Abstract
The non-ABC transport protein RalBP1 has been shown to be overexpressed in various cancer cell lines and implicated in the process of metastasis formation, but its expression in tissue samples and prognostic significance has not been shown. In this study matched tumor-mucosa tissue samples from 78 CRC patients were investigated. The RalBP1 mRNA and protein levels were quantified by real-time quantitative PCR (qPCR) and ELISA. RalBP1 was found to be overexpressed in tumor at the mRNA level both overall (p = 0.027), and for stages I (p = 0.024), II (p = 0.038) and IV (p = 0.004). At the protein level, RalBP1 was only significantly overexpressed in stage IV patients (p = 0.018). Expression of RalBP1 mRNA and protein were inversely correlated (r = 0.4173; p = 0.0004). Multivariate Cox regression analysis including sex, age, stage, grade, and nodal status as covariates showed that overexpression of RalBP1 protein, but not mRNA, was an independent predictor of both decreased disease free survival (p = 0.016, RR = 6.892) and overall survival (p = 0.039, RR = 5.986). These results suggest that RalBP1 protein is an independent predictor of poor survival and early relapse for CRC patients. Owing to its multifunctional intermediary role in cell survival, chemotherapeutic resistance, and metastasis formation, RalBP1 represents a promising novel therapeutic target.
Keywords: colorectal cancer, metastasis, prognosis, RalBP1, survival
Introduction
RalA and RalB are nearly identical proteins (85% amino acid identity) within the Ras family of monomeric G proteins which, in addition to normal cellular functions, contribute to cancer cell migration, chemotherapy resistance, invasion and metastasis.4-9 The best-characterized Ral effector is Ral-binding protein (RalBP1), whose association with Ral-mediated tumorigenesis has previously been suggested.10 RalBP1 is a multifunctional membrane protein that has been implicated in cancer cell proliferation,11 radiation and chemoresistance,12-16 and ligand dependent receptor internalization.17-19 Recent studies have shown that overexpression of RalBP1 enhanced migration and invasion of fibrosarcoma cells,20 whereas depletion inhibited tumor growth and metastasis formation in prostate and bladder metastasis models.11 Depletion or inhibition of RalBP1 increased apoptosis in cultured cells from various malignancies, such as small cell lung cancer, non-small cell lung cancer, melanoma, ovarian cancer, prostate cancer, lymphoma, and myeloid leukemia.21 Xenograft models in which RalBP1 was depleted by antisense caused regression of commercial cell lines of lung and colon cancer.21 In summary, RalBP1 may be a promising therapeutic target for cancer therapy owing to its intermediary signaling role in affecting cellular migration, metastasis formation, and chemotherapeutic resistance. The objective of the present study is to establish the relative baseline RalBP1 expression levels in matched human CRC and normal mucosa tissue.
Results
Quantitative RT-PCR Analysis for RalBP1 mRNA Expression
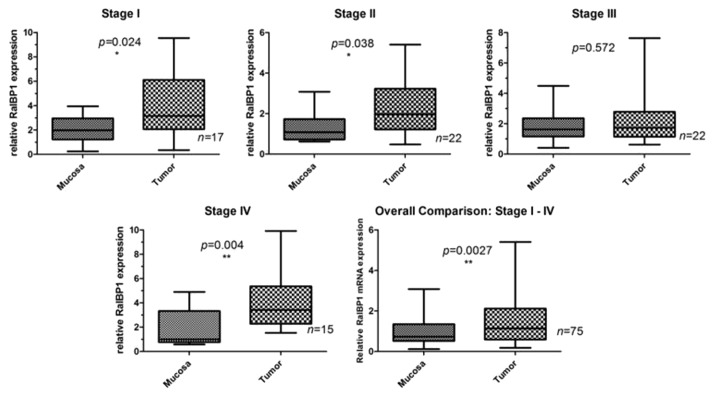
Quantitative RT-PCR (qPCR) analysis showed mRNA expression for RalBP1 in all tumor and mucosa samples; however, marked differences in mRNA levels were seen between normal mucosa and tumor samples for RalBP1 (Fig. 1). Pairwise comparison demonstrated that 70.0% (53/76) overall, 82.4% (14/17) in stage I, 63.6% (14/22) in stage II, 54.5% (12/22) in stage III, and 86.7% (13/15) of patients in stage IV overexpressed RalBP1 in tumor as compared with mucosa. Significantly higher RalBP1 mRNA levels were observed in tumor than in normal mucosa for patients in UICC stage I (p = 0.024), stage II (p = 0.038), and stage IV (p = 0.004). When grouping all stages, RalBP1 mRNA was significantly overexpressed in tumor when compared with normal mucosa (p = 0.027).
