Abstract
A number of interrelated factors can affect the precision and accuracy of lung nodule size estimation. To quantify the effect of these factors, we have been conducting phantom CT studies using an anthropomorphic thoracic phantom containing a vasculature insert to which synthetic nodules were inserted or attached. Ten repeat scans were acquired on different multi-detector scanners, using several sets of acquisition and reconstruction protocols and various nodule characteristics (size, shape, density, location). This study design enables both bias and variance analysis for the nodule size estimation task. The resulting database is in the process of becoming publicly available as a resource to facilitate the assessment of lung nodule size estimation methodologies and to enable comparisons between different methods regarding measurement error. This resource complements public databases of clinical data and will contribute towards the development of procedures that will maximize the utility of CT imaging for lung cancer screening and tumor therapy evaluation.
OCIS codes: (110.6955) Tomographic imaging, (110.1758) Computational imaging, (110.7440) X-ray imaging, (110.6880) Three-dimensional image acquisition
1. Introduction
Technological advances in computer tomography (CT) over the last decade have enabled the acquisition of thin (less than 1mm), near isotropic thoracic scans in a single breath hold. These advances can potentially improve temporal CT analysis so that the detection of small (less than 1cm in diameter) lung nodules can be obtained and changes in nodule size can be assessed early. These improvements have led to an improved ability to both diagnose disease and to characterize the response of tumors to therapy so that the proper treatment for individual patients can be administered. Change in lesion size is one approach for estimating drug response, however it assumes that bias (difference between measured and true size) is constant along temporal scans. If this assumption does not hold, then evaluating the change in measured lesion size between scans can be problematic. An approach that could quantify size that is specific to the CT hardware and scan parameters would be preferable in order to correctly account for these biases.
Currently, lung nodule size is typically assessed using the RECIST criteria [1,2], which are based on the measurement of the maximum diameter of a nodule from a single slice. The RECIST criteria suffer from certain limitations [3], the most important of which is the assumption of nodule sphericity. Volumetric assessment of nodule size has been investigated as an approach that is better suited to the true representation of lung nodule shape due to its use of 3D data. However, a number of factors can affect the precision and accuracy of volumetric CT for the estimation of lung nodule size, as was summarized in recent review articles [4,5]. These factors include acquisition and reconstruction parameters, nodule characteristics, and the performance and usage of measurement tools. We have been conducting phantom studies to quantify the effect of such factors with an overall goal of developing methods to account for errors in lung nodule volumetry. Phantom studies provide a framework in which the true size, shape, and location of nodules is known, allowing for bias analysis. Moreover, they allow for the acquisition of multiple scans required for variance analysis, which would be more difficult to acquire in human studies because of the additional radiation exposure to patients. Additionally phantom data sets may serve as a means for directly comparing the performance of different nodule sizing algorithms on a fixed set of phantom nodules [6].
In order for findings from phantom studies to be applicable to clinical data, phantoms should somehow be representative of actual lung nodules, and should also incorporate the complexity of the surrounding background and interfering vessel structures. State-of-the-art thoracic phantoms incorporating lung vasculature are now available and, while their characteristics do not completely match those of a clinical scan, they are much better suited for synthesizing the variability inherent in the vascular nature of the lung field. Synthetic nodules can be attached to the vasculature of such a thoracic phantom to approximate the complexity of clinical nodules; studies have shown that the performance of segmentation algorithms is significantly worse for cases of attached nodules [7–9]. Synthetic nodules with irregular shapes and margins, as well as heterogeneous densities that mimic non-solid nodules can be manufactured. Scans of these anthropomorphic phantoms and synthetic nodules can be used in the development and evaluation of methodologies for nodule size estimation.
Our phantom studies have employed an anthropomorphic thoracic phantom with a vasculature insert to which synthetic nodules with characteristics (nodule size, shape, density) that span the range of clinical nodules can be attached or inserted. The thoracic phantom has been scanned using multiple scanners and imaging protocols to examine the effects of scanner model, acquisition, and reconstruction parameters on lung nodule size estimation. Each imaging protocol and nodule layout has been scanned repeatedly (10 repeats) to enable variance analysis in addition to bias analysis.
Using a systematic approach to probe the factors that may affect the precision and accuracy in lung nodule estimation, we have collected over 5000 phantom CT scans. This data is in the process of becoming publicly available via the National Biomedical Imaging Archive (NBIA) as a resource for the development and assessment of nodule size estimation methodologies and to enable developers to perform comparisons between different methods regarding measurement error. This resource complements public databases of clinical data sets such as the Reference Image Database for Evaluation of Response (RIDER) and the Lung Image Database Consortium (LIDC) databases, created by the National Cancer Institute (NCI) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB).
