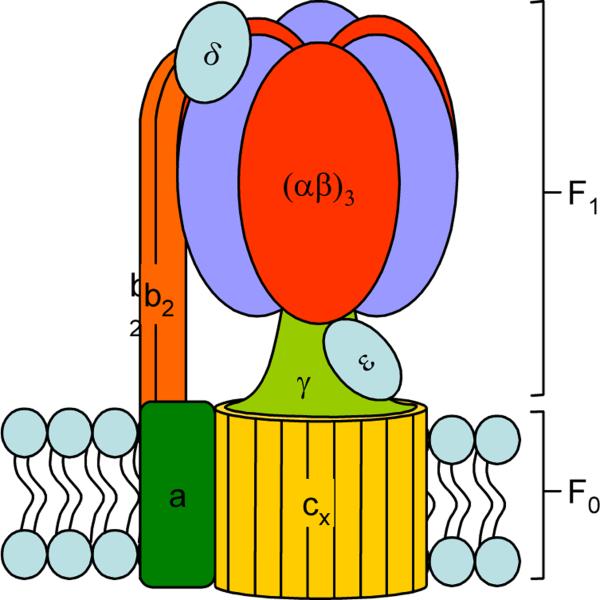
Figure 1. Subunit Arrangement of ATP Synthase.
ATP synthase is a membrane-bound protein found in chloroplasts, mitochondria and bacteria. It is responsible for using the energy stored in an electrochemical proton gradient to synthesize ATP from ADP and Pi and consists of two major moieties—F1 and F0. F1 consists mainly of soluble proteins with the subunit composition α3β3γδε and contains 3 catalytic sites (one on each β subunit). F0 has the subunit composition ab2cx (x ranges from 10–15 and depends on the species) and contains a membrane-bound ion channel.
During ATP synthesis, rotation of the asymmetric axle (γε) within the core of the catalytic hexamer (α3β3) causes a change in the binding properties of the active site which promotes binding of the substrates, synthesis and release of the product. This rotation is caused by proton flux through a proton channel (formed by subunits a and c) that causes a stepwise protonation of the each c subunit in the oligomeric ring.
Table 1. Crystallographic Parameters.
Data collection and crystallographic analysis. The numbers in parentheses refer to the higher resolution bin (3.60–3.45 Å).
| Space Group | C2 |
|---|---|
| Unit Cell | a=145 Å b=97 Å c=127 Å, β = 106° |
| Resolution | 50–3.45 Å (3.60–3.45 Å) |
| Multiplicity | 3.4 (2.9) |
| Completeness | 92.6% (65.1%) |
| I/σ | 15.7 (2.3) |
| Rsym | 0.074 (0.353) |
Acknowledgements
This research was supported by NIH, proposal GM081490-01. We thank Robert Lawrence for critical reading of the manuscript and helpful discussion.