Figure 1. Summary of RalBP1 mRNA expression in normal mucosa and tumor by both stage and overall. mRNA levels are expressed as a relative ratio according to the Equation 2-ΔΔCT.
ELISA analysis for RalBP1 protein expression
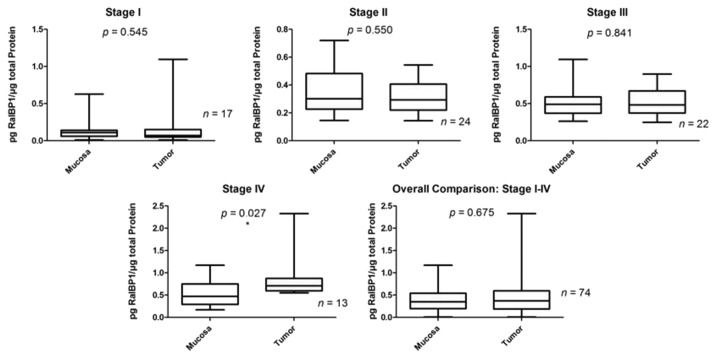
All of the tumor and mucosa samples were found to be positive for RalBP1 measured by ELISA. Pairwise comparison demonstrated that 50.0% (37/74) overall, 17.6% (3/17) in stage I, 37.5% (9/24) in stage II, 54.5% (12/22) in stage III, and 100% (13/13) of patients in stage IV overexpressed RalBP1 in tumor as compared with mucosa. Significantly higher RalBP1 protein levels were observed in tumor than in normal mucosa only for stage IV patients (p = 0.018, Fig. 2).
Figure 2. Summary of RalBP1 protein expression in normal mucosa and tumor by both stage and overall by ELISA method.
RalBP1 expression relationship
By crosstable-calculation (χ2-test), we found RalBP1 protein overexpression to have a significant correlation with nodal status (p = 0.044), UICC stage (p < 0.001), recurrence (p < 0.001), and death (p = 0.001, Table 1). There were no significant correlations between RalBP1 mRNA overexpression and any of the clinicopathologic parameters.
Table 1. Correlation between RalBP1 expression and clinical and pathologic parameters (X2-test).
| Characteristics | RalBP1 mRNA |
RalBP1 Protein |
||||
|---|---|---|---|---|---|---|
| Overexpressed | Not overexpressed | p value | Overexpressed | Not overexpressed | p value | |
|
Gender |
|
|
0.512 |
|
|
0.876 |
|
Male |
31 |
16 |
|
22 |
23 |
|
|
Female |
22 |
7 |
|
14 |
12 |
|
|
Age |
|
|
0.454 |
|
|
0.701 |
|
> Mean |
28 |
15 |
|
20 |
22 |
|
|
≤ Mean |
25 |
8 |
|
16 |
13 |
|
|
Lymph node status |
|
|
0.355 |
|
|
0.044 |
|
N0 |
31 |
11 |
|
15 |
26 |
|
|
N1 |
12 |
4 |
|
11 |
5 |
|
|
N2 |
9 |
8 |
|
9 |
4 |
|
|
N3 |
1 |
0 |
|
1 |
0 |
|
|
Differentiation status |
|
|
0.791 |
|
|
0.305 |
|
Well |
1 |
1 |
|
0 |
2 |
|
|
Moderate |
36 |
16 |
|
23 |
23 |
|
|
Poor |
16 |
6 |
|
13 |
10 |
|
|
UICC Stage |
|
|
0.207 |
|
|
< 0.001 |
|
I |
14 |
3 |
|
3 |
12 |
|
|
II |
14 |
8 |
|
9 |
14 |
|
|
III |
12 |
10 |
|
12 |
9 |
|
|
IV |
13 |
2 |
|
12 |
0 |
|
|
Recurrencea |
|
|
0.633 |
|
|
< 0.001 |
|
Yes |
16 |
5 |
|
18 |
2 |
|
|
No |
37 |
18 |
|
18 |
33 |
|
|
Deatha |
|
|
0.536 |
|
|
0.001 |
|
Yes |
42 |
16 |
|
21 |
33 |
|
| No | 11 | 7 | 15 | 2 | ||
UICC, Unio Internationalis Contra Cancrum; statistically significant p values are highlighted in bold; arecurrence or death at any time
Spearman correlation coefficient analysis showed a statistically significant direct correlation between mRNA Cp (crossing point) values and tissue protein levels for both tumor (r = 0.4173; p = 0.0004) and mucosa (r = 0.2933; p = 0.0160).