2. Database Description
2.1 Anthropomorphic phantom
The anthropomorphic thoracic phantom (Kyotokagaku Incorporated, Tokyo, Japan) employed in this study is shown in Fig. 1 , along with the vasculature insert. The phantom was manufactured using urethane and epoxy resins to mimic soft-tissue and bone respectively. To date, we have acquired scans with nodules attached to the vasculature as well as scans with nodules inserted without attachment to vessels by placing them in low-density foam. The phantom does not contain lung parenchyma so the space within the vascular structure is filled with air.
Fig. 1.
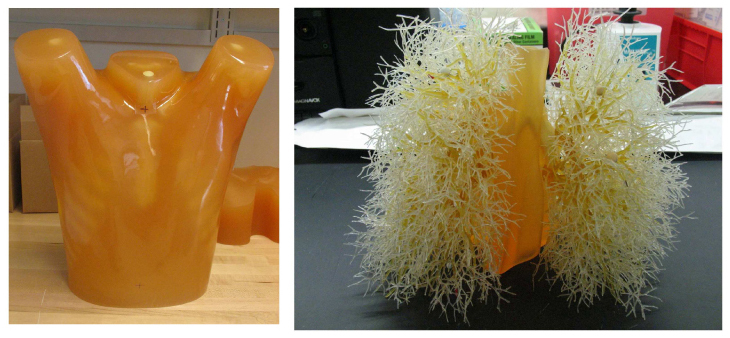
Photograph of the exterior shell of the thoracic phantom (left) and the vasculature insert (right).
2.2 Synthetic lung nodules
The set of synthetic lung nodules used in this study were independently manufactured by Kyotokagaku Incorporated (Japan) and Computerized Imaging Reference Systems (CIRS, Norfolk, VA). They consisted of objects varying in size (5, 8, 10, 12, 20, 40 mm), shape (spherical, elliptical, lobulated, spiculated, and irregular), and density (−800, −630, −300, −10, + 20, and + 100 HU). Each synthetic nodule was made from a material of relatively uniform density; however different materials were used for nodules of different densities. For the Kyotokagaku nodules, foamed urethane resin was used to construct the −800 and −630 HU nodules, and urethane resins along with hydro-apatite was used for the + 100HU nodules. Lathing was used to shape the −800 and −630HU nodules whereas the + 100HU were shaped using a molding process. For the CIRS nodules, epoxy resin material loaded with different fillers was used to achieve the target HU for the various lung nodules, and molding was used to shape them. Figure 2 shows samples of synthetic nodules in various sizes and shapes.
Fig. 2.
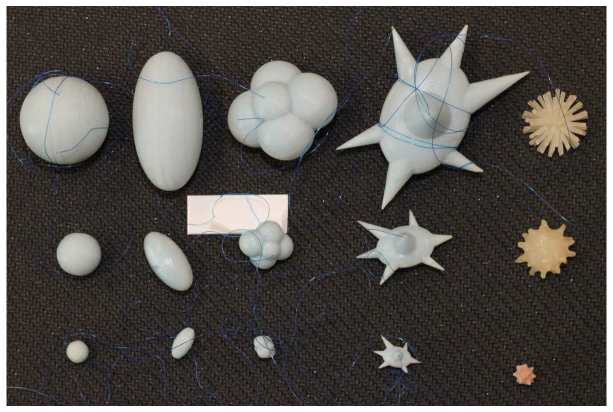
: Photograph of the different types of synthetic nodules used in this study. Each column contains nodules in three sizes, and each row contains the five nodule shape categories. From left to right, spherical, elliptical, lobulated, spiculated, and irregular shapes are shown. The three sizes shown here (with the exception of the irregular shapes on the right) were manufactured to have the equivalent nominal volumes of spherical nodules with diameters of 5, 10, and 20 mm.
Eight different layouts of nodules were specified by placing them in pre-marked positions within the phantom vasculature, where they were either attached to vessels or suspended in foam (non-attached configuration). Care was taken to maintain constant positioning of the nodules when a particular layout was scanned multiple times or using different protocols. For that purpose, vessels on which nodules were attached were color coded. Table 1 tabulates the nodule configuration for each layout in terms of nodule positioning, size, shape, and density. Figure 3 shows an example diagram of one layout used in this study.
