Abstract
Anti-neoplastic potential of calorie restriction or ligand-induced activation of peroxisome proliferator activated receptors (PPARs) has been demonstrated in multiple studies; however, mechanism(s) by which tumor cells respond to these stimuli remain to be elucidated. One of the potent agonists of PPARα, fenofibrate, is a commonly used lipid-lowering drug with low systemic toxicity. Fenofibrate-induced PPARα transcriptional activity is expected to shift energy metabolism from glycolysis to fatty acid β-oxidation, which in the long-term, could target weak metabolic points of glycolysis-dependent glioblastoma cells. The results of this study demonstrate that 25 μM fenofibrate can effectively repress malignant growth of primary glial tumor cells and glioblastoma cell lines. This cytostatic action involves G1 arrest accompanied by only a marginal level of apoptotic cell death. Although the cells treated with 25 μM fenofibrate remain arrested, the cells treated with 50 μM fenofibrate undergo massive apoptosis, which starts after 72 h of the treatment. This delayed apoptotic event was preceded by FoxO3A nuclear accumulation, FoxO3A phosphorylation on serine residue 413, its elevated transcriptional activity and expression of FoxO-dependent apoptotic protein, Bim. siRNA-mediated inhibition of FoxO3A attenuated fenofibrate-induced apoptosis, indicating a direct involvement of this transcription factor in the fenofibrate action against glioblastoma. These properties of fenofibrate, coupled with its low systemic toxicity, make it a good candidate in support of conventional therapies against glial tumors.
Keywords: fenofibrate, PPARα, FoxO3A, glioblastoma, energy metabolism
Introduction
Despite recent advances in cancer therapy, there is an enormous need for new therapeutic approaches against CNS malignances. Recent reports suggest a beneficial role for lipid-lowering drugs, fibrates and statins in anticancer treatment.1-7 For example, a ten-year mortality study of over 7,722 French subjects treated with different fibrates revealed that the use of these drugs is associated with a significantly lower total mortality and reduced probability of death from cancer.8 In cell culture and in animal studies, various members of the fibrate family, which are synthetic agonists of peroxisome proliferator activated receptor α (PPARα), were shown to exert interesting anticancer properties. Clofibrate attenuated human ovarian cancer cell proliferation in vitro and in mouse xenograft model;9,10 growth arrest and apoptotic response were observed in glioblastoma cells treated with gemfibrozil;11 and fenofibrate induced apoptosis and decreased proliferation rate in endometrial cancer cells4 and in human and mouse medulloblastoma cell lines.12 In addition, fenofibrate sensitized melanoma cells to staurosporin2 and inhibited motility and invasiveness of human glioblastoma cell lines.13 In spite of these multiple observations, molecular mechanism(s) by which fenofibrate targets cancer cells are not well defined, and a possibility of PPARα-independent action is gaining attention. With respect to these mechanisms, we have previously reported highly encouraging in vivo effects of fenofibrate oral administration against melanoma lung metastases in which anticancer effect was associated with the inhibition of the constitutively active in these tumor cells, Akt.2,14 Other examples of a possible PPARα-independent action include fenofibrate-induced attenuation of the IGF-IR signal transduction,12,13 inhibition of endothelial cell growth and associated attenuation of angiogenesis;3,15 effects on mitochondrial respiration;16,17 and inhibition of cell motility and gap-junction intercellular coupling in prostate cancer.18
In our current study, we propose a new mechanism that links the pro-apoptotic action of fenofibrate with FoxO3A transcriptional activity in human glioblastoma cells. FoxO3A is a member of an evolutionary conserved class of transcription factors, the forkhead-box O (FoxO), which have been shown to play an important role in the control of various cellular processes, such as cell death, cell cycle progression, metabolism and longevity.19,20 Recent studies implicate FoxO3A in cellular responses to environmental stress, where it can play a crucial role as a molecular switch between detoxification and induction of cell death.21 FoxO3A undergoes intensive posttranslational modifications, including phosphorylation, acetylation and ubiquitynation, which affect its subcellular localization and transcriptional activity.20,21 Here, we demonstrate that glioblastoma cells responded to the fenofibrate treatment by G1 cell cycle arrest, with only a moderate effect on cell survival during first 48 h of the treatment. However, prolonged exposure to fenofibrate triggered massive apoptosis of the tumor cells. In our experimental setting, this delayed apoptotic event was preceded by nuclear retention and serine phosphorylation of FoxO3A (pS413), which resulted in FoxO-dependent expression of pro-apoptotic protein Bim. Since other agonists of PPARα were much less effective, and siRNA against PPARα only partially rescued glioblastoma cell from the treatment, we concluded that both PPARα-dependent and -independent mechanisms might contribute to the fenofibrate-induced activation of FoxO3A/Bim apoptotic axis.
