Abstract
Striatal-enriched phosphatase (STEP) is a non-receptor tyrosine phosphatase that is specifically expressed in neurons of the central nervous system. STEP regulates the activity of several effector molecules involved in synaptic plasticity and neuronal cell survival, including mitogen-activated protein kinases (MAPKs), Src family kinases and N-methyl-D-aspartic acid (NMDA) receptors. The critical role of STEP in regulating these effectors requires that its activity be tightly regulated. Previous studies demonstrated that the activity of STEP is regulated through reversible phosphorylation of a serine residue within the kinase interacting motif (KIM), by cAMP-dependent protein kinase A (PKA). Here we show that STEP is endogenously phosphorylated at two additional sites located within the kinase specificity sequences (KIS). Basal activity of ERK and p38 MAPKs plays an important role in the phosphorylation of these two sites. Dephosphorylation of these two sites leads to poly-ubiquitination and proteolytic degradation of STEP. Conversely, the proteasome inhibitors MG-132 and epoxomicin can stabilize STEP. The active form of STEP is more susceptible to degradation than the inactive form. Taken together our results establish that ubiquitin-dependent proteolysis could be a novel mechanism for terminating the activity of STEP irreversibly.
Keywords: tyrosine phosphatase, striatal enriched phosphatase (STEP), kinase specificity sequence (KIS), kinase interacting motif (KIM), ubiquitination, proteasomal degradation
INTRODUCTION
Protein tyrosine phosphatases (PTPs) are a large family of structurally diverse proteins that, together with tyrosine kinases, regulate the tyrosine phosphorylation state of many important signaling molecules. It is now apparent that PTPs exhibit exquisite substrate specificity and are critical regulators of a wide array of cellular signaling pathways [1, 2]. However an important area in the field that remains to be investigated is the characterization of mechanisms by which the activity of PTPs themselves may be regulated.
STEP (striatal-enriched phosphatase, also known as PTPN5) is an intracellular PTP that is exclusively expressed in the central nervous system whose specific targets and functions are beginning to emerge. STEP is preferentially expressed in neurons of the basal ganglia, hippocampus, cortex and related structures [3, 4]. The STEP-family of PTPases includes both membrane associated (STEP61) and cytosolic (STEP46) variants that are formed by alternative splicing of a single gene [5]. STEP61 differs from STEP46 by the presence of an additional 172 amino acids at its N-terminus [6]. STEP along with two other PTPs, PTPRR and HePTP belongs to a family of PTPs that contains a highly conserved 16-amino acid substrate-binding domain termed the kinase interacting motif (KIM domain) [7]. A regulatory serine residue lies in the middle of the KIM domain (ser 49 in STEP46/ser 221 in STEP61) and dephosphorylation of this residue renders STEP active in terms of its ability to bind to its substrates. Phosphorylation of ser 49/ser 221 is mediated by dopamine/D1 receptor dependent activation of the cAMP/PKA pathway [8] while dephosphorylation is mediated by glutamate/NMDA receptor induced activation of the Ca2+ dependent phosphatase, calcineurin [9].
In its active form STEP can modulate synaptic plasticity by regulating the activity of extracellular regulated kinase 1/2 (ERK1/2), a key protein involved in memory formation [10, 11]. Active STEP can also modulate NMDA receptor-dependent long term potentiation by interfering with NMDA receptor trafficking to synaptic membrane, possibly through regulation of the upstream kinase Fyn [12] and tyrosine dephosphorylation of NR2B-NMDA receptor subunits [13]. Several studies also indicate a role of active STEP in neuroprotection through its regulation of p38 mitogen-activated protein kinase [14, 15].
Although reversible phosphorylation of ser49/ser 221 within the KIM domain appears to play an important regulatory function additional mechanisms may also exist for stricter control of STEP activity. It has been reported that there is a second conserved domain carboxy-terminal to the KIM domain present in STEP, PTP-SL and HePTP. This domain has been termed as the kinase specificity sequence (KIS domain) [16]. The aim of the present study was to investigate the role of the KIS domain in regulating the function of STEP. Our results show that in intact cells STEP is phosphorylated at two specific sites within the KIS domain. Dephosphorylation of these two sites leads to degradation of STEP and the active form of STEP is specifically targeted for degradation by ubiquitination.
EXPERIMENTAL
Materials
STEP61 and STEP46 cDNAs were cloned into the pcDNA 3.1/V5-His mammalian expression vector (Invitrogen). Deletion and point mutants of STEP61 and STEP46 were obtained by polymerase chain reaction (PCR) based site-directed mutagenesis. All mutations were verified by nucleotide sequencing. Hela, Cos-7 and N2A cells were obtained from ATCC. Antibodies used were as follows: monoclonal anti-STEP from Novus Biologicals, monoclonal anti-V5 and anti-V5-HRP from Invitrogen, anti-β-tubulin from Sigma-Aldrich and anti-ubiquitin (FK2) from Biomol International. All secondary antibodies were from Cell Signaling. Proteasome inhibitor MG-132 and epoxomicin were from Boston Biochemicals. Protein G Sepharose was from GE Healthcare. 32P-labelled sodium orthophosphate and Easy Tag Express 35S Protein Labeling Mix was obtained from Perkin Elmer. All other reagents were from Sigma-Aldrich.
Cell culture and stimulation
Hela, Cos-7 and N2A cells were grown in Dulbeco’s Modified Essential Medium (DMEM) containing 1X antibiotics-antimycotic and 10% fetal calf serum, at 37°C in a humidified atmosphere consisting of 5% CO2 and 95% air. Cells were transiently transfected with 2 μg of each DNA using Lipofectamine 2000 (Invitrogen). After 18 hr cells were treated with or without MG-132 (25 μM and 50 μM) and epoxomicin (2.5μM, 5 μM and 10 μM) for 6 and 9 hr at 37°C. Cells were either processed for immunoprecipitation or lysed in SDS-containing buffer. Immune-complexes and lysates were then subjected to SDS-PAGE and immunoblot analysis. In some experiments PD98059 (20 μM) and SB203580 (20 μM) were added 6 hr after transfection. Cells were harvested after another 6 hr and processed for SDS-PAGE and immunoblot analysis.
Immunoprecipitation
Cells were lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 50 mM NaF, 10 mM Na4P2O7, 1 mM Na3VO4, 0.1% NP-40 and protease inhibitor cocktail (Boehringer). Lysates were centrifuged for 10 min at 14,000 rpm to remove insoluble material, and then pre-cleared with protein G-sepharose for 1 hr. For immunoprecipitation of STEP or ubiquitin, samples were incubated overnight with anti-V5 or anti-ubiquitin antibody. The immune complexes were incubated with 30 μl of protein G-sepharose for 2 hr at 4°C. Beads were collected by centrifugation at 1000 rpm for 2 min and washed five times with lysis buffer. Proteins were eluted using SDS-sample buffer and processed for SDS-PAGE and immunoblot analysis with antibodies as described in the individual experiments.
Immunoblotting
Equal amount of protein lysates, as estimated using bicinchoninic acid protein assay kit (Pierce Biotechnology), were resolved by 8% SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membranes were blocked for 1 hr at room temperature with 5% non-fat dry milk and then incubated for 1 hr at room temperature or at 4°C overnight with the appropriate primary antibody. Horseradish peroxidase coupled to anti-rabbit or anti-mouse IgG that were raised in goat were used as secondary antibodies. Immune complexes were detected on X-ray film after treatment with West Pico super signal chemiluminescence reagent (Pierce Biotechnology).
Phosphatase activity assay
Hela cells were transfected with V5-tagged STEP46 S49A or one of its mutant. After 24 hr cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl and 1% Triton-X-100. Half of the lysate was processed for immunoblot analysis of immunoprecipitated STEP and the remaining half was used for phosphatase assay. STEP46 was immunoprecipitated using anti-V5 antibody. The immune complexes bound to protein G beads were then washed three times in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl and 0.1% Triton-X-100 and then once in a buffer containing 30 mM HEPES, pH 7.0 and 120 mM NaCl. For para-nitrophenylphosphate (pNPP) assay the beads were incubated in 100 mu;l reaction mixture containing 30 mM HEPES, pH-6.0; 120 mM NaCl and 10 mM pNPP for 30 min at 30°C. The reactions were terminated by addition of 900 μl of 0.2 N NaOH. Phosphatase activity of the STEP46 mutants was measured by colorimetric quantitation of the formation p-nitrophenolate at 410 nm using a spectrophotometer. The values obtained were normalized for the amount of STEP immunoprecipitated [17].