Table 1. Summary of nodule placement and description for each nodule layout.
| Nodule layout | Vessel attachment | Nodule placement and description | |||||
|---|---|---|---|---|---|---|---|
| Left lung | Right lung | ||||||
| Size (mm)a | Shape | Density (HU) | Size (mm)a | Shape | Density (HU) | ||
| 1 | attached | 5,8,10 | spherical | −630 | 5,8,10 | spherical | −800 |
| 2 | attached | 5,10,12 | irregular | −300, + 20 | 5,8,10 | spherical | + 100 |
| 3 | attached | 5,8,10, 20, 40 | spherical | −630 | 5,8,10, 20, 40 | spherical | + 100 |
| 4 | attached | 10, 20 | elliptical lobulated, spiculated | −630 | 10, 20 | elliptical lobulated, spiculated | + 100 |
| 5 | attached | 40 | spiculated | −630 | 40 | spiculated | + 100 |
| 6 | attached | 5,8,10, 20, 40 | spherical | −10 | 20, 40 | spherical | −630, + 100 |
| 7 | attached | 10, 20 | elliptical lobulated, spiculated | −10 | 5,8 | elliptical lobulated, spiculated | −10 |
| 8 | non-attached | 5,8,10 | spherical | −630, + 100 | 5,8,10 | spherical | −800, −10 |
aNodule sizes in the figure correspond to equivalent diameters of spheres with the same nominal volume as the nodule. Volume measurements of each nodule will be provided along with location coordinates in a document accompanying the data sets.
Fig. 3.
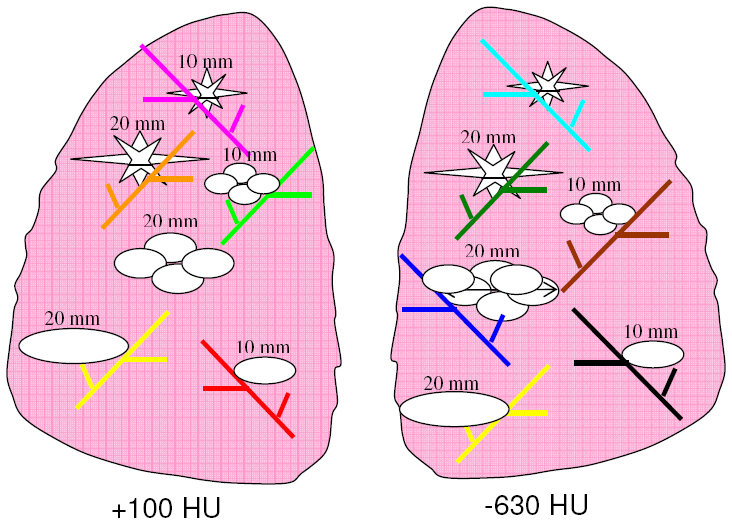
An example layout (Layout #4) indicating the positioning of each nodule along with information on the size, shape, and density of each nodule in the layout. Vessel branches within the anthropomorphic phantom were color coded in order to map nodules to specific positions within the phantom’s vasculature structure in a reproducible manner. Nodule sizes in the figure correspond to equivalent diameters of spheres with the same nominal volume as the nodule.
Future additions to the set of nodules (currently under construction) will include heterogeneous objects (i.e., a 5 mm, −630 HU object, enclosed in a 10 mm, −10 HU object). Different combinations of sizes and densities are being manufactured to more closely mimic non-solid or part-solid nodules as well as nodules surrounded by inflammation or with necrotic centers.
A key component of this CT lung phantom project is that it provides the ability to compare the estimated nodule size with the known true size or reference gold standard. As part of our project, volume was used as a surrogate measure of size. The true volume estimate of each synthetic nodule was derived from weight and density measures. Both the CIRS and Kyotokagaku nodules were accompanied by density measures. Nodule weights were measured in our lab using a precision scale of 0.1 mg tolerance (Adventurer Pro AV 2646, Ohaus Corp, Pine Brook, NJ). Three repeat weight measurements were made and then averaged to produce a final estimated weight for each nodule. The resulting estimates of true volume are provided along with location coordinates in a document accompanying the data sets.
2.3 Scan acquisition and reconstruction parameters
The phantom was scanned using a Philips 16-row scanner (Mx8000 IDT, Philips Healthcare, Andover, MA) and a Siemens 64-row scanner (Somatom 64, Siemens Medical Solutions USA, Inc., Malvern, PA). Scans were acquired with varying combinations of exposure, pitch, and slice collimation, and were reconstructed with varying combinations of slice thicknesses and reconstruction kernels. Examples of such scans are shown in Figs. 4 and 5 . Ten exposures were acquired for each imaging protocol. The phantom position was not changed during the 10 repeat exposures; however it was repositioned between different imaging protocols or different nodule layouts.