Results
Fenofibrate mediated inhibition of cell proliferation and survival.
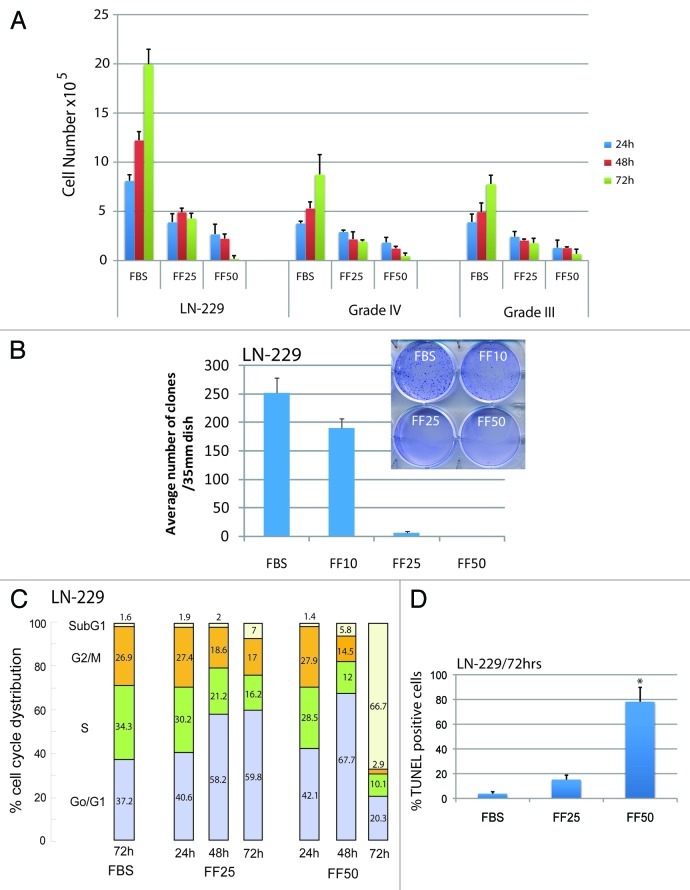
We have previously reported that 25 ∝M fenofibrate attenuated growth of mouse and human medulloblastoma cell lines in anchorage independence12 and inhibited glioblastoma cell motility and invasiveness without major effects on cell survival.13 We have also noticed that prolonged cell exposure to fenofibrate often triggers massive apoptosis in monolayer cultures of different tumor cells. To further investigate this delayed pro-apoptotic action of fenofibrate, we tested human glioblastoma cell line, LN-229, which is PPARα-positive, responds to IGF-1 stimulation and expresses wild type PTEN.13 Results in Figure 1A (first panel) demonstrate that the exponential growth of LN-229 cells is effectively inhibited in the presence of 25 ∝M fenofibrate (FF25), and this reversible cytostatic effect (the cells exposed to 25 ∝M fenofibrate for 48 h are able to resume proliferation after removal of the drug) was accompanied by only a minimal cell loss. In contrast, LN-229 cells exposed to 50 ∝M fenofibrate (FF50), caused, in addition to growth retardation at 48 h, a severe cell loss at 72 h. To verify that the observed anticancer effects were not restricted to this particular glioblastoma cell line, we tested primary tumor cells isolated from brain biopsies of patients diagnosed with grade III and IV Astrocytic tumors. As shown in the second and third panel of Figure 1A, the expected increase in cell number in control cultures (10% FBS) was effectively blocked by 25 ∝M fenofibrate, and, similar to the observed effect on LN-229, these primary tumor cells were practically eliminated after 72 h of the exposure to 50 ∝M fenofibrate. In addition, both 25 and 50 ∝M fenofibrate treatment resulted in a complete inhibition of LN-229 clonogenic growth, further supporting anticancer potential of this common lipid-lowering drug (Fig. 1B).
