Abstract
Small GTPases regulate a wide range of homeostatic processes such as cytoskeletal dynamics, organelle homeostasis, cell migration and vesicle trafficking, as well as in pathologic conditions such as carcinogenesis and metastatic spreading. Therefore, it is important to understand the regulation of small GTPase signaling, but this is complicated by the fact that crosstalk exists between different GTPase families and that we have to understand how they signal in time and space. The Golgi apparatus represents a hub for several signaling molecules and its importance in this field is constantly increasing. In this review we will discuss small GTPases signaling at the Golgi apparatus. Then, we will highlight recent work that contributed to a better understanding of crosstalk between different small GTPase families, with a special emphasis on their crosstalk at the Golgi apparatus. Finally, we will give a brief overview of available methods and tools to investigate spatio-temporal small GTPase crosstalk.
Keywords: Arf, Cdc42, Golgi apparatus, Rac, Ras, small GTPases, spatio-temporal signaling
Introduction: Small GTPases and the Golgi Apparatus
Small GTPases are a family of GTP-binding proteins with a molecular weight in the range of ~21 kDa (hence their designation as small). Small GTPases are involved in many different cellular processes relevant for several homeostatic, developmental and pathologic conditions. They are used by the cell to regulate protein synthesis, to transduce signals from the plasma membrane in response to external stimuli, to regulate the cytoskeleton, to regulate trafficking of proteins and ligands, as well as many other processes. Small GTPases cycle between an inactive (GDP-bound) and an active (GTP-bound) state. Activation of the GTPase is mediated via a guanine-nucleotide exchange factor (GEF) and inactivation is mediated by a GTPase-activating protein (GAP). In this current review we will focus on four of the major families of small GTPases; namely, Ras, Rho, Rab and Arf, and their roles at the Golgi apparatus.
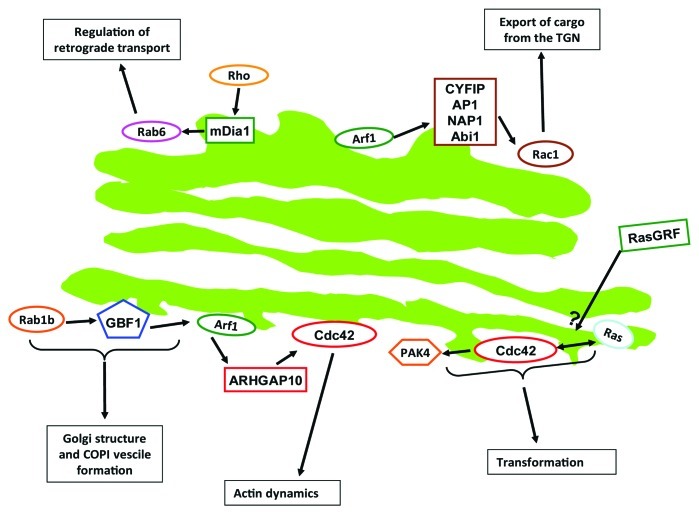
The Golgi apparatus is a membrane-bound organelle composed of stacks of flattened cisternae located in close proximity to the centrosome in the juxtanuclear region of mammalian cells. This organization is specific to mammalian cells. The Golgi is best known for its roles in post-translational modification (e.g., glycosylation) and sorting of proteins.1 However, the Golgi is increasingly viewed as hub for cellular signaling molecules, and by spatially organizing signaling pathways the Golgi contributes to the biological outcome of signal transduction.2 In this review, we will give an overview on what is known about signaling of the above mentioned GTPases and their regulators (GEFs and GAPs) to and from the Golgi and then we will discuss evidence for crosstalk between different GTPase families occurring at the Golgi. Considering the enormity of the field, this review does not intend to give a detailed description of a certain type of GTPase or on its crosstalk with other GTPases. The aim here is to give a broad overview over the extent of crosstalk that small GTPases exert at the Golgi. Furthermore, we will also give a short description of methods that are used to study spatiotemporal signaling of small GTPases. A schematic of some of the crosstalk routes is shown in Figure 1 and key crosstalk routs are listed in Table 1.
Figure 1. Schematic of some of the crosstalk routs described in this review.
Table 1. Main crosstalk routes described in this review.
| Families involved in the interaction |
Direction and Quality of interaction |
Ref |
| Ras – Rho |
Ras→ Cdc42→ transformation Ras→ Cdc42→ PAK4→ transformation RasGRF –I Cdc42 Ras Rac1→ PLCγ1→ RasGRP1→ Ras |
52
53 54 58 |
| Arf – Rho |
Arf →COPI Cdc42 Arf1→ Rac1→ TGN carrier formation ARAP1 –I Arf1 Cdc42 Arf1→ ARHGAP21 –I Cdc42 |
20
,
21
61 29 27 |
| Arf – Rab Rho – Rab |
Rab1b→ GBF1→ Arf1 Rab11→ Rab8→ TGN to cilia transport Arf4 Rho→ mDia→ Rab6→ retrograde transport |
78
83 , 84 88 |
Ras GTPases
Ras GTPases regulate various aspects of cell biology like cell proliferation, apoptosis, senescence and differentiation. One of the core signaling pathways activated by Ras is the Raf-MEK-ERK mitogen-activated protein kinase cascade (MAPK cascade) by which Ras is able to signal to targets in the cytosol as well as in the nucleus. In mammals, there are three genes for Ras: K-Ras, H-Ras and N-Ras, which all become farnesylated at their C-terminal CAAX motif, an event that occurs at the cytosolic face of the endoplasmic reticulum, allowing their targeting to various membranes. N-Ras and H-Ras are further palmitoylated at the Golgi and this mediates their association with the inner leaflet of the plasma membrane. K-Ras uses a poly-lysine region for plasma membrane targeting.3,4 H-Ras and N-Ras cycle between the Golgi and the plasma membrane and this cycling is regulated by a depalmitoylation-repalmitoylation process.5 While palmitoylation occurs mainly at the Golgi, depalmitoylation was shown to occur virtually everywhere in the cell.5,6 When a Ras protein loses its palmitoyl moiety, it goes unspecifically to all endomembranes because the farnesyl group mediates a weak affinity to cellular membranes. Because palmitoylation occurs only at the Golgi, Ras is kinetically trapped, which explains why a significant pool of Ras has been observed at the Golgi apparatus in several studies.5,6
Over a decade ago, Ras was shown not only to localize, but also to signal from endomembranes.7 The biologic response elicited by Ras was dependent on the cellular location from where the Ras signal was initiated.7,8 The fact that Ras may signal from endomembranes was met with such excitement that it was considered equal to “life on Mars”9. However, it also provoked questions on the molecular determinants that regulate Ras activation on the Golgi, or in other words, are there GEFs that are specifically responsible for activation of Ras on the Golgi? The main known Ras-GEFs are the Son Of Sevenless proteins (Sos1 and Sos2), the Ras guanosine nucleotide releasing factors (GRFs) and Ras guanosine nucleotide releasing proteins (GRPs). All these GEFs have a common Cdc25 domain, which is responsible for the GDP to GTP exchange in Ras GTPases. In addition to the Cdc25 domain, Sos and GRF have a DH-domain (Dbl-Homology domain), that is typically found in GEFs activating Rho family GTPases. As we will discuss below, Sos and RasGRF are capable of mediating crosstalk between Ras and Rho GTPases due to the fact that they have both types of GEF domains.
