Abstract
The compaction of genomic DNA into chromatin has profound implications for the regulation of key processes such as transcription, replication and DNA repair. Nucleosomes, the repeating building blocks of chromatin, vary in the composition of their histone protein components. This is the result of the incorporation of variant histones and post-translational modifications of histone amino acid side chains. The resulting changes in nucleosome structure, stability and dynamics affect the compaction of nucleosomal arrays into higher-order structures. It is becoming clear that chromatin structures are not nearly as uniform and regular as previously assumed. This implies that chromatin structure must also be viewed in the context of specific biological functions.
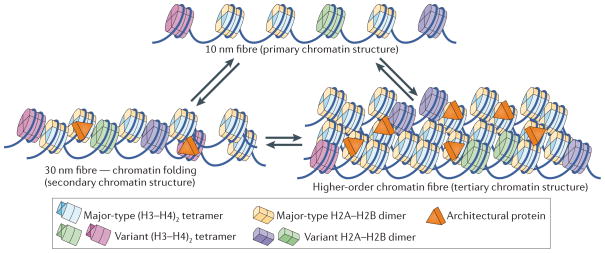
Genomic DNA in eukaryotic cells ranging from yeasts to humans is packaged with an equal mass of protein to form chromatin. Through various mechanisms, the organized and compacted genome is made accessible for readout by the complex machinery involved in gene transcription, DNA replication and DNA repair. The repeating unit of chromatin is the nucleosome, which is formed by wrapping ~145–147 bp of DNA around a histone octamer core1. Nucleosomes are connected by short DNA segments (termed ‘linker DNA’) into nucleosomal arrays, which undergo short-range interactions with neighbouring nucleosomes to form chromatin fibres. Subsequent fibre–fibre interactions contribute to the high degree of compaction observed in the condensed chromosome2. The beads-on-a-string organization of individual nucleosomes (which vary in the DNA sequence that is being organized, as well as in the amino acid sequence and combinations of post-translational modifications (PTMs) of the histones) can be termed the ‘primary structure’ of chromatin, which in turn defines ‘secondary’ and ‘tertiary’ higher-order chromatin structures3 (FIG. 1).
Figure 1. Primary, secondary and tertiary structure of chromatin.
The primary structure is shown as nucleosomal arrays consisting of nucleosomes with canonical histones (shown in light blue and yellow) and combinations of different histone variants (shown in green, purple and light blue). Nucleosomes with canonical or histone variants may vary in the degree of post-translational modifications (PTMs; such as acetylation, methylation, phosphorylation, ubiquitylation and sumoylation), generating the possibility for nucleosomes with a large number of different ‘colours’. Histone variants and PTMs may affect nucleosome structure and dynamics. The spacing between nucleosomes may vary on the basis of the underlying sequence, action of chromatin-remodelling enzymes and DNA binding by other factors (for example, transcription activators). Short-range nucleosome–nucleosome interactions result in folded chromatin fibres (secondary chromatin structure, lower left panel). Fibre–fibre interactions, which are defined by long-range interactions between individual nucleosomes, are also affected by the primary structure of chromatin fibres, including PTMs, histone variants and spacing of nucleosomes. Secondary and tertiary structures are stabilized by architectural proteins, such as linker histone H1, methyl-CpG-binding protein 2 (MeCP2), heterochromatin protein 1 (HP1), high mobility group (HMG) proteins, poly(ADP-ribose) polymerase 1 (PARP1), myeloid and erythroid nuclear termination stage-specific protein (MENT), Polycomb group proteins and many others. Transitions between the different structural states are indicated by double arrows; these may be regulated by changes in patterns of PTMs, binding or displacement of architectural proteins, exchange of histone variants and chromatin-remodelling factors.
Given the ever-increasing number of histone variants and PTMs that are being identified, and considering that each nucleosome contains two copies of each histone, the number of theoretically possible variations in chromatin primary structure is astronomical. In addition to variations of the components of the nucleosomes themselves, architectural chromatin proteins (ACPs) and nucleosome-binding proteins (including those that specifically recognize modified histones), histone chaperones and ATP-dependent chromatin remodellers also affect chromatin structure at all levels. Changes to chromatin structure can apply to the nano-scale, for example by establishing the local structure of an active promoter, or to the micro-scale, in which case megabases of DNA are organized into specialized structures such as the centromere and surrounding constitutive heterochromatin.
There is now a large collection of high-resolution nucleosome crystal structures from different species, showing PTMs, histone variants and nucleosomes in complex with nuclear proteins (reviewed in REF. 4). Single-molecule approaches have led to exciting insights into the dynamic properties of nucleosomes that were not apparent from the crystal structures5. It is now clear that the various crystal structures represent just one possible state of the nucleosome, and that the incorporation of PTMs and histone variants has the potential to shift the equilibrium between different structural states. This variability affects the compaction of the chromatin fibre and the interaction of nucleosomes with non-histone proteins. Numerous in vitro studies have addressed the effect of PTMs and histone variants on chromatin condensation, and several (sometimes contradictory) experimental and computational models for higher-order structure have been proposed. High-resolution sequencing techniques have allowed the mapping of nucleosome position over entire genomes to near base-pair resolution6, and exciting (if controversial) progress has been made in predicting nucleosome position from DNA sequence alone7. Here we summarize evidence for the dynamic structural states of nucleosomes that result from changes in nucleosome primary structure and discuss how the variability of nucleosome structural states in turn influences the folding of chromatin into more compacted states. We also review recent work that challenges the long-held notion of a hierarchical organization of chromatin.
Nucleosome structure and stability
In the ‘canonical nucleosome structure’, which is assembled from major-type, unmodified histones and a DNA fragment of defined length and sequence, 147 base pairs of DNA form a tight, two-turn ‘superhelix’ around a wedge-shaped compact histone octamer composed of two copies each of the histones H2A, H2B, H3 and H4 (FIG. 2). The nucleosome is stabilized by a multitude of protein–protein interactions within the histone octamer and by numerous electrostatic and hydrogen bonds between protein and DNA over its entire length1,8,9. The vast majority of DNA–histone interactions are between structured regions of the histones and DNA10, whereas the flexible histone tails (the sites of most PTMs) extend away from nucleosomal DNA and are mainly involved in interaction with neighbouring nucleosomes or with nuclear factors11.
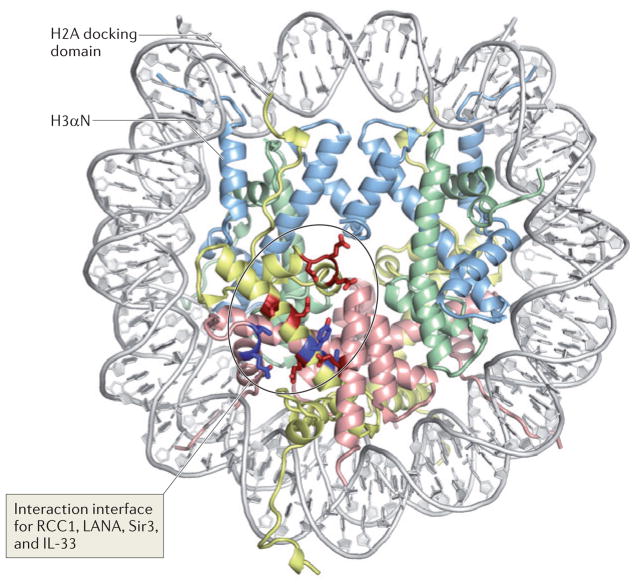
Figure 2. Nucleosome structure and the acidic patch: a common interaction interface for many nucleosome-interacting proteins.
The structure of the nucleosome (Protein Data Bank code 1AOI) is viewed down the superhelical axis of the DNA. Histones H3, H4, H2A and H2B are shown in light blue, green, yellow and red, respectively. The figure indicates the amino-terminal α-helix of H3 (H3αN), which organizes the penultimate 10 bp of the DNA, and the carboxy-terminal end of the H2A docking domain. Acidic residues on H2A and H2B (the ‘acidic patch’) that are involved in the interaction with the H4 tail and with nucleosome-interacting proteins (such as the latency-associated nuclear antigen (LANA) peptide, interleukin-33 (IL-33), regulator of chromosome condensation 1 (RCC1), silent information regulator 3 (Sir3) and high mobility group nucleosome-binding domain-containing protein 2 (HMGN2)) are indicated in bright red; additional residues that are implicated in the interaction interfaces with the proteins listed above are shown in dark blue. Note that the number of total histone residues implicated in all of these protein–protein interfaces is relatively small, and that all cluster in a contained region on the surface of the histone octamer. In the absence of these factors, the interaction of the H4 tail from a neighbouring particle with the acidic patch mediates nucleosome–nucleosome interactions, thereby promoting chromatin folding. By using similar interactions with acidic patch on the nucleosome, the proteins listed above may compete with the H4 tail and modulate chromatin structure.