RNA isolation and cDNA synthesis
Total RNA was isolated using RNA easy kits (Qiagen), from Hela cells transiently transfected for 24 hr with either STEP46 S49A or its mutant STEP46 S49A/T59A/S72A. The quality of the RNA was analyzed by electrophoresis on 1% agarose gels. 2 μg of each RNA was used for cDNA synthesis with Superscript III cDNA synthesis kit (Invitrogen), using oligo dT. The following primers were used for subsequent PCR based amplification (16, 20, 25 and 30 cycles) of STEP46 (Forward primer – 5′ ATGGAGGAGAAGGTAGAGGATGAC 3′; Reverse primer – 5′ CTCTGAGGACTGGAGGGTCAGCTG 3′). Amplification (16, 20, 25 and 30 cycles) of β-actin (Forward primer – 5′ GCTCGTCGTCGACAACGGCT 3′; Reverse primer – 5′ CAAACATGATCTGGGTCATCTTCTC 3′) was performed as a control to show that equal amounts of starting total RNA were present in each sample. The PCR products were analyzed by agarose gel electrophoresis.
Pulse chase experiments
Hela cells expressing STEP46 S49A or STEP46 S49A/T59A/S72A were washed twice with warm PBS and then incubated for 30 min at 37°C in Met/Cys free DMEM supplemented with 5% dialyzed fetal calf serum. Cells were then incubated with 35S protein labeling mix (0.1 mCi per plate) in fresh Met/Cys free DMEM supplemented with 5% dialyzed fetal calf serum for 45 min. The radiolabel was then removed by washing the cells two times with PBS followed by incubation at 37°C in warm DMEM supplemented with 10% fetal calf serum and 2 mM each of cysteine and methionine for the specified time periods. The medium was then removed, and the cells were rinsed twice with ice-cold PBS. Cells were then lysed in a buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 50 mM NaF, 10 mM Na4P2O7, 1 mM Na3VO4, 0.1% NP-40 and protease inhibitor cocktail (Boehringer) and then processed for immunoprecipitation.
32P labeling of cells
Hela cells expressing wild type STEP46 or one of its mutants were grown for 24 hr in the presence of DMEM containing 1X antibiotics-antimycotic and 10 % fetal calf serum. The cells were then grown in phosphate free DMEM containing 0.5% serum for 18 hr and then incubated with 32P sodium phosphate (0.3 mCi per plate) for additional 6 hr in the same medium. The labeling buffer was then removed, and the cells were rinsed twice with ice-cold PBS (pH 7.4). Cells were then lysed and processed for immunoprecipitation as described above.
Statistical analysis
Densitometric analysis of the 35S- and 32P-labeled STEP bands in the autoradiogram as well as the levels of STEP and ubiquitin proteins detected in immunoblots was performed using Image J software. Statistical comparison was done using one-way analysis of variance (ANOVA, Bonferroni’s multiple comparison test) and differences were considered significant when p < 0.05.
RESULTS
STEP is phosphorylated endogenously at SP/TP sites in the KIS domain
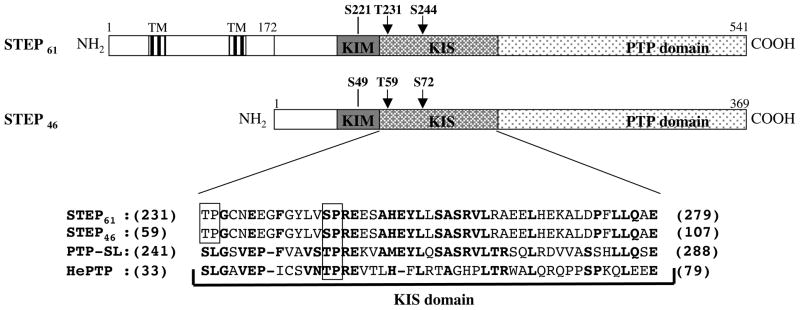
STEP61 and STEP46 both include the KIM and KIS domains and the phosphatase motif (PTP domain) (Fig. 1). Both STEP61 and STEP46 are phosphorylated endogenously at a consensus PKA phosphorylation site within the KIM domain (ser 221 in STEP61/ser 49 in STEP46) [8]. Additionally, a serine and a threonine residue (T231/T59 and S244/S72) are present in the KIS domain within the consensus sequences of proline directed kinases (SP/TP sites; Fig. 1). Initial experiments explored the possibility of endogenous phosphorylation of these two sites in Hela cells. Cells were transfected with either V5-tagged wild type STEP46 (STEP46 WT) or its mutant that cannot be phosphorylated endogenously at the KIM domain serine residue (V5-tagged STEP46 S49A) and then labeled with 32P-orthophosphate for 6 hr. STEP was then immunoprecipitated using anti-V5 antibody, resolved on SDS-PAGE, transferred to PVDF membrane and processed for autoradiography. A phospho-protein of ~50 kDa was detected by autoradiography in cell lysates transfected with STEP46 WT (Fig. 2A, upper panel, lane 2). Immunoblot analysis identified the protein as STEP46 (Fig. 2A, lower panel, lane 2). This is consistent with earlier findings that STEP is endogenously phosphorylated at the PKA site within the KIM domain [8]. Consistent with this idea, mutation of this site (STEP46 S49A) greatly reduced, but did not abolish phosphorylation of the mutant form (Figure 2A). Thus a phospho-protein of similar molecular weight was observed in cell lysates transfected with STEP46 S49A, although the level was low and the band position was slightly lower than that observed with STEP46 WT (Fig. 2A, lane 3). Such a downward shift in mobility has been reported in earlier studies of STEP46 following dephosphorylation or mutation of the PKA site [9, 13, 14]. Phosphorylation of mutant STEP46 (STEP46 S49A) implies that in addition to the PKA site within the KIM domain STEP46 must be endogenously phosphorylated at other sites.
Fig. 1.
Schematic diagram of STEP61 and STEP46. The illustration indicates the positions of the transmembrane domains (TMs), the kinase-interacting motif (KIM), the PKA phosphorylation site in the KIM domain, the kinase specificity sequences (KIS) and catalytic domain (PTP domain) of STEP61 and STEP46. The arrows indicate the positions of potential proline-directed kinase phosphorylation sites in both isoforms (SP/TP sites). Also shown are the sequence alignment of the KIS domains of STEP61 (231–279), STEP46 (59–107), PTP-SL (241–288) and hHePTP (33–79). The conserved residues are in bold. The boxes show the location of the SP/TP sites.
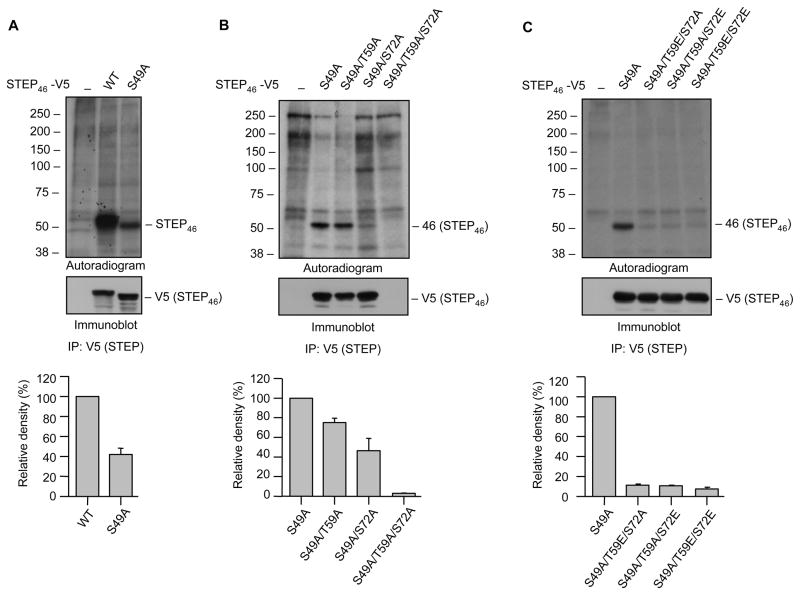
Fig. 2.