Kaplan-Meier Survival Curves
Survival curves were compared via the log rank test, and the corresponding p values are printed on each graph (Figs. 3 and 4). Survival probabilities are comparable for patients with and without mRNA overexpression (DFS: p = 0.595; OS: p = 0.490). Patients with protein overexpression have lower survival probabilities at each time‐point (DFS: p < 0.001; OS: p = 0.002). As expected, more advanced colorectal cancer stages have lower survival rates (DFS: p < 0.001; OS: p < 0.001). When analyzing prognostic factors for disease-free and OS, univariate analysis using the log-rank test confirmed well-known prognostic parameters such as lymph node status (p < 0.001), UICC tumor stage (p < 0.001) and grade (p = 0.026, Table 2) to be of prognostic relevance in our patient cohort. Regarding RalBP1 expression, median DFS and OS were significantly reduced in tumors with protein overexpression. In a multivariate analysis based on the Cox proportional hazards regression model, we tested the independent predictive value for all relevant clinical and pathological parameters and RalBP1 expression. Lymph node status was excluded for its linear depending covariance with tumor stage. The group of patients who overexpressed RalBP1 protein expression had significantly worse DFS (p = 0.016, HR = 6.892) and OS (p = 0.039, HR = 5.986) at each time point, whereas mRNA expression had no effect on DFS or OS (Table 3).
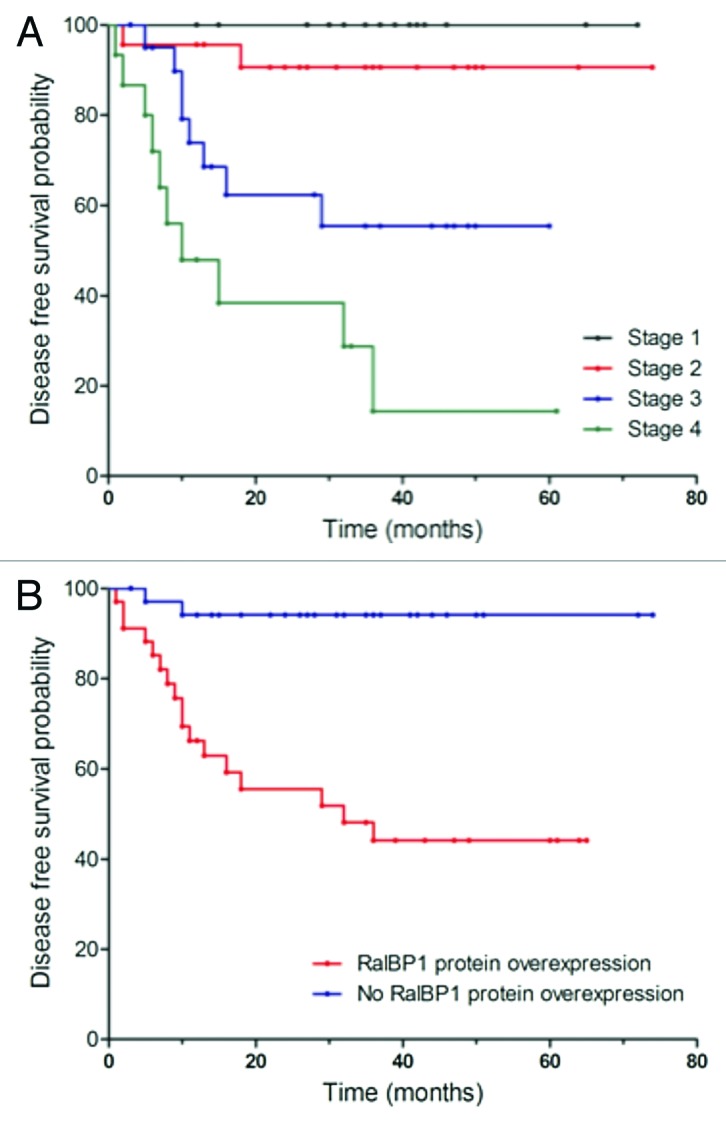
Figure 3. Univariate analysis (log-rank test, Kaplan-Meier curves) of statistically significant prognostic parameters for disease free survival in colorectal cancer. (A) Stage; (B) RalBP1 protein overexpression.