Figure 1. Evaluation of glioblastoma cells growth in vitro. (A) Human glioblastoma cell line, LN-229, and two primary cells from patients diagnosed with grade III and grade IV Astrocitic tumors were cultured in 10%FBS (FBS) in the presence or absence of 25 (FF25) and 50μM (FF50) fenofibrate. The cells were plated at 5 × 105/35mm dish and the number of cells was evaluated at 24, 48 and 72 h. The control cells (FBS) were treated with equal volume of DMSO (vehicle) used to dissolve fenofibrate. Viable cells were counted by utilizing GUAVA easyCyte 8HT flowcytometer and ViaCount assay (Millipore). The data represents average number of cells from three separate experiments in triplicate (n = 9) ± SD (B) Inhibition of clonogenic growth by fenofibrate. The cells were plated at the clonal density, 1 × 103/35 mm dish, in the presence of 10%FBS ± fenofibrate used at 10, 25 and 50 μM. The treatment was applied every 3 d over a period of 2 weeks by changing fresh media with indicated concentrations of fenofibrate. Three plates for each condition were used in a single experiment and the experiment was repeated 3 times (n = 9). Note that the number of clones decreased sharply with the increasing concentration of fenofibrate. (C) Flowcytometry-based analysis of cell cycle distribution of exponentially growing LN-229 cells in 10%FBS, and following the treatment with 25 and 50 μM fenofibrate. The aliquots of cells collected at 24, 48 and 72 h of the treatment were labeled with propidim iodine (PI) and were analyzed by GUAVA easyCyte 8HT flowcytometer using CellCycle software included in GuavaSoft 1.1 (Millipore). The diagram is a representative experiment of cell cycle distribution, which was repeated at least three times. (D) Apoptotic cell death by TUNEL assay. The TUNEL staining, specific for apoptosis related DNA damaged, was performed on untreated (FBS) and fenofibrate (25 μM, 50 μM) treated cells at 72 h time point using TUNEL assay reagents (Roche) and Guava/Express plus software. The data are presented as average % of TUNEL positive cells with standard deviation. * Indicates a value statistically different from FBS (p ≤ 0.05). Statistical significance between two measurements was determined with the two-tailed Student’s t-test.
Flowcytometric analysis of the cell cycle, depicted in Figure 1C, demonstrated 37.2% of cells in G0/G1, 34.3% in S, 26.9% in G2/M and 1.6% in sub-G1 phase in control cultures of LN-229 cells in the presence of 10% FBS. The treatment with 25 ∝M fenofibrate applied for 72 h resulted in G1 arrest, which was accompanied by proportional decreases in S and G2/M content, and moderate accumulation of the cells in sub-G1 (dead cells). Quantitatively, 59.8% of cells were in G0/G1, 16.2% in S, 17.0% in G2/M and 7% in sub-G1. In the presence of 50 ∝M fenofibrate, the cells demonstrated more advance G0/G1 arrest at 48 h (67.7%), followed by extensive cell death at 72 h of treatment (sub-G1 = 66.7%). The TUNEL analysis performed at the same time revealed that the sub-G1 cells are indeed apoptotic (Fig. 1D).