Endogenous phosphorylation of STEP at SP/TP sites in the KIS domain. Hela cells were transiently transfected with (A) V5-tagged STEP46 (WT) or its mutant, STEP46 S49A that cannot be phosphorylated at the Ser residue in the KIM domain, (B) V5-tagged STP46 S49A or one of its mutants where Thr 59 and/or Ser 72 in the KIS domain were converted to alanine (S49A/T59A, S49A/S72A, S49A/T59A/S72A) to render them non-phosphorylatable and (C) V5-tagged STEP46 S49A or one of its mutants where Thr 59 and/or Ser 72 in the KIS domain were converted to glutamic acid (T59E/S72A, T59A/S72E, T59E/S72E) to mimic phosphorylated form. 24 hr after transfection cells were labeled with 32P-orthophosphate. STEP was immunoprecipitated from cell lysates using anti-V5 antibody, resolved on SDS-PAGE, transferred to PVDF membrane and processed for autoradiography (upper panel) and immunoblot analysis (lower panel). The line on the right side of each representative autoradiograph (upper panel) indicates the position of the phosphorylated protein band corresponding to STEP46. The molecular weight standards are indicated in kilodaltons on the left side of each panel. The phosphorylated bands were identified as STEP by probing the membrane with anti-STEP antibody (lower panel). Immunoprecipitation with protein-G sepharose alone was used as control (lane 1). Quantitation of phosphorylated STEP46 was performed by computer-assisted densitometry and Image J analysis. Values are mean ± SEM (n = 3).
To determine whether the SP/TP sites in the KIS domain (Thr 59 and Ser 72) are endogenously phosphorylated Hela cells were transfected with either V5-tagged STEP46 S49A or one of its mutants (S49A/T59A, S49A/S72A or S49A/T59A/S72A). Cells were labeled with 32P-orthophosphate and then processed for immunoprecipitation of STEP and analysis by autoradiography and immunoblotting as described above. Figure 2B shows that the level of expression of STEP46 S49A, STEP46 S49A/T59A and STEP46 S49A/S72A remained unaltered (lower panel, lanes 2–4), but a significant reduction in phosphorylation was observed following mutation of either Thr 59 (upper panel, lane 3; 74.9% ± 3.6) or Ser 72 (upper panel, lane 4; 46.4 ± 10.2). These results suggest preferential phosphorylation of Ser 72 as compared to Thr 59. Interestingly, in the mutant where both Thr 59 and Ser 72 were converted to alanine (STEP46 S49A/T59A/S72A), to mimic the non-phosphorylatable form, no specific band was observed either in the autoradiogram (Fig. 2B, upper panel, lane 5) or in the immunoblot (Fig. 2B, lower panel, lane 5), suggesting that mutation of both Thr 59 and Ser 72 may lead to loss of expression of STEP. To further test whether phosphorylation of both these sites is essential for maintaining the STEP expression, additional mutants were constructed where one site in the KIS domain was converted to glutamic acid to mimic the phosphorylated form, and the other site was converted to alanine to make it non-phosphorylatable (S49A/T59E/S72A and S49A/T59A/S72E). These results were compared with mutants where both sites were converted to glutamic acid (S49A/T59E/S72E) to mimic the phosphorylated form. Figure 2C shows that expression of both the phosphorylatable (STEP46 S49A) and the phospho-mimicking forms (S49A/T59E/S72A, S49A/T59A/S72E and S49A/T59E/S72E) were stable (Fig. 2C, lower panel, lanes 2–5), even though only STEP46 S49A could be phosphorylated (Fig. 2C, upper panel, lane 2). Taken together these data indicates that both Thr 59 and Ser 72 are endogenously phosphorylated in intact cells and that phosphorylation of at least one of these sites is necessary for maintaining expression of STEP46 that is dephosphorylated at the serine residue within the KIM domain.
Dephosphorylation of the SP/TP sites in the KIS domain does not affect STEP’s association with its substrate or phosphatase activity
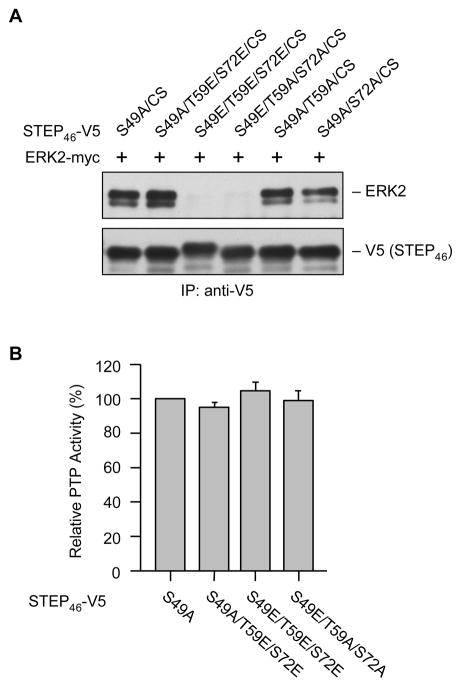
Dephosphorylation of STEP46 at the serine residue within the KIM domain (Ser 49) allows STEP to bind to its substrates for subsequent dephosphorylation [9]. To examine the consequence of phosphorylation-dephosphorylation of the KIS domain sites on the association of STEP with its downstream target ERK2 (extracellular regulated kinase 2), we generated a dominant negative variant of STEP (STEP46 S49A/C300S) where the KIM domain serine residue was converted to alanine (S49A) so that it can bind constitutively its substrate(s) and the regulatory cysteine residue in the phosphatase domain was converted to serine (C300S) so that it is enzymatically inactive (phosphatase dead form). Hela cells were co-transfected with myc-ERK2 and V5-STEP46 S49A/C300S or one of its mutants (S49A/T59E/S72E/C300S, S49E/T59E/S72E/C300S, S49E/T59A/S72A/C300S, S49A/T59A/C300S or S49A/S72A/C300S). STEP was immunoprecipitated using anti-V5 antibody and co-immunoprecipitation of ERK2 was determined by immunoblotting with anti-ERK2 antibody. The results indicate that in intact cells the STEP mutants, where the KIM domain serine residue was mutated to alanine, were able to interact with ERK2. In these mutants conversion of the KIS domain sites either to alanine (non-phosphorylatable form) or to glutamic acid (phospho-mimicking form) did not affect STEP’s interaction with ERK2 (Fig. 3A, lanes 1, 2, 5 and 6). However conversion of the KIM domain regulatory serine residue to glutamic acid (phospho-mimicking form) inhibited STEP’s interaction with ERK2, irrespective of the phosphorylation status of the KIS domain sites (Fig. 3A, lanes 3 and 4). To determine whether mutation of the KIS domain sites either to alanine or to glutamic acid has any effect on the phosphatase activity of STEP, Hela cells were transfected with V5-STEP46 S49A or one of its mutants (S49A/T59E/S72E, S49E/T59E/S72E, S49E/T59A/S72A). Tyrosine phosphatase activity of immunoprecipitated STEP was measured using pNPP as a substrate and the values were normalized for the amount of STEP immunoprecipitated. The results show that there is no change in phosphatase activity of STEP46 when the phosphorylation sites in the KIS domain was converted to either glutamic acid or alanine (Fig. 3B). Taken together these findings suggest that phosphorylation-dephosphorylation of the SP/TP sites in the KIS domain does not affect the intrinsic catalytic activity or the interaction of active STEP with its downstream target.
Fig. 3.
Dephosphorylation of SP/TP sites in the KIS domain does not affect the association of STEP with ERK2 or its phosphatase activity. Hela cells were transfected with (A) myc-tagged ERK2 and V5-tagged STEP46 S49A/C300S or one of its mutants (S49A/T59E/S72E/C300S, S49E/T59E/S72E/C300S, S49E/T59A/S72A/C300S, S49A/T59A/C300S, S49A/S72A/C300S) and (B) V5-tagged STEP46 S49A or one of its mutants (S49A/T59E/S72E, S49E/T59E/S72E, S49E/T59A/S72A). (A, B) STEP46 was immunoprecipitated using anti-V5 antibody. (A) Immune-complexes were processed for SDS-PAGE and immunoblot analysis with anti-ERK2 antibody to evaluate the association of ERK2 with STEP mutants (upper panel). Blots were reprobed with anti-V5 antibody (lower panel). (B) PTP activity was assayed using pNPP as a substrate. Quantitative measurement of the formation of para-nitrophenolate is represented as mean ± SEM (n = 4).
