Abstract
Rho GTPases undergo ubiquitylation and degradation via the ubiquitin-proteasome pathway. We now report in the November issue of Developmental Cell that the E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of GTP-bound Rac1. Depletion of HACE1 leads to an increase of Rac1 activity. We have proposed that HACE1 limits Rac1 activity in cells, a regulation that is usurped by some pathogenic bacteria for efficient invasion of host cell monolayers. We here review these findings in parallel with the regulation of RhoA by the ubiquitin and proteasome system (UPS) and discuss the impact of these regulations on the capacity of Rho GTPases to signal.
Keywords: CNF1, GTPase, HACE1, HECT, Rac, RhoA, Smurf1, toxin, ubiquitin
Introduction
Protein degradation represents the most reliable way to dampen the amplitude and duration of signal transduction pathways.1 For instance, protein degradation by ubiquitin-mediated targeting to the proteasomal machinery plays a crucial role in cell signaling, notably in pathways that control actin cytoskeleton dynamics2,3 and during host pathogen interactions.4 The ubiquitylation reaction consists of the covalent attachment of ubiquitin, an 8-kDa polypeptide, to lysine residues on the target.1 This involves a cascade of transfer reactions between ubiquitin-carrier proteins. Among these factors, the E3 ubiquitin-ligases confer the specificity to the reaction by binding distinctively a panel of target proteins.1 Additional molecules of ubiquitin can be subsequently attached to one of the seven lysines of the previously cross-linked ubiquitin molecule, leading to the formation of various types of poly-ubiquitin chains, notably Lysine-48 (K48)-poly-ubiquitylation for substrate targeting to proteasomal destruction. Posttranslational modifications of proteins by mono- or multi- and poly-ubiquitylation form a repertoire of modifications allowing specific interactions with multiple families of ubiquitin-binding domain containing proteins.5 This system of regulation is completed by the action of de-ubiquitylating enzymes.6 Ubiquitylation is now viewed as a system of molecular barcodes controlling protein sorting at the membrane, as well as local and/or temporal activation of proteins and protein inhibition by degradation.5,7 It is now firmly established that ubiquitylation of small GTPases, notably that of Rho proteins, controls their capacity to signal. Determination of the cellular actors responsible for ubiquitylation of Rho proteins has started to unveil some functions and the importance of this type of posttranslational modification with regard to the canonical GTPase-based spatio-temporal regulation (Fig. 1).
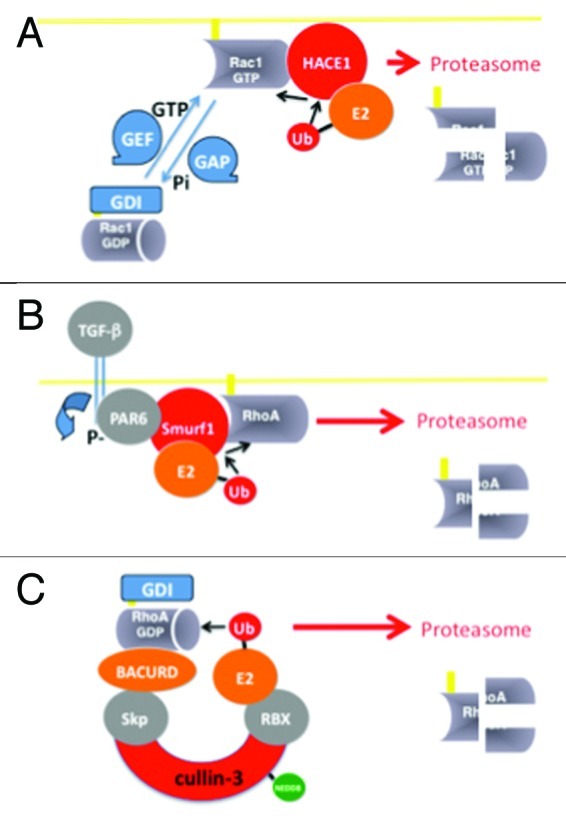
Figure 1. Rho protein ubiquitylation. (A) HACE1 HECT-domain containing protein E3 ubiquitin-ligase binds preferentially to the GTP-bound form of Rac1, and to an ubiquitin conjugated to an E2 enzyme. Ubiquitin molecules are next transferred to a conserved cysteine residue (C876 of HACE1) of the HECT-domain prior to conjugation on Rac1 to form K48-poly-ubiquitin chain, a signal for targeting to the proteasomal machinery. (B) RhoA is a target of Smurf1 HECT-domain containing E3 ubiquitin-ligase. (C) The GDP-bound form of RhoA is recognized by a multi-subunit E3 ubiquitin-ligase homologous to SCF (for Skp1-Cullin-1 and RING-finger), which contains Cullin-3, as scaffold protein, and the BTB-domain containing protein BACURD for specific binding to RhoA. This allows a direct conjugation of ubiquitin to RhoA for subsequent proteasomal destruction.