Next we asked whether the observed cytostatic and cytotoxic effects of fenofibrate depend on the activation of PPARα. Results in Figure 2A demonstrate over 90% decrease in PPARα protein levels when LN-229 cells were incubated with 100 nM PPARα siRNA for 48 h (Fig. 2A), and the siRNA treatment partially rescued LN-229 cells from both fenofibrate-induced cell cycle arrest (Fig. 2B) and cytotoxicity (Fig. 2C). Quantitatively, we observed over 5-fold decrease in cell number between fenofibrate-treated and untreated cells, when the experiment was performed in the presence of control (off-target) siRNA against nuclear lamin (NL). This strong cytostatic effect of fenofibrate was diminished by almost 3-fold (from 5-fold decrease to 1.8-fold decrease) when the experiment was performed in the presence of the on-target siRNA against PPARα (Fig. 2B). In respect to fenofibrate-induced cytotoxicity, we have employed a flow cytometry-based JC-1/7-AAD assay. The cells were labeled with JC-1 fluorescent dye in which a gradual increase in JC-1 green fluorescence and loss of orange fluorescence represents compromised mitochondrial membrane potential. By double labeling the same cells with 7-Aminoactinomycin D (7-AAD), we were also able to distinguish the population of dead cells in the same experiment.22 Results depicted in Figure 2C demonstrate 81.6 ± 9.5% of cells with severely compromised cell membrane integrity when 50 ∝M fenofibrate treatment was applied in the presence of control siRNA against nuclear lamin (FF50 + NLsiRNA). This cytotoxic effect was reduced by an almost 2.5-fold (decrease from 81.6% to 33.74% of dead cells) when the fenofibrate treated cultures were pre-incubated with 100 nM siRNA against PPARα, confirming a partial involvement of this nuclear receptor in the fenofibrate action against glioblastoma. Surprisingly, two other potent agonists of PPARα, gemfibrozil and WY14643, did not induce any significant changes in cell viability even when used at 100 ∝M concentration (Fig. 2D), which, in contrast to siRNA data, suggest PPARα-independent mode of the fenofibrate action. Alternatively, LN-229 cells could be simply resistant to gemfibrozil and WY14643. However, we have previously reported that LN-229 express PPARα protein at a level comparable to other tumor cell lines examined,12,13 and gemfibrozil, similarly to fenofibrate, activated PPAR-responsive elements in these cells (not shown). Therefore, there is still an unidentified and unique property of fenofibrate, which seems to synergize with a PPARα-dependent mechanism for its anticancer action.
Figure 2. Evaluation of PPARα involvement in fenofibrate effects against glioblastoma cell growth and survival. (A) Western blot analysis showing PPARα protein levels in LN-229 cells incubated with 100 nM of OFF-TARGET siRNA against nuclear lamins (NL: 100 nM), which efficiently downregulated nuclear lamines without affecting PPARα or Grb-2 13; and with 100 and 200 nM of ON-TARGRT plus SMARTpool siRNA against human PPARα (Thermo Scientific). (B) Evaluation of LN-229 cell number following 72 h of a continuous cell growth in 10% FBS (FBS), in which the treatment with 25 μM fenofibrate (FF25) was evaluated in the presence of 100 nM of NL-siRNA or PPARα-siRNA. The data are presented as an average of cell number at 72 h with standard deviation. *Indicates a value statistically different from FBS+NL-siRNA; ** Indicates a value statistically different from FBS+PPARα-siRNA (p ≤ 0.05). Statistical significance between two measurements was determined with the two-tailed Student’s t-test. (C and D) Flowcytometry-based evaluation of cell death using 7-AAD red fluorescent dye from Guava EasyCyte MitoPotential Kit. Exponentially growing LN-229 cells, in 10% FBS, were treated with 100 nM of indicated siRNAs in the presence or absence of 50 μM fenofibrate (C) or with three different PPARα agonists including fenofibrate, gemfibrozli and Wy14643 at the indicated concentrations and time points (D). Data represent average % of cell death from at least three independent experiments with standard deviation.
Fenofibrate affects FoxO3A subcellular localization and activity.