Variations in the protein composition of the nucleosome
Histone variants are encoded by genes that are distinct from the genes encoding their major-type counterparts. Unlike major-type histones, variants are expressed throughout the cell cycle and are incorporated into nucleosomes in DNA replication-independent pathways (reviewed in REF. 12) that often involve specific histone chaperones and ATP-dependent chromatin remodelling factors (see below). Histone variants replace missing histones or are specifically recruited to precise genomic locations. Most histone variants have been identified for histones H2A and H3 (BOX 1). Histone variants are highly conserved between different species, indicating that they have evolved to fulfil important functions that cannot be accomplished by their major-type counterparts, as has been demonstrated for H2A.Z13 and CenH3 (REFS 14,15). By contrast, H2A.Bbd-like histone variants, including human H2A.Bbd16 and mouse H2A.Lap1 (REF. 17) are evolving rapidly to carry out tissue- and even cell-specific transcription roles in the testes and brain.
Box 1. Histone variants for histones H2A and H3.
H2A.Z
This is found in virtually all eukaryotes. Despite intense studies, its functions remain enigmatic. Roles in establishing a poised or active RNA polymerase II promoter architecture, and more generally in gene activation and silencing, have been described. It appears to be of particular importance at developmental genes in embryonic stem cells and is thus essential for early development, chromosome stability and centromere function.
MacroH2A
This is a vertebrate-specific H2A variant with a large carboxy-terminal ‘macrodomain’ that is connected to the histone fold region through a flexible linker. It exists in several splice variants that exhibit different functions. It is enriched on the mammalian inactive X-chromosome.
H2A.Bbd
This is a human-specific H2A variant that is expressed in the testes and the brain. Its function is largely unknown.
H2A.Lap1
This is the mouse isoform of H2A.Bbd, and is likewise expressed in the testes and brain. It is involved in the temporal and spatial activation of testis-specific genes.
H2A.X
H2A.X has an SQ C-terminal motif that becomes Ser phopshorylated at sites of double-strand DNA breaks.
H3.3
H3.3 replaces H3 at regions of active transcription. Like most histone variants, it is incorporated in a replication-independent manner. It is involved in gene activation as well as in heterochromatin formation.
CenH3
The centromeric H3 histone variant (CenH3, but specifically referred to as Cse4 in yeast, CENPA in humans and mice, and Cid in Drosophila melanogaster) is essential for assembly of the proteinaceous kinetochore to which the spindle microtubules attach during mitosis and meiosis.
PTMs are small, chemical modifications to amino acid side chains18 that are added and removed post-translationally by a multitude of highly specific enzymes, with numerous complex and very specific biological outcomes. Histone-modifying enzymes are recruited through diverse mechanisms: for example, site-specific DNA-binding factors19, co-activators and repressors20, RNA polymerase II21 or preceding histone modifications22. Virtually all known types of protein modifications are found on histones, and new types of modifications as well as new locations on histone proteins are discovered at a rapid pace23, although in many instances their biological significance awaits confirmation.
Despite the absence of site-specific interactions between histones and DNA bases, DNA sequence affects nucleosome structure through its sequence-encoded propensity for being distorted into the tight superhelical conformation that is dictated by the histone octamer; these properties are also thought to affect nucleosome positioning. This is a key parameter, as the DNA sequence of each nucleosome is unique.
Numerous X-ray crystallographic studies show that the overall architecture of the nucleosome is remarkably unaffected by variations in histone sequence, PTMs and DNA sequence (reviewed in REFS 4,24). Nevertheless, crystallographic studies allude to effects on nucleosome stability and/or dynamics. First, consistent differences are found at the small interface at which two H2A–H2B dimers interact, when H2A is replaced with either H2A.Z or macroH2A25. Second, an ‘acidic patch’ formed by several amino acids from H2B and from the docking domain of H2A (FIG. 2) is expanded in nucleosomes containing H2A.Z without causing effects on DNA interaction but with significant consequences for chromatin folding (see below). Third, minor variations in local DNA geometry have been observed when comparing crystal structures of nucleosomes with DNA binding ligands or with different DNA sequences4.
In most structures, the ends of the DNA are tightly organized through base-stacking interactions between neighbouring nucleosomes26. The notable exception is a nucleosome in which the variant CenH3 replaces H3 (BOX 1). In this structure, the 13 terminal base pairs of DNA are not visible owing to weakened interactions between DNA and the histone octamer at entry and exit sites of the nucleosome; consequently, the crystal packing does not involve the DNA ends27. Single-molecule DNA unzipping experiments confirmed that octameric CenH3 nucleosomes from yeast bind the terminal DNA segments much more weakly than their major-type counterparts28. This may not be a unique property of centromeric H3, as the histone variants H2A.Bbd29,30 and H2A.Lap1 (REF. 17) also form nucleosomes in which the terminal DNA segments are only loosely organized. However, when compared with chicken erythrocyte nucleosomes, nucleosomes with certain plant H2B and H2A variants appear to protect additional linker DNA31,32.
Nucleosome stability
Changes in the chemical composition of nucleosome components can also affect the overall stability of the nucleosome (that is, the number and strength of protein–protein and protein–DNA interactions) without changing its overall structure. When considering nucleosome stability, it must be remembered that nucleosomes are modular, multi-component assemblies, and thus their thermodynamic stability can be defined in a number of ways33. Quantitative descriptions of the binding constants of the (H3–H4)2 tetramer to DNA and of the H2A–H2B dimer to the (H3–H4)2 tetramer–DNA complex under physiological conditions are essential to understand how modifications of histones modulate DNA accessibility (see, for example, REF. 34). These parameters also provide insights into the pathways by which nucleosomes are assembled and disassembled (FIG. 3a). Recent in vivo work suggests that histones turn over in a replication- and transcription-independent manner, and at a much more rapid rate than previously anticipated35,36; this, in turn, is at least in part determined by the intrinsic stability of the nucleosomes themselves. Several studies have investigated the stability of H2A.Z nucleosomes in response to variations in ionic strength, and some interesting effects have been identified37. Additionally, variant nucleosomes are shown to differ in their ability to be remodelled by the various ATP-dependent chromatin-remodelling factors38–40.
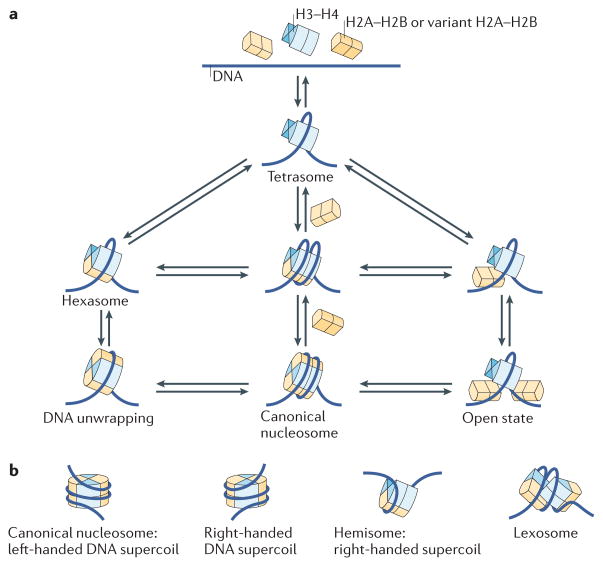
Figure 3. The many structural states of the nucleosome.
a | Structural states of the nucleosome that are likely to be interchangeable. These include the tetrasome, which is formed by the wrapping of ~80 bp DNA around an (H3–H4)2 tetramer. Hexasomes (nucleosomes lacking one H2A–H2B heterodimer) are intermediate states during nucleosome assembly or disassembly, as well as during transcription of nucleosome templates by RNA polymerase II. Both hexasomes and fully formed nucleosomes may undergo spontaneous structural transitions that are characterized either by the transient release of the DNA ends (DNA breathing) or by a transient opening of the interface between histone subcomplexes (open state). Some of the specific states may be favoured by DNA sequence, histone variant incorporation or post-translational modifications (PTMs). Histone variants are likely to be incorporated by similar pathways. b | Nucleosomes may also exist in alternative states that vary in the direction of the handedness of the DNA superhelix (left-handed versus right-handed) or in the stoichiometry (hemisome) and structural states (lexosome) of histones. These alternative structures, if they do indeed exist in vivo, would affect DNA accessibility and the interaction of nuclear factors with chromatin.