We want to thank Dr. Dmitrii Vavilin for the reverse-phase HPLC analysis of the pigment content of the crystals. We would also like to thank the beamline scientists at the ALS and APS, especially James Holton and George Meigs at ALS 8.3.1 and Corie Ralston at the Berkeley Center for Structural Biology (ALS). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the US Department of Energy under contract No. DEAC02- 05CH11231 at Lawrence Berkeley National Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Bottcher B, Graber P. The structure of the H(+)-ATP synthase from chloroplasts and its subcomplexes as revealed by electron microscopy. Biochim Biophys Acta. 2000;1458:404–16. doi: 10.1016/s0005-2728(00)00090-6. [DOI] [PubMed] [Google Scholar]
- (2).Dickson VK, Silvester JA, Fearnley IM, Leslie AGW, Walker JE. On the structure of the stator of the mitochondrial ATP synthase. Embo Journal. 2006;25:2911–2918. doi: 10.1038/sj.emboj.7601177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Devenish RJ, Prescott M, Roucou X, Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim Biophys Acta. 2000;1458:428–42. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- (4).Weber J. ATP synthase--the structure of the stator stalk. Trends Biochem Sci. 2007;32:53–6. doi: 10.1016/j.tibs.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fromme P, Boekema EJ, Graber P. Isolation and Characterization of a Supramolecular Complex of Subunit-Iii of the Atp-Synthase from Chloroplasts. Zeitschrift Fur Naturforschung C-a Journal of Biosciences. 1987;42:1239–1245. [Google Scholar]
- (6).Herrmann RG, Steppuhn J, Herrmann GS, Nelson N. The nuclear-encoded polypeptide Cfo-II from spinach is a real, ninth subunit of chloroplast ATP synthase. FEBS Lett. 1993;326:192–8. doi: 10.1016/0014-5793(93)81789-3. [DOI] [PubMed] [Google Scholar]
- (7).Boyer PD. The binding change mechanism for ATP synthase--some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–50. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- (8).Borsch M, Diez M, Zimmermann B, Reuter R, Graber P. Stepwise rotation of the gamma-subunit of EF(0)F(1)-ATP synthase observed by intramolecular single-molecule fluorescence resonance energy transfer. FEBS Lett. 2002;527:147–52. doi: 10.1016/s0014-5793(02)03198-8. [DOI] [PubMed] [Google Scholar]
- (9).Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–8. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- (10).Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–50. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- (11).Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–41. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- (12).Bowler MW, Montgomery MG, Leslie AG, Walker JE. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 A resolution. J Biol Chem. 2007;282:14238–42. doi: 10.1074/jbc.M700203200. [DOI] [PubMed] [Google Scholar]
- (13).Shirakihara Y, Leslie AG, Abrahams JP, Walker JE, Ueda T, Sekimoto Y, Kambara M, Saika K, Kagawa Y, Yoshida M. The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer. Structure. 1997;5:825–36. doi: 10.1016/s0969-2126(97)00236-0. [DOI] [PubMed] [Google Scholar]
- (14).Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. Embo J. 2006;25:5433–42. doi: 10.1038/sj.emboj.7601410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Del Rizzo PA, Bi Y, Dunn SD. ATP synthase b subunit dimerization domain: a right-handed coiled coil with offset helices. J Mol Biol. 2006;364:735–46. doi: 10.1016/j.jmb.2006.09.028. [DOI] [PubMed] [Google Scholar]
- (16).Dmitriev O, Jones PC, Jiang WP, Fillingame RH. Structure of the membrane domain of subunit b of the Escherichia coli F0F1 ATP synthase. Journal of Biological Chemistry. 1999;274:15598–15604. doi: 10.1074/jbc.274.22.15598. [DOI] [PubMed] [Google Scholar]
- (17).Groth G, Pohl E. The structure of the chloroplast F1-ATPase at 3.2 A resolution. J Biol Chem. 2001;276:1345–52. doi: 10.1074/jbc.M008015200. [DOI] [PubMed] [Google Scholar]
- (18).Groth G. Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc Natl Acad Sci U S A. 2002;99:3464–8. doi: 10.1073/pnas.052546099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Vik SB, Ishmukhametov RR. Structure and function of subunit a of the ATP synthase of Escherichia coli. J Bioenerg Biomembr. 2005;37:445–9. doi: 10.1007/s10863-005-9488-6. [DOI] [PubMed] [Google Scholar]