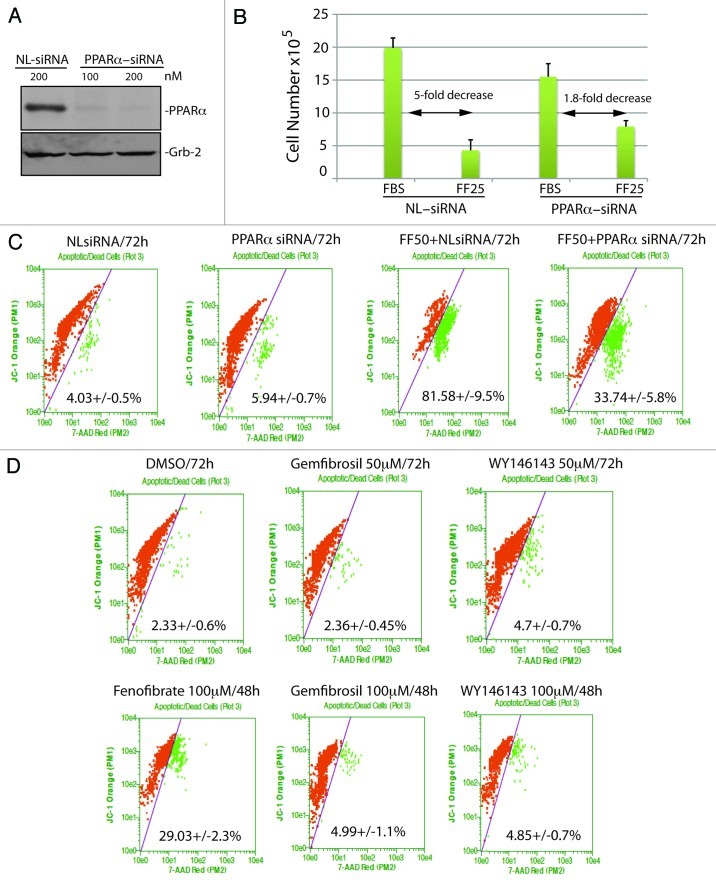
Since fenofibrate has been shown to inhibit constitutively active Akt in melanoma cell lines,2 attenuated IGF-I-induced Akt phosphorylation in LN-229 cells,13 and activation of Akt by growth factors is a known negative regulator of FoxO3A,21,23 we asked if apoptotic cell death observed in fenofibrate-treated glioblastoma cells depend on FoxO3A. Results in Figure 3A demonstrate immunofluorescent detection of FoxO3A (green fluorescence) in exponentially growing LN-229 cells (10% FBS) and in cells treated with 25 and 50 ∝M fenofibrate for 24 h. The co-localization between FoxO3A specific (green fluorescence) and DAPI (nuclear blue fluorescence) demonstrate a significant 3-fold increase in FoxO3A nuclear accumulation in the presence of 50 ∝M fenofibrate. In contrast, control cells and cells treated with 25 ∝M fenofibrate did not show any significant changes in FoxO3A nuclear content. We have confirmed this nuclear accumulation of FoxO3A by utilizing subcellular fractionation and western blot analysis, in which Lamin A/C and GAPDH were used as markers of nuclear and cytosolic fraction, respectively (Fig. 3B and C). In addition, fenofibrate-induced nuclear accumulation of FoxO3A was accompanied by a transient increase in serine phosphorylation of FoxO3A (pS413), detected 24 h following the treatment (Fig. 3D). This specific phosphorylation was not observed in cells treated with 25 ∝M fenofibrate, and we did not observe any detectible changes in FoxO3A acetylation (not shown). Importantly, these fenofibrate-induced FoxO3A modifications resulted in a 4-fold increase in FoxO transcriptional activity, detected by luciferase-based reporter construct containing the region from Fas-ligand promoter with three canonical forkhead responsive elements (Fig. 3E).