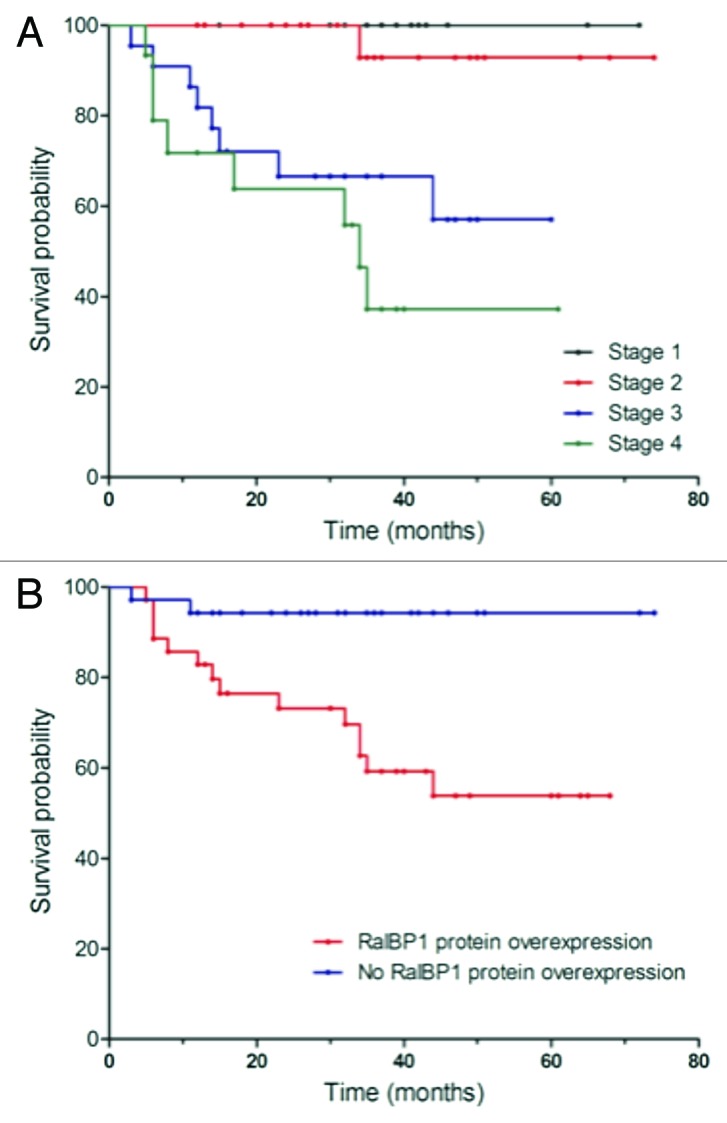
Figure 4. Univariate analysis (log-rank test, Kaplan-Meier curves) of statistically significant prognostic parameters for overall survival in colorectal cancer. (A) Stage; (B) RalBP1 protein overexpression.
Table 2. Univariate analysis (log-rank test) of prognostic parameters in colorectal cancer for progression-free survival and overall survival.
| Characteristics | Cases | Progression free survival | Overall survival | ||
|---|---|---|---|---|---|
|
Time (months) |
p value |
Time (months) |
p value |
||
|
Gender |
|
|
0.921 |
|
0.947 |
|
Male |
48 |
38.4 |
|
29.2 |
|
|
Female |
30 |
30.0 |
|
23.6 |
|
|
Age |
|
|
0.062 |
|
0.375 |
|
> Mean |
45 |
34.2 |
|
12.9 |
|
|
≤ Mean |
33 |
31.2 |
|
30.0 |
|
|
Lymph node status |
|
|
< 0.001 |
|
< 0.001 |
|
Negative |
44 |
33.3 |
|
30.3 |
|
|
Positive |
34 |
31.0 |
|
22.0 |
|
|
Differentiation status |
|
|
0.025 |
|
0.120 |
|
Well/Moderate |
55 |
|
|
27.3 |
|
|
Poor |
23 |
|
|
25.7 |
|
|
UICC stagea |
|
|
< 0.001 |
|
< 0.001 |
|
II |
24 |
34.0 |
|
17.3 |
|
|
III |
22 |
33.4 |
|
21.1 |
|
|
IV |
15 |
25.1 |
|
17.9 |
|
|
RalBP1 mRNA |
|
|
0.490 |
|
0.595 |
|
Overexpressed |
53 |
38.0 |
|
28.6 |
|
|
Normal expression |
23 |
26.5 |
|
14.4 |
|
|
RalBP1 Protein |
|
|
0.002 |
|
< 0.001 |
|
Overexpressed |
36 |
10.7 |
|
9.9 |
|
| Normal expression | 35 | 33.7 | 22.8 | ||
UICC, Unio Internationalis Contra Cancrum; a there were no stage I patients who experienced either recurrence or death at time of last follow-up; statistically significant p values are highlighted in bold
Table 3. Multivariate analysis (Cox proportional hazards regression model) of prognostic parameters in colorectal cancer for progression-free survival and overall survival.