Alternative nucleosome structures
These findings are consistent with the notion put forward by the late Jonathan Widom that the terminal DNA segments in all nucleosomes undergo spontaneous unwrapping and rewrapping41,42 (FIG. 3a). Restriction enzyme analysis, quantitative measurements of the interaction of proteins with nucleosomal DNA, fluorescence resonance energy transfer (FRET) and high-speed atomic force microscopy (high-speed AFM) have shown that nucleosomal DNA partially and rapidly dissociates from the histone octamer in a reversible manner41–44. This concept has wide-ranging implications for the regulation of nuclear processes, as it provides an explanation for how DNA-binding factors access their site within the context of chromatin. The rates and extent of unwrapping are affected by DNA sequence as well as by variations in histones (especially histones H2A and H3, as these contact the terminal DNA segments; FIG. 2). For example, acetylation of histone H3 Lys56 (H3K56) leads to increased rates of site exposure45, whereas PTMs such as acetylation of H3K115 and H3K122 near the nucleosomal dyad have distinctly different effects46. As pointed out above, the histone variants H2A.Bbd and CenH3 do not tightly organize the terminal DNA segments and thus are more likely to exist in an ‘open state’. The organization of nucleosomes into chromatin fibres only results in modest changes in the rates of DNA exposure, suggesting that nucleosomes in chromatin fibres also undergo transient DNA breathing43.
More recently, single-molecule FRET approaches led to the identification of an open state of the nucleosome during salt-dependent nucleosome assembly and disassembly47,48 (FIG. 3a). This state is characterized by a partial disruption of the interface between the H2A–H2B dimer and the (H3–H4)2 tetramer while the H2A–H2B dimers remain attached to the DNA. Thermodynamic considerations predict that this state is significantly populated under physiological conditions. A recent high-speed AFM study of single nucleosomes showed that DNA not only fluctuates at the entry and exit sites but also that nucleosomes can undergo spontaneous sliding, ‘splitting’ and complete dissociation without the involvement of additional proteins such as ATP-dependent chromatin-remodelling factors44.
Independent evidence for alternative nucleosome structures in solution, at concentrations that are more physiological than those used in single-molecule or AFM experiments, has been provided by small angle X-ray scattering (SAXS)28,49,50. SAXS data are consistent with nucleosomal states involving partially dissociated peripheral DNA and/or with the structural states described in FIG. 3a. Distinct differences are observed between nucleosomes reconstituted with various DNA sequences and between nucleosomes reconstituted with different histone variants. As SAXS measures the bulk of all molecules in solution, equilibria between open and closed states are consistent with the observed results.
Several accounts of more extreme, non-canonical nucleosome structures have been reported over the years (see REFS 51,52 for recent reviews and FIG. 3b for cartoon representations of some of the proposed models). These include still controversial accounts of centromeric nucleosomes containing right-handed DNA wrapping as opposed to the conventional left-handed path53, perhaps with less than a full complement of histones54,55,56–58. An extreme type of open nucleosome structure, the lexosome (which is characterized by the splitting of the two halves of the (H3–H4)2 tetramer), was proposed in 1991 (REF. 59), but no further reports have confirmed this phenomenon. Non-canonical nucleosome structures are not likely to exist in equilibrium with the canonical nucleosome structure.
Although the biological significance of the structural transitions described above still awaits confirmation, it is likely that non-canonical nucleosomes exist in vivo. These might be transient — for example, as the intermediates of assembly and disassembly processes34,60 — or they might occur during transcription61. More permanent changes may result from the incorporation of histone variants (see, for example, REFS 53,54). There is also recent and mounting evidence for non-canonical nucleosomes that protect less than the requisite 147 base pairs at specific regions of the genome in vivo (see, for example, REFS 62–64).
New tools to sequence the primary structure of chromatin
Together, these examples illustrate how intrinsic modifications to components of the nucleosome affect its dynamic properties. With the advent of even more refined techniques to study nucleosome structure (for example, REFS 65–67), paired with the ability to generate ‘designer nucleosomes’ bearing all kinds of combinations of PTMs, histone variants and DNA sequence68–72, we are likely to see a more systematic investigation of biologically relevant modifications on nucleosome structure. It is intuitively obvious that alternative nucleosome structures potentially affect all interactions that they are engaged in, especially the formation of higher-order chromatin structures. Additionally, they may contribute to the regulation of DNA accessibility through differences in their ability to be remodelled by histone chaperones and ATP-dependent chromatin remodelling factors (see below).
Nucleosome positioning
The specific positioning of key regulatory nucleosomes around promoter regions and transcription start sites, often to base-pair accuracy, is an important component of gene regulation, as is the localized absence of nucleosomes. Recent technological advances have allowed the mapping of nucleosomes throughout entire genomes with very high resolution, and attempts have been made to derive a ‘nucleosome positioning code’ that may exclusively be embedded in the DNA sequence73,74. Although opinions still diverge on whether DNA sequence alone determines nucleosome position (reviewed in REF. 75), it is clear that other nuclear factors, such as ATP-dependent remodelling factors or pre-bound proteins, also contribute by creating local boundaries74,76–81. To what level the outcome is determined by one or the other is likely to be context dependent. One must also keep in mind that nucleosome position is not absolute but subject to change in time through spontaneous movement or through the action of remodelling factors and polymerases. In light of the growing ensemble of alternative nucleosome structures52 (FIG. 3), a pertinent question in interpreting the wealth of mapping data is also how inclusive our definition of a nucleosome should be63,64. Nucleosome mapping experiments provide a snapshot through a large population of cells, and thus important information on the longevity of mapped positions is lost. An elegant approach to provide this missing information comes through the development of a method to directly measure histone turnover35.
ATP-dependent chromatin remodelling factors
ATP-dependent chromatin remodelling factors are large, macromolecular machines that use energy from ATP hydrolysis to change chromatin structure through sliding, disassembling or otherwise restructuring nucleosomes (reviewed in REF. 82). Accordingly, the biological outcomes of remodelling activity are varied. For example, the nucleosome sliding activity of remodellers (particularly of the ISWI family) alters the spacing of nucleosomes, which is important for chromatin assembly and folding into higher-order structures after DNA replication83–85. However, localized nucleosome sliding can lead to the exposure or occlusion of DNA elements, which may result in transcription activation or repression. SWI/SNF family remodellers are more disruptive, and their activity includes nucleosome sliding, displacement of H2A–H2B dimers and/or complete removal of nucleosomes86–88. Members of this family are usually associated with transcription activation.
The activities of many remodellers are affected by the presence of histone variants. MacroH2A and H2A.Bbd reduce the efficiency of the SWI/SNF family of remodelling factors39,89, whereas H2A.Z stimulates remodelling by the ISWI family38. The H2A variant effects on remodelling are at least partly associated with the sequence difference in the docking domain/acidic patch of H2A (FIG. 2), which is required for remodelling activities by these factors90.
The ATP-dependent SWR1 complex specifically mediates the exchange of H2A with H2A.Z at specific genomic loci91–93. This happens in a stepwise manner94, consistent with the pathways shown in FIG. 3a. The INO80 complex in turn removes H2A.Z from ‘wrong’ locations95. ATP-dependent remodellers also contribute to constraining centromeric protein A (CENPA) to the centromere96. Incorporation, and perhaps also removal, of either histone variant is aided by specific histone chaperones such as yeast Chz1 (REF. 97) and suppressor of chromosome mis-segregation protein 3 (Scm3; known as Holliday junction recognition protein (HJURP) in humans), which distinguish them from their major-type counterparts.
Many ATP-dependent chromatin-remodelling enzymes functionally interact with the activities involved in the PTMs of histones. Some PTMs promote ATP-dependent chromatin remodelling by creating the binding sites for remodellers98,99. For instance, acetylation of nucleosomes aids the recruitment of SWI/SNF remodellers (through their acetyl-group-binding bromo-domain) and increases remodelling efficiency. Acetylation of H2A or H4 tails stimulates the SWR1-mediated exchange of H2A with H2A.Z100,101. By contrast, ISWI and chromodomain-helicase-DNA-binding protein 1 (CHD1) remodelling activities are inhibited by acetylation of the nucleosomes99,102.
Chromatin secondary structure
Linear arrays of nucleosomes are folded and compacted into three-dimensional (3D) assemblages of higher-order structures. To better describe higher-order chromatin structures, secondary chromatin structures have been defined as those structures that arise from the folding of an individual array (the primary structure) to produce a defined fibre (for example, the 30 nm fibre). Subsequent intermolecular interactions between secondary chromatin structures produce large-scale configurations (tertiary structures; FIG. 1).
Given the fundamental role of chromatin higher-order structures in regulating all DNA- and chromosome- dependent processes, a major goal has been to elucidate the 3D arrangement of long nucleosomal arrays in vivo at interphase, and their transition to the highly compacted form at metaphase. Because of the complexity of the problem and limitations of current microscopic approaches, it is not surprising that progress has been slow and interpretation of the results has been controversial. Significant advances have come from a reductionist approach in which defined in vitro chromatin is assembled from purified DNA and histones. Under physiological salt conditions, an array of nucleosomes folds into a secondary chromatin structure with the same hydrodynamic shape as the 30 nm fibre, and a population of these fibres self-associates to form highly condensed tertiary structures103. Although valuable information on the principles governing chromatin compaction has been obtained from these in vitro experiments, it must be pointed out that in recent years the very existence of large sections of 30 nm fibres in living cells has been called into question104–106.