Dephosphorylation of active STEP at the SP/TP sites in the KIS domain leads to its loss of expression
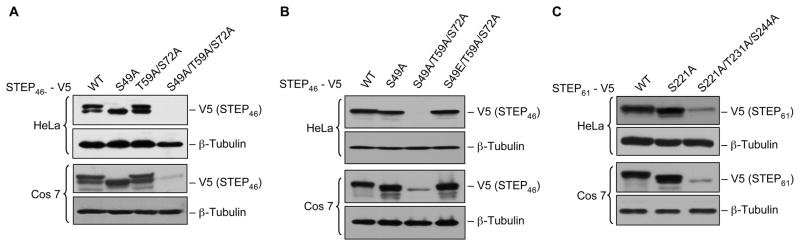
To determine whether the active form of STEP46 is more susceptible to destabilization, either STEP46 WT or one of its mutant (S49A, T59A/S72A or S49A/T59A/S72A) was transfected into both Hela and Cos-7 cells and expression levels were analyzed by immunoblot analysis of total cell lysates. Figure 4A shows that expression levels of STEP46 WT (inactive form), STEP46 S49A (active form) and STEP46 T59A/S72A (inactive form dephosphorylated at the two phosphorylation sites in the KIS domain) remained unaltered (lanes 1–3). However there was a significant loss in expression of the STEP46 S49A/T59A/S72A, the active form of STEP dephosphorylated at the two phosphorylation sites within the KIS domain (Fig. 4A, lane 4). Additional evidence that the active STEP is more susceptible to destabilization was obtained from mutants that mimic the inactive form of STEP and dephosphorylated at the two phosphorylation sites in the KIS domain (STEP46 S49E/T59A/S72A). Immunoblot analysis showed that there was no significant loss in expression of the inactive mutant of STEP46, even in the complete absence of KIS domain phosphorylation (Fig. 4B, lane 4).
Fig. 4.
Dephosphorylation of active STEP at the SP/TP sites in the KIS domain results in loss of expression. Hela and Cos-7 cells were transiently transfected with (A) V5-tagged STEP46 (WT) or one of its mutants (S49A, T59A/S72A or S49A/T59A/S72A), (B) V-tagged-STEP46 (WT), -STEP46 S49A or one of its mutants (S49A, S49A/T59A/S72A or S49E/T59A/S72A) and (C) V5-tagged STEP61 (WT) or one of its mutants (S221A or S221A/T231A/S244A). Expression of the STEP variants was analyzed by probing total cell lysates with anti-V5 antibody. Equal protein loading was confirmed by re-probing the blots using a polyclonal antibody against endogenously expressed β-tubulin.
Further analysis was also carried out with STEP61 WT variant (inactive form) and its corresponding mutants S221A (active form) and S221A/T231A/S244A (active form dephosphorylated at the two phosphorylation sites within the KIS domain). As shown in Figure 4B the expression profile of STEP61 WT and STEP61 S221A remained unaltered in both Hela and Cos-7 cells (lanes 1–2). However, a significant loss in expression of STEP61 S221A/T231A/S244A was observed in both the cell types (Fig. 4B, lane 3), which is consistent with our findings in STEP46.
To evaluate whether the loss of expression of active STEP following mutation of the sites in the KIS domain may be attributed to reduced transcription Hela cells were transiently transfected with STEP46 S49A or STEP46 S49A/T59A/S72A. Total RNA isolated from the cell lysates were used for cDNA synthesis and subsequent PCR amplification (16, 20, 25 and 30 cycles) with primers against STEP46 as described in the Experimental Procedures. Analysis of PCR products by agarose gel electrophoresis showed a single band in the gel demonstrating that the amplification was RNA specific. Additionally there were no differences in the amounts of PCR products between STEP46 S49A and STEP46 S49A/T59A/S72A in any of the cycles tested (see Supplemental data, Fig. S1). Amplification of the PCR product using primers specific for β-actin confirmed that equal amounts of cDNA were amplified in each sample. Taken together the findings of Figures 4 and S1 demonstrate that the loss of expression of active STEP observed in both Hela and Cos-7 cells (Fig. 4) was not regulated by transcription but rather the active form of STEP appears susceptible to destabilization following dephosphorylation of the SP/TP sites within the KIS domain.
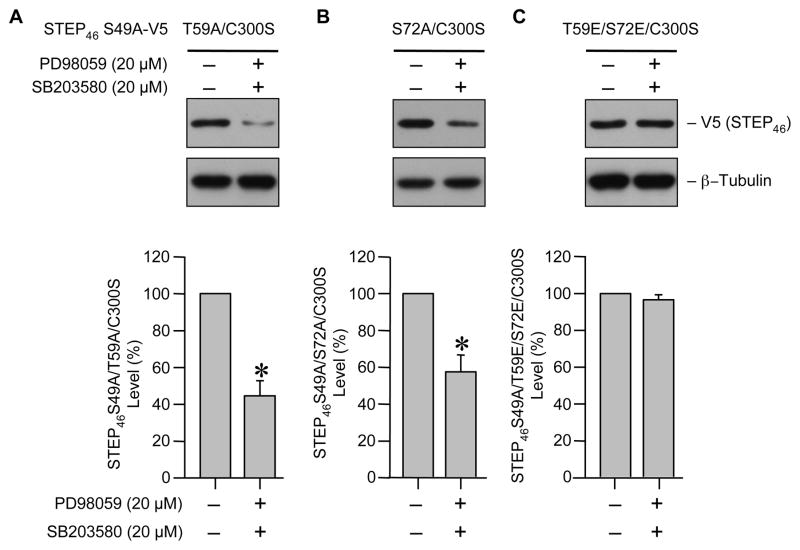
SP/TP sites are putative mitogen activated protein kinase (MAPK) target sites and two members of this family, ERK and p38 MAPKs are known to interact with STEP [9, 14]. To examine whether ERK and p38 MAPK dependent phosphorylation of the two SP/TP sites in the KIS domain leads to stabilization of STEP several additional mutants were constructed using STEP46 S49A/C300S, the dominant negative form that can bind constitutively to its substrates, as the template. In the first two mutants one of the phosphorylation sites in the KIS domain was converted to alanine to render it non-phosphorylatable while the other remained accessible to phosphorylation by endogenous kinases (STEP46 S49A/T59A/C300S and STEP46 S49A/S72A/C300S). In the third mutant both the KIS domain sites were converted to glutamic acid to mimic the phosphorylatable form (STEP46 S49A/T59E/S72E/C300S). Hela cells expressing these mutants were treated with pharmacological inhibitors of both ERK (PD98059, 20 μM) and p38 (SB203580, 20 μM) MAPKs [18, 19]. Immunoblot analysis of total lysates shows a significant decrease in the expression of STEP mutants, in the presence of MAPK inhibitors, where either Thr 59 or Ser 72 was accessible to phosphorylation. (Fig. 5A and B, upper panel, lane 2). However the expression of the phospho-mimicking form remained unaltered in the absence or presence of MAPK inhibitors (Fig. 5C, upper panel). Thus it is likely that the basal activity of either ERK or p38 MAPK is essential for phosphorylation of Thr 59 and Ser 72 to maintain the stability of STEP. Inhibition of ERK or p38 MAPKs individually did not significantly affect the level of expression of the STEP mutants (data not shown) indicating that the loss of activity of one of the MAPKs may be compensated by the other.
Fig. 5.
Phosphorylation of SP/TP sites in the KIS domain is mediated by ERK and p38 MAPKs. Hela cells expressing (A) STEP46 S49A/T59A/C300S, (B) STEP46 S49A/S72A/C300S or (C) STEP46 S49A/T59E/S72E/C300S were treated with PD98059 (20 μM) and SB203580 (20 μM) 6 hr after transfection. Cells were harvested after another 6 hr and processed for SDS-PAGE and immunoblot analysis with anti-V5 antibody (upper panel). Total tubulin was also analyzed to indicate equal protein loading (lower panel). Steady-state expression level of STEP mutants was quantified by computer-assisted densitometry and Image J analysis. Values represent mean ± SEM of measurements (n = 3). * Indicate significant difference from untreated control (p < 0.05).