Fig. 4.
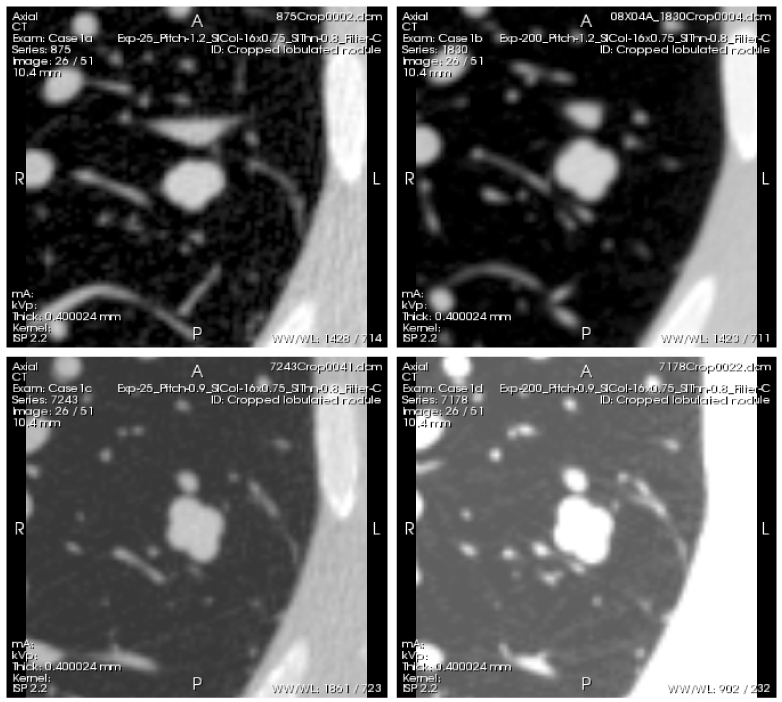
Example scans of a lobulated nodule with −10HU density and 10mm diameter (nominal volume equal to that of a 10 mm diameter sphere), acquired with the following 4 different protocols: Top left (Case 1a)- low exposure (25 mAs), 1.2 pitch. Top right (Case 1b)- high exposure (200 mAs), 1.2 pitch. Bottom left (Case 1c)- low exposure (25m As), 0.9 pitch. Bottom right (Case 1d)- high exposure (200m As), 0.9 pitch. Reconstructed slice thickness was 0.8 mm for all scans. The series of scans can be viewed by clicking on the Interactive Science Publishing (ISP) hyperlink ( View 1).
Fig. 5.
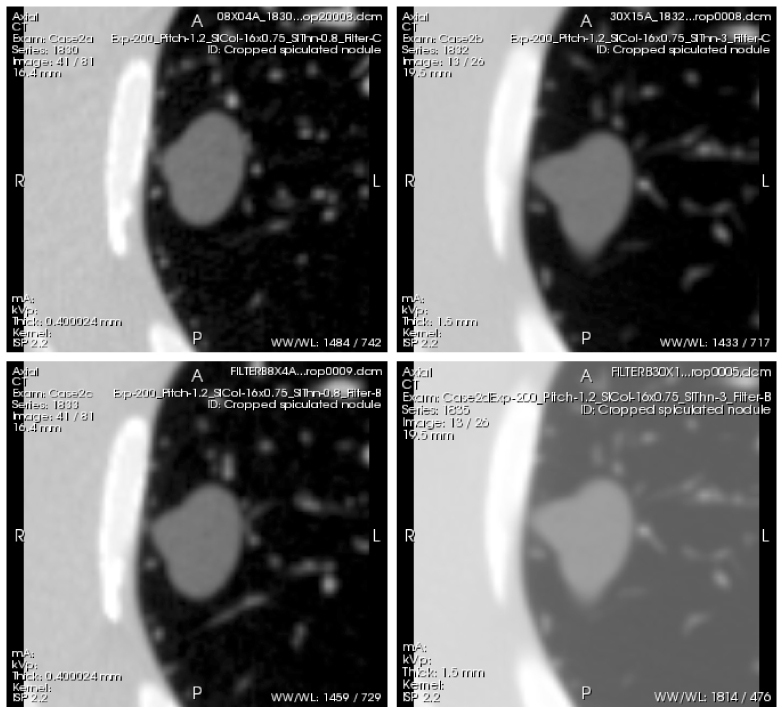
Example scans of a spiculated nodule of −630HU density and 20 mm diameter (nominal volume equal to that of a 20 mm diameter sphere), acquired with the following 4 protocols: Top left (Case 2a)- thin slice thickness (0.8 mm), detail reconstruction kernel (BF60). Top right (Case 2b)- thick slice thickness (3.0mm), detail reconstruction kernel (BF60). Bottom left (Case 2c)- thin slice thickness (0.8 mm), medium reconstruction kernel (BF40). Bottom right (Case 2d)- thick slice thickness (3.0 mm), medium reconstruction kernel (BF40). All scans were acquired with a high exposure (200 mAs) and 1.2 pitch. The whole series of each scan can be viewed by clicking on the Interactive Science Publishing (ISP) hyperlink ( View 2).