Zigzag or solenoid? Two models for the 30 nm fibre
Despite years of effort, the structure of the 30 nm fibre has not been resolved. Two competing models, the solenoid and zigzag arrangement of nucleosomes, have been proposed on the basis of in vitro data2 (FIG. 4). In the ‘one-start’ solenoid model, consecutive nucleosomes interact with each other and follow a helical trajectory with bending of linker DNA107. In the ‘two-start’ zigzag structure, two rows of nucleosomes form a two-start helix so that alternate nucleosomes (for example, N1 and N3) become interacting partners, with relatively straight linker DNA108 (FIG. 4). Twisting or coiling of the two stacks can produce different forms of the zigzag model.
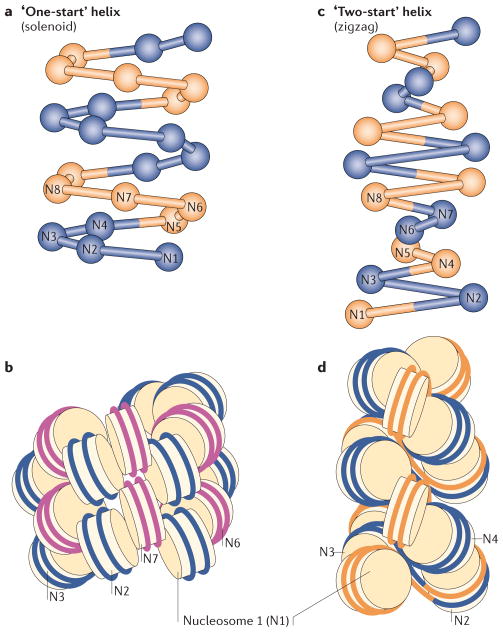
Figure 4. Two models for chromatin secondary structure.
The solenoid model is characterized by interactions between consecutive nucleosomes (n, n + 1; a,b), whereas the zigzag model implies interactions between alternate nucleosomes (n, n + 2; c,d). The alternative nucleosomes are numbered from N1 to N8. In the solenoid model proposed by Rhodes and colleagues111, the 30 nm chromatin fibre is an interdigitated one-start helix in which a nucleosome in the fibre interacts with its fifth and sixth neighbour nucleosomes. Alternative helical gyres are coloured blue and magenta (b). In the zigzag model, the chromatin fibre is a two-start helix in which nucleosomes are arranged in a zigzag manner such that a nucleosome in the fibre binds to the second neighbour nucleosome108. Alternative nucleosome pairs are coloured blue and orange (d). The two models also differ in the trajectory and degree of bending of the DNA that connects two nucleosomes (linker DNA). Figure is reproduced, with permission, from REF. 160 © (2011) Elsevier.
The main reason the structure of the 30 nm structure has not been resolved is that its highly compacted nature prevents the path of the DNA from being visualized by electron microscopy109. Crosslinking studies and the ultimate crystallization of a tetranucleosome array (albeit with a very short 167 bp nucleosome repeat length (NRL)) have provided the strongest evidence to date that the 30 nm fibre adopts a zigzag structure108,110. However, the biological relevance of this structure is questioned given that the short repeat length and the absence of linker histone H1 is not typical for higher eukaryotes111,112. Linker histone H1 (for which several variants and post-translationally modified versions have been identified) binds tightly to nucleosomes with linker DNA113,114 and stabilizes the higher-order structure of chromatin111. Although its biological role is still controversial115, it is clear that most nucleosomes in metazoans are bound by H1. Cryo-electron microscopy on long regular nucleosomal arrays with defined and constant repeat length, and in the presence of linker histone, resulted in a very tight compaction of reconstituted chromatin fibres, especially for repeats above 207 bp109,116. At that time, this was interpreted as evidence for a multiple-start interdigitated solenoid model111 because the observed high nucleosome packing ratio was not compatible with two-start zigzag or classic one-start solenoidal models. A more recent molecular modelling study showed that these compact structures were also consistent with a set of multi-start chromatin fibre models with extended nucleosome linker DNA117.
A recent study using electron microscopy-assisted nucleosome capture (EMANIC) may have finally resolved this issue, at least for in vitro-assembled chromatin118. In this approach, a limited number of inter-nucleosomal contacts within condensed chromatin are fixed by formaldehyde. Chromatin is then decondensed in low salt, and transmission electron microscopy is applied to quantitate the nature of nucleosome–nucleosome contacts. This study demonstrated that there is not one uniform type of helical fibre organization but rather conformational heterogeneity of nucleosome interactions (hence ‘heteromorphic’ fibre). Second, although the fibres showed a predominantly two-start organization, the structures were interspersed with partially bent linker DNA where interactions between consecutive nucleosomes typical of one-start solenoids occurred.
Innovative modelling and simulation approaches have been applied to explore the structure of the 30 nm fibre under a wide range of parameters, including variations in nucleosome shape, linker length, strength of tail-mediated interactions and salt conditions and in the presence and absence of linker histones117,119–123. Remarkably, mesoscopic modelling revealed the same heteromorphic 30 nm fibre structure obtained through EMANIC118. A heteromorphic fibre with predominant two-start (zigzag) type interspersed with one-start conformations was energetically more favourable than uniform zigzag or solenoid conformations under conditions that promoted the most compact folding (that is, the presence of linker histone and Mg2+ counter ions). These results suggest that linker length could influence the chromatin fibre organization: short to medium NRLs (173–209 bp) favoured chromatin fibre condensation into irregular zigzag structures, whereas solenoid features were viable for longer NRLs (218–226 bp)121. Experimentally, the NRL was shown to affect chromatin folding of the arrays with short NRLs (165–177 bp) but not medium NRLs (188–209 bp)124. On the basis of the compaction behaviour of reconstituted chromatin112 and meso-scale modelling of oligo-nucleosomes, it has been argued that in short to long NRLs (173–209 bp), linker histones promote stiffening of linker DNA. The formation of the DNA stem prevents DNA bending, thereby destabilizing solenoid-like features121. On nucleosomal arrays with even longer NRLs, linker histones cannot prevent bending of linker DNA in their middle section125,126.
Most of the in vitro studies discussed above were done on homogenous nucleosomal arrays with a regular repeat length and exquisitely accurate positioning of nucleosomes. However, the very technological advance that enabled these studies may also distort the results towards a highly defined architecture that probably does not accurately reflect the in vivo structure of an ‘average’ chromatin fibre with variable DNA sequence, repeat length and heterogeneous histone populations with different PTMs. Rather than a ‘universal’ structure of chromatin, multiple conformations may exist depending on the physiological context (for example, the transcriptional state, cell cycle stage, developmental stage and response to environmental signals and damage). All of these impart their own specific requirements for a unique chromatin structure to control genome function. Therefore, understanding chromatin structure is exceedingly more complex than previously assumed, and it is unlikely that a ‘one-fits-all’ solution to the problem can be found.
Histone tail meets acidic patch
Despite the apparent controversy, much has been learnt about the molecular determinants of chromatin folding in vitro. Various crystal structures of nucleosomes give insights into the variability of nucleosome–nucleosome packing1,110,127; two examples are shown in FIG. 5. Central to all of these interactions is the acidic patch on histone H2A–H2B (FIGS 2,5). In vitro neutralization of just three acidic residues within the acidic patch on the nucleosome inhibited inter-nucleosome interactions and the formation of the 30 nm fibre, demonstrating the importance of this patch in promoting nucleosome–nucleosome interactions128. Conversely, neutralization of additional acidic amino acid residues within this patch inhibited rather than facilitated interactions between nucleosomes129. Therefore, the relative strength of the interaction between adjacent nucleosomes in the chromatin fibre may be a balance between attractive and repulsive interactions (FIG. 5). A comprehensive histone mutation approach in yeast revealed that specific acidic amino residues within this patch are essential for cell survival130. This same study also showed that specific amino acid residues that contribute to transcriptional activation, transcriptional silencing or DNA repair and DNA replication cluster in distinct regions on the histone octamer surface.