The description of Ras signaling from endomembranes raised new questions that now need to be taken into account when studying endomembrane signaling of any small GTPase. We will frame two questions here, the answer to which is, in our opinion, crucial when studying spatial signaling of small GTPases.
(1) Is there a GEF, a GAP or any kind of anchor on the examined cellular compartment capable of modulating small GTPase signaling? In fact, if there is nothing to module GTPase activity on a specific cellular compartment, then it is likely that any detectable signaling cascade taking place there is only an echo originating from another cellular compartment. Answering this question provides sufficient grounds to state that a certain GTPase is signaling from a specific cellular location. After this question is answered, the following question becomes important:
(2) Is endomembrane-originated signaling associated with any specific biological response?
As will become evident from what we highlight below, these two questions are partially answered for Ras signaling at the Golgi. In lymphocytes, activation of Ras on the Golgi is mediated by RasGRP1.10 In a study by Caloca et al.,11 Ras at the Golgi was only activated by RasGRP1, contrary to Ras at the plasma membrane or on the endoplasmic reticulum, which was also activated by other GEFs.11 Other factors that contribute to the specificity of Ras signaling at the Golgi are scaffold proteins that regulate the Raf-MEK-ERK cascade that signals downstream of Ras. This was investigated using Ras constructs that were artificially hooked to different subcellular locations and revealed that, depending on the origin of Ras signaling, different scaffolds were utilized by the Raf-MEK-ERK cascade. This differential scaffold use was then linked to differential signaling of Ras to cytosolic and nuclear targets.12 Despite the apparent richness of evidence for Ras signaling at the Golgi, this fact is not beyond doubt. A recent study used fluorescent sensors to monitor the spatio-temporal activity of Ras in MDCK cells.13 While activation of Ras at the plasma membrane was quick and transient, the activity at the Golgi was slightly delayed and sustained. Using a combination of live imaging and modeling, the authors concluded that the Golgi does not contain appreciable amounts of GEFs and GAPs for Ras. Therefore, it was concluded that the active pool of Ras at the Golgi is an “echo” of Ras that is activated at the plasma membrane and the endoplasmic reticulum.13 However, the latter finding does not fit well with the finding that Ras-GEFs (e.g., RasGRP1) have been found at the Golgi. It could well be that this is simply due to the usage of different cells lines, but this has to be resolved in future studies. Therefore, despite some controversy, the question whether regulators of GTPase activity are present at the Golgi is at least partially answered.
The next question is whether there is a site specific action of Ras. A striking example of the biological effect deriving from Ras signaling at the Golgi was provided by Daniels et al.14 They generated OVA peptide variants with different affinities to the T-cell receptor and studied the ability of those ligands to induce positive or negative selection of thymic lymphocytes. Positive selectors resulted in a substantial increase in the number of CD8 single positive cells, whereas negative selectors did not. In cells treated with negative selectors, the Ras GEF, RasGRP1, Ras and its effector Raf1 were recruited on the plasma membrane. On the other hand, positive selectors led to a colocalization of these proteins at the Golgi. The ERK response elicited by positive selectors (i.e., that originating from the Golgi) was more sustained compared with the ERK response elicited by negative selectors (i.e., that originating from the plasma membrane). This difference in kinetics could be due to the fact that the milieu at the plasma membrane imposes a more stringent negative regulation on Ras than the Golgi. The final result of this study is that the origin of the ERK cascade determines the selection outcome of T-lymphocytes.14
Because the two questions we framed above seem to be at least partially answered, it appears that the Golgi is a bona fide site for Ras signaling. Nevertheless, there still is some controversy and the search for biological functions that this pool of Ras may exert is an active field of research. While the two questions are partially answered for Ras, this is not the case for another family of small GTPases, which we will highlight in the next section.
Rho GTPases
Rho GTPases are a family of small G-proteins composed of over 20 members that function as key regulators of the cytoskeleton, thereby modulating cell migration, neurite growth, vesicle trafficking and cytokinesis.15 The three main groups within the Rho family are Rho, Rac and Cdc42. In addition, to their large number, Rho family GTPases have more than 60 different GEFs and at least 70 GAPs, thus indicating the wide range of potential actions they may exert. In addition to GEFs and GAPs, there are three classes of RhoGDIs (Rho guanine-nucleotide dissociation inhibitors) that act as negative regulators of Rho GTPases. When a Rho GTPase is in its inactive (GDP-bound) state, it is bound to a GDI that acts as a chaperone, preventing premature uncontrolled activation of the GTPase.16 The GDI-bound pool is thought to account for at least 95% of the total pool of Rho family proteins.17,18
Rho GTPases, and in particular Cdc42, have been shown to play an active role at the Golgi. Cdc42 was shown to localize to the Golgi,19 and by interaction with the γCOP subunit of the coatomer,20 regulates transport between the Golgi and the endoplasmic reticulum. This very likely involved active Cdc42 signaling as the effect of Cdc42 on Golgi-to-ER transport was dependent on the Cdc42 effector N-WASP.21 In order to elucidate the spatio-temporal dynamics of Rho GTPase signaling, considerable effort was invested into the development of fluorescent sensors that report the subcellular location where the Rho GTPase is active.18,22,23 These sensors are mostly based on fluorescence resonance energy transfer (FRET; see later). Using different kinds of probes, Cdc42 was shown to be active at the Golgi.24,25 An open question (as in the case of Ras) is whether the active Cdc42 observed at the Golgi is the result of local activation. In support of this, the Golgi protein GM130 was shown to interact with Tuba, a Cdc42 GEF.26 Knockdown of GM130 reduced the levels of active Cdc42, thus providing the first evidence that Cdc42 might be activated locally at the Golgi.26 However, the pool of Tuba at the Golgi was very faint and other studies (including our unpublished observations) indicate other subcellular locations for Tuba than the Golgi. Therefore, more work is needed to finally elucidate whether Golgi membranes provide the environment for local activation of Cdc42. The Golgi is well known for its active role in cell migration. Whether the Golgi plays a primary role in cell migration, i.e. whether the Golgi is sufficient to induce cell migration, remains unclear. As Cdc42 is a master regulator of cell migration, a Golgi-based activation cascade for Cdc42 would further support the primary role of this organelle in directional cell motility.
Coming back to the two key questions related to spatial signaling of small GTPases (see above), we find that although some Rho GEFs and GAPs have been found at the Golgi,27-29 their presence has not always been linked directly to signaling of any Rho family GTPases member at this cellular location. Thus, we do not know whether the Golgi provides the environment for modulation of Rho GTPase signaling. Furthermore, whether Golgi-localized Rho GTPases exert any specific cellular function has never been investigated. This research area is currently heavily investigated and we expect that future results will help to provide answers to yet unresolved questions.
ARF GTPases
The ADP-ribosylation factor (Arf) family is a group of G proteins implicated in the control of membrane traffic and organelle architecture. In mammals, there are six Arfs (numbered from 1–6), divided into three classes: class I, composed of Arf 1 and 3, class II, composed of Arf 4 and 5, and class III, composed only of Arf6, the most divergent protein of this group.30 In humans, Arf2 is identical to Arf4. The Arf family also includes Sar1 and more than 20 Arf-like proteins (ARLs).
