Eukaryotic genomes are organized into functional domains that facilitate the fundamental nuclear processes of transcription, replication, and DNA repair. An appreciation of chromosome structure effects on gene expression dates back over 70 years to Muller's discovery of Drosophila mutants with a mosaic (red/white) eye-color phenotype, caused by translocation of an eye-color gene from an open (euchromatic) region to a position near condensed (heterochromatic) chromatin. Many different examples of silent chromatin are now known, which can encompass a few thousand base pairs of DNA, as in mating-type gene silencing in the budding yeast Saccharomyces cerevisiae, or whole chromosomes, as in mammalian X-inactivation. These now classic examples of gene silencing may be related to important developmental gene repression phenomena in metazoa, exemplified by the Polycomb system in Drosophila that controls the inheritance of homeotic gene expression states (see refs. 1 and 2 for recent reviews). However, despite their central importance in chromosome biology, we still lack a solid biochemical understanding of any system of regional chromatin silencing. It is thus particularly noteworthy that the report from Tanner et al. (3) in this issue of PNAS provides the first detailed biochemical description of a novel, evolutionarily conserved enzyme (Sir2) required for silencing in yeast. This milestone provides a good opportunity to summarize our current view of silencing, recount a fascinating tale of discovery, and consider how this remarkable new enzyme may motivate future studies.
Extensive genetic and biochemical analysis of mating-type gene silencing and a related, but metastable, form of repression near telomeres in yeast have led to a working molecular model (reviewed in ref. 4 and diagramed in Fig. 1). Mating-type gene silencer elements or telomeres act as cis initiation sites for the polymerization of SIR (silent information regulator) protein complexes onto neighboring nucleosomes. Silencers recruit SIR proteins through the combined action of different DNA-binding factors: the origin recognition complex (ORC) and either or both of two multifunctional regulators, repressor/activator protein 1 (Rap1p) and ARS binding factor 1 (Abf1p). At telomeres, the TG1–3 repeat binding protein Rap1 is essential for Sir protein recruitment, but recent studies show that the Ku heterodimer also plays an important role in promoting SIR recruitment, which is counteracted by other factors (5). The subsequent assembly of Sir2/3/4 complexes along the adjacent nucleosomes is thought to be driven by a network of interactions between the SIR proteins (6) and a set of critical contacts between both Sir3p and Sir4p and the N-terminal tails of the core histones H3 and H4 (7). These SIR-histone interactions are thought to provide the glue that prevents access to the underlying DNA by factors involved in transcription, repair, and recombination, although the structure and dynamics of silent chromatin are still poorly understood. Nevertheless, the intimate involvement of the highly conserved core histone tails in yeast silencing suggests that global repression mechanisms in more complex eukaryotes may be related at some fundamental level to the yeast system.
Figure 1.
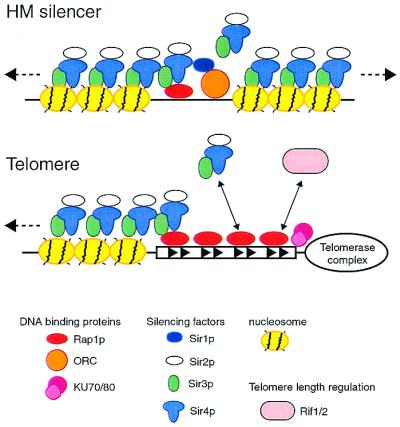
Molecular model for silencing at HM mating-type loci and telomeres. Silencing is initiated by DNA-binding proteins [Rap1p and origin recognition complex (ORC), or Rap1p and Ku] that cooperate to recruit SIR protein complexes. These histone-interacting SIR complexes then assemble along adjacent nucleosomal DNA, masking or in some other way altering the nearby chromatin. At telomeres, where silencing is unstable, or variegated, recruitment of SIRs is negatively regulated by the RIF proteins
One feature of silent chromatin in yeast that is clearly common to many other forms of repressed chromatin is the relative hypoacetylation of the core histone tails in comparison to open or active chromatin (8). How, then, does silent chromatin become underacetylated? Is it assembled from hypoacetylated histone precursors during replication and kept in this state by the action of SIR proteins, or are the SIR proteins directly involved in an active deacetylation process? Early work from James Broach and coworkers (8) suggested that the latter possibility might be correct. They showed that overexpression of Sir2p leads to global histone deacetylation and suggested that Sir2p might be a histone deacetylase. Shortly thereafter, a flood of studies demonstrated the central importance of reversible histone acetylation in gene regulation. However, it also became clear that Sir2p is not homologous to known deacetylases, and no such activity could then be demonstrated for Sir2p, for reasons that will become clear below. Nevertheless, the discovery that Sir2p is a member of an evolutionarily conserved family of proteins, and that yeast alone has five Sir2p family members, served to focus attention on this protein (9). Furthermore, the fact that bacteria have Sir2-like proteins, but lack histones and silent chromatin, suggested that Sir2p might have a general function, not strictly related to gene silencing.