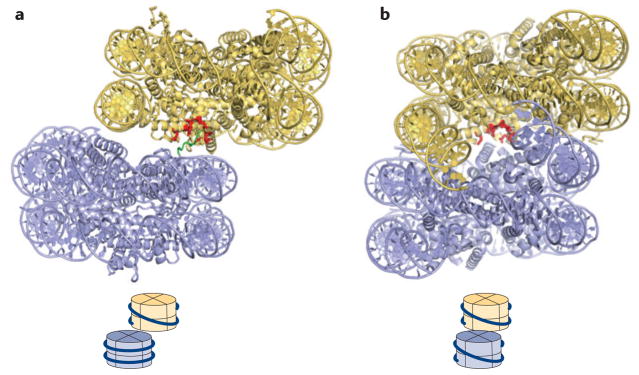
Figure 5. Nucleosome–nucleosome interactions mediated by histone tails and the nucleosomal surface.
a | Nucleosome–nucleosome interactions in the crystal structures of unconnected 147 bp nucleosomes (Protein Data Bank (PDB) code 1AOI).b | Nucleosome– nucleosome interactions, as seen in the low-resolution tetranucleosome structure of four connected nucleosomes (PDB code 1ZBB; only two nucleosomes are shown). The resolution of this structure does not reveal the location of the H4 tail. A cartoon at the bottom of each panel shows the relative arrangement of the two nucleosomes with respect to each other. Residues constituting the acidic patch are shown in red, and the H4 tail (in a) is shown in green; it is not resolved in the structures shown in b.
There is evidence that the amino-terminal tail of histone H4 originating from an adjacent nucleosome interacts with the acidic patch to mediate nucleosome–nucleosome interactions108 (FIG. 5) and that this interaction is disrupted by the acetylation of H4K16 (REF. 102). A thorough analysis of the effect of acetylation and Gln mutation of Lys residues 5, 8, 12 and 16 and different cations led to the proposal that the H4 tail also simultaneously interacts with a site on histone H2B131. Whereas acetylation of the H4 tail inhibits chromatin compaction, dimethylation and trimethylation of Lys20, a mark for constitutive heterochromatin, promotes intra-nucleosome interactions and the formation of the 30 nm fibre132. The H4 tail can also interact with the acidic patch on its own nucleosome133, and it has been proposed that this interaction may indirectly stabilize the wrapping of DNA at the entry and exit points128. The tails of the other histones also contribute in the following order of importance: H4 >H3 > H2A/H2B133–135. Of note is the unusual carboxy-terminal linker domain of macroH2A that promotes extreme chromatin compaction in vitro136. The less important role of the other histone tails, especially of H3, in directly mediating chromatin compaction enables them to participate in other functions, such as the recruitment of a large variety of effector proteins through their modification status.
Local modulation of the 30 nm fibre
Adding to the notion that a uniform chromatin secondary structure would rarely exist in a living cell, various subtle changes to the structure and/or composition of a nucleosome can have a profound effect on the extent of chromatin compaction and the types of structures assembled. For example, histone H2B ubiquitylation disrupts local and higher-order chromatin compaction137. Also of note are minor alterations to the acidic patch region generated by the replacement of H2A with its variant forms. H2A.Z promotes internucleosome interactions and the formation of the 30 nm fibre (compared to H2A), and this ability is dependent on the extended acidic patch of H2A.Z (remarkably, just two amino acid residues) in vitro138. Significantly, this same region of H2A.Z is required for survival in Drosophila melanogaster13. Histone H2A variants with a partially neutralized acidic patch have also been identified. The human histone H2A. Bbd variant139 lacks three of the six H2A acidic amino acid residues and, consequently, H2A.Bbd nucleosome arrays cannot fold128. Intriguingly, the major amino acid residue differences in H2A.Z, H2A.Bbd and its mouse homologue, H2A.Lap1, that are responsible for changes in the dynamics of the chromatin folding pathway all lie in the acidic patch of H2A.
H4 tail mimicry
A recently defined class of chromatin-binding proteins can mimic the ability of the H4 tail to dock onto the acidic patch, thereby potentially modulating chromatin structure. The 20 N-terminal amino acids of the Kaposi’s sarcoma-associated herpesvirus protein latency-associated nuclear antigen (LANA) mediate the attachment of the viral genome to chromosomes by specifically binding to the acidic patch of nucleosomes140. This mechanism of interaction is also used by the cytokine interleukin-33 (IL-33), an abundant chromatin- associated factor in endothelial cells that is involved in regulating transcription. Like LANA, IL-33 binds to chromatin by specifically interacting with the acidic patch141. The crystal structure of regulator of chromosome condensation 1 (RCC1) bound to the nucleosome demonstrated an interaction between a surface loop in RCC1 and the acidic patch and neighbouring residues via an extensive network of hydrogen bonds and van der Waals contacts142. Silent information regulator 3 (Sir3) also contacts the same area on the nucleosome143 (FIG. 2), with intriguing implications for possible higher-order structure organization. A recent tour-de-force in NMR has added high mobility group nucleosome-binding domain-containing protein 2 (HMGN2) to this group of proteins144. Finally, the extended acidic patch of H2A.Z enhances the chromatin-binding affinity of heterochromatin protein 1α (HP1α; also known as CBX5)138. Thus, it appears that the region containing the acidic patch has evolved as the hub for nucleosome-interacting proteins, in addition to being key for chromatin condensation.
Chromatin tertiary structures
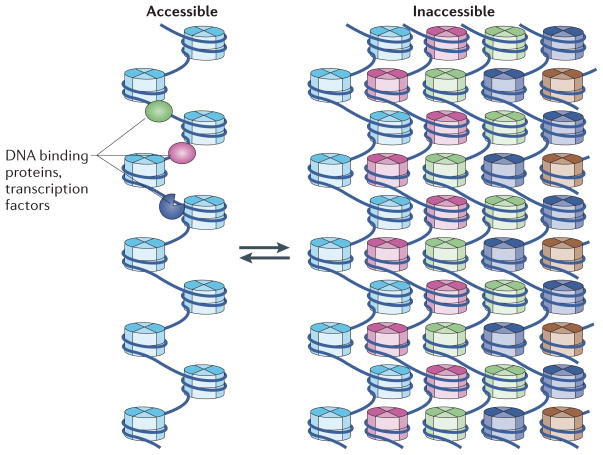
The most easily visualized chromatin structures are metaphase chromosomes at the end point of chromatin compaction in dividing cells. The tertiary structure of nucleosomes in metaphase chromosomes must be constrained and nonrandom to a high degree to allow for the rod-like structures with reproducible lengths and diameters, and unique banding patterns. The fundamental question is whether long nucleosomal arrays actually fold and compact in a reproducible manner during the formation of metaphase chromosomes. The textbook view is that chromosomes form from the ordered hierarchical coiling of the 30 nm fibre into sequential higher-order stages of condensation, with each more compacted stage dependent on the coiling or folding of the chromatin fibre of the previous stage. However, recently the existence of the 30 nm fibre has been questioned104–106, as neither it nor any other type of ordered secondary structure has been observed reproducibly in mammalian cells. Alternative models have been proposed that involve a less-ordered chromosome condensation process105. One plausible scenario is that the physical arrangement of the metaphase chromosome is similar to a ‘molten globule’ or ‘polymer melt’ state2,105,145 arising from the interdigitation of over-crowded and irregularly folded nucleosomal arrays (FIG. 6). Electron spectroscopic imaging of interphase chromatin appears to show a mesh of 10 nm fibres105. A criterion for any model for the metaphase chromosome is that the compaction ratio must increase by 10 nucleosomes per 10 nm146,147, and such interdigitation allows these packing ratios to be reached109. Although the polymer melt model may explain how chromatin fibres interact and compact, it cannot account for the rod-like structures that occur at metaphase. Other chromatin-organizing and chromatin-compacting proteins, such as topoisomerases and condensins, are likely to have an important role at this final organizational level, and so this issue is far from being resolved.
Figure 6. A model for chromatin tertiary structure by interdigitation of nucleosomal arrays.
The dynamic motion of arrays, possibly aided by nucleosome remodelling factors and histone-modifying enzymes, enables partial decompaction by the ‘unpeeling’ of an unfolded array (light blue) from the large-scale interdigitated tertiary chromatin fibre. As a result, this ‘string of pearls’ (shown on the left) slides out of the compact array and becomes more accessible as it is removed from the interdigitated stacks (shown on the right). This unpeeling process from the surface of the fibre facilitates transcription factor access. The fractal nature of the interdigitated chromatin fibre enables access to even DNA that is buried deep within the fibre.
An important aspect of the higher organization of chromatin is that specific nucleosomes may be distant with respect to their primary structure but may be within interacting distance in the context of higher-order 3D (or tertiary) structures, in analogy with protein folding (FIG. 6). For example, analysis of extended chromatin fibres (~50–100 times their normal interphase length) revealed that 10–40 kb regions of CenH3- and H2A-containing nucleosomes are interspersed with similar-sized domains of dimethylated H3 and H2A.Z. Upon folding of the fibre at metaphase, CenH3- and H2A-containing nucleosomes and dimethylated H3- and H2A.Z-containing nucleosomes occupied distinct 3D locations148. Ultimately, if the 3D path of DNA can be elucidated for a chromosome or a chromosome domain, it may then be possible to superimpose linear epigenetic maps onto this 3D structure to fully understand how the epigenetic code links chromatin structure and function.