Arf GTPases contribute to the structural integrity of the Golgi, which is most evident when cells are treated with the fungal metabolite brefeldin A (BFA), which inhibits Arf GEFs, by binding to their Sec7 domain. Treatment with BFA leads to a rapid disassembly of the Golgi and its fusion with the ERGIC and the ER.31,32 At least three members, namely Arf1, Arf4 and Arf5 have been shown to regulate budding of COPI vesicles,33 which are the major carriers mediating Golgi-to-ER trafficking. Arf family GTPases seem to exert their function cooperatively, as knockdown of a single Arf isoform has little or no appreciable effect. Depletion of Arf1 and Arf4 was shown to regulate the integrity of the Golgi and the ERGIC and to regulate trafficking from pre-Golgi compartments.34,35 Arf GTPases also have well appreciated roles in endocytosis, with Arf6 being the best characterized among endocytic Arf family members that exerts its biological role mainly in the endocytic pathway and has little direct roles on the Golgi.36
Arls arose early in the evolution and are functionally related to Arfs. Some Arl GTPases (like Arl1) regulate the recruitment of GRIP-domain Golgins to the TGN and mediate TGN localization of Arf interacting proteins like Arfaptins that regulate formation of tubules and vesicles from the TGN. Other Arls, like Arl2, regulate assembly of microtubules at the centrosome and therefore contribute to Golgi positioning.37 Several Arls are necessary for intraflagellar transport and ciliogenesis.37-39 Other Arls are involved in maintaining Golgi structure, like Arl3 which localizes to the Golgi, its knockdown leads to strong fragmentation of this organelle.37
Rab GTPases
Rab GTPases are the largest family of Ras-related proteins, with 11 components in yeast and at least 60 in mammals. The various members of the Rab GTPase family play a key role in regulating membrane trafficking at different locations of the endomembrane system. While Arf GTPases control vesicle biogenesis, Rab GTPases are important for directed carrier movement and tethering at the target membrane.40 TRAPP-I, a multiprotein complex that functions as a GEF for Rab1, is involved in tethering and homotypic fusion of COPII vesicles and thereby regulates cargo transport between ER and Golgi.41 Besides being so numerous, Rab GTPases have an equally large set of regulators (GEFs and GAPs). In order to elucidate which of the 60 Rabs function at the Golgi, Haas et al.42 screened 38 Rab-GAP proteins for their effect on Golgi morphology. Overexpression of two GAPs, RN-tre and TBC1D20, was found to disrupt both the Golgi and protein transport.42 RN-tre is a GAP for Rab43 and regulates endosome-to-Golgi transport. TBC1D20 is a GAP for Rab1 and regulates transport from the ER to the Golgi. The functional significance of this finding was later underscored by the observation that disruption of the Golgi with RN-tre and TBC1D20 inhibited secondary envelopment of herpes simplex virus 1 and thereby inhibited viral production.43 Another Rab isoform that is important at the Golgi is Rab6, which regulates retrograde (Golgi-to-ER) transport in a COPI-independent manner.44 Rab6 localizes mainly to the TGN and this specific localization requires Rab33.45,46 Other Rabs that have been shown to act at the Golgi are Rab34 that controls intra-Golgi protein transport,47 Rab13 that controls trafficking between the TGN and the endocytic pathway48 and Rab8, which controls exit from the TGN toward lysosomes.49 Overall, the above-mentioned Rabs, but also others, are important regulators of the functional organization of the Golgi.
Crosstalk on the Golgi Between Small GTPases
Crosstalk between Ras family and Rho family GTPases
As mentioned above, Ras has its main role in the control of cell proliferation and differentiation. Rho GTPases seem to be more prominently involved in regulating cell migration and cell shape.15 Thus, it is conceivable that cell fate decisions are controlled by a balance between these two families. For instance, PC12 cells may (depending on stimulus) either maintain a proliferative state, where they exhibit a round morphology, or they may cease proliferation to differentiate into a neuron-like morphology.50 Therefore, crosstalk between Ras and Rho GTPases appears self-evident and necessary.
Evidence for such a crosstalk came from a work performed in yeast. Using two different mutants of Ras1 (the only Ras form present in yeast), it was shown that a plasma membrane restricted Ras1 supported mating via the Byr2 MAPK pathway, while an endomembrane restricted Ras1 did not.51 However, Ras1 restricted to endomembranes supported elongated cell morphology by activating Cdc42, an effect not observed with plasma membrane Ras1.51 This example nicely illustrates crosstalk between Ras and Cdc42 in a manner dependent on a specific spatial location of Ras. This work was later extended to mammalian cells, where it was shown that efficient transformation by Ras requires interaction of Ras and Cdc42 at endomembranes.52 This interaction did occur on endosomes, and not on the Golgi, but it nevertheless tempts to speculate on the possibility of a physical and/or functional interaction of Cdc42 and Ras at the Golgi. Such an assumption seems safe when taking an earlier observation into account where it was shown that Cdc42 not only is present at the Golgi, but also actively signals at this location as it activated its effector protein PAK4 at the Golgi.53 Active PAK4 was important for Ras-mediated transformation, thus implying a functional crosstalk between Cdc42 and Ras signaling at the Golgi.53 Because of the relevance of this crosstalk for cell transformation, it appears worthwhile to test whether this crosstalk plays a role in cancer specimens. Overall, these papers place Cdc42 downstream of Ras signaling (Table 1). Recently, RasGRF was proposed to act as a physical link that mediates crosstalk between Ras and Cdc42.54 There, RasGRF was shown to bind both Cdc42 and Ras. However, while RasGRF functions as a GEF for Ras, it is unable to activate Cdc42. Accordingly, knockdown of RasGRF2 was shown to induce Cdc42 activity.55 The inhibitory effect of RasGRF on Cdc42 translated into an inhibition of cell migration and invasion. However, it remains to be determined how RasGRF balances its activities toward Ras and Cdc42. In addition, no information about the subcellular location was provided. Because there is evidence for localization of RasGRF at endomembranes, we may speculate that the RasGRF-mediated crosstalk between Ras and Cdc42 might occur (at least partially) on the Golgi. Thus, while the above-mentioned studies placed Ras upstream of Cdc42, this study places them at the same level and inversely controlled by RasGRF.
Crosstalk was also demonstrated between Ras and Rac. The Rac effector PAK was shown to activate Raf and MEK1, both downstream targets of Ras.56 Ras was also shown to bind to and positively modulate Tiam1, a GEF for Rac.57 So, in general, Rac was placed downstream of Ras, which is analogous to Cdc42. However, unlike with Cdc42, no information is available on the spatial organization of the Ras-Rac crosstalk. In one study, not only an inverted hierarchy of Ras-Rac crosstalk was found, but also some evidence pointing to an involvement of the Golgi. In T lymphocytes, Rac appeared to increase the activity of phsopholipase-gamm1, which in turn led to formation of diacylglycerol and the activation of the Ras GEF, RasGRP1,58 and thus Ras was downstream of Rac. As mentioned above, RasGRP1 was shown to mediate Golgi-based activation of Ras in T-lymphocytes58 and therefore it appears likely that this crosstalk takes place at the Golgi.