An important clue to the biochemical function of Sir2-like proteins came from work in the bacteria Salmonella typhimurium, where it was shown that a Sir2p homolog (CobB) could substitute for the loss of CobT, a known phosphoribosyltransferase enzyme (10). This led to the idea that Sir2p family proteins might act as (mono) ADP-ribosylating enzymes, and evidence in support of this idea was soon provided by Frye (11), who showed that a human Sir2p homolog could transfer label from NAD to BSA. Shortly thereafter, Moazed and colleagues (12) showed that yeast Sir2p itself has ADP-ribosyltransferase activity, and that this activity is important for its in vivo silencing function. These studies led to the suggestion that ADP ribosylation of histones might be Sir2p's critical silencing function, although there was no evidence for ribosylated histones in silent chromatin.
The story of Sir2's enzymatic function might have ended here, but for the fact that two other groups either failed to detect a Sir2 ribosyltransferase activity or noted something peculiar about Sir2 reaction products. In the first case, Rolf Sternglanz and colleagues (13) decided to examine more closely the interaction between Sir2 and NAD. They noted that Escherichia coli DNA ligase, an enzyme that uses NAD as a cofactor to join DNA ends, also will catalyze the reversible cleavage of NAD. Based on this analogy, they reasoned that Sir2 family proteins might be able to catalyze the transfer of label from nicotinamide to NAD in a similar exchange reaction. In a recent report they showed that Sir2p and its yeast family member Hst2p can indeed catalyze such an exchange reaction. Significantly, neither protein alone could do the job, but instead required the presence of other proteins, in this case histones isolated from chicken red blood cells. In an additional series of elegantly designed experiments, Sternglanz and colleagues (13) went on to demonstrate directly that Sir2 family proteins are in fact NAD-dependent deacetylases. The same conclusion was reached by Guarente and coworkers (14), who noticed that a weak Sir2p ribosyltransferase activity appeared to be stimulated by acetylated histone tail peptides. When they examined the products of this reaction, they realized that the histone peptides had not been ADP-ribosylated, but instead had been deacetylated by Sir2p, in an NAD-dependent manner. Finally, Boeke and colleagues (15) described a robust NAD-dependent histone deacetylase activity in yeast extracts that depends on the SIR2 homolog HST2. These striking discoveries put the prescient observations of Broach and colleagues (8) back in the spotlight and suggested that the important role of Sir2p in silencing might be the deacetylation of histone tails. However, these new studies (13–15) left the nature and possible biological role of Sir2p's ribosyltransferase activity unresolved.
The report by Tanner et al. (3) goes a long way toward resolving this controversy and provides the first detailed biochemical view of a novel and remarkably complex enzyme. Through a careful kinetic analysis of products from a reaction with the Sir2p homolog Hst2p, those authors first showed that there is a strict 1:1 correspondence between the amount of NAD consumed and the quantity of deacetylated histone H3 peptide produced. This tight coupling of nicotinamide formation and peptide deacetylation was established further by measuring the NAD-concentration dependence of both reactions. The almost unavoidable conclusion from this analysis is that Sir2-like enzymes perform a novel reaction in which NAD cleavage and deacetylation are tightly coupled.