Figure 3. Effects of fenofibrate on subcellular localization and transcriptional activity of FoxO3A. (A) Quantification of FoxO3A nuclear localization in exponentially growing LN-229 cells (FBS) in presence of 25 μM (FF25) and 50 μM (FF50) fenofibrate. Nuclear localization of FoxO3A was calculated from the entire volume of the nucleus (DAPI labeled) by utilizing Mask Operation included in SlideBook5 software. The data represents average number of voxels per nucleus ± SD *Indicates values significantly different from FBS (p ≤ 0.05). Lower panel: representative immunofluorescent images of nuclear FoxO3A. The cells treated with 50 μM fenofibrate show over 30% increase in FoxO3A nuclear co-localization. (B) Western blot to detect FoxO3A-regulated pro-apoptotic gene, Bim, was performed using protein extracts from exponentially growing LN-229 cells (10% FBS), and from the same cells exposed to 25 and 50 μM fenofibrate for 48 h. Grb-2 was used as a loading control. (C) Densitometric analysis of the experiment depicted in Panel B by utilizing EZQuant-Gel 2.17 (EZQuant Biology Software Solutions). The data represent average values from three independent blots with standard deviation. FoxO3A values were normalized either with cytosolic marker GAPDH or with nuclear marker, Lamin A/C. (D) Western blot to detect FoxO3A phosphorylation on serine residue (S413), using anti-pS413-FoxO3A antibody. Total proteins were extracts from exponentially growing LN-229 cells (10% FBS), at indicated time points following the treatment with 50 μM fenofibrate. Grb-2 was used as a loading control. (E) FoxO3A transcriptional activity was evaluated in