Crosstalk between Arf family and Rho family GTPases
Golgi architecture is controlled by Arf GTPases. On the other hand, positioning of the Golgi is dependent on the cytoskeleton, which is controlled by Rho GTPases. Based on these two facts, it appears likely that the Golgi is controlled by crosstalk between Arf and Rho family GTPases appears. Crosstalk between these two families also appears to occur at the level of carrier formation at both the cis- and the trans-face of the Golgi. Formation of COPI vesicles on the cis-side of the Golgi was shown to be dependent on Arf family GTPases, namely Arf1, Arf4 and Arf5.33 However, Cdc42 was reported to interact with the coatomer subunit γCOP and to itself control retrograde Golgi-to-ER transport.20,21 Therefore, Arf GTPases and Cdc42 seem to cooperate at the level of COPI vesicle trafficking. Also, with regard to carrier formation at the TGN, there seems to be crosstalk between Arf and Rho family GTPases. In mammalian cells, trafficking of specific cargoes (like the mannose-6-phosphate receptors) from the TGN is dependent on clathrin and the AP-1 adaptor.59 Also, actin dynamics were shown to control this trafficking step.60 Anitei et al.61 showed that Arf1 recruits a complex, composed of clathrin heavy chain, the adaptor protein AP-1, CYFIP (cytoplasmic fragile-X mental retardation interacting protein), NAP1 and Abi1.61 This Arf1-dependent multi-protein complex serves as a platform for the recruitment of Rac1 and its GEF protein, β-PIX, leading to N-WASP mediated actin polymerization.61 This facilitates formation of tubules that will give rise to carriers that mediate export of cargo from the TGN. Thus, Rac1 functions downstream of Arf1 during formation of TGN carriers (Table 1).
ArfGAP1 was the first GAP identified for an Arf GTPase and it was shown to localize to the Golgi.62 Besides promoting GTP hydrolysis in Arf1, it has been implicated in the regulation of COPI vesicle biogenesis and in protein transport from the ER to the cell surface.63,64 When a truncation mutant of ArfGAP1 (lacking the N-terminal 141 amino acids) was expressed, cells displayed impaired flattening and spreading, suggesting a defect in the actin cytoskeleton.65 Mechanistically, this truncated mutant of ArfGAP1, suppressed the activity of Rac1 and thereby exerted its negative effect on actin nucleation, which translated into an inhibition of cell migration.65 Thus, ArfGAP1 could principally mediate crosstalk between Arf1 and Rac1 and thereby regulate actin polymerization. A potential role for Arf1 in actin biogenesis appears likely as Arf1 stimulates the production of PI(4,5)P2, a lipid that is known to stimulate actin nucleation. This study based its conclusion primarily on overexpression of a truncated mutant of ArfGAP1. Therefore, more experiments under more physiological conditions are needed to test whether such a crosstalk is physiologically relevant. This could, for instance, be achieved by monitoring Rac1 activity at the Golgi using FRET reporter probes (see later) and by testing whether ArfGAP1 has any effect on local Rac1 activity.
ARAP1 is another Arf-GAP protein that was shown to mediate crosstalk between Arf and Rho family GTPases.66 Endogenous ARAP1 localized to the Golgi apparatus and displayed GAP activity not only toward Arf1 and Arf5, but also toward Rho and Cdc42.66 The GAP activity toward Arf1 and Arf5 was dependent on phosphatidylinositol (3,4,5) trisphosphate. Overexpression of ARAP1 disrupted Golgi morphology, most likely due to its negative effect on Arf1. Interestingly, despite the fact that ARAP1 had a GAP activity toward Rho GTPases, its overexpression resulted in elevated Cdc42 activity, which was associated with the morphological changes in the actin cytoskeleton.66 Thus, ARAP1 forms an intersection point for signaling pathways regulating membrane remodeling and actin. An important point that needs to be addressed here is whether the effect of ARAP1 on Arf is causally linked to its effect on Rho and Cdc42 and vice versa. In addition, it remains to be determined whether the effects of ARAP1 on Cdc42 take place at the Golgi. This appears likely, because ARAP1 itself localizes to the Golgi and its overexpression leads to formation of plasma membrane blebs that contain Golgi markers.66 Nevertheless, this point has to be formally proven and could be addressed using the FRET reporter probes for Cdc42.
A further, more direct example for a crosstalk between Arf1 and Cdc42 at the Golgi is represented by the finding that the Cdc42 GAP protein, ARHGAP21 (also referred to as ARHGAP10), is recruited to the Golgi in a manner dependent on Arf1.27 At the Golgi, ARHGAP21 regulates Cdc42 activity and thereby the Arp2/3 complex and actin dynamics. The control of actin dynamics implies that ARHGAP21 has a role in cell migration, which was later shown to be the case.67 Thus, ARHGAP21 represents an example how Arf1 would directly recruit a regulator of Cdc42 to the Golgi.
Finally, we will discuss a very recent work that strongly suggests that crosstalk between Arf GTPases and Rac2 might exist at the Golgi and although it is speculative, we think that it merits being discussed here. PLD2 (phospholipase D2), which was shown to be located on endomembranes in a juxtanuclear region, was shown to act as a GEF for Rac2.28 PLD2 is the first phospholipase shown to act as a GEF, but its specificity toward different Rho GTPases has not yet been tested.28 PLD isoforms were shown to be activated by different Arf family members68 and in particular Arf4 was shown to activate PLD2 in response to mitogenic stimulation.69 Arf4 localizes to the Golgi and is involved in COPI vesicle production33,35 and therefore PLD2 might represent a molecule that mediates crosstalk between Arf4 and Rac2. The question arises now of whether Arf4 controls Golgi localization of PLD2 and by this might control Rac2 signaling at the Golgi. Whether this is the case, and what the potential functional implications of such a crosstalk mean, remain to be determined. Of note, is that PLD2 controls the Golgi localization of the adaptor protein Grb2, together with Sos and Ras, in response to EGF stimulation.70 Therefore, PLD2 might represent a molecular switchboard that links signaling of three different small GTPases at the Golgi (namely Ras, Cdc42 and Arf4). Further work is required to elucidate this exciting possibility.
Crosstalk between Arf family and Ras family GTPases
As mentioned above, the Golgi seems to represent a bona fide site for Ras signaling, and many functions of this organelle are controlled by Arf GTPases. Nevertheless, evidence for crosstalk between these two families is sparse. Mor et al.71 showed that recruitment of RasGRP1 to specific cellular locations was dependent on PLD and its ability to produce phosphatidic acid, which in turn is converted to diacylglycerol, which activates RasGRP1. Inhibition of PLD resulted in the inhibition of Ras at the plasma membrane and at the Golgi.71 As already mentioned above, PLD can be activated by several Arf68,69 and Rho GTPases.68 Thus, we may speculate that Arfs recruit and activate PLD at the Golgi, which leads to recruitment of RasGRP1 and Ras activation. In agreement with this, it was recently shown that depletion of Arf1 attenuates ERK activation by adrenergic signaling.72 This could at least partially be due to altered recruitment of RasGRP1 to the Golgi, which reduces Ras signaling to ERK.