3. Results and Discussion
Table 2 summarizes the acquired CT scan data as of May 2010 by specifying the layout, scanner, and imaging parameters for each data set. We have obtained 5400 scans and currently, 480 reconstructed CT scans are available (Nodule Layout #1) and can be accessed through the National Biomedical Imaging Archive (NBIA) (https://imaging.nci.nih.gov). NBIA is a searchable repository of in vivo images managed by the National Cancer Institute, that provides the biomedical research community, industry, and academia with access to image archives to be used in the development and validation of analytical software tools that support lesion detection and classification, accelerated diagnostic imaging decision, and quantitative imaging assessment of drug response. As mentioned previously, the specific nodules (size, shape, density, attachment) within each layout are tabulated in Table 1. As an example, Fig. 6 shows slice data obtained from a high exposure (200 mAs), thick slice (3.0 mm) scan of Layout 4.
Table 2. Summary of reconstructed CT data sets: a description of the individual nodule layouts are provided in Table 1.a .
| Nodule Layout, Scanner | Exposure (mAs) | Slice collimation (mm) | Slice overlap | Pitch | Recon. Slice thickness (mm) | Recon. Kernels | # of sets |
|---|---|---|---|---|---|---|---|
| 1, S1 | 20, 50, 100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail | 480 |
| 2, S1 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 720 |
| 3, S1 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 720 |
| 3, S2 | 20,100,200 | 64x0.6 | 0%, 50% | 0.9,1.2 | 0.75,1.5,3 | detail, medium | 720 |
| 4, S2 | 20,100,200 | 16x0.75, (16x1.5) | 0%, 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 720 |
| 5, S2 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 360 |
| 6, S2 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 360 |
| 7, S2 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 720 |
| 8, S2 | 20,100,200 | 16x0.75, (16x1.5) | 50% | 0.9,1.2 | 0.75,1.5,3 (2,3,5) | detail, medium | 720 |
| TOTAL | 5400 |
aS1: 16-row Philips Mx8000 IDT (Philips Healthcare, Andover, MA), S2: 64-row Siemens Somatom Definition (Siemens Medical Solutions USA, Inc., Malvern, PA).
Fig. 6.
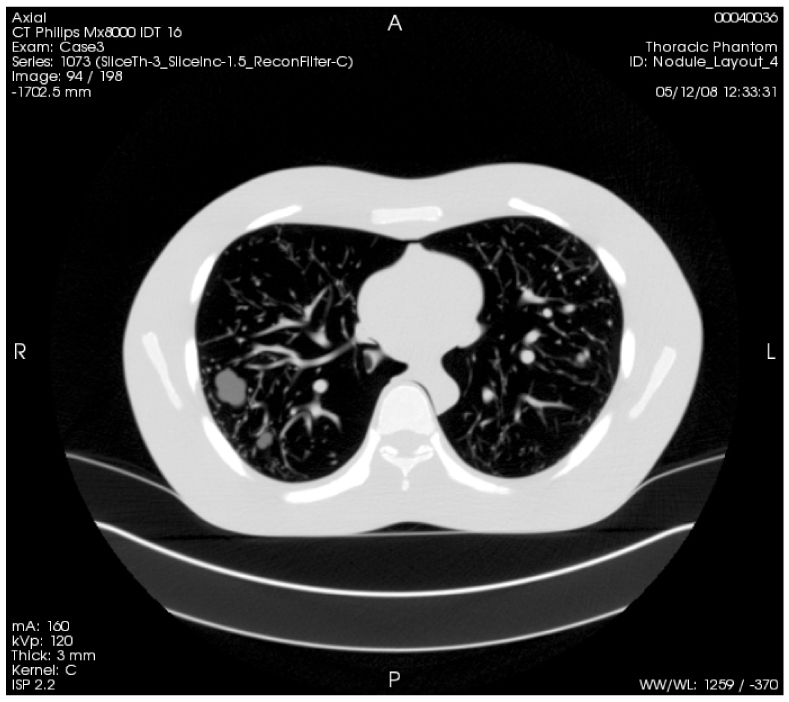
Example series of Layout 4, acquired with a 100mAs exposure and reconstructed to 3mm slices. The whole series can be viewed by clicking on the Interactive Science Publishing (ISP) hyperlink ( View 3).