Rho GTPases have a crucial role in controlling virtually all actin-dependent cellular processes.8,9 The importance of this family of proteins is further established by links unveiled between their dysregulation and several human diseases such as inflammatory and neurological disorders, as well as cancer.10-12 Moreover, this class of proteins is the target of potent virulence factors of highly pathogenic bacteria responsible for infectious diseases of major public health threat and cost, such as Clostridium difficile responsible for antibiotic-induced diarrhea and pseudomembranous colitis, as well as uropathogenic Escherichia coli (UPEC) a leading cause of urinary tract infection and sepsis.13,14
The crucial function of Rho proteins is achieved through their ability to orchestrate the timely local organization of complex arrays of protein-protein interactions.8 Most Rho proteins bind guanosine 5′-diphosphate (GDP) and guanosine 5′-triphosphate (GTP) guanine nucleotides, as well as hydrolyze GTP into GDP.15 Several residues, including the glutamine-61 of Rac1 (Q63 in RhoA), are essential for hydrolysis of the gamma-phosphate of GTP. Transitions between guanine nucleotide-bound forms of Rho proteins produce conformational changes in two flexible regions referred to as the switch I and II regions. Binding to GTP allows the switch I region to bind to and activate downstream effector proteins. Transition between both GTP/GDP forms of Rho is catalyzed by a large number of proteins containing GEF (Guanine nucleotide exchange factors) or GAP (GTPase activating protein) domains.9 Most of these regulatory proteins contain additional functional domains and thus most likely act as regulatory-effectors. Finally, the targeting of Rho proteins to cellular membranes for their activation is regulated by guanine nucleotide dissociation inhibitor (GDI) factors, which retain the GDP-bound GTPase in the cytosol.16 Together, this complex GTPase-based spatiotemporal regulation of small GTPases satisfactorily accounts for their ability to signal. Nevertheless, studies have clearly established that Rho proteins are also subjected to several posttranslational modifications such as ubiquitylation.17 This now raises the question of the implication of these additional modes of regulation in the control of the activity of small GTPases and their possible diversion in human pathologies.