There is also more evidence for crosstalk between Arf and Ras families. A protein called PDEδ (phosphodiesterase 6delta) was shown to act as a solubilizing factor for prenylated proteins of the Ras superfamily.73 PDEδ was earlier shown to interact with Arl2 and Arl3 in a GTP-dependent manner.74 Later, it was shown that interaction of GTP-loaded Arl2 and Arl3 is capable of releasing farnesylated cargo from PDEδ 73. Another study then showed that PDEδ binds and solubilizes farnesylated Ras and enhances their diffusion in the cytoplasm.75 The same work showed that this enhances kinetic trapping of depalmitoylated Ras at the Golgi.75 Altogether, Arl2 and Arl3 seem to control Ras subcellular trafficking and therefore act as upstream factors regulating Ras signaling. Thus, although the evidence for crosstalk between the Arf and Ras family is sparse, the available data place Ras downstream of Arf family GTPases.
Crosstalk between Arf family and Rab family GTPases
The Arf and Rab family of GTPases are mainly involved in the generation, transport, tethering and fusion of vesicles, as well as in the maintenance of organelle structure. Therefore, it is not surprising to find that crosstalk between these GTPases is involved in the control of vesicular transport at various stages of the secretory pathway. This was noted when Alvarez et al.76 expressed an inactive mutant of Rab1b which led to disassembly of the Golgi. Because a constitutively active mutant of Rab1b protected the Golgi against brefeldin A, it was suggested that Rab1b was involved in Arf1-dependent COPI recruitment to the Golgi.76 Later, this was further shown to be mediated by an interaction between Rab1 and GBF1, a GEF protein for Arf1.77 When Rab1b was depleted using siRNA, the recruitment of GBF1 to the Golgi was impaired. A more recent work uncovered the mechanistic details of Rab1b-dependent GBF1 recruitment to the Golgi. There, it was observed that expression of a constitutive active mutant of Rab1b increased the levels of PI4P at the Golgi and that the presence of this lipid is required for recruitment of GBF1.78 A model was proposed where Rab1b leads to activation of PI4KIIIα, which leads to synthesis of PI4P and in consequence the recruitment of GBF1.78 Future work will be needed to determine how Rab1b activates PI4KIIIα. Altogether, Arf1 functions downstream of Rab1b in the control of Golgi structure and trafficking events at and to the cis-Golgi.
Crosstalk between Rab and Arf GTPases also occurs at the trans-Golgi. Rab6 and Arl1 were proposed to cooperate in the recruitment of the large coiled-coil protein GCC185 to the TGN.79 GCC185 is required for the Rab9-dependent transport of mannose-6-phosphate from late endosomes to the TGN.80,81 Thus, the golgin, GCC185, appears to be an effector of two small GTPases that act in concert to control endosome-to-Golgi transport. While this appears appealing, the model is mainly based on in vitro interactions. Moreover, another paper claimed that endogenous GCC185 localizes normally in cells where Rab6 and Arl1 were depleted using siRNA.82 This work casts doubt on the relevance of the previous finding. However, it has to be stressed that siRNA-mediated depletion is a knock-down and not a knockout of the protein of interest. Thus, it could be that the remaining levels of Rab6 and Arl1 are sufficient to control the localization of GCC185. More work is needed to resolve this issue.
Crosstalk between Rab and Arf family GTPases at the TGN also exists in the context of ciliary transport. Arf4 and Rab11 were shown to form a ternary complex with the effector protein FIP3 and the Arf GAP protein, ASAP1.83 This complex was shown to mediate targeting of rhodopsin to cilia. Interestingly, Rab11 was required for recruitment of Rabin8, a GEF for Rab8, to the TGN.84 Taken together, a working model may be proposed where Rab11 and Arf4 act in concert to recruit and activate Rab8, which in turn control the trafficking of TGN carriers to cilia membrane.
Crosstalk between Rho family and Rab family GTPases
Positioning as well as structural integrity of the Golgi are dependent on microtubules (MTs). This becomes evident when cells are treated with the MT-depolymerizing drug nocodazole, which leads to fragmentation of the Golgi into several ministacks.85 Removal of the drug leads to rapid recovery of the Golgi in a biphasic process.86 In contrast to MTs, the role of actin at the Golgi is less clear, but recent work showed that the Golgi protein GOLPH3 interacts with MYO18A (an unconventional myosin), thus linking the Golgi to the actin cytoskeleton.87 Zilberman et al.88 studied the effect of Rho activation on Golgi morphology. Expression of active Rho, or its pharmacological activation, led to dispersion of the Golgi. This effect was dependent on Rho-mediated activation of mDia1 and was accompanied by formation of F-actin patches at the Golgi.88 Another important observation was that activation of the Rho-mDia1 pathway enhanced the formation of Golgi-derived Rab6-positive vesicles, which were found to transiently become positive for mDia1.88 This work implicates crosstalk between Rho and Rab6 in the regulation of Golgi structure and retrograde transport.
Methods to Study Spatio-Temporal Signaling of Small GTPases
Biochemical approaches
Initially, the activation of small GTPases was studied using biochemical approaches by performing pull-down experiments. There, an effector fused typically to GST was used as a bait to bind the active form of the small GTPase of interest in a total cell lysate. This method was first developed for Ras89 and subsequently adapted for almost all other small GTPases. These experiments proved to be very valuable for initially characterizing the temporal activation patterns of small GTPases. However, biochemical approaches fail to provide insight into spatial activation pattern of small GTPases. Another problem with certain types of GTPases was the small pool of the active form. For instance, in the case of Rho GTPases, even the most potent stimuli will activate, on average 5%, of the total pool of Rho GTPases in the cell.18 Thus, changes might not become visible due to a poor signal to noise ratio. Therefore, there was a need to develop novel approaches to study small GTPases.
Overexpression of small GTPases
As evident from the sections above, the spatial aspect of signaling has to be taken into account in order to fully understand how a signaling pathway works. Given that pull-down approaches are not useful for this purpose, a specific small GTPase can be visualized in the cell by expressing a fluorescently tagged version of it. However, overexpression of a small GTPase might lead to erratic results that have to be treated with caution. This is, for instance, important for Rho family GTPases, which are usually bound to Rho-GDI proteins in the inactive state. Overexpression of a Rho GTPase will over-saturate the GDI binding capacity and thus the overexpressed protein will more likely to be found in its active. In addition, by out-competing other Rho family members, additional erratic results might occur.90 Furthermore, to study the regulation of a small GTPase, it can be useful to overexpress one of its regulators (GEFs and GAPs) to see where it localizes. A problem occurring when overexpressing a GEF or a GAP is that this overexpressed regulatory protein will overwhelm all the other endogenous regulators, thus creating an environment that could be not representative of the physiological situation.