Regarding the currently available data from the two scanners, the same nodules were scanned with both scanners, albeit in different layouts. For example all spherical nodules of Layouts 1, 2, and 6 that were scanned using the Philips scanners were incorporated in Layout 3 that was scanned using the Siemens scanner. Naturally, direct comparisons must take into account the fact that nodules were positioned differently within the layouts.
The database described in this manuscript can serve as a publicly available resource for a number of different applications in the field of thoracic CT imaging. Primarily, it is well-suited for the development and assessment of methodologies for lung nodule size estimation. Both bias and variance analysis of nodule sizing can be obtained using this phantom database because the reference standard (i.e., true nodule size) and repeat exposures for each configuration are included in the database. This is the main advantage of such phantom data over clinical data, where the true size or extent of a nodule is unknown and repeat scans are difficult to justify due to the radiation exposure. The phantom database can serve in a complementary role to existing or developing clinical databases such as the Reference Image Database for the Evaluation of Response (RIDER) (https://wiki.nci.nih.gov/display/Imaging/RIDER).
Limitations of our phantom study include a lack of complexity in the lung field (i.e., only air surrounding the synthetic vasculature) and nodule shapes, the simplicity of the human anatomy (e.g., lack of bronchial pathway structures and heart details), and some inaccuracies in CT values of soft tissue compared to patient scans.
Although the phantom lacks the complexity of human lung anatomy, the wide range in size, shape, and density of the synthetic nodules and the presence of the vasculature structure can be useful in providing important information on the performance of and comparisons across nodule size estimators. A number of studies have used phantoms for lung nodule measurement but all have used their own phantoms, making it difficult to compare results across methodologies; this database provides a common framework for such comparisons. It has already been employed in two projects, namely VOLCANO’09 (http://www.via.cornell.edu/challenge/) and BIOCHANGE (http://www.itl.nist.gov/iad/894.05/biochange2008/Biochange2008-webpage.htm). The VOLCANO’09 competition is part of the Second International Workshop of Pulmonary Image Analysis whose goal was to compare the outcomes of various algorithms measuring the change in volume of pulmonary nodules in CT scans using a common data set and performance evaluation method. Data from our phantom CT collection (repeat scans of a 10 mm nodule reconstructed using 1.5 and 3.0 mm slice thicknesses) were included in the competition data set along with clinical data provided by the Weill Medical College of Cornell University. Preliminary results [10] showed no significant differences between different size change estimation methods but significant disagreement between different volume estimation methods. The authors comment that the latter may have occurred due to a bias in volume measurement between methods that might be neutralized when computing size change. However, it still needs to be examined whether the aforementioned bias is constant between scans or not. For instance, certain algorithms might overestimate the volume of small nodules and underestimate the volume of larger nodules, resulting in a small size change when in fact a nodule may have grown significantly. The BIOCHANGE 2008 project was organized by the National Institute of Standards and Technology (NIST) as a benchmarking pilot study of lung CT change measurement algorithms and computer-aided diagnosis tools. In related work Kinnard et al. [9] reported on volumetric analysis of phantom data extracted from this database comparing different 3D segmentation algorithms. The development of this data collection and lessons learned from its usage will hopefully inspire the development of publicly available data for other diseases/modalities to provide a common, reference set for developing and/or assessing medical imaging software
Another use of the phantom CT database is the optimization of lung nodule estimation methodologies. Recent work by co-authors of this study [11,12] included the development of a matched-filter approach for the estimation of lung nodules. The matched filter minimized a cost function between the lung nodule to be measured and a bank of simulated 3D nodule templates. The simulated templates were generated using a model of the helical MDCT imaging system, which included a forward projection and filtered back projection-based image reconstruction and derived simulated reconstructed data of nodule objects (templates) of varying size. The templates were then matched to CT data of the target nodules in order to derive the estimate of the scanned nodule size. The phantom CT database was used to compare cost functions and assess the performance of this method. Results demonstrated the effect of vessel attachments, nodule characteristics, and imaging protocols on volumetric precision and accuracy, supporting the value of the database to provide lower bounds on performance. Another approach would be to develop algorithms on clinical data and test it on the phantom CT data collection for assessment based on known truth. Such an assessment would provide a lower bound on performance and provide information regarding bias errors that would be unattainable using clinical data.