Tanner et al. then proceeded to examine the reaction in more detail, expecting to verify acetate as one of its products. Instead they found that the cleaved acetyl group is transferred to a molecule whose mass matches that predicted for acetyl-ADP ribose. Because many NAD-dependent enzymes are believed to form an oxo-carbenium ADP-ribose cation through nicotinamide release, the authors suggest that the acetyl product in their reactions is 1-O-acetyl-ADP ribose, an apparently novel compound. They propose two alternative mechanisms to explain the formation of this product (see figure 4 of their paper) that cannot be distinguished at present. Finally, Tanner et al. also suggest a plausible explanation for the ADP-ribosylation activity of Sir2 family proteins, which in any event appears to be considerably less robust than its deacetylation activity, and is in fact undetectable under their conditions. They point out that ADP-ribose transfer to protein might be a side reaction resulting from an uncoupling of the intrinsic reaction in the presence of extremely high concentrations of an alternative acceptor, such as reactive protein side chains.
The present study provokes a number of intriguing biological questions. The first one raised by Tanner et al. is: why do Sir2 family enzymes couple deacetylation to the production of 1-O-acetyl-ADP ribose? One interesting possibility they suggest is that this molecule serves as an effector of other related enzymatic processes. An equally pressing question relates to the significance of the NAD dependence of the Sir2 family deacetylase activity. A recent report (15) shows that mutation of the yeast NPT1 gene, which is required for NAD synthesis through a salvage pathway, reduces both intracellular NAD levels and silencing. This raises the possibility that silencing might be modulated in wild-type cells by physiological conditions that alter the cellular energy state. Significantly, very recent observations by Guarente and colleagues (16, 17) show that caloric restriction in yeast increases lifespan through a SIR2- and NPT1-dependent pathway. Taken together, these observations suggest the intriguing possibility that Sir2p is a molecular link in a pathway connecting the cellular energy state to chromatin effects that control lifespan. Another interesting question is whether the free energy of NAD cleavage is used by Sir2p to perform work, for example in some form of nucleosome remodeling, or in the assembly of a functional Sir2/3/4 complex (18). What is known about the biological role of other Sir2 family members? Genetic studies in yeast (9) and the existence of cytoplasmic Sir2 family members and homologs in bacteria and Archaea (19) all point to diverse functions and deacetylase targets. This fact alone suggests that reversible acetylation, now a well-characterized histone modification, may be a much more general protein regulatory mechanism than previously appreciated. Characterizing the mechanisms that determine the target specificity of Sir2 family enzymes will be an important challenge. In this regard, it should be pointed out that there is still no direct evidence that histones are the actual (or only) Sir2p substrate, nor is a role for ADP ribosylation ruled out. Sir2p itself poses another interesting problem, because it carries out a set of still poorly understood, but clearly Sir3- and Sir4-independent functions in the nucleolus related to suppression of recombination, transcriptional silencing, and aging (reviewed in ref. 17). Recent studies indicate that Sir2p's nucleolar role can be genetically separated from its telomeric and mating-type silencing functions (20), but the mechanisms underlying its targeting to different sites are still not clear. What is clear, though, is that the discovery of a novel Sir2 family enzymatic function will motivate intensive research into these and many other interesting biological questions.
Acknowledgments
I thank G. Cuperus for comments, N. Roeggli for the figure, and the Swiss National Fund and the Canton of Geneva for support.
Footnotes
See companion article on page 14178.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011506198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011506198
References
- 1.Henikoff S. Biochim Biophys Acta. 2000;1470:O1–O8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs J J, van Lohuizen M. Semin Cell Dev Biol. 1999;10:227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- 3.Tanner K G, Landry J, Sternglanz R, Denu J M. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. . (First Published December 5, 2000; 10.1073/pnas.250422697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grunstein M. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 5.Mishra K, Shore D. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- 6.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 7.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 8.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 10.Tsang A W, Escalante-Semerena J C. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 11.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 12.Tanny J C, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 13.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. . (First Published May 16, 2000; 10.1073/pnas.110148297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 15.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wol-berger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S J, Defossez P A, Guarente L. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 17.Guarente L. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 18.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afshar G, Murnane J P. Gene. 1999;234:161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 20.Cuperus G, Shafaatian R, Shore D. EMBO J. 2000;19:2641–2642. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]