Fluorescent biosensors
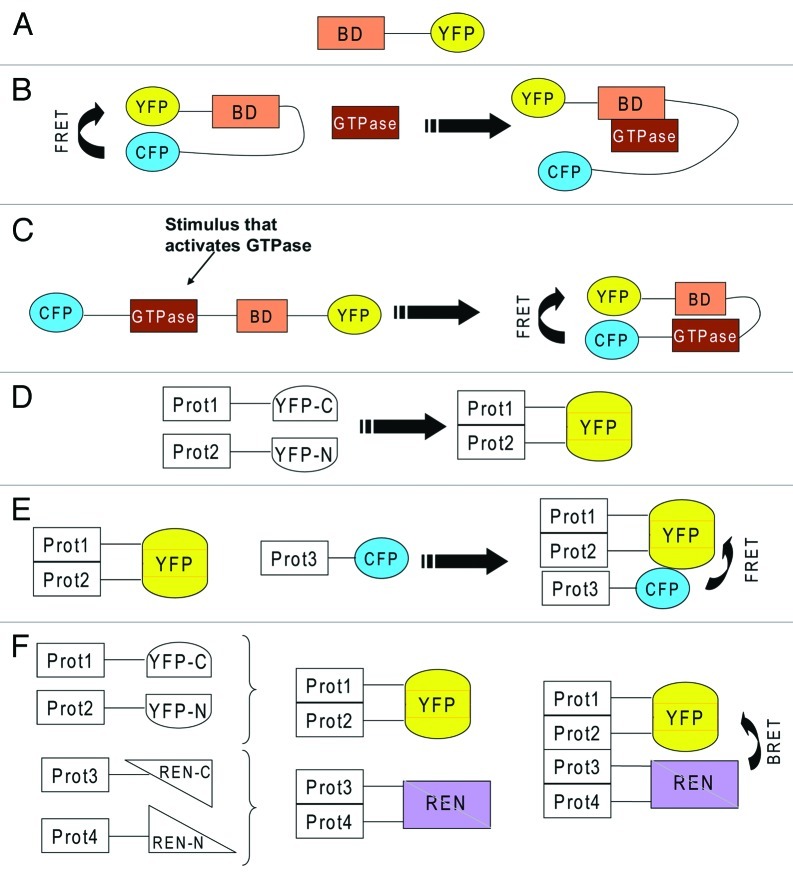
simple overexpression of fluorescently tagged GTPases, or of their regulators, does not appear to be the best method of choice. Thus, the development of reporters that visualize the sub-cellular location of the active form of the small GTPase is certainly a promising alternative. The simplest form of this reporter contains a GTPase effector domain coupled to a fluorescent molecule. This reporter will only be enriched at a certain cellular location when the GTPase is in its active form (Fig. 2A). This approach has, for instance, been used to visualize compartmentalized Ras signaling.7 Overexpression of such a reporter can lead to further problems. In the case of Ras, it can induce apoptosis by sequestering and blocking the endogenous Ras signaling.91 This is typically counteracted by co-overexpress of the sensor together with a Ras protein.
Figure 2. (A) YFP fused to a binding domain (BD) that recognizes an active GTPase. Because of the binding domain, if taken from an effector of a small GTPase, this reporter will specifically bind a certain type of GTPase. (B) BD sandwiched between CFP and YFP, which exhibit FRET because they are in close proximity. An incoming active GTPase squeezes in to bind to BD and displaces CFP and YFP, which leads to loss of FRET. (C) the GTPase and a BD from its effector are sandwiched between CFP and YFP. Activation of the GTPase will lead to binding of BD to the active GTPase which leads to closure of the reporter which allows FRET between CFP and YFP. (D) two proteins (Prot1 &2) are fused to two severed halves of YFP (YFP-C and YFP-N). Interaction of the two proteins will lead to complementation of YFP, which starts to fluoresce. (E) the interaction of two proteins is detected via YFP complementation. A third protein (Prot3) also joins the complex and this is visualized by FRET between CFP and YFP. (F) the interaction of two proteins (Prot 3&4) is detected via YFP complementation. The interaction of two further proteins (Prot 3&4) is detected via complementation of two severed halves of renilla luciferase (REN). The formation of a tetrameric complex is visualized by BRET.
Other reporters are based on FRET and do not only report on the localization of a GTPase but also on its activation status.18 Some probes contain the GTPase plus an effector protein sandwiched between two fluorescent proteins. In this case, activation of the GTPase will lead to an increase of FRET. Other probes contain an effector domain sandwiched between two fluorescent proteins in such a way that there is constitutive FRET. When an active GTPase encounters this probe it squeezes in, which leads to a loss of FRET (Fig. Two B&C). For a more detailed review on these and other reporters, the reader is referred to another recently published review.18 The use of these reporters has provided valuable insight into spatio-temporal small GTPase signaling. However, the results obtained with probes for Rho family members have to be taken with caution for the same reason mentioned above, namely that some of these reporters contain a full length small GTPase that is not subject to regulation by Rho-GDI. Therefore, a major improvement would be to design probes that are regulated by Rho-GDI in order to perform experiments that are closer to the physiologic situation. Further room for improvement would be to modulate the backbone of these probes to enhance the efficiency of FRET, and such an attempt has been published recently.92
As discussed above, there are numerous examples for crosstalk between GTPases, but little attention has been given to the spatial aspect of this crosstalk. Studying crosstalk requires methods to visualize protein-protein interactions. FRET provides a method to visualize protein-protein interactions in living cells.
Protein complementation
a method to study interactions of proteins, and therefore crosstalk, is the protein complementation assay (PCA). As illustrated in Figure 2D, two proteins are fused to severed fragments of YFP. Interaction of these proteins will lead to a complementation of the two halves of YFP, which can be then detected microscopically. YFP-PCA can then be combined with FRET in order to detect a ternary complex (Fig. 2E). Complementation may also be performed using severed halves of luciferase. Interaction between luciferase and YFP can be detected using BRET (bioluminescence resonance energy transfer). This allows, in principle, to detect a tetrameric complex (Fig. 2F). Another advantage of the YFP-PCA system is that it can be used to screen for interaction partners.93,94 Complementation of the YFP protein is irreversible and therefore, once a complex has formed it will not dissociate. This is, on one hand, an advantage as it visualizes weak and transient interactions. On the other hand, irreversible linking of two proteins is also often considered a disadvantage, as it does not allow the examination of the dynamics of a system. In contrast to YFP-PCA, complementation of luciferase is reversible and allows us to study the dynamics of a system. A disadvantage is that it does not easily allow visualization of the subcellular location of this interaction. Thus, ideally, complementation of YFP and luciferase should be done in parallel. For a more detailed review about protein complementation, the reader is referred to other recent reviews.95
Overall, with all these methods in hand (together with necessary improvements), we will be able in the future to not only better characterize old interactions, but also uncover new signaling crosstalk at different subcellular locations.
Concluding Remarks
Most attention in past decades of research has focused on the temporal regulation of signaling. However, the spatial regulation is increasingly recognized to contribute immensely to the outcome of signaling. Thus, a major task for the future is to dissect the anatomy of small GTPase crosstalk in time and space. This requires a stronger awareness of the importance of the spatial aspect when studying signaling. In addition, an improvement of the available tools will certainly facilitate progress in this field as well as yield data that better reflect the physiologic situation. It is becoming increasingly appreciated that the control of cell fate and various homeostatic and pathologic processes cannot be understood by focusing on single signaling pathways, but it is rather the result of crosstalk between signaling pathways. Moreover, crosstalk has to be studied with a special attention to where it occurs in the cell. With this review, we hope to have increased the awareness of the importance of crosstalk between small GTPases. We have focused this review on crosstalk at the Golgi, but we emphasize that in the future there will be an increasing need to compare signaling crosstalk between different locations. It is conceivable that the development of improved therapeutic strategies for many pathologic conditions, like cancer and its metastatic spreading, will be facilitated by the dissection of signaling networks in time and space and by understanding their biological impacts.