| |
Progression Free Survival |
Overall Survival |
||||
|---|---|---|---|---|---|---|
| Parameter | Hazard ratio | 95% CI | p value | Hazard ratio | 95% CI | p value |
|
Gender |
0.566 |
0.217–1.476 |
0.2441 |
0.491 |
0.172–1.406 |
0.1853 |
|
Age |
0.998 |
0.957–1.040 |
0.9207 |
0.965 |
0.921–1.011 |
0.1301 |
|
Differentiation status |
0.825 |
0.318–2.138 |
0.6921 |
0.582 |
0.204–1.656 |
0.3104 |
|
UICC stage |
8.811 |
1.847–42.047 |
0.0063 |
17.348 |
1.953–33.125 |
0.0105 |
|
RalBP1 mRNA expression |
0.709 |
0.233–2.160 |
0.5449 |
0.276 |
0.083–0.920 |
0.0797 |
| RalBP1 protein expression | 6.892 | 1.429–33.249 | 0.0162 | 5.986 | 1.094–32.766 | 0.0391 |
UICC, Unio Internationalis Contra Cancrum; CI, confidence intervals; statistically significant p values are highlighted in bold
Discussion
Multivariate analysis found that overexpression of RalBP1 protein was an independent predictor of both decreased DFS and OS in CRC patients. To our knowledge, our results are the first to show that RalBP1 protein expression provides prognostic information for CRC patients. From a functional point of view, these results could be explained by the intermediary signaling role that RalBP1 plays in metastasis formation, and its protective role against oxidative stress metabolites and xenobiotics.
A number of studies have demonstrated the role of RalBP1 in cellular migration and metastasis. Wu et al. showed that RalBP1 depletion not only inhibited cell migration, but also inhibited metastasis formation in an experimental metastasis model of bladder cancer.11 RalBP1 has also been shown to act upon CDK1 (cdc42), a protein known to be involved in regulating cell cycle progression and migration.27 After binding to CDK1, RalBP1 dissociates from the cell membrane to act as a motor for spindle movement at the mitotic spindle.28,29 Our data demonstrated a significant correlation with RalBP1 protein overexpression, increasing stage, and nodal positivity; providing further evidence for the role of RalBP1 in invasion and metastasis formation.
RalBP1 is an ATP-dependent non-ABC transporter which actively transports chemotherapeutic agents, in addition to anionic metabolites like glutathione-conjugates of electrophiles (GS-E).35,36 Studies performed in RalBP1 −/− mice showed a significant increase in the concentration of aldehydes, lipid hydroperoxides, and alkenals in tissues as a consequence of RalBP1 loss.36 This loss translates into greater sensitivity to xenobiotic toxins including traditional chemotherapeutic agents, which are substrates of RalBP1, as well as other alkylating agents and platinum-coordinates that are metabolized to GS-E. Induction of RalBP1 thus results in cellular resistance to apoptosis by metabolizing and excluding stress metabolites at a higher rate.37
Although RalBP1 mRNA was overexpressed in tumor vs. mucosa, it was not a significant predictor of either disease-free or overall survival. Indeed, there was an inverse correlation demonstrated between RalBP1 mRNA and protein expression. Furthermore, as RalBP1 mRNA transcript amounts decreased with increasing stage (data not shown), RalBP1 protein amounts increased as stage increased. Oncogenes have been demonstrated to co-opt the translational process during transformation, and interestingly RalBP1 has been linked to translational regulation of some mRNAs.38 Panner et al. demonstrated that RalA binding of RalBP1 induced suppression of S6 kinase and the translation of the antiapoptotic FLICE-like inhibitory protein. Further research will be needed to determine the mechanism by which the translation of RalBP1 begins to occur at an accelerated rate during tumorigenesis.
In conclusion, overexpression of the RalBP1 protein is an independent predictor of poor survival and early relapse for CRC patients. Owing to its multifunctional intermediary role in cell survival, chemotherapeutic resistance, and metastasis formation, RalBP1 represents a promising novel therapeutic target.