Dephosphorylation of the SP/TP sites in the KIS domain results in ubiquitin-mediated proteasomal degradation of active STEP
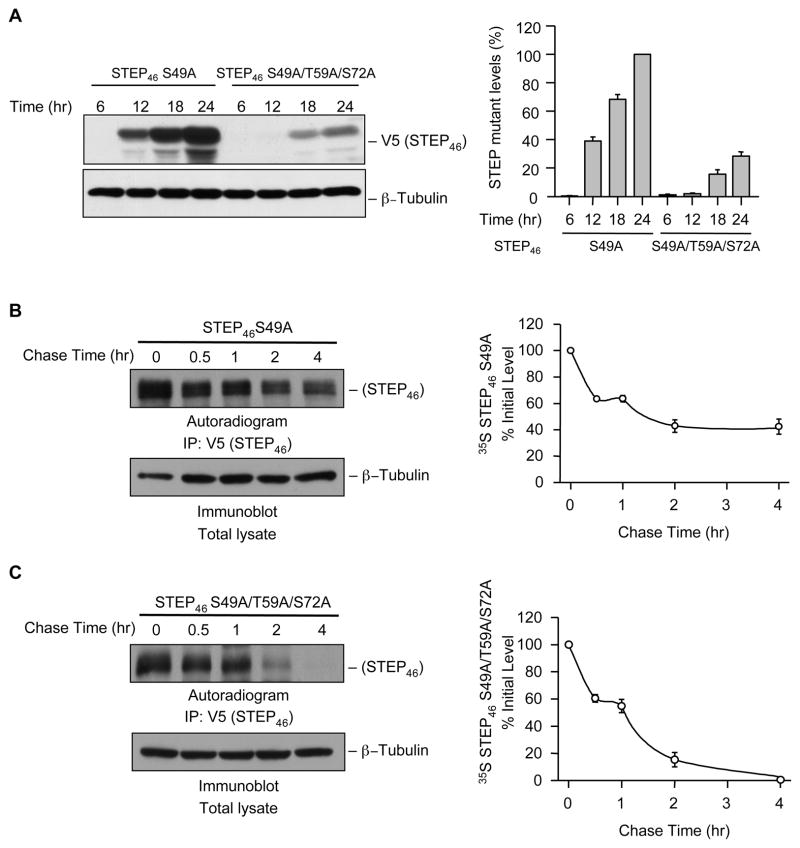
We next determined the time course of changes in the expression of active STEP and its mutant form that cannot be phosphorylated endogenously at the SP/TP sites within the KIS domain. Hela cells were transiently transfected with V5-tagged STEP46 S49A or -STEP46 S49A/T59A/S72A. At specific time points after transfection (6, 12, 18 and 24 hr) cells were harvested, lysed and processed for immunoblot analysis. As shown in Figure 6A expression of STEP46 S49A increased substantially with time from 12 hr onwards. However there was a significant delay and dramatic reduction in the expression of STEP46 S49A/T59A/S72. A very low level of expression of STEP46 S49A/T59A/S72 was observed at 18 and 24 hr.
Fig. 6.
Phosphorylation of Thr 59 and Ser 72 regulates the stability of STEP. (A) V5-tagged STEP46 S49A or its mutant STEP46 S49A/T59A/S72A was expressed in Hela cells. The expression of STEP over time (6, 12, 18 and 24 hr) was analyzed by immunoblot analysis of total cell lysates with anti-V5 antibody. Level of expression of STEP mutants was also quantified by densitometry and presented in the bar graph. (B) Hela cells expressing V5-tagged STEP46 S49A or its mutant STEP46 S49A/T59A/S72A were subjected to pulse-chase experiments as described in Experimental Procedures. STEP was immunoprecipitated from equal amount of cell lysates using anti-V5 antibody, resolved on SDS-PAGE and processed for autoradiography (upper panel). Equal amount of protein from the cell lysates were also processed for immunoblot analysis with anti-tubulin antibody (lower panel). Quantification of the 35S-labeled STEP band in the autoradiograph was done by computer assisted densitometry and Image J analysis. The representative autoradiograph and the corresponding graph show the time course of degradation of STEP46 and STEP46 S49A/T59A/S72A. Values are mean ± SEM (n = 3).
Having established that the expression of active STEP is decreased when dephosphorylated at Ser 59 and Thr 72 residues in the KIS domain we hypothesized that preventing phosphorylation of these two sites may destabilize the active form of the protein. To test this hypothesis, we conducted pulse-chase experiments using Hela cells transiently transfected with either STEP46 S49A or STEP46 S49A/T59A/S72A. Newly synthesized mutant STEP proteins were metabolically labeled with 35S-Met/Cys protein labeling mix for 45 min (pulse) and the labeled products were then followed (chase) by replacing the medium with non-radioactive medium containing an excess of unlabeled Met/Cys. The degradation pattern of 35S-labeled STEP46 S49A and STEP46 S49A/T59A/S72A was monitored by immunoprecipitating them from cell lysates obtained at various time points during the chase. Analysis of the immunoprecipitates by SDS-PAGE and autoradiography showed a significant increase in the rate of degradation of active STEP46 S49A/T59A/S72A (Figure 6B and C), where the phosphorylation sites within the KIS domain were mutated to alanine (non-phosphorylatable form).
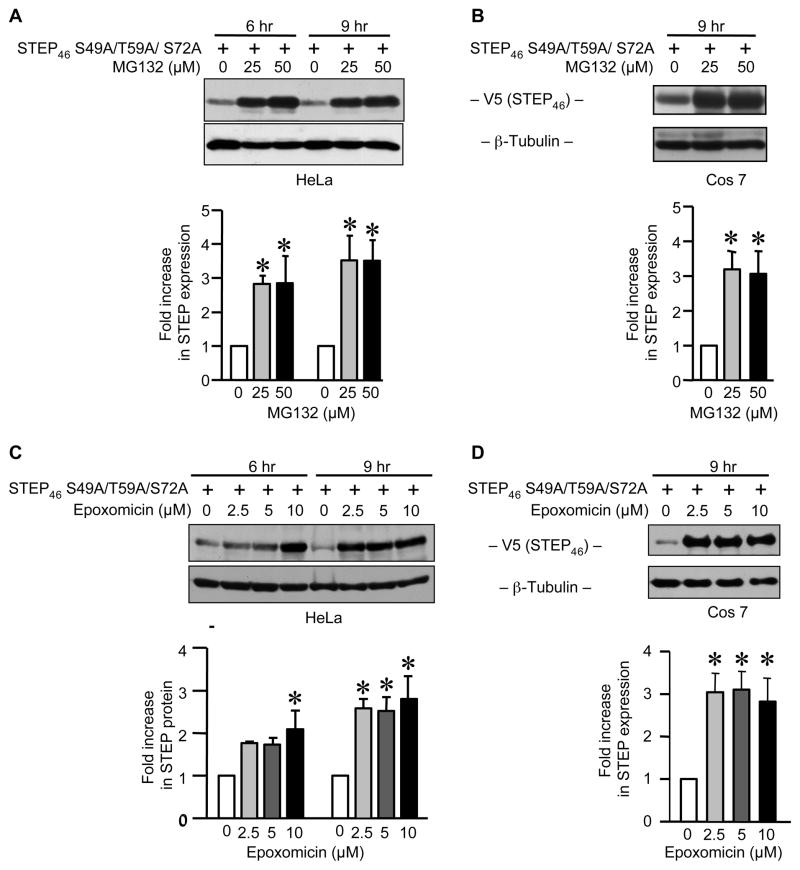
To clarify the mechanism(s) by which the active form of STEP is degraded following dephosphorylation of Thr 59 and Ser 72 in the KIS domain we tested the effects of two well established proteasome inhibitors, MG-132 and epoxomicin [20, 21]. Hela and Cos-7 cells were transiently transfected with V5-tagged STEP46 S49A/T59A/S72A and then treated with one of the inhibitors (MG-132, 25 – 50 μM; epoxomicin, 2.5 – 10 μM) for 6 or 9 hr. Figure 7 shows that both inhibitors effectively prevented STEP degradation. Exposure to 25 μM MG-132 for 6 or 9 hr was sufficient to enhance the expression of STEP46 S49A/T59A/S72A in Hela cells (~ 3 fold increase, Fig. 7A). In Cos-7 cells the level of STEP expression also increased (~3–3.5 folds) following 9 hr treatment with 25 μM MG-132 (Fig. 7B). Epoxomicin exerted similar effects at a minimum concentration of 2.5 μM (9 hr treatment), in both Hela and Cos-7 cells (Fig. 7C and D). These findings are consistent with a primary role for proteasomal degradation of active STEP.