Footnotes
Previously published online: www.landesbioscience.com/journals/smallgtpases/article/19842
References
- 1.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–32. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farhan H, Rabouille C. Signalling to and from the secretory pathway. J Cell Sci. 2011;124:171–80. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- 3.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–77. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 4.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 5.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–52. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 6.Rocks O, Gerauer M, Vartak N, Koch S, Huang Z-P, Pechlivanis M, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141:458–71. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–50. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 8.Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol Biol Cell. 2008;19:4776–84. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Fiore PP. Signal transduction: life on Mars, cellularly speaking. Nature. 2003;424:624–5. doi: 10.1038/424624a. [DOI] [PubMed] [Google Scholar]
- 10.Bivona TG, Pérez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, et al. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–8. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 11.Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J Biol Chem. 2003;278:33465–73. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- 12.Casar B, Arozarena I, Sanz-Moreno V, Pinto A, Agudo-Ibáñez L, Marais R, et al. Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol Cell Biol. 2009;29:1338–53. doi: 10.1128/MCB.01359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorentzen A, Kinkhabwala A, Rocks O, Vartak N, Bastiaens PIH. Regulation of Ras localization by acylation enables a mode of intracellular signal propagation. Sci Signal. 2010;3:ra68. doi: 10.1126/scisignal.20001370. [DOI] [PubMed] [Google Scholar]
- 14.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, et al. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–9. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 15.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–35. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 16.Boulter E, Garcia-Mata R. RhoGDI: A rheostat for the Rho switch. Small Gtpases. 2010;1:65–8. doi: 10.4161/sgtp.1.1.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren X-D, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–85. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci. 2010;123:1841–50. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 19.Erickson JW, Zhang C, Kahn RA, Evans T, Cerione RA. Mammalian Cdc42 is a brefeldin A-sensitive component of the Golgi apparatus. J Biol Chem. 1996;271:26850–4. doi: 10.1074/jbc.271.43.26850. [DOI] [PubMed] [Google Scholar]
- 20.Wu WJ, Erickson JW, Lin R, Cerione RA. The gamma-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature. 2000;405:800–4. doi: 10.1038/35015585. [DOI] [PubMed] [Google Scholar]
- 21.Luna A, Matas OB, Martínez-Menárguez JA, Mato E, Durán JM, Ballesta J, et al. Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell. 2002;13:866–79. doi: 10.1091/mbc.01-12-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiyokawa E, Aoki K, Nakamura T, Matsuda M. Spatiotemporal regulation of small GTPases as revealed by probes based on the principle of Förster Resonance Energy Transfer (FRET): Implications for signaling and pharmacology. Annu Rev Pharmacol Toxicol. 2011;51:337–58. doi: 10.1146/annurev-pharmtox-010510-100234. [DOI] [PubMed] [Google Scholar]
- 23.Welch CM, Elliott H, Danuser G, Hahn KM. Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol. 2011;12:749–56. doi: 10.1038/nrm3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–91. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–9. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 26.Kodani A, Kristensen I, Huang L, Sütterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol Biol Cell. 2009;20:1192–200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois T, Paléotti O, Mironov AA, Fraisier V, Stradal TEB, De Matteis MA, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–64. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 28.Mahankali M, Peng H-J, Henkels KM, Dinauer MC, Gomez-Cambronero J. Phospholipase D2 (PLD2) is a guanine nucleotide exchange factor (GEF) for the GTPase Rac2. Proceedings of the National Academy of Sciences 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, et al. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9:109–19. doi: 10.1016/S1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Kelly WG, Logsdon JM, Jr., Schurko AM, Harfe BD, Hill-Harfe KL, et al. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 2004;18:1834–50. doi: 10.1096/fj.04-2273com. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263:18545–52. [PubMed] [Google Scholar]
- 32.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–13. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popoff V, Langer JD, Reckmann I, Hellwig A, Kahn RA, Brügger B, et al. Several ADP-ribosylation factor (Arf) isoforms support COPI vesicle formation. J Biol Chem. 2011;286:35634–42. doi: 10.1074/jbc.M111.261800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Tekaya H, Kahn RA, Hauri H-P. ADP ribosylation factors 1 and 4 and group VIA phospholipase A₂ regulate morphology and intraorganellar traffic in the endoplasmic reticulum-Golgi intermediate compartment. Mol Biol Cell. 2010;21:4130–40. doi: 10.1091/mbc.E10-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volpicelli-Daley LA, Li Y, Zhang C-J, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol Biol Cell. 2005;16:4495–508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweitzer JK, Sedgwick AE, D'Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Seminars in Cell &. Dev Biol. 2011;22:39–47. doi: 10.1016/j.semcdb.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol Biol Cell. 2006;17:2476–87. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J Cell Biol. 2010;189:1039–51. doi: 10.1083/jcb.200912001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiens CJ, Tong Y, Esmail MA, Oh E, Gerdes JM, Wang J, et al. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J Biol Chem. 2010;285:16218–30. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1:E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- 41.Yu S, Satoh A, Pypaert M, Mullen K, Hay JC, Ferro-Novick S. mBet3p is required for homotypic COPII vesicle tethering in mammalian cells. J Cell Biol. 2006;174:359–68. doi: 10.1083/jcb.200603044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas AK, Yoshimura S-i, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 43.Zenner HL, Yoshimura S-i, Barr FA, Crump CM. Analysis of Rab GTPase-activating proteins indicates that Rab1a/b and Rab43 are important for herpes simplex virus 1 secondary envelopment. J Virol. 2011;85:8012–21. doi: 10.1128/JVI.00500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, et al. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–30. doi: 10.1038/15658. [DOI] [PubMed] [Google Scholar]
- 45.Starr T, Sun Y, Wilkins N, Storrie B. Rab33b and Rab6 are functionally overlapping regulators of Golgi homeostasis and trafficking. Traffic. 2010;11:626–36. doi: 10.1111/j.1600-0854.2010.01051.x. [DOI] [PubMed] [Google Scholar]
- 46.Young J, Stauber T, del Nery E, Vernos I, Pepperkok R, Nilsson T. Regulation of microtubule-dependent recycling at the trans-Golgi network by Rab6A and Rab6A’. Mol Biol Cell. 2005;16:162–77. doi: 10.1091/mbc.E04-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldenberg NM, Grinstein S, Silverman M. Golgi-bound Rab34 is a novel member of the secretory pathway. Mol Biol Cell. 2007;18:4762–71. doi: 10.1091/mbc.E06-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nokes RL, Fields IC, Collins RN, Fölsch H. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J Cell Biol. 2008;182:845–53. doi: 10.1083/jcb.200802176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Toro D, Alberch J, Lázaro-Diéguez F, Martín-Ibáñez R, Xifró X, Egea G, et al. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20:1478–92. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci U S A. 1976;73:2424–8. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Onken B, Wiener H, Philips MR, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci U S A. 2006;103:9045–50. doi: 10.1073/pnas.0603318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng C-M, Li H, Gasman S, Huang J, Schiff R, Chang EC. Compartmentalized Ras proteins transform NIH 3T3 cells with different efficiencies. Mol Cell Biol. 2011;31:983–97. doi: 10.1128/MCB.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callow MG, Clairvoyant F, Zhu S, Schryver B, Whyte DB, Bischoff JR, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–8. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- 54.Calvo F, Sanz-Moreno V, Agudo-Ibáñez L, Wallberg F, Sahai E, Marshall CJ, et al. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–26. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 55.Calvo F, Sanz-Moreno V, Agudo-Ibáñez L, Wallberg F, Sahai E, Marshall CJ, et al. RasGRF suppresses Cdc42-mediated tumour cell movement, cytoskeletal dynamics and transformation. Nat Cell Biol. 2011;13:819–26. doi: 10.1038/ncb2271. [DOI] [PubMed] [Google Scholar]