Materials and Methods
Patients and samples
Matched tissue samples (histologically proven) of normal colon mucosa and tumor were obtained from the surgical specimens of 78 patients with curatively resected (R0) primary sporadic colon adenocarcinomas treated between February 2004 and April 2007 at the Department of Surgery, University of Heidelberg in accordance with the ethics committee. Tissue samples were frozen in liquid nitrogen immediately after surgical removal and maintained at 80°C until RNA extraction. Clinical and pathological data were documented prospectively, and entered into a specific tumor registry at the time of surgery and at each follow-up. The patients’ ages ranged from 39 to 90 y (mean age, 65 y). Median follow-up time of patients alive at last follow-up was 35 mo.3 The tumors were staged according to the Unio Internationalis Contra Cancrum (UICC) system with the following distribution: 21.8% stage I (17/78), 30.8% stage II (24/78), 28.2% stage III (22/78), and 19.2% stage IV (15/78) patients.
RNA extraction and qPCR
Fresh tumor and mucosa tissue were frozen in RNAlater Reagent (Invitrogen) and stored at -80°C. Approximately 20 mg of tissue was used for each RNA extraction. SV total RNA isolation (Promega) was performed, followed by ImProm-II reverse transcription (Promega) for cDNA synthesis. The cDNA was amplified with real-time quantitative polymerase chain reaction (qPCR) and was performed with the LightCycler 2.0 Real-Time PCR system (Roche Applied Science). qPCR conditions for RalBP1 were as follows: initial denaturation for 5 min at 95°C, 35 cycles (95°Cfor 30 sec, annealing at 55°C for 45 sec, 30 sec at 72°C), followed by 15 min at 72°C; incubated with appropriate forward (5′- TCTATAGTGCTCAGCCCAAC-3′) and reverse (5′-ATCGCAGAGGTTTCATCAC-3′) primers (Invitrogen). cDNA was also amplified with β-actin primers (ACTB) forward (5′-ATGTGGCCGAGGACTTTGATT-3′) and reverse (5′-AGTGGGGTGGTTTTAGGATG-3′) (Invitrogen), serving as an internal control. Threshold cycle (Ct) and melting curves were acquired. Only genes with clear and single melting peaks were taken for further data analysis. Samples with irregular melting peaks were excluded from the calculation. The threshold was set manually, using identical threshold levels for one gene in all analyzed samples. Reaction efficiency was established for each set of primers, after quantification of four different dilutions of a reference cDNA.
Analysis of RalBP1 expression using the 2 -ΔΔCt method
Details of the 2 -ΔΔCt method have been previously described.23,24 Briefly, the mean target gene mRNA expression level for the three mRNA measurements was calculated. The 2-ΔΔCt method was used to calculate relative changes in gene expression determined from real-time quantitative PCR experiments. In the present study, the data are presented as the fold change in target genes RalBP1 expression in tumors normalized to the internal control gene (ACTB) and relative to the normal control (matched normal as calibrator). Results of the qPCR data were represented as threshold cycle (Ct) values, where Ct was defined as the threshold cycle number of PCR at which amplified product was first detected. There is an inverse correlation between Ct and amount of target: lower amounts of target correspond to a higher Ct value. The average Ct was calculated for both the target genes and ACTB and the ΔCt was determined as (the mean of the triplicate Ct values for the target gene) minus (the mean of the triplicate Ct values for ACTB). The ΔΔCt represented the difference between the paired tissue samples, as calculated by the formula ΔΔCt = (ΔCt of tumor - ΔCT of normal). The differential expression in the target gene of a tumor sample compared with the normal counterpart was expressed as 2-ΔΔCt.23,24
Protein extraction and ELISA
Tissue extracts were prepared from frozen tissues by a standard extraction protocol.25 The protein content of cell lysates and tissue extracts was determined using the Lowry protein assay (Sigma). The enzyme-linked immunosorbent assay (ELISA)Kit for human RalBP1 was purchased from USCN Life Science, Inc. (E97265Hu) and performed as indicated by the manual. The standard was reconstituted and a dilution series was made with the standard serving as a blank. The blank and samples were loaded and incubated for 2 h at 37°C. Every item of the standard dilution series, the blank and samples were loaded as duplicates. The liquid of each well was removed and a detection reagent was added and incubated for 1 h at 37°C. The liquids were removed and washed (Wash Solution and aqua dest) 3 times after the incubation was finished. A second detection reagent was added to each well and incubated for 30 min at 37°C, after which the samples were washed 5 times as described above. The detection procedure continued by adding 90 µl Substrate Solution to each well and incubation for 15–20 min at 37°C. The microplate was immediately measured by 450 nm in a standard ELISA microplate reader.