Ubiquitin-modification of Rho Proteins and proteasomal degradation
It is now recognized that in addition to the C-terminal prenylation of Rho proteins for anchoring at the membrane, the activity of Rho GTPases in cells is also modulated by several posttranslational modifications such as ubiquitylation, sumoylation, serotonylation and phosphorylation.17 A microbial centric view indicates that bacteria have recognized for a long time the benefit of modifying small Rho GTPases by posttranslational reactions in order to corrupt host innate responses and successfully invade their host.13,18 These modifications target either the switch-I or -II domain of Rho proteins to block or activate their downstream signaling, respectively.13 The Cytotoxic Necrotizing Factor-1 (CNF1) toxin, for example, is a paradigmatic toxin targeting Rho proteins for activation.14 This toxin is synthesized by a large number of uropathogenic Escherichia coli, a pathogen responsible for urinary tract infections and sepsis.14,19 CNF1 activates Rho proteins by modification of the glutamine 61 of Rac1 (Q63 for RhoA). Indeed, CNF1 catalyzes the deamidation of this glutamine residue into a glutamic acid.20,21 This type of modification impairs the intrinsic and GAP-stimulated GTPase activity of Rac1.22 Consequently, Rac1 remains in an activated GTP-bound form. Two reports in 2002 have demonstrated that once Rac1 is activated, it is ubiquitylated for targeting to proteasomal degradation.3,23 Indeed, the persistent activation of Rac1, resulting from mutations in position Q61 as well as G12, sensitizes this GTPase to K48-poly-ubiquitylation, thereby reducing its stability. Sensitization of Rac1 to ubiquitylation also occurs upon its activation by the GEF domain of Dbl, or following stimulation of the HGF (Hepatocyte Growth Factor) receptor.3,24 We have established a direct correlation between the strength of activation of Rac1 (Rac1-WT < E61 < V12 < L61) and its ubiquitylation efficiency.3,25 Poly-ubiquitylation of Rac1 occurs at cellular membranes and requires functional caveolin-1.25,26 Based on these data it was assumed that activation of Rac1 at the membrane sensitizes this GTPase to ubiquitin-mediated proteasomal degradation. Complementary to these findings it was suggested that RhoA and CDC42 are also targeted to UPS once they are activated3
Ubiquitylation of RhoA for local inhibition
A major turn in deciphering the function of Rho GTPase regulation by UPS came from the discovery that RhoA is a substrate of the C2-WW-HECT domains containing Smurf1 E3 ubiquitin-ligase, a regulator of the TGF-β signaling.2 This study established that the Rac1 and CDC42-effector complex PAR6/PKCzeta controls the local recruitment of Smurf1 in cell protrusions for targeting of RhoA to UPS. Smurf1 binds to the nucleotide-free form of RhoA, known to interact with GEF for GTP loading. Based on these findings, it was assumed that UPS-mediated degradation of RhoA occurs prior to its activation (GTP loading) by GEFs in order to prevent the inhibitory effect that RhoA signaling would have on the formation of membrane protrusions induced by Rac1 and CDC42. Complementary to these findings, it was later shown that Smurf1 also controls the degradation of RhoA for disruption of tight junctions during epithelial-to-mesenchymal transition (EMT) promoted by TGF-β in mammary gland epithelial cells.27 Stimulation of TGF-β receptors induces a recruitment of PAR6 and its phosphorylation by the TGF-β receptor-2 subunit. This allows the recruitment of Smurf1 and consequently the poly-ubiquitylation of RhoA at two acceptor lysines located at the N-terminal part of RhoA. These studies concluded that UPS-mediated degradation of RhoA by Smurf1 likely triggers its local inactivation. Thus in this case, the degradation of RhoA seems to substitute for or act in concert with GAP regulators to provide a tight spatial regulation of Rho signaling. Still, the localization of Rho ubiquitylation remains to be monitored. Smurf1 also participates to the degradation of the cellular pool of RhoA once activated by CNF1.28 Thus, Smurf1 likely participates in the downregulation of RhoA prior to and after its sustained activation. More recently, it was shown that the E3 ubiquitin-ligase activity of Smurf1 is stimulated by release of an autoinhibition between Smurf1-homodimers achieved upon APC/Cdh1 interaction.29 This interaction is independent of APC/Cdh1 E3 ubiquitin-ligase activity, which in other respects plays a key role in mitotic exit and G1/S transition. Consistent with above findings, forced expression of Cdh1, together with RhoA, leads to a poly-ubiquitylation and downregulation of RhoA.29