In addition to lung nodule size estimation, the phantom CT scan database can be used for a number of other applications in thoracic CT imaging such as the analysis of helical CT noise to derive useful properties such as noise correlation. Understanding noise properties is necessary for developing signal-detection theoretic estimators that make optimal use of the deterministic and stochastic processes of the image formation process. The database provides a large number of regions of interest from CT scans acquired with multiple imaging parameters that can be used for noise analysis. Related work and references on this topic are presented in the recent study by Zeng et al [13]. Other applications may include the development and evaluation of 3D algorithms for segmenting the lung field or the lung vasculature.
Future work will target more realistic phantoms to better mimic the x-ray attenuation coefficients of the human thorax. Regarding synthetic nodules, we are in the process of manufacturing and scanning non-solid nodules. Despite their clinical importance [14] only a small number of authors have reported volumetric measurement results on non-solid or partly solid nodules. The presence of a common collection of non-solid nodules in a phantom study would likely be valuable. Moreover, our plans include the acquisition of phantom CT data at multiple sites and using scanners from all major CT vendors. Additionally, findings from upcoming studies related to the impact of different factors on volumetric and size change estimation might indicate a need to adjust the range of an imaging parameter (such as adjusting the range in pitch settings) in order to better determine its influence. Regarding the types of data we have acquired so far, future acquisition will include scanning of nodules with small differences in diameter (8, 9, and 10 mm) positioned at the same location, in order to enable change analysis.
4. Conclusions
We have collected multiple CT data sets using a factorial design across image acquisition parameters and nodule characteristics using a well-characterized phantom and a variety of imaging platforms. Repeat phantom studies provide a framework in which the true size, shape, and location of nodules are known, allowing for both bias and variance analysis. The data is being released to the public through NIBA in stages (https://imaging.nci.nih.gov). It is a resource to examine the impact of CT acquisition parameters and data analysis approaches on the accuracy and precision of tumor size estimation and to facilitate the development of procedures to maximize the utility of CT imaging for lung cancer screening and tumor therapy evaluation. The database complements public databases of clinical data and provides a framework for comparisons between different nodule size estimators in terms of measurement error.
Acknowledgments
This research was funded though a Critical Path grant from the U.S. Food and Drug Administration. The intramural research program of the National Institute of Biomedical Imaging and Bioengineering and the National Cancer Institute through IAG no. 224-07-6030 provided partial support for this work. Phantom scans on the Siemens Somatom Definition were conducted at the Center for Clinical Imaging Research in the Mallinckrodt Institute of Radiology (MIR) at the Washington University School of Medicine in St. Louis, MO. The authors would like to thank Bruce Whiting of MIR for his assistance. Phantom scans on the Philips IDT Mx8000 were supported in part by the Center for Interventional Oncology at the National Institutes of Health (NIH) and an Interagency Agreement between the NIH and the United States Food and Drug Administration (FDA). Finally, the authors would like to thank Wei-Chung Cheng for his assistance with photographs. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Footnotes
Data sets associated with this article are available at http://hdl.handle.net/10376/1538. Links such as “View 1” that appear in figure captions and elsewhere will launch custom data views if ISP software is present.