Fig. 7.
Proteasomal inhibitors MG-132 or epoxomicin increases the steady-state level of active STEP dephosphorylated at the SP/TP sites in the KIS domain. (A and C) Hela and (B and D) Cos-7 cells were transiently transfected with V5-tagged STEP46 S49A/T59A/S72A cDNA. 18 hr after transfection cells were treated with different doses of MG-132 or epoxomicin: (A) 0, 25 or 50 μM of MG-132 for 6 and 9 hr; (B) 0, 25 or 50 μM of MG-132 for 9 hr; (C) 0, 2.5, 5.0 or 10 μM of epoxomicin for 6 and 9 hr; (D) 0, 2.5, 5.0 or 10 μM of epoxomicin for 9 hr. Equal amount of total cellular protein from each sample was resolved on SDS-PAGE and processed for immunoblot analysis with anti-V5 antibody. The membranes were re-probed with anti-β-tubulin antibody to ensure equal protein loading. The amount of STEP46 S49A/T59A/S72A protein was quantified by densitometry and presented in the bar graphs. Values represent mean ± SEM of measurements (n = 3). * Indicate significant difference from untreated control (p < 0.05).
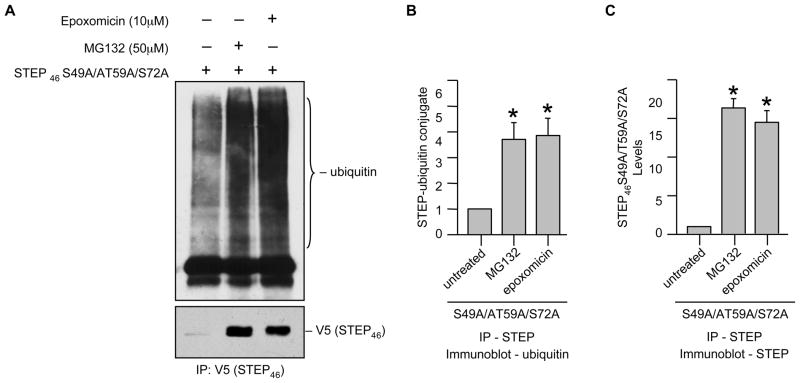
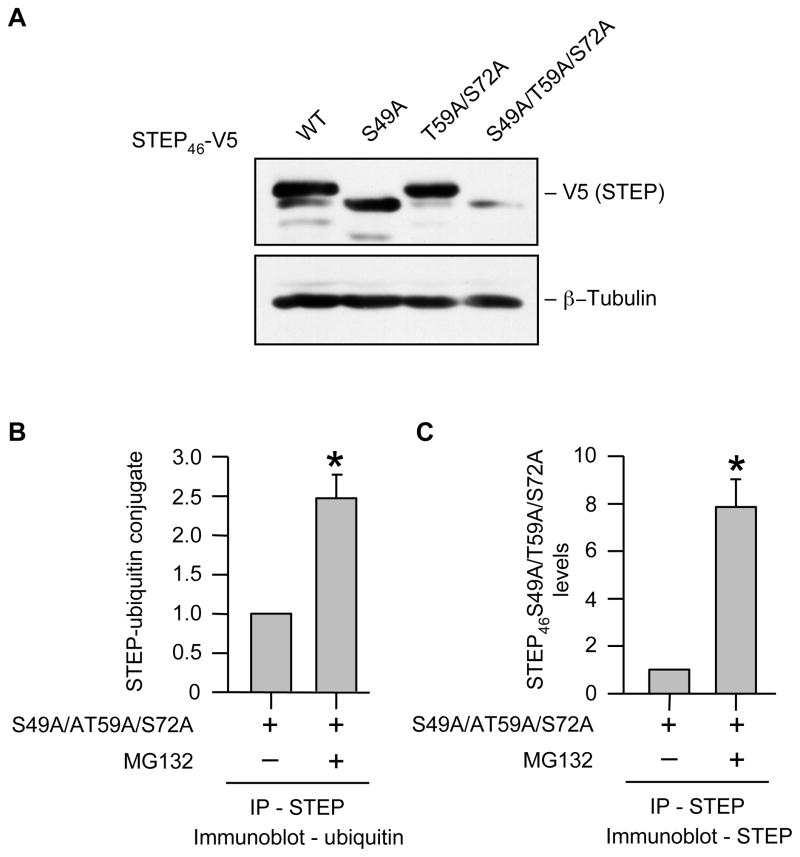
Ubiquitination is an important first step in proteasomal degradation and it is well established that when a substrate protein is marked with multiple ubiquitin peptides (generally four or more), this complex can be recognized by proteasomes resulting in degradation [22]. We next tested whether STEP is ubiquitinated prior to degradation. Hela cells were transfected with V5-tagged STEP46 S49A/T59A/S72A that is susceptible to degradation and treated with MG-132 or epoxomicin. STEP was immunoprecipitated and immune complexes were processed for immunoblotting with an anti-ubiquitin antibody. The representative immunoblot and the corresponding mean data shows that in the absence of MG-132 or epoxomicin, most of the STEP46 S49A/T59A/S72A was degraded and accumulation of ubiquitinated STEP46 S49A/T59A/S72A was minimal (Fig. 8A, lower panel, lane 1 and Fig. 8C). In the presence of MG-132 or epoxomicin the level of expression of STEP46 S49A/T59A/S72A increased significantly as compared to the untreated control (Fig. 8A, lower panel, lanes 2–3 and Fig. 8C) and formed a stable complex with ubiquitin as evident from co-immunoprecipitation of ubiquitin along with STEP (Fig. 8A, upper panel, lanes 2–3 and Fig. 8B). In a reverse set of experiments ubiquitin was immunoprecipitated and co-immunoprecipitation of STEP with ubiquitin was analyzed using anti-V5 antibody. A considerable increase in co-immunoprecipitation of STEP46 S49A/T59A/S72A along with ubiquitin was observed in cells treated with MG-132 or epoxomicin (data not shown), consistent with our hypothesis that ubiquitin-mediated proteasomal degradation plays a critical role in regulation of STEP levels.
Fig. 8.
Ubiquitination of active STEP dephosphorylated at the KIS domain sites in Hela cells. Hela cells were transiently transfected with V5-tagged STEP46 S49A/T59A/S72A cDNA. 18 hr after transfection cells were treated with MG-132 (50 μM) or epoxomicin (10 μM) for 6 hr. (A) STEP was immunoprecipitated from equal amount of total cell lysates using anti-V5 antibody. (A) Immune complexes were resolved on SDS-PAGE and processed for immunoblot analysis with anti-ubiquitin (upper panel) and anti-V5 (lower panel) antibodies. Quantification of (B) co-immunoprecipitated ubiquitin in complex with STEP and (C) total immunoprecipitated STEP46 S49A/T59A/S72A was performed by computer-assisted densitometry and Image J analysis. Values are mean + SEM (n = 3). *Indicate significant difference from untreated control (p < 0.05).
Since STEP is predominantly expressed in neurons, in a final set of experiments we examined whether the mechanisms described above for degradation of active STEP also apply following expression in the more neuron-like N2A cells (mouse neuroblastoma cells) that do not express STEP endogenously. Cells were transfected with STEP46 WT or its mutants (S49A, T59A/S72A or S49A/T59A/S72A) and cell lysates were processed for immunoblot analysis with anti-V5 antibody. Consistent with our observation in Hela and Cos-7 cells, a loss in stability/expression was only observed with the STEP46 S49A/T59A/S72A mutant in N2A cells (Fig. 9A, lane 4). Furthermore, treatment of cells expressing V5-tagged STEP46 S49A/T59A/S72A with MG-132 followed by immunoprecipitation of STEP and immunoblot analysis with anti-ubiquitin antibody imply a similar dependence on ubiquitin-mediated proteasomal degradation. Accordingly, a significant increase in STEP46 S49A/T59A/S72A level (Fig. 9B) as well as STEP46 S49A/T59A/S72A-ubiquitin complex formation (~ 2.5 fold) was seen in the presence of MG-132 (Fig. 9C).
Fig. 9.