- 56.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–38. doi: 10.1016/S0092-8674(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 57.Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, et al. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621–5. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- 58.Zugaza JL, Caloca MJ, Bustelo XR. Inverted signaling hierarchy between RAS and RAC in T-lymphocytes. Oncogene. 2004;23:5823–33. doi: 10.1038/sj.onc.1207768. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–12. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 60.Salvarezza SB, Deborde S, Schreiner R, Campagne F, Kessels MM, Qualmann B, et al. LIM kinase 1 and cofilin regulate actin filament population required for dynamin-dependent apical carrier fission from the trans-Golgi network. Mol Biol Cell. 2009;20:438–51. doi: 10.1091/mbc.E08-08-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anitei M, Stange C, Parshina I, Baust T, Schenck A, Raposo G, et al. Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat Cell Biol. 2010;12:330–40. doi: 10.1038/ncb2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 63.Beck R, Adolf F, Weimer C, Bruegger B, Wieland FT. ArfGAP1 activity and COPI vesicle biogenesis. Traffic. 2009;10:307–15. doi: 10.1111/j.1600-0854.2008.00865.x. [DOI] [PubMed] [Google Scholar]
- 64.Reiterer V, Maier S, Sitte HH, Kriz A, Rüegg MA, Hauri HP, et al. Sec24- and ARFGAP1-dependent trafficking of GABA transporter-1 is a prerequisite for correct axonal targeting. J Neurosci. 2008;28:12453–64. doi: 10.1523/JNEUROSCI.3451-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siu KY, Yu MK, Wu X, Zong M, Roth MG, Chan HC, et al. The non-catalytic carboxyl-terminal domain of ARFGAP1 regulates actin cytoskeleton reorganization by antagonizing the activation of Rac1. PLoS One. 2011;6:e18458. doi: 10.1371/journal.pone.0018458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, et al. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9:109–19. doi: 10.1016/S1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 67.Bigarella CLo, Borges L, Costa FF, Saad STO. ARHGAP21 modulates FAK activity and impairs glioblastoma cell migration. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 2009; 1793:806. [DOI] [PubMed]
- 68.Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–16. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S-W, Hayashi M, Lo J-F, Yang Y, Yoo J-S, Lee J-D. ADP-ribosylation factor 4 small GTPase mediates epidermal growth factor receptor-dependent phospholipase D2 activation. J Biol Chem. 2003;278:2661–8. doi: 10.1074/jbc.M205819200. [DOI] [PubMed] [Google Scholar]
- 70.Di Fulvio M, Frondorf K, Henkels KM, Lehman N, Gomez-Cambronero J. The Grb2/PLD2 interaction is essential for lipase activity, intracellular localization and signaling in response to EGF. J Mol Biol. 2007;367:814–24. doi: 10.1016/j.jmb.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mor A, Campi G, Du G, Zheng Y, Foster DA, Dustin ML, et al. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat Cell Biol. 2007;9:713–9. doi: 10.1038/ncb1592. [DOI] [PubMed] [Google Scholar]
- 72.Dong C, Li C, Wu G. Regulation of α(2B)-adrenergic receptor-mediated extracellular signal-regulated kinase 1/2 (ERK1/2) activation by ADP-ribosylation factor 1. J Biol Chem. 2011;286:43361–9. doi: 10.1074/jbc.M111.267286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ismail SA, Chen Y-X, Rusinova A, Chandra A, Bierbaum M, Gremer L, et al. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7:942–9. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- 74.Hanzal-Bayer M, Linari M, Wittinghofer A. Properties of the interaction of Arf-like protein 2 with PDEdelta. J Mol Biol. 2005;350:1074–82. doi: 10.1016/j.jmb.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 75.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, et al. The GDI-like solubilizing factor PDE[delta] sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol 2011; advance online publication. [DOI] [PubMed]
- 76.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–27. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–10. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumaresq-Doiron K, Savard M-F, Akam S, Costantino S, Lefrancois S. The phosphatidylinositol 4-kinase PI4KIIIalpha is required for the recruitment of GBF1 to Golgi membranes. J Cell Sci. 2010;123:2273–80. doi: 10.1242/jcs.055798. [DOI] [PubMed] [Google Scholar]
- 79.Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–98. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy JV, Burguete AS, Sridevi K, Ganley IG, Nottingham RM, Pfeffer SR. A functional role for the GCC185 golgin in mannose 6-phosphate receptor recycling. Mol Biol Cell. 2006;17:4353–63. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–73. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 82.Houghton FJ, Chew PL, Lodeho S, Goud B, Gleeson PA. The localization of the Golgin GCC185 is independent of Rab6A/A’ and Arl1. Cell. 2009;138:787–94. doi: 10.1016/j.cell.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 83.Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–92. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knödler A, Feng S, Zhang J, Zhang X, Das A, Per Ã. ¤nen J, et al. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proceedings of the National Academy of Sciences 2010; 107:6346-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rogalski AA, Singer SJ. Associations of elements of the Golgi apparatus with microtubules. J Cell Biol. 1984;99:1092–100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller PM, Folkmann AW, Maia ARR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–80. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dippold HC, Ng MM, Farber-Katz SE, Lee S-K, Kerr ML, Peterman MC, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–51. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, et al. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–11. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taylor SJ, Shalloway D. Cell cycle-dependent activation of Ras. Current biology: CB 1996; 6:1621. [DOI] [PubMed]
- 90.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, et al. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat Cell Biol. 2010;12:477–83. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Augsten M, Pusch R, Biskup C, Rennert K, Wittig U, Beyer K, et al. Live-cell imaging of endogenous Ras-GTP illustrates predominant Ras activation at the plasma membrane. EMBO Rep. 2006;7:46–51. doi: 10.1038/sj.embor.7400560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, Kamioka Y, et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647–56. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nyfeler B, Reiterer V, Wendeler MW, Stefan E, Zhang B, Michnick SW, et al. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J Cell Biol. 2008;180:705–12. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reiterer V, Nyfeler B, Hauri H-P. Role of the lectin VIP36 in post-ER quality control of human α1-antitrypsin. Traffic. 2010;11:1044–55. doi: 10.1111/j.1600-0854.2010.01078.x. [DOI] [PubMed] [Google Scholar]
- 95.Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. Protein-Fragment Complementation Assays for Large-Scale Analysis, Functional Dissection and Dynamic Studies of Protein–Protein Interactions in Living Cells. 2011:395-425. [DOI] [PubMed] [Google Scholar]
- 96.Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9:2221–35. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]