Statistical Analysis
All statistical analyses were performed using Statistical Analysis Systems (SAS) (SAS Corp, NC). The t-test was used to compare all continuous parameters from normal vs. tumor samples, and the χ2 test was used for comparison of discrete parameters. The relationship between RalBP1 protein and mRNA expression levels within the same samples was examined using the Spearman correlation coefficient analysis. The Kaplan-Meier method was used to estimate the survival probability, and the log-rank test was used to compare the survival curves between groups. Independent predictive factors affecting survival were analyzed by the Cox multivariate proportional hazards regression model with stepwise and enter models, respectively. All p values were two-sided and considered statistically significant if p < 0.05. Overall survival (OS) was defined as the time interval between the date of surgery and the date of death or last follow-up. Disease free survival (DFS) was defined as the time interval between the date of surgery and the date of disease recurrence or death from any cause, whichever came first, or date of last follow-up evaluation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/20087
References
- 1.Camonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–32. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–25. doi: 10.1016/S0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 3.van Dam EM, Robinson PJ. Ral: mediator of membrane trafficking. Int J Biochem Cell Biol. 2006;38:1841–7. doi: 10.1016/j.biocel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Rossé C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–34. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 6.Yin J, Pollock C, Tracy K, et al. Activation of the RalGEF/Ral Pathway Promotes Prostate Cancer Metastasis to Bone. Mol Cell Biol. 2007. [DOI] [PMC free article] [PubMed]
- 7.Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–94. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Tchevkina E, Agapova L, Dyakova N, Martinjuk A, Komelkov A, Tatosyan A. The small G-protein RalA stimulates metastasis of transformed cells. Oncogene. 2005;24:329–35. doi: 10.1038/sj.onc.1208094. [DOI] [PubMed] [Google Scholar]
- 9.Ward Y, Wang W, Woodhouse E, Linnoila I, Liotta L, Kelly K. Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol Cell Biol. 2001;21:5958–69. doi: 10.1128/MCB.21.17.5958-5969.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–45. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 2010;12:1003–12. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awasthi S, Singhal SS, Yadav S, Singhal J, Drake K, Nadkar A, et al. RLIP76 is a major determinant of radiation sensitivity. Cancer Res. 2005;65:6022–8. doi: 10.1158/0008-5472.CAN-05-0968. [DOI] [PubMed] [Google Scholar]
- 13.Awasthi S, Singhal SS, Sharma R, Zimniak P, Awasthi YC. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RALBP1): a novel link between G-protein and tyrosine kinase signaling and drug resistance. Int J Cancer. 2003;106:635–46. doi: 10.1002/ijc.11260. [DOI] [PubMed] [Google Scholar]
- 14.Awasthi S, Singhal SS, Singhal J, Cheng J, Zimniak P, Awasthi YC. Role of RLIP76 in lung cancer doxorubicin resistance: II. Doxorubicin transport in lung cancer by RLIP76. Int J Oncol. 2003;22:713–20. [PubMed] [Google Scholar]
- 15.Awasthi S, Singhal SS, Singhal J, Yang Y, Zimniak P, Awasthi YC. Role of RLIP76 in lung cancer doxorubicin resistance: III. Anti-RLIP76 antibodies trigger apoptosis in lung cancer cells and synergistically increase doxorubicin cytotoxicity. Int J Oncol. 2003;22:721–32. [PubMed] [Google Scholar]
- 16.Stuckler D, Singhal J, Singhal SS, Yadav S, Awasthi YC, Awasthi S. RLIP76 transports vinorelbine and mediates drug resistance in non-small cell lung cancer. Cancer Res. 2005;65:991–8. [PubMed] [Google Scholar]
- 17.Morinaka K, Koyama S, Nakashima S, Hinoi T, Okawa K, Iwamatsu A, et al. Epsin binds to the EH domain of POB1 and regulates receptor-mediated endocytosis. Oncogene. 1999;18:5915–22. doi: 10.1038/sj.onc.1202974. [DOI] [PubMed] [Google Scholar]
- 18.Nakashima S, Morinaka K, Koyama S, Ikeda M, Kishida M, Okawa K, et al. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–42. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jullien-Flores V, Mahé Y, Mirey G, Leprince C, Meunier-Bisceuil B, Sorkin A, et al. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J Cell Sci. 2000;113:2837–44. doi: 10.1242/jcs.113.16.2837. [DOI] [PubMed] [Google Scholar]
- 20.Coon BG, Burgner J, Camonis JH, Aguilar RC. The epsin family of endocytic adaptors promotes fibrosarcoma migration and invasion. J Biol Chem. 2010;285:33073–81. doi: 10.1074/jbc.M110.124123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singhal SS, Awasthi YC, Awasthi S. Regression of melanoma in a murine model by RLIP76 depletion. Cancer Res. 2006;66:2354–60. doi: 10.1158/0008-5472.CAN-05-3534. [DOI] [PubMed] [Google Scholar]
- 22.Singhal SS, Singhal J, Yadav S, Dwivedi S, Boor PJ, Awasthi YC, et al. Regression of lung and colon cancer xenografts by depleting or inhibiting RLIP76 (Ral-binding protein 1) Cancer Res. 2007;67:4382–9. doi: 10.1158/0008-5472.CAN-06-4124. [DOI] [PubMed] [Google Scholar]
- 23.ABI. User bulletin No. 2ABI prism 7700 Sequence Detection System. 1997. Relative quantitation of gene expression.