Ubiquitin-mediated proteasomal degradation of active STEP in neuronal cells. (A) Neuronal origin N2A cells were transiently transfected with V5-tagged STEP46 WT or one of its mutants (S49A, T59A/S72A or S49A/T59A/S72A). Expression of STEP mutants was analyzed by probing total cell lysates with anti-V5 antibody. Equal protein loading was confirmed by reprobing the blots with anti-β-tubulin antibody. (B, C) N2A cells were transiently transfected with V5-tagged STEP46 S49A/T59A/S72A. 18 hr after transfection cells were treated with MG-132 (50 μM) for 6 hr. STEP was immunoprecipitated using anti-V5 antibody, immune complexes were resolved on SDS-PAGE and processed for immunoblot analysis with anti-ubiquitin and anti-V5 antibodies. Quantification of (B) co-immunoprecipitated ubiquitin in complex with STEP and (C) total immunoprecipitated STEP46 S49A/T59A/S72A was performed by computer-assisted densitometry and Image J analysis. Values are mean + SEM (n = 3). *Indicate significant difference from untreated control (p < 0.05).
DISCUSSION
The current study highlights the role of KIS domain in regulation of the level of active STEP. We identified two SP/TP sites in the KIS domain whose phosphorylation is mediated primarily through the basal activity of ERK and p38 MAPKs. Dephosphorylation of these two sites selectively results in ubiquitin-mediated degradation of the active form of STEP. The effects of proteasome inhibitors also strongly implicate a role of the proteasomal machinery in degradation of active STEP. Previous studies have demonstrated that a major switch in the ability of STEP to bind to its substrate(s) involves reversible phosphorylation and dephosphorylation of a serine residue within the KIM domain [8, 9]. The specific proteolysis of the active form of STEP, as demonstrated in this report, reveals a previously unidentified cellular mechanism for attenuating STEP activity irreversibly.
Our findings indicate that the Ser residue within the KIM domain (Ser 49) is not the only site involved in phosphorylation of STEP in intact cells. Thr and Ser residues (Thr 59 and Ser 72) present within the consensus sequences of proline-directed kinases in the KIS domain are also phosphorylated endogenously in STEP. The fact that mutation of S72A in the active form of STEP (STEP46 S49A) results in ~55% reduction in active STEP phosphorylation, while mutation of T59A results in ~25% decrease may suggest that Ser 72 is a major phosphorylation site in addition to Ser 49. Alternatively, it is also possible that phosphorylation of Thr 59 in intact cells is enhanced only when Ser 72 is phosphorylated. Several studies have also suggested that Ser/Thr Kinases have a preference for phosphorylating Ser residues, while most Ser/Thr phosphatases shows a striking bias towards dephosphorylating phospho-Thr residues [23]. This relative biasness may also account for the over-representation of Ser residue phosphorylation in intact cells. Nevertheless, phosphorylation of either of the two sites appears to play a central role in stabilization of STEP.
Ubiquitin is a small protein of 76 amino acids that marks a protein for degradation by the proteasome pathway [24, 25]. The formation of the polyubiquitin-protein complex is an ATP-dependent enzymatic process that requires the concerted efforts of E1, E2 and E3 ubiquitin ligases [26]. E3 ligases control the specificity of target protein selection and therefore are key to controlling target protein degradation. While ubiquitination is involved in many cellular processes, only relatively recently has its involvement in neuronal function begun to be investigated [27–29]. Phosphorylation is the most common mechanism by which a stable protein can become a target for ubiquitination and subsequent degradation in neurons. For example, phosphorylation of p35 (a neuronal specific activator of cyclin-dependent kinase 5; Cdk5) by Cdk5 kinase results in its degradation by an ubiquitin-mediated pathway [30]. Similarly, tyrosine phosphorylation of the NR2B-NMDA receptor in a Fyn-kinase dependent manner is also thought to result in ubiquitination and down-regulation of NMDA receptor function [31]. However, phosphorylation can also have an opposing effect of stabilizing a short-lived protein. In vertebrate neurons, NMDA receptor-dependent phosphorylation of c-jun renders the transcription factor resistant to ubiquitin-mediated degradation and activates its ability to bind to DNA and initiate transcription [32]. A key finding of the current study is that endogenous phosphorylation of Thr 59 and Ser 72 residues within the KIS domain plays an important role in the stabilization of STEP.
Consistent with the requirement of multiple-site phosphorylation to maintain the stability of STEP, a single mutation of either of the two sites alone did not affect the stability of the protein. It is important to note that the requirement of multi-site phosphorylation as the driving force for ubiquitination of a protein is well established and is thought to increase the sensitivity of a protein for ubiquitin-mediated degradation [33–36]. Recent studies have also demonstrated a role for multi-site phosphorylation in the protection of a protein from ubiquitin-mediated degradation [37], however the underlying mechanism by which this occurs is not yet well understood. One possible explanation for this phenomenon is that more than one kinase is involved in the phosphorylation of the two sites within the KIS domain in STEP and that dephosphorylation of a single site is not sufficient for ubiquitination and degradation. In this way, multi-site phosphorylation could serve to set a higher threshold for susceptibility to degradation.
Ubiquitination is increasingly recognized as a post-translational modification that, along with phosphorylation, is likely to be an important regulator of synaptic plasticity [29]. Recent evidence suggests that at glutamatergic synapses in the brain, activation of NMDA receptors can alter synaptic protein composition in an ubiquitin-proteasome dependent manner [38]. In particular, post-synaptic density-95 (PSD-95), a scaffolding protein that regulates NMDA-receptor dependent synaptic plasticity through GluR trafficking [39–41] has been shown to be a target for degradation by the ubiquitin-proteasome system. Ubiquitination of PSD-95 by ubiquitin ligase Mdm2, and its subsequent degradation, is regulated by the Ca2+ dependent phosphatase calcineurin [42]. STEP is part of the NMDA receptor complex [43] and NMDA receptor mediated activation of STEP is also regulated by calcineurin [9]. Since active STEP is more susceptible to degradation this leads us to speculate that calcineurin may also play a role in the degradation of active STEP, thereby limiting the duration of STEP signaling following its activation. This negative regulation may be one mechanism by which neurons may avert the patho-physiological consequences of over activation of STEP.
Several lines of evidence suggest that dysfunction of the proteasomal degradation machinery plays an important role in the pathogenesis of neurodegenerative disorders [44]. Aberrant accumulation of ubiquitinated proteins due to decline in proteasomal function has been reported in both Alzheimer’s and Parkinson’s disease [45–47]. Accumulating evidence from both animal models and human post-mortem studies suggests that progressive increase in the generation of reactive oxygen and reactive nitrogen species may contribute to failure of proteasomal system, leading to accumulation of ubiquitinated proteins involved in the pathology of these disorders [48, 49]. In this context a recent study reported elevated levels of STEP61 in the cortex of aged transgenic AD model mice (Tg2576) and in human AD brains [50]. The study also showed that in the AD mice model the elevated levels of STEP are ubiquitinated and associated with an increase in tyrosine phosphatase activity. Thus, it appears that impairment of the proteasomal degradation machinery to remove STEP-ubiquitin conjugates may lead to the accumulation of active STEP61 in AD. Consistent with this view our study indicates that active STEP is more susceptible to degradation and that inhibition of the proteasomal machinery leads to accumulation of active STEP-ubiquitin conjugates. It is possible that such accumulation of active STEP may contribute, at least in part, to the pathophysiology of AD. Results from previous studies found that Aβ-induced activation of STEP leads to dephosphorylation of NR2B-NMDA receptor subunits and is responsible for endocytosis of functional NMDA receptors [13]. This raises the possibility that disruption of normal NMDAR trafficking as a result of Aβ-mediated accumulation of active STEP may contribute to the synaptic dysfunction as observed in AD.
Thus, taken together with the previous findings, the work presented here suggest that the KIS domain plays a role in maintaining the stability of STEP, and furthermore that dephosphorylation of the two SP/TP sites within the KIS domain is essential for the targeted degradation of active STEP by the ubiquitin-proteasomal pathway. Such degradation of active STEP may be essential for the normal turn over of the protein. However failure of the proteasomal degradation machinery and subsequent accumulation of active STEP-ubiquitin conjugate may contribute to the patho-physiology of a range of neurodegenerative disorders.
Supplementary Material
Acknowledgments
We would like to thank Dr. William Shuttleworth, University of New Mexico for his helpful comments.
FUNDING
This work was supported by grants from the National Institute of Health [grant number NS059962] to Paul, S.