References
- 1.Therasse P., Arbuck S. G., Eisenhauer E. A., Wanders J., Kaplan R. S., Rubinstein L., Verweij J., Van Glabbeke M., van Oosterom A. T., Christian M. C., Gwyther S. G., “New guidelines to evaluate the response to treatment in solid tumors,” J. Natl. Cancer Inst. 92(3), 205–216 (2000). 10.1093/jnci/92.3.205 [DOI] [PubMed] [Google Scholar]
- 2.Eisenhauer E. A., Therasse P., Bogaerts J., Schwartz L. H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., Rubinstein L., Shankar L., Dodd L., Kaplan R., Lacombe D., Verweij J., “New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1),” Eur. J. Cancer 45(2), 228–247 (2009). 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 3.Jaffe C. C., “Measures of response: RECIST, WHO, and new alternatives,” J. Clin. Oncol. 24(20), 3245–3251 (2006). 10.1200/JCO.2006.06.5599 [DOI] [PubMed] [Google Scholar]
- 4.Gavrielides M. A., Kinnard L. M., Myers K. J., Petrick N., “Noncalcified lung nodules: volumetric assessment with thoracic CT,” Radiology 251(1), 26–37 (2009). 10.1148/radiol.2511071897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNitt-Gray M. F., Bidaut L. M., Armato. III S. G., Meyer C. R., Gavrielides M. A., Fenimore C. P., McLennan G., Petrick N., Zhao B., Reeves A. P., Beichel R., Kim H.-J. G., Kinnard L., “CT assessment of response to therapy: tumor volume change measurement, truth data and error,” Translational Oncol. 2, 216–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer C. R., Armato. III S. G., Fenimore C. P., McLennan G., Bidaut L. M., Barboriak D. P., Gavrielides M. A., Jackson E. F., McNitt-Gray M. F., Kinahan P. E., Petrick N., Zhao B., “Quantitative Imaging to assess tumor response to therapy: common themes, of measurement, truth data, and error sources,” Translational Oncol. 2, 198–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das M., Ley-Zaporozhan J., Gietema H. A., Czech A., Mühlenbruch G., Mahnken A. H., Katoh M., Bakai A., Salganicoff M., Diederich S., Prokop M., Kauczor H. U., Günther R. W., Wildberger J. E., “Accuracy of automated volumetry of pulmonary nodules across different multislice CT scanners,” Eur. Radiol. 17(8), 1979–1984 (2007). 10.1007/s00330-006-0562-1 [DOI] [PubMed] [Google Scholar]
- 8.Kostis W. J., Reeves A. P., Yankelevitz D. F., Henschke C. I., “Three-dimensional segmentation and growth-rate estimation of small pulmonary nodules in helical CT images,” IEEE Trans. Med. Imaging 22(10), 1259–1274 (2003). 10.1109/TMI.2003.817785 [DOI] [PubMed] [Google Scholar]
- 9.Kinnard L. M., Gavrielides M. A., Myers K. J., Zeng R., Peregoy J., Pritchard W., Karanian J., Petrick N., “Volume Error Analysis for Lung Nodules Attached to Bronchial Vessels in an Anthropomorphic Thoracic Phantom,” Proc. SPIE 6915, 69152Q (2008). 10.1117/12.773039 [DOI] [Google Scholar]
- 10.A. P. Reeves, A. C. Jirapatnakul, A. M. Biancardi, T. V. Apanasovich, C. Schaefer, J. J. Bowden, M. Kietzmann, R. Korn, M. Dillmann, Q. Li, J. Wang, J. H. Moltz, J. Kuhnigk, T. Hayashi, X. Zhou, H. Fujita, T. Duindam, B. v. Ginneken, R. Avila, J. P. Ko, K. Melamud, H. Rusinek, R. Wiemker, G. Soza, C. Tietjen, M. Thorn, M. F. McNitt-Gray, Y. Valenciaga, M. Khatonabadi, Y. Kawata, and N. Niki, “The VOLCANO'09 challenge: Preliminary results,” in Second International Workshop of Pulmonary Image Analysis, (2009), pp. 353–364. [Google Scholar]
- 11.Gavrielides M. A., Zeng R., Kinnard L. M., Myers K. J., Petrick N., “A template-based approach for the analysis of lung nodules in a volumetric CT phantom study,” Proc. SPIE 7260, 726009 (2009). 10.1117/12.813560 [DOI] [Google Scholar]
- 12.Gavrielides M. A., Zeng R., Kinnard L. M., Myers K. J., Petrick N., “Information-theoretic approach for analyzing bias and variance in lung nodule size estimation with CT: a phantom study,” IEEE Trans. Med. Imaging. in press) [DOI] [PubMed] [Google Scholar]
- 13.Zeng R., Petrick N., Gavrielides M. A., Myers K. J., “Approximations of noise structures in helical multi-detector CT scans: application to lung nodule volume estimation,” Proc. SPIE 7624, 762415 (2010). 10.1117/12.844216 [DOI] [Google Scholar]
- 14.Henschke C. I., Yankelevitz D. F., Mirtcheva R., McGuinness G., McCauley D., Miettinen O. S., ELCAP Group , “CT screening for lung cancer: frequency and significance of part-solid and nonsolid nodules,” AJR Am. J. Roentgenol. 178(5), 1053–1057 (2002). [DOI] [PubMed] [Google Scholar]