Tertiary chromatin structure: hierarchy or anarchy?
To directly distinguish whether the same nucleosomes (that is, the same DNA sequences) are always positioned in the same 3D space within a metaphase chromosome or whether there is an element of disorganization and randomness implied by the molten globule model, artificial segments of chromosomes were created and tagged by the insertion of multiple copies of the lac operator sequence149. No reproducibility in the lateral positions of the tagged sequences was observed in mitotic chromosomes. Moreover, positioning of these sequences differed even between sister chromatids. This argues against a reproducible hierarchical folding model for the formation of a metaphase chromosome but instead suggests a disordered and random aspect to the compaction of chromatin at metaphase, at least at some stage of the condensation process. Therefore, both at the local level of the chromatin fibre and at the global level of the chromosome, structural uniformity is not a dominant feature of chromatin.
Transcription in a compact environment
What is the correlation between chromatin compaction and transcription? In vivo, it has been shown that actively transcribed genes exist in a chromatin state that is up to 25 times more compact than the 30 nm fibre, suggesting that transcription occurs within condensed large-scale tertiary chromatin fibres at interphase150. Perhaps counter-intuitively, an interdigitated state may in fact facilitate greater access of transcription factors to DNA via the dynamic movement of unfolded nucleosome fibres (FIG. 6). In such an arrangement, DNA would more often be exposed than in highly folded secondary structures such as the 30 nm fibre, which has been shown to be highly refractory to transcription in vitro128. Moreover, the location of transcribed nucleosome arrays on the surface of tertiary chromatin structures, aided by specific histone modifications and ATP-dependent chromatin-remodelling machines151, would further enhance the accessibility of the transcriptional machinery (perhaps by being able to peel away from the large-scale interdigitated tertiary chromatin fibre)152–154. Supporting this notion is the in vitro finding that promoting fibre–fibre interactions (while simultaneously inhibiting 30 nm fibre formation) is positively correlated with transcriptional activation128. Thus, even the long-held view that chromatin must be significantly decondensed for transcription to occur is being challenged.
ACPs also contribute to the modulation of nucleosome and chromatin structure through diverse mechanisms (reviewed in REF. 155). ACPs modulate the structural properties of chromatin to create functionally unique, higher-order chromatin structures. All known ACPs are involved in silencing gene expression by sterically hindering access to nucleosomal and linker DNA. The most intensively studied ACPs include HP1, methyl-CpG-binding protein 2 (MeCP2) and the Polycomb group complex. HP1 binds nucleosomes as a tetramer156 and enhances the interactions between nucleosomes and the formation of compacted chromatin secondary structures. MeCP2 and the Polycomb complex organize nucleosomes in their own distinct ways to generate functional unique compacted structures157,158. Therefore, it appears that the type of repressive chromatin structure assembled may vary depending on the specific transcription networks that need to be repressed.
Final perspective
An emerging key feature of chromatin is that small changes in chromatin primary structure yield dramatic effects in its large-scale organization. Numerous and often subtle changes by PTMs, histone variants and DNA sequence can significantly affect nucleosome shape and stability and its protein surface. These alterations are subsequently amplified via altered intra- and internucleosome interactions within a chromatin fibre to produce vastly different chromatin higher-order structures, which in turn define function. Adding to this complexity of structural regulation are architectural chromatin-binding proteins and a plethora of protein (and RNA) ‘readers’ and ‘writers’ of PTMs.
Together, the emerging data forces us to re-evaluate the notion of the nucleosome and chromatin as highly defined structural states, and calls into question the long-held truism that the chromatin fibre must be decondensed for transcription to occur. Rather, we have to consider a continuum of various inter-convertible states at all levels of condensation. One probable function of histone variants, PTMs, histone chaperones and ATP-dependent chromatin-remodelling factors may be to ease transitions between these many states. Although valuable information can be obtained from the investigation of defined model systems of chromatin higher-order structures, the challenge of the future is not to elucidate the universal structure (or structures) of chromatin at various levels of condensation but to define the type of structure that performs a particular function in a cell. This will require the development of new imaging technologies that can visualize individual nucleosomes (including their modification state) in real time, together with multi-pronged approaches that also incorporate 1D and 3D genome-wide mapping techniques (for example, soft X-ray tomography) and overall chromatin condensation (see, for example, REF. 159). The continued development of approaches to study the dynamics of nucleosome and chromatin structure in solution will be critical to provide the molecular basis of how different specialized chromatin structures form.
Acknowledgments
The authors dedicate this contribution to the late Jonathan Widom in acknowledgement of his pioneering work on chromatin structure and dynamics. Work in the authors’ laboratories is supported by the US National Institutes of Health (grant GM088409 to K.L.) and the National Health and Medical Research Council (grants 471422, 1009850 and 1009851 to D.J.T.). K.L. is supported by the Howard Hughes Medical Institute. We thank U. M. Muthurajan for help with figures 2 and 5, and S. Grigoryev for discussion.
Glossary
- Post-translational modifications
(PTMs). Chemical modifications added post-translationally (and reversibly) to many histone amino acid side chains by highly specific enzymes. PTMs have important functions in the regulation of transcription and DNA repair
- Architectural chromatin proteins
(ACPs). Abundant nuclear proteins that interact with nucleosomes and influence the three-dimensional arrangement of nucleosomal arrays
- Histone chaperones
A diverse group of nuclear proteins that prevent the aggregation of folded histones with DNA during the assembly of nucleosomes. They are also implicated in the transport of histones into the nucleus
- Centromere
The most constricted and compacted region of a chromosome. Spindle fibres attach to the centromere to equally partition newly replicated sister chromatids between daughter cells during cell division
- Sumoylation
A post-translational modification that is involved in various cellular processes, such as nuclear–cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress and progression through the cell cycle. It involves the covalent attachment of small ubiquitin- like modifier (SUMO) proteins to other proteins to modify their function
- ATP-dependent chromatin remodelling factors
Large macromolecular complexes that use the energy from ATP hydrolysis to slide, disassemble or otherwise structurally alter nucleosomes
- Fluorescence resonance energy transfer
(FRET: also known as Förster resonance energy transfer). Distance-dependent energy transfer between two chromophores, commonly used to measure conformational changes in a single molecule or to measure interactions between different molecules
- High-speed atomic force microscopy
(High-speed AFM). A high-resolution type of scanning probe microscopy with a demonstrated resolution on the order of fractions of a nanometre. High-speed AFM allows direct visualization of dynamic structural changes and dynamic processes of functioning biological molecules in physiological solutions at high spatiotemporal resolution
- Transient DNA breathing
A transient structural state of the nucleosome characterized by the dissociation of the 10–20 penultimate base pairs of DNA from the histone octamer, leading to ‘transient site exposure’
- Open state of the nucleosome
A transient structural state of the nucleosome characterized by the opening of the interface between histone H2A–H2B dimers and (H3–H4)2 tetramers
- Small angle X-ray scattering
(SAXS). A technique in which the elastic scattering of X-rays by a sample is recorded at very low angles. Unlike X-ray crystallography, this technique does not require crystals, and is used to determine the maximum dimensions and overall shape of a macromolecule or a macromolecular complex
- ISWI
The name is derived from its founding member, the Drosophila melanogaster protein imitation switch (ISWI). This family of ATP-dependent chromatin remodellers includes SWI/SNF, ISWI, CHD and INO80 in eukaryotes
- Electron microscopy-assisted nucleosome capture
(EMANIC). A technique whereby nucleosomal arrays or whole cells are subject to controlled formaldehyde crosslinking such that only a few of the nucleosome–nucleosome contacts become covalently linked. Subsequently, the arrays are allowed to disperse in low salt and are imaged by electron microscopy
- Mesoscopic modelling
Pertains to the resolution of the computational approach being the intermediate between the atomic and macroscopic scale. Over a number of years, mesoscopic models of chromatin have been developed that alleviate the prohibitive computational demands of atomistic simulations but incorporate the key features of the chromatin fibre, thereby making it amenable to large-scale simulations
- Ubiquitylation
The covalent attachment of a small protein domain (ubiquitin) to cellular proteins. Monoubiquitylation is a common histone modification; polyubiquitylation (the attachment of multiple molecules of ubiquitin) is usually a marker for intracellular protein transport and degradation. It is found in all cells of higher organisms.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Karolin Luger’s homepage: http://lugerlab.org/
David J. Tremethick’s homepage: http://jcsmr.anu.edu.au/research/genome-biology
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. This is the first structure of the nucleosome that detailed the interactions between DNA and histones as well as among histone proteins at nearly atomic resolution. [DOI] [PubMed] [Google Scholar]
- 2.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock CL, Dimitrov S. Higher-order structure of chromatin and chromosomes. Curr Opin Genet Dev. 2001;11:130–135. doi: 10.1016/s0959-437x(00)00169-6. [DOI] [PubMed] [Google Scholar]