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual, Ed. 1. Cold Spring Harbor Laboratory Press, New York, 1989. [Google Scholar]
- 26.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J Cell Biol. 2006;174:877–88. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, et al. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–7. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 28.Rossé C, L’Hoste S, Offner N, Picard A, Camonis JH. RLIP, an effector of the Ral GTPases, is a platform for Cdk1 to phosphorylate epsin during the switch off of endocytosis in mitosis. J Biol Chem. 2003;278:30597–604. doi: 10.1074/jbc.M302191200. [DOI] [PubMed] [Google Scholar]
- 29.Singhal SS, Yadav S, Vatsyayan R, Chaudhary P, Borvak J, Singhal J, et al. Increased expression of cdc2 inhibits transport function of RLIP76 and promotes apoptosis. Cancer Lett. 2009;283:152–8. doi: 10.1016/j.canlet.2009.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–15. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awasthi S, Singhal SS, Singhal J, Cheng J, Zimniak P, Awasthi YC. Role of RLIP76 in lung cancer doxorubicin resistance: II. Doxorubicin transport in lung cancer by RLIP76. Int J Oncol. 2003;22:713–20. [PubMed] [Google Scholar]
- 32.Stuckler D, Singhal J, Singhal SS, Yadav S, Awasthi YC, Awasthi S. RLIP76 transports vinorelbine and mediates drug resistance in non-small cell lung cancer. Cancer Res. 2005;65:991–8. [PubMed] [Google Scholar]
- 33.Awasthi S, Singhal SS, Srivastava SK, Zimniak P, Bajpai KK, Saxena M, et al. Adenosine triphosphate-dependent transport of doxorubicin, daunomycin, and vinblastine in human tissues by a mechanism distinct from the P-glycoprotein. J Clin Invest. 1994;93:958–65. doi: 10.1172/JCI117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singhal SS, Sehrawat A, Sahu M, Singhal P, Vatsyayan R, Rao Lelsani PC, et al. Rlip76 transports sunitinib and sorafenib and mediates drug resistance in kidney cancer. Int J Cancer. 2010;126:1327–38. doi: 10.1002/ijc.24767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Awasthi S, Cheng J, Singhal SS, Saini MK, Pandya U, Pikula S, et al. Novel function of human RLIP76: ATP-dependent transport of glutathione conjugates and doxorubicin. Biochemistry. 2000;39:9327–34. doi: 10.1021/bi992964c. [DOI] [PubMed] [Google Scholar]
- 36.Awasthi S, Singhal SS, Sharma R, Zimniak P, Awasthi YC. Transport of glutathione conjugates and chemotherapeutic drugs by RLIP76 (RALBP1): a novel link between G-protein and tyrosine kinase signaling and drug resistance. Int J Cancer. 2003;106:635–46. doi: 10.1002/ijc.11260. [DOI] [PubMed] [Google Scholar]
- 37.Cheng JZ, Sharma R, Yang Y, Singhal SS, Sharma A, Saini MK, et al. Accelerated metabolism and exclusion of 4-hydroxynonenal through induction of RLIP76 and hGST5.8 is an early adaptive response of cells to heat and oxidative stress. J Biol Chem. 2001;276:41213–23. doi: 10.1074/jbc.M106838200. [DOI] [PubMed] [Google Scholar]
- 38.Panner A, Nakamura JL, Parsa AT, Rodriguez-Viciana P, Berger MS, Stokoe D, et al. mTOR-independent translational control of the extrinsic cell death pathway by RalA. Mol Cell Biol. 2006;26:7345–57. doi: 10.1128/MCB.00126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]