Ubiquitin-mediated control of Rac1 and RhoA stability and activity
We had previously established that ubiquitin-mediated degradation of RhoA or Rac1 is specifically impaired or inefficient in different immortalized and cancer cell lines, suggesting that several enzymes specifically target Rho protein members to ubiquitin-mediated proteasomal degradation.28 Consistent with these findings, Smurf1 targets RhoA but not Rac1 or CDC42. By conducting a RNAi-based screen of the subgroup of E3 ubiquitin-ligase containing Homologous to E6-associated protein C-Terminus (HECT)-domains, we were able to unveil the key function of HACE1 in the degradation of Rac1.30 This established Rac1 as a first target of HACE1 E3 ubiquitin-ligase activity, a likely important finding considering the protective role of HACE1 in tumorigenesis. In humans, the epigenetic silencing of hace1 has been linked to Wilm’s tumors.31 Genetic inactivation of hace1 in mice induces the development of spontaneous, late-onset cancer of a large variety of origin.33 HACE1 tumor suppressor activity has been linked to its E3 ubiquitin-ligase activity, responsible for indirect degradation of Cyclin-D1 in response to cell stress in order to control anchorage-dependent growth and cell cycle progression.33 Thus, it is interesting to speculate that the tight control of cell cycle requires Rac1 regulation by HACE1, considering for instance that Rac1 controls cyclin-D1 levels for anchorage-dependent cell cycle progression.32 In our recent study, we showed that HACE1 depletion had no effect on RhoA or CDC42 stability. In vitro and in vivo studies show a 2-fold preferential binding of HACE1 to GTP-bound Rac1.30 This suggests that Rac1 binds directly to HACE1 once it is activated. Biochemical assays conducted in vitro suggest a strong specificity of the ubiquitin-ligase activity of HACE1 toward the active GTP-bound form of Rac1.30 Collectively, these findings suggest that the binding of Rac1-GTP to HACE1 stimulates its ubiquitin-ligase activity. Functional studies have also allowed us to determine that HACE1 acts by reducing cellular levels of active Rac1. Both activation and degradation of Rac1 confer advantages to pathogenic bacteria. Proteasomal degradation, by limiting the cellular level of activation of Rac1, allows efficient disruption of intercellular junctions, triggers a polarization of epithelial cells that adopt a migratory phenotype as well as confers epithelial cells efficient phagocytic properties.3 Bacteria likely benefit from these regulations to breach cellular barriers and penetrate into cells to form intracellular bacterial colonies. Consistent with these previous findings, both Rac1 and HACE1 appear to be required for efficient bacterial internalization into endothelial cell monolayers.30 Rac1 degradation may also potentially limit the inflammation resulting from its activation by CNF1, a host response detrimental to bacterial infection.34,35
RhoA shows a high turnover, as compared to Rac1 which is a stable protein.36 The multi-subunit E3 Cullin3-RING ubiquitin-ligase (C3RL), homologous to SCF (Skp1-Cullin-1-F-box protein complex), in association with the BTB-containing substrate binding factor BACURD forms a complex identified as the machinery controlling the degradation of GDP-bound RhoA36 (Fig. 1). CRLs are activated and sorted outside the nucleus upon cross-linking of NEDD8 ubiquitin-like molecule to the cullin subunits.37,38 NEDD8 is targeted for inactivation by several bacterial toxins, notably Cif of enteropathogenic E. coli.39 As a consequence, the resulting default in CRLs activation leads to a massive increase of cellular levels of RhoA, but not RhoB, RhoC or Rac1, and subsequently to formation of thick actin cables.39 Moreover the RNAi-mediated depletion of BACURD produces an increase of cellular levels of RhoA and actin cable formation.36 These findings suggest that in cultured cells the levels of active RhoA/Rac1 are limited by the amount of total GTPases available as substrate for relatively constant GEF/GAP-regulated cycles of GTP loading and hydrolysis.
Rac1 is likely a target of more than one E3 ubiquitin-ligase, as shown for RhoA. A recent study shows that Rac1 binds and is poly-ubiquitylated on its lysine-147 by the Ring-domain containing inhibitors of apoptosis proteins (IAPs) E3 ubiquitin-ligases, c-IAP1 and XIAP.40 The binding of these IAPs to Rac1 is nucleotide-independent. Downregulation of c-IAP1 or XIAP leads to an increase of Rac1 level and activity in cells, promoting cell elongation and migration.