- 4.Tan S, Davey CA. Nucleosome structural studies. Curr Opin Struct Biol. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. This article summarizes the structure of all nucleosomes determined up to 2011 and reviews the structure of the nucleosome in complex with proteins and other small molecules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Killian JL, Li M, Sheinin MY, Wang MD. Recent advances in single molecule studies of nucleosomes. Curr Opin Struct Biol. 2012;22:80–87. doi: 10.1016/j.sbi.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nature Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. A good review of a whole-genome analysis of nucleosome positioning, chromatin remodelling factors, and the interplay between nucleosome positioning and DNA sequence. The Review also discusses the organization of nucleosomes around the promoter and transcription start sites, and the consequences of this for transcription regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan N, Hughes TR, Lieb JD, Widom J, Segal E. Contribution of histone sequence preferences to nucleosome organization: proposed definitions and methodology. Genome Biol. 2010;11:140. doi: 10.1186/gb-2010-11-11-140. A discussion of the controversies surrounding the effect of histone sequence preferences on nucleosome organization in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 9.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 11.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends Genet. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- 14.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 15.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chadwick BP, Willard HF. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soboleva TA, et al. A unique H2A histone variant occupies the transcriptional start site of active genes. Nature Struct Mol Biol. 2012;19:25–30. doi: 10.1038/nsmb.2161. This study identified a unique testis histone H2A variant (H2A.Lap1) that occupies chromatin at the transcription start site of active genes in a differentiation-specific manner. Enrichment of H2A.Lap1 at transcription start sites positively correlates with active gene transcription as well as H2A.Z occupancy, which is a mark of genes poised for expression or active genes. H2A.Lap1 lacks the acidic patch domain and consequently folds into less-compacted chromatin than chromatin with regular histone H2A, suggesting its role in transcription activation. [DOI] [PubMed] [Google Scholar]
- 18.Andrews AJ, Luger K. Histone modifications: chemistry and structural consequences. In: Begley TP, editor. Wiley Encyclopedia of Chemical Biology. Vol. 1. Wiley; 2009. pp. 275–284. [Google Scholar]
- 19.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownell JE, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. This is the first demonstration that the transcription co-activator/adaptor Gcn5 complex is a histone acetyltransferase, directly linking chromatin modifications with transcription regulation. It was proposed that the bromodomains anchor Gcn5 to chromatin. Numerous subsequent studies have shown that bromodomains are acetyl-group-binding domains that recruit and stabilize the binding of different chromatin factors, including histone acetyltransferases, to chromatin. [DOI] [PubMed] [Google Scholar]
- 21.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 22.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dechassa ML, Luger K. In: Genome Organization and Function in the Cell Nucleus. Rippe K, editor. Wiley-VCH; 2011. pp. 55–87. [Google Scholar]
- 25.Chakravarthy S, Bao Y, Roberts VA, Tremethick D, Luger K. Structural characterization of histone H2A variants. Cold Spring Harb Symp Quant Biol. 2004;69:227–234. doi: 10.1101/sqb.2004.69.227. [DOI] [PubMed] [Google Scholar]
- 26.Suto RK, et al. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- 27.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. The crystal structure of human CENPA nucleosomes, showing that the 13 bp DNA at the entry and exit sites of the nucleosome is only loosely bound. [DOI] [PubMed] [Google Scholar]
- 28.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nature Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao Y, et al. Nucleosomes containing the histone variant H2A.Bbd organize only 118 base pairs of DNA. EMBO J. 2004;23:3314–3324. doi: 10.1038/sj.emboj.7600316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier T, et al. Histone variant H2ABbd confers lower stability to the nucleosome. EMBO Rep. 2004;5:715–720. doi: 10.1038/sj.embor.7400182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsey GG, Thompson P. S(T)PXX motifs promote the interaction between the extended N-terminal tails of histone H2B with “linker” DNA. J Biol Chem. 1992;267:14622–14628. [PubMed] [Google Scholar]
- 32.Lindsey GG, Orgeig S, Thompson P, Davies N, Maeder DL. Extended C-terminal tail of wheat histone H2A interacts with DNA of the “linker” region. J Mol Biol. 1991;218:805–813. doi: 10.1016/0022-2836(91)90268-b. [DOI] [PubMed] [Google Scholar]
- 33.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 34.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA Interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. A highly innovative approach to address nucleosome dynamics in vivo. The approach is based on co-translational incorporation of the Met analogue azidohomoalanine (Aha) into newly expressed proteins, and the subsequent modification of Aha by biotin. The affinity-purified micrococcal-nuclease-digested chromatin was then analysed with tiling microarrays. Nucleosomes in active genes, epigenetic regulatory elements and origins of replication were found to undergo fast turnover. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Hoch DA, Stratton JJ, Gloss LM. Protein-protein Forster resonance energy transfer analysis of nucleosome core particles containing H2A and H2A.Z. J Mol Biol. 2007;371:971–988. doi: 10.1016/j.jmb.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman JA, Garlick JD, Kingston RE. Chromatin remodeling by imitation switch (ISWI) class ATP-dependent remodelers is stimulated by histone variant H2A.Z. J Biol Chem. 2010;285:4645–4651. doi: 10.1074/jbc.M109.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang EY, et al. MacroH2A allows ATP-dependent chromatin remodeling by SWI/SNF and ACF complexes but specifically reduces recruitment of SWI/SNF. Biochemistry. 2008;47:13726–13732. doi: 10.1021/bi8016944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angelov D, et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 41.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 42.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. Evidence that nucleosomal DNA spontaneously unwraps and re-wraps, resulting in ‘transient site exposure’ of nucleosomal DNA. This and other work from this team demonstrates that the nucleosome is a dynamic structure that regulates the accessibility of underlying DNA. [DOI] [PubMed] [Google Scholar]
- 43.Poirier MG, Bussiek M, Langowski J, Widom J. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyagi A, Ando T, Lyubchenko YL. Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry. 2011;50:7901–7908. doi: 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 45.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon M, et al. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci USA. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bohm V, et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. Evidence for an intermediate open state of the nucleosome where the H2A–H2B dimers dissociate from the tetramer while still associated with the DNA. This might be an intermediate during nucleosome disassembly, histone exchange and other processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of nucleosome invasion by DNA binding proteins. J Mol Biol. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mangenot S, Leforestier A, Vachette P, Durand D, Livolant F. Salt-induced conformation and interaction changes of nucleosome core particles. Biophys J. 2002;82:345–356. doi: 10.1016/S0006-3495(02)75399-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, van der Woerd MJ, Muthurajan UM, Hansen JC, Luger K. Biophysical analysis and small-angle X-ray scattering-derived structures of MeCP2-nucleosome complexes. Nucleic Acids Res. 2011;39:4122–4135. doi: 10.1093/nar/gkr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavelle C, Prunell A. Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle. 2007;6:2113–2119. doi: 10.4161/cc.6.17.4631. [DOI] [PubMed] [Google Scholar]
- 52.Zlatanova J, Bishop TC, Victor JM, Jackson V, van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 53.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 56.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camahort R, et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 58.Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen TA, Smith MM, Le SY, Sternglanz R, Allfrey VG. Nucleosome fractionation by mercury affinity chromatography, Contrasting distribution of transcriptionally active DNA sequences and acetylated histones in nucleosome fractions of wild-type yeast cells and cells expressing a histone H3 gene altered to encode a cysteine 110 residue. J Biol Chem. 1991;266:6489–6498. [PubMed] [Google Scholar]
- 60.Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol Cell. 2011;43:638–648. doi: 10.1016/j.molcel.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kireeva ML, et al. Nucleosome remodeling induced by RNA polymerase II. Loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 62.Floer M, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kent NA, Adams S, Moorhouse A, Paszkiewicz K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 2011;39:e26. doi: 10.1093/nar/gkq1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henikoff JG, Belsky JA, Krassovsky K, Macalpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci USA. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chien FT, van Noort J. 10 years of tension on chromatin: results from single molecule force spectroscopy. Curr Pharm Biotechnol. 2009;10:474–485. doi: 10.2174/138920109788922128. [DOI] [PubMed] [Google Scholar]
- 66.Ando T, et al. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. 2008;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 67.Panchenko T, et al. Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc Natl Acad Sci USA. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 69.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. A comprehensive description of the in vitro preparation of nucleosomes from recombinant histone proteins and DNA. [DOI] [PubMed] [Google Scholar]
- 70.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nature Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. An alternative approach that involves the co-translational incorporation of acetyl-Lys into any position in a protein in Escherichia coli. [DOI] [PubMed] [Google Scholar]
- 71.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahto SK, Howard CJ, Shimko JC, Ottesen JJ. A reversible protection strategy to improve Fmoc-SPPS of peptide thioesters by the N-acylurea approach. Chembiochem. 2011;12:2488–2494. doi: 10.1002/cbic.201100472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nature Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trifonov EN. Cracking the chromatin code: precise rule of nucleosome positioning. Phys Life Rev. 2011;8:39–50. doi: 10.1016/j.plrev.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 76.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaplan N, et al. Nucleosome sequence preferences influence in vivo nucleosome organization. Nature Struct Mol Biol. 17:918–920. doi: 10.1038/nsmb0810-918. author reply 920–922 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z, et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pugh BF. A preoccupied position on nucleosomes. Nature Struct Mol Biol. 2010;17:923. doi: 10.1038/nsmb0810-923. [DOI] [PubMed] [Google Scholar]