- (20).Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- (21).Girvin ME, Rastogi VK, Abildgaard F, Markley JL, Fillingame RH. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- (22).Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- (23).Stahlberg H, Muller DJ, Suda K, Fotiadis D, Engel A, Meier T, Matthey U, Dimroth P. Bacterial Na+-ATP synthase has an undecameric rotor. Embo Reports. 2001;2:229–233. doi: 10.1093/embo-reports/kve047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Seelert H, Poetsch A, Dencher NA, Engel A, Stahlberg H, Muller DJ. Structural biology - Proton-powered turbine of a plant motor. Nature. 2000;405:418–419. doi: 10.1038/35013148. [DOI] [PubMed] [Google Scholar]
- (25).Pogoryelov D, Yu JS, Meier T, Vonck J, Dimroth P, Muller DJ. The c(15) ring of the Spirulina platensis F-ATP synthase: F-1/F-0 symmetry mismatch is not obligatory. Embo Reports. 2005;6:1040–1044. doi: 10.1038/sj.embor.7400517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Turina P, Samoray D, Graber P. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1-liposomes. Embo Journal. 2003;22:418–426. doi: 10.1093/emboj/cdg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fromme P. Die ATP-synthase aus Chloroplasten: Biochemische Untersuchungen zur Struktur and kinetische Messungen zum Mechanismus des Enzyms. Dissertation, Technical University Berlin. 1988:1–248. [Google Scholar]
- (28).Wellburn A. R. a. L., H. Formulae and program to determine total carotenoids and chlorophylls A and B leaf extracts in different solvents. Vol. II. The Hague; The Netherlands: 1984. [Google Scholar]
- (29).Fischer S, Graber P. Comparison of Delta pH- and Delta phi-driven ATP synthesis catalyzed by the H+-ATPases from Escherichia coli or chloroplasts reconstituted into liposomes. Febs Letters. 1999;457:327–332. doi: 10.1016/s0014-5793(99)01060-1. [DOI] [PubMed] [Google Scholar]
- (30).Richard P, Rigaud JL, Graber P. Reconstitution of CF0F1 into liposomes using a new reconstitution procedure. Eur J Biochem. 1990;193:921–5. doi: 10.1111/j.1432-1033.1990.tb19418.x. [DOI] [PubMed] [Google Scholar]
- (31).Vavilin DV, Vermaas WF. Regulation of the tetrapyrrole biosynthetic pathway leading to heme and chlorophyll in plants and cyanobacteria. Physiol Plant. 2002;115:9–24. doi: 10.1034/j.1399-3054.2002.1150102.x. [DOI] [PubMed] [Google Scholar]
- (32).Xu H, Vavilin D, Vermaas W. The presence of chlorophyll b in Synechocystis sp. PCC 6803 disturbs tetrapyrrole biosynthesis and enhances chlorophyll degradation. J Biol Chem. 2002;277:42726–32. doi: 10.1074/jbc.M205237200. [DOI] [PubMed] [Google Scholar]
- (33).Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallographica Section D-Biological Crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- (34).Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (35).Collaborative Computational Project, N. The Ccp4 Suite - Programs for Protein Crystallography. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- (36).Fromme P, Witt HT. Improved isolation and crystallization of Photosystem I for structural analysis. Biochimica Et Biophysica Acta-Bioenergetics. 1998;1365:175–184. [Google Scholar]
- (37).Zouni A, Jordan R, Schlodder E, Fromme P, Witt HT. First photosystem II crystals capable of water oxidation. Biochimica Et Biophysica Acta-Bioenergetics. 2000;1457:103–105. doi: 10.1016/s0005-2728(00)00100-6. [DOI] [PubMed] [Google Scholar]
- (38).Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science. 2003;302:1009–14. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- (39).Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b(6)f complex. Nature. 2003;426:413–8. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- (40).Dashdorj N, Zhang H, Kim H, Yan J, Cramer WA, Savikhin S. The single chlorophyll a molecule in the cytochrome b6f complex: unusual optical properties protect the complex against singlet oxygen. Biophys J. 2005;88:4178–87. doi: 10.1529/biophysj.104.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000;7:1055–61. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- (42).Evron Y, Johnson EA, McCarty RE. Regulation of proton flow and ATP synthesis in chloroplasts. J Bioenerg Biomembr. 2000;32:501–6. doi: 10.1023/a:1005669008974. [DOI] [PubMed] [Google Scholar]
- (43).Avenson TJ, Cruz JA, Kramer DM. Modulation of energy-dependent quenching of excitons in antennae of higher plants. Proc Natl Acad Sci U S A. 2004;101:5530–5. doi: 10.1073/pnas.0401269101. [DOI] [PMC free article] [PubMed] [Google Scholar]