Other potential functions of Rho protein ubiquitylation
One interesting question is whether ubiquitylation might confer new biochemical properties to Rac1. A two-hybrid screen in yeast for ub-Rac1 chimera interacting proteins allowed us to isolate Tollip (Toll and IL1-Receptor interacting protein).41 Tollip comprises an ubiquitin binding domain and a TBD-domain for interaction with the clathrin-interacting protein Tom1.42 Tollip, Tom1 and clathrin localize at the site of internalized bacteria.41 These factors are required for efficient bacterial entry into cells intoxicated by CNF1, or expressing active Rac1, via a β-1 integrin pathway.41 Both Rac1 and ubiquitylated-Rac1 bind to Tollip.41 These findings raise the interesting question of whether the ubiquitylation of Rac1 might modulate Tollip interactions. For example, it has been reported that ubiquitin and Tollip bind to an overlapping region of the GAT-domain of Tom1 in a mutually exclusive manner.42 The lysine 147 of Rac1 is most likely the acceptor site of ubiquitin.43 This lysine is located in the vicinity of a polybasic stretch of amino-acids residues at the C-terminal part of Rac1. This region is crucial for binding to several regulatory proteins as well as for the proper localization of Rac1 at the membrane. In line with this, it has been suggested that ubiquitylation might be involved in Rac1 dynamics in peripheral membrane ruffles and sorting of Rac1 in endocytic vesicles.26,44 Further work will likely decipher the importance of Rac1 ubiquitylation not only as a restrictive regulation of its signaling, but also as a mean to modulate its interactions with effectors and with the membrane.
Acknowledgments
Our laboratory is supported by institutional funding from INSERM, grants from Fondation Infectiopôle Sud, the Agence Nationale de la Recherche (ANR-11BSV300401), and the Association pour la Recherche sur le Cancer (ARC SFI 20111203659 and ARC SFI 20111203671).
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/19221
References
- 1.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 2.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, et al. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–9. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 3.Doye A, Mettouchi A, Bossis G, Clément R, Buisson-Touati C, Flatau G, et al. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–64. doi: 10.1016/S0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- 4.Munro P, Flatau G, Lemichez E. Bacteria and the ubiquitin pathway. Curr Opin Microbiol. 2007;10:39–46. doi: 10.1016/j.mib.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–71. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 9.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 10.Bokoch GM. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 2005;15:163–71. doi: 10.1016/j.tcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Nadif Kasri N, Van Aelst L. Rho-linked genes and neurological disorders. Pflugers Arch. 2008;455:787–97. doi: 10.1007/s00424-007-0385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Aktories K. Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol. 2011;9:487–98. doi: 10.1038/nrmicro2592. [DOI] [PubMed] [Google Scholar]
- 14.Lemonnier M, Landraud L, Lemichez E. Rho GTPase-activating bacterial toxins: from bacterial virulence regulation to eukaryotic cell biology. FEMS Microbiol Rev. 2007;31:515–34. doi: 10.1111/j.1574-6976.2007.00078.x. [DOI] [PubMed] [Google Scholar]
- 15.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 16.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–63. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Visvikis O, Maddugoda MP, Lemichez E. Direct modifications of Rho proteins: deconstructing GTPase regulation. Biol Cell. 2010;102:377–89. doi: 10.1042/BC20090151. [DOI] [PubMed] [Google Scholar]