- 82.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 83.Corona DF, et al. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nature Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sala A, et al. Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 2011;30:1766–1777. doi: 10.1038/emboj.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bruno M, et al. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dechassa ML, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Doyen CM, et al. Dissection of the unusual structural and functional properties of the variant H2A.Bbd nucleosome. EMBO J. 2006;25:4234–4244. doi: 10.1038/sj.emboj.7601310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shukla MS, et al. The docking domain of histone H2A is required for H1 binding and RSC-mediated nucleosome remodeling. Nucleic Acids Res. 2011;39:2559–2570. doi: 10.1093/nar/gkq1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kobor MS, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:e131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 93.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luk E, et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. An intriguing mechanistic study on the histone variant-specific histone exchange by the ATP-dependent chromatin-remodelling enzyme SWR1 through a heterotypic nucleosome. Hyperstimulation of the ATPase activity of SWR1 by heterotypic nucleosomes promotes the exchange of the second H2A.Z–H2B dimer to form homotypic H2A.Z–H2B dimer-containing nucleosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gkikopoulos T, et al. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. EMBO J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luk E, et al. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 98.Hassan AH, Neel KE, Workman JL. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 99.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 αN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu PY, Levesque N, Kobor MS. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem Cell Biol. 2009;87:799–815. doi: 10.1139/O09-062. [DOI] [PubMed] [Google Scholar]
- 101.Altaf M, Auger A, Covic M, Cote J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol. 2009;87:35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 102.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. This is the first description of the profound effect of a PTM on chromatin higher-order structure and ATP- dependent chromatin remodelling of nucleosomes. [DOI] [PubMed] [Google Scholar]
- 103.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 104.Nishino Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. Using cryo-electron microscopy and X-ray scattering, the authors show that mitotic chromosomes are mainly composed of irregularly folded nucleosome fibres with a diameter not more than 11 nm. The report suggests that irregular chromatin organization allows dynamic states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fussner E, Ching RW, Bazett-Jones DP. Living without 30nm chromatin fibers. Trends Biochem Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. A thought-provoking review on chromatin compaction. [DOI] [PubMed] [Google Scholar]
- 106.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 107.Kruithof M, et al. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nature Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. [DOI] [PubMed] [Google Scholar]
- 108.Dorigo B, et al. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 109.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. Electron microscopy measurements of fully folded fibres with various linker DNA lengths in the presence of linker histones. Two distinict folded chromatin structures were observed. Nucleosomal arrays with 10–40bp linker DNA produced fibres that were 33nm in diameter, whereas arrays with 50–70bp linker DNA produced fibres that were 44nm in diameter. On the basis of the electron microscopy data, the authors proposed an interdigitated one-start helix model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. A low-resolution crystal structure of an oligonucleosome. The structure of tetranucleosomes with 20 bp linker DNA suggests a two-start model for the 30 nm chromosome fibre. [DOI] [PubMed] [Google Scholar]
- 111.Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 112.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci USA. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hieb AR, D’Arcy S, Kramer MA, White AE, Luger K. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res. 2012;40:e33. doi: 10.1093/nar/gkr1045. Approaches to measure affinities between histones, nucleosomes and nuclear factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caterino TL, Hayes JJ. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fan Y, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 116.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 117.Wong H, Victor JM, Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. The two-start zigzag topology and the type of linker DNA bending that defines solenoid models may be simultaneously present in a structurally heteromorphic chromatin fibre with a uniform 30nm diameter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Depken M, Schiessel H. Nucleosome shape dictates chromatin fiber structure. Biophys J. 2009;96:777–784. doi: 10.1016/j.bpj.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perisic O, Collepardo-Guevara R, Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stehr R, Kepper N, Rippe K, Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys J. 2008;95:3677–3691. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc Natl Acad Sci USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Correll SJ, Schubert MH, Grigoryev SA. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012;30:2416–2426. doi: 10.1038/emboj.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kelbauskas L, Yodh J, Woodbury N, Lohr D. Intrinsic promoter nucleosome stability/dynamics variations support a novel targeting mechanism. Biochemistry. 2009;48:4217–4219. doi: 10.1021/bi900476t. [DOI] [PubMed] [Google Scholar]
- 126.Kelbauskas L, Woodbury N, Lohr D. DNA sequence-dependent variation in nucleosome structure, stability, and dynamics detected by a FRET-based analysis. Biochem Cell Biol. 2009;87:323–335. doi: 10.1139/O08-126. [DOI] [PubMed] [Google Scholar]
- 127.White CL, Suto RK, Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nature Struct Mol Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. This study demonstrates that the acidic patch is a key macromolecular determinant on the surface of the nucleosome. The acidic patch is required for modulating the extent of chromatin compaction, and it couples this to transcriptional repression. [DOI] [PubMed] [Google Scholar]
- 129.Chodaparambil JV, et al. A charged and contoured surface on the nucleosome regulates chromatin compaction. Nature Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Matsubara K, Sano N, Umehara T, Horikoshi M. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells. 2007;12:13–33. doi: 10.1111/j.1365-2443.2007.01031.x. [DOI] [PubMed] [Google Scholar]
- 131.Allahverdi A, et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu X, et al. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nature Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tse C, Hansen JC. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 135.Zheng C, Hayes JJ. Intra- and inter-nucleosomal protein-DNA interactions of the core histone tail domains in a model system. J Biol Chem. 2003;15:15. doi: 10.1074/jbc.M302817200. [DOI] [PubMed] [Google Scholar]
- 136.Muthurajan UM, McBryant SJ, Lu X, Hansen JC, Luger K. The linker region of macroH2A promotes self-association of nucleosomal arrays. J Biol Chem. 2011;286:23852–23864. doi: 10.1074/jbc.M111.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fierz B, et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nature Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2AZ alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 139.Chadwick BP, Willard HF. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Barbera AJ, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;5762:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 141.Roussel L, Erard M, Cayrol C, Girard JP. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A-H2B acidic pocket. EMBO Rep. 2008;9:1006–1012. doi: 10.1038/embor.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. An intriguing model for chromatin higher-order structure formation by Sir3. The structure of the Sir3 BAH domain–nucleosome complex revealed the extensive interactions of Sir3 with surfaces of the nucleosome. The structural data explain the numerous genetic mutations reported previously and provide a model of how interactions of the BAH domain with the nucleosome surface lead to silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kato H, et al. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc Natl Acad Sci USA. 2011;108:12283–12288. doi: 10.1073/pnas.1105848108. NMR and mutational analyis reveal that HMGN2 interacts with the acidic patch of the nucleosome and nucleosomal DNA at the entry and exit regions. The work demonstrates a possible mechanism for how HMGN2 regulates chromatin structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Daban JR. Physical constraints in the condensation of eukaryotic chromosomes, Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry. 2000;39:3861–3866. doi: 10.1021/bi992628w. [DOI] [PubMed] [Google Scholar]
- 147.Daban JR. High concentration of DNA in condensed chromatin. Biochem Cell Biol. 2003;81:91–99. doi: 10.1139/o03-037. [DOI] [PubMed] [Google Scholar]
- 148.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Strukov YG, Belmont AS. Mitotic chromosome structure: reproducibility of folding and symmetry between sister chromatids. Biophys J. 2009;96:1617–1628. doi: 10.1016/j.bpj.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 152.Dietzel S, et al. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- 153.Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- 154.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.McBryant SJ, Adams VH, Hansen JC. Chromatin architectural proteins. Chromosome Res. 2006;14:39–51. doi: 10.1007/s10577-006-1025-x. [DOI] [PubMed] [Google Scholar]
- 156.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. An intriguing model for the spreading of HP1 to promote the formation of heterochromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Georgel PT, et al. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 158.Grau DJ, et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.McDermott G, Le Gros MA, Larabell CA. Visualizing cell architecture and molecular location using soft X-ray tomography and correlated cryo-light microscopy. Annu Rev Phys Chem. 2011;63:225–239. doi: 10.1146/annurev-physchem-032511-143818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]