Abbreviations used
- STEP
striatal-enriched phosphatase
- NMDA
N-methyl-D-aspartic acid
- KIM
kinase interacting motif
- KIS
kinase specificity sequence
- ERK
extracellular regulated kinase
- PKA
protein kinase A
- Ser
serine
- Thr
threonine
- PCR
polymerase chain reaction
References
- 1.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21:7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tonks NK, Neel BG. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr Opin Cell Biol. 2001;13:182–195. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 3.Lombroso PJ, Naegele JR, Sharma E, Lerner M. A protein tyrosine phosphatase expressed within dopaminoceptive neurons of the basal ganglia and related structures. J Neurosci. 1993;13:3064–3074. doi: 10.1523/JNEUROSCI.13-07-03064.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulanger LM, Lombroso PJ, Raghunathan A, During MJ, Wahle P, Naegele JR. Cellular and molecular characterization of a brain-enriched protein tyrosine phosphatase. J Neurosci. 1995;15:1532–1544. doi: 10.1523/JNEUROSCI.15-02-01532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bult A, Zhao F, Dirkx R, Jr, Raghunathan A, Solimena M, Lombroso PJ. STEP: a family of brain-enriched PTPs. Alternative splicing produces transmembrane, cytosolic and truncated isoforms. EJCB. 1997;72:337–344. [PubMed] [Google Scholar]
- 6.Bult A, Zhao F, Dirkx R, Jr, Sharma E, Lukacsi E, Solimena M, Naegele JR, Lombroso PJ. STEP61: a member of a family of brain-enriched PTPs is localized to the endoplasmic reticulum. J Neurosci. 1996;16:7821–7831. doi: 10.1523/JNEUROSCI.16-24-07821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pulido R, Zuniga A, Ullrich A. PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul S, Snyder GL, Yokakura H, Picciotto MR, Nairn AC, Lombroso PJ. The Dopamine/D1 receptor mediates the phosphorylation and inactivation of the protein tyrosine phosphatase STEP via a PKA-dependent pathway. J Neurosci. 2000;20:5630–5638. doi: 10.1523/JNEUROSCI.20-15-05630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul S, Nairn AC, Wang P, Lombroso PJ. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat Neurosci. 2003;6:34–42. doi: 10.1038/nn989. [DOI] [PubMed] [Google Scholar]
- 10.Paul S, Olausson P, Venkitaramani DV, Ruchkina I, Moran TD, Tronson N, Mills E, Hakim S, Salter MW, Taylor JR, Lombroso PJ. The striatal-enriched protein tyrosine phosphatase gates long-term potentiation and fear memory in the lateral amygdala. Biol Psychiatry. 2007;61:1049–1061. doi: 10.1016/j.biopsych.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen TH, Liu J, Lombroso PJ. Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J Biol Chem. 2002;277:24274–24279. doi: 10.1074/jbc.M111683200. [DOI] [PubMed] [Google Scholar]
- 13.Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, Choi EY, Nairn AC, Salter MW, Lombroso PJ, Gouras GK, Greengard P. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 14.Poddar R, Deb I, Mukherjee S, Paul S. NR2B-NMDA receptor mediated modulation of the tyrosine phosphatase STEP regulates glutamate induced neuronal cell death. J Neurochem. 2010;115:1350–1362. doi: 10.1111/j.1471-4159.2010.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Kurup P, Zhang Y, Goebel-Goody SM, Wu PH, Hawasli AH, Baum ML, Bibb JA, Lombroso PJ. Extrasynaptic NMDA receptors couple preferentially to excitotoxicity via calpain-mediated cleavage of STEP. J Neurosci. 2009;29:9330–9343. doi: 10.1523/JNEUROSCI.2212-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munoz JJ, Tarrega C, Blanco-Aparicio C, Pulido R. Differential interaction of the tyrosine phosphatases PTP-SL, STEP and HePTP with the mitogen-activated protein kinases ERK1/2 and p38alpha is determined by a kinase specificity sequence and influenced by reducing agents. Biochem J. 2003;372:193–201. doi: 10.1042/BJ20021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deb I, Poddar R, Paul S. Oxidative stress-induced oligomerization inhibits the activity of the non-receptor tyrosine phosphatase STEP61. J Neurochem. 2011;116:1097–1111. doi: 10.1111/j.1471-4159.2010.07165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang PY, Liu P, Weng J, Sontag E, Anderson RG. A cholesterol-regulated PP2A/HePTP complex with dual specificity ERK1/2 phosphatase activity. EMBO J. 2003;22:2658–2667. doi: 10.1093/emboj/cdg255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan PM, Lim L, Manser E. PAK is regulated by PI3K, PIX, CDC42, and PP2Calpha and mediates focal adhesion turnover in the hyperosmotic stress-induced p38 pathway. J Biol Chem. 2008;283:24949–24961. doi: 10.1074/jbc.M801728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito Y, Tsubuki S, Ito H, Kawashima S. The structure-function relationship between peptide aldehyde derivatives on initiation of neurite outgrowth in PC12h cells. Neurosci Lett. 1990;120:1–4. doi: 10.1016/0304-3940(90)90153-z. [DOI] [PubMed] [Google Scholar]
- 21.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 23.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 24.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci USA. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499–504. doi: 10.1016/s1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- 27.Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14:1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- 28.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, Goodman CS. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 29.Hegde AN, DiAntonio A. Ubiquitin and the synapse. Nat Rev. 2002;3:854–861. doi: 10.1038/nrn961. [DOI] [PubMed] [Google Scholar]
- 30.Patrick GN, Zhou P, Kwon YT, Howley PM, Tsai LH. p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. J Biol Chem. 1998;273:24057–24064. doi: 10.1074/jbc.273.37.24057. [DOI] [PubMed] [Google Scholar]
- 31.Jurd R, Thornton C, Wang J, Luong K, Phamluong K, Kharazia V, Gibb SL, Ron D. Mind bomb-2 is an E3 ligase that ubiquitinates the N-methyl-D-aspartate receptor NR2B subunit in a phosphorylation-dependent manner. J Biol Chem. 2008;283:301–310. doi: 10.1074/jbc.M705580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Musti AM, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 33.Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 34.Meimoun A, Holtzman T, Weissman Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 36.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Kim HS, Lee SJ, Kim KT. Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. J Cell Sci. 2007;120:2259–2271. doi: 10.1242/jcs.03468. [DOI] [PubMed] [Google Scholar]
- 38.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 39.Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci. 2004;24:916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 41.Bingol B, Schuman EM. A proteasome-sensitive connection between PSD-95 and GluR1 endocytosis. Neuropharmacology. 2004;47:755–763. doi: 10.1016/j.neuropharm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 42.Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelkey KA, Askalan R, Paul S, Kalia LV, Nguyen TH, Pitcher GM, Salter MW, Lombroso PJ. Tyrosine phosphatase STEP is a tonic brake on induction of long-term potentiation. Neuron. 2002;34:127–138. doi: 10.1016/s0896-6273(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 44.Miller RJ, Wilson SM. Neurological disease: UPS stops delivering! Trends Pharmacol. Sci. 2003;24:18–23. doi: 10.1016/s0165-6147(02)00011-1. [DOI] [PubMed] [Google Scholar]
- 45.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 46.van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, Sluijs JA, Koycu S, Ramdjielal RD, Salehi A, Martens GJ, Grosveld FG, Peter J, Burbach H, Hol EM. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer’s and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 47.McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat Rev. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- 48.Gu Z, Nakamura T, Lipton SA. Redox reactions induced by nitrosative stress mediate protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Mol Neurobiol. 2010;41:55–72. doi: 10.1007/s12035-010-8113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith MA, Rottkamp CA, Nunomura A, Raina AK, Perry G. Oxidative stress in Alzheimer’s disease. Biochem Biophys Acta. 2000;1502:139–144. doi: 10.1016/s0925-4439(00)00040-5. [DOI] [PubMed] [Google Scholar]
- 50.Kurup P, Zhang Y, Xu J, Venkitaramani DV, Haroutunian V, Greengard P, Nairn AC, Lombroso PJ. Abeta-mediated NMDA receptor endocytosis in Alzheimer’s disease involves ubiquitination of the tyrosine phosphatase STEP61. J Neurosci. 2010;30:5948–5957. doi: 10.1523/JNEUROSCI.0157-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.