- 18.Boquet P, Lemichez E. Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 2003;13:238–46. doi: 10.1016/S0962-8924(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 19.Dubois D, Delmas J, Cady A, Robin F, Sivignon A, Oswald E, et al. Cyclomodulins in urosepsis strains of Escherichia coli. J Clin Microbiol. 2010;48:2122–9. doi: 10.1128/JCM.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–9. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 21.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–33. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 22.Lerm M, Selzer J, Hoffmeyer A, Rapp UR, Aktories K, Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1: activation of c-Jun N-terminal kinase in HeLa cells. Infect Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerm M, Pop M, Fritz G, Aktories K, Schmidt G. Proteasomal degradation of cytotoxic necrotizing factor 1-activated rac. Infect Immun. 2002;70:4053–8. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch EA, Stall J, Schmidt G, Chavrier P, D’Souza-Schorey C. Proteasome-mediated degradation of Rac1-GTP during epithelial cell scattering. Mol Biol Cell. 2006;17:2236–42. doi: 10.1091/mbc.E05-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doye A, Boyer L, Mettouchi A, Lemichez E. Ubiquitin-mediated proteasomal degradation of Rho proteins by the CNF1 toxin. Methods Enzymol. 2006;406:447–56. doi: 10.1016/S0076-6879(06)06033-2. [DOI] [PubMed] [Google Scholar]
- 26.Nethe M, Anthony EC, Fernandez-Borja M, Dee R, Geerts D, Hensbergen PJ, et al. Focal-adhesion targeting links caveolin-1 to a Rac1-degradation pathway. J Cell Sci. 2010;123:1948–58. doi: 10.1242/jcs.062919. [DOI] [PubMed] [Google Scholar]
- 27.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–9. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 28.Boyer L, Turchi L, Desnues B, Doye A, Ponzio G, Mege JL, et al. CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1-/- cells. Mol Biol Cell. 2006;17:2489–97. doi: 10.1091/mbc.E05-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan L, Zou W, Gao D, Inuzuka H, Fukushima H, Berg AH, et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Mol Cell. 2011;44:721–33. doi: 10.1016/j.molcel.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–65. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms’ tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–74. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- 32.Mettouchi A, Klein S, Guo W, Lopez-Lago M, Lemichez E, Westwick JK, et al. Integrin-specific activation of Rac controls progression through the G(1) phase of the cell cycle. Mol Cell. 2001;8:115–27. doi: 10.1016/S1097-2765(01)00285-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Anglesio MS, O’Sullivan M, Zhang F, Yang G, Sarao R, et al. The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat Med. 2007;13:1060–9. doi: 10.1038/nm1621. [DOI] [PubMed] [Google Scholar]
- 34.Munro P, Flatau G, Doye A, Boyer L, Oregioni O, Mege JL, et al. Activation and proteasomal degradation of rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J Biol Chem. 2004;279:35849–57. doi: 10.1074/jbc.M401580200. [DOI] [PubMed] [Google Scholar]
- 35.Boyer L, Magoc L, Dejardin S, Cappillino M, Paquette N, Hinault C, et al. Pathogen-derived effectors trigger protective immunity via activation of the Rac2 enzyme and the IMD or Rip kinase signaling pathway. Immunity. 2011;35:536–49. doi: 10.1016/j.immuni.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–55. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20:2326–33. doi: 10.1128/MCB.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer L, Travaglione S, Falzano L, Gauthier NC, Popoff MR, Lemichez E, et al. Rac GTPase instructs nuclear factor-kappaB activation by conveying the SCF complex and IkBalpha to the ruffling membranes. Mol Biol Cell. 2004;15:1124–33. doi: 10.1091/mbc.E03-05-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–8. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, et al. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visvikis O, Boyer L, Torrino S, Doye A, Lemonnier M, Lorès P, et al. Escherichia coli producing CNF1 toxin hijacks Tollip to trigger Rac1-dependent cell invasion. Traffic. 2011;12:579–90. doi: 10.1111/j.1600-0854.2011.01174.x. [DOI] [PubMed] [Google Scholar]
- 42.Katoh Y, Shiba Y, Mitsuhashi H, Yanagida Y, Takatsu H, Nakayama K. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J Biol Chem. 2004;279:24435–43. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 43.Visvikis O, Lorès P, Boyer L, Chardin P, Lemichez E, Gacon G. Activated Rac1, but not the tumorigenic variant Rac1b, is ubiquitinated on Lys 147 through a JNK-regulated process. FEBS J. 2008;275:386–96. doi: 10.1111/j.1742-4658.2007.06209.x. [DOI] [PubMed] [Google Scholar]
- 44.Nethe M, Hordijk PL. The role of ubiquitylation and degradation in RhoGTPase signalling. J Cell Sci. 2010;123:4011–8. doi: 10.1242/jcs.078360. [DOI] [PubMed] [Google Scholar]