fenofibrate treated (FF25 and FF50) LN-229 cells by utilizing three copies of FoxO3A consensus sequences from the Fas ligand promoter and dual-Firefly/Renilla luciferase reporter system. Data are presented as mean ± SD calculated from two experiments in triplicates (n = 6). *Indicates value statistically significantly different (p ≤ 0.05) from control (FBS; cells treated with vehicle only). Statistical significance between two measurements was determined with the two-tailed Student’s t-test.
Fenofibrate-induced FoxO3A-Bim apoptotic pathway.
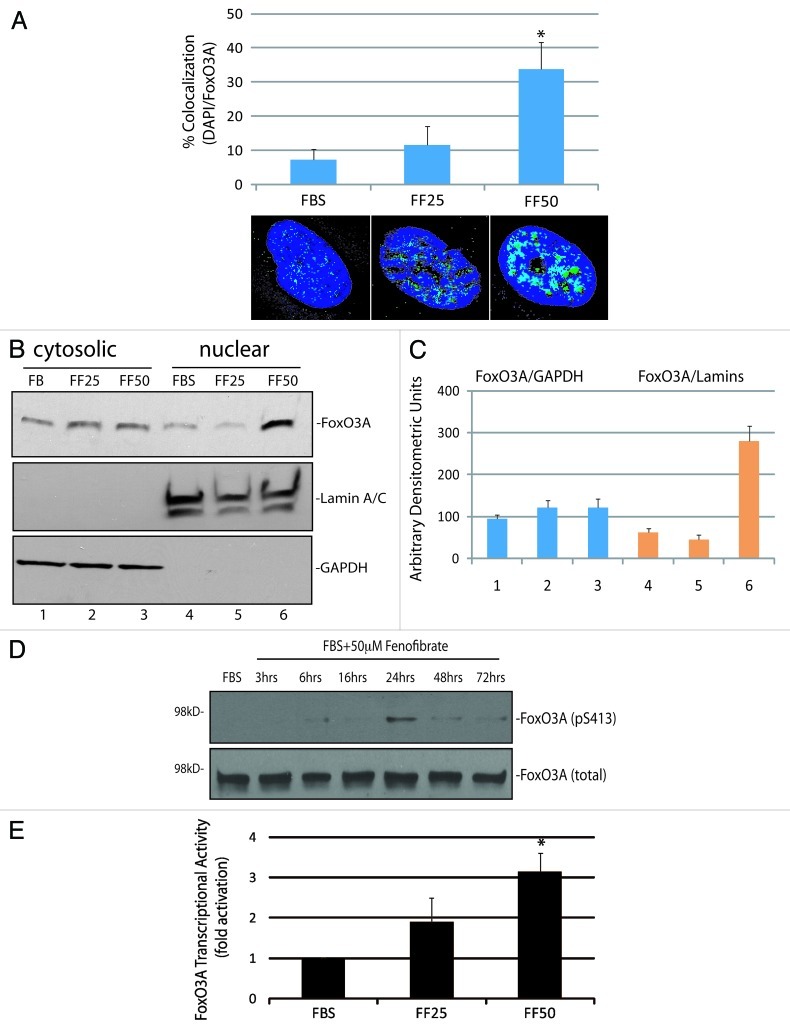
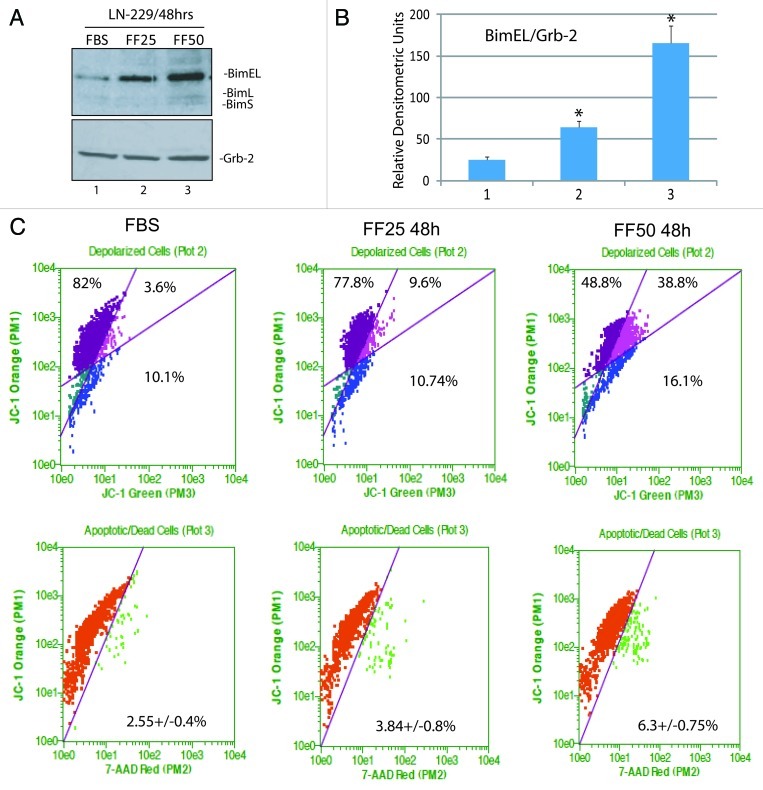
Among a large number of FoxO3A-dependent genes, there are several which are directly involved in promoting apoptosis.20,21,24 In particular, FoxO3A is known to bind and activate the promoter of pro-apoptotic gene, Bim.25-27 Our preliminary real-time PCR screening of the LN-229 cells treated with 50 ∝M fenofibrate revealed high levels of Bim mRNA (data not shown). The western blot in Figure 4A demonstrates a strong increase in BimEL protein level, and detectable bands corresponding to BimL and BimS isoforms following 48 h cell incubation with 50 ∝M fenofibrate.
Figure 4. Fenofibrate-induced activation of pro-apoptotic protein, Bim. (A) Western blot showing FoxO3A-dependent pro-apoptotic gene, Bim, and Grb-2 (loading control) in LN-229 cells cultured in 10% FBS and in cells treated with 25 and 50 μM fenofibrate for 48 h. Note that anti-Bim antibody recognizes three forms of Bim protein: BimEL, BimL and BimS. (B) Densitometric analysis of the experiment depicted in Panel A by utilizing EZQuant-Gel 2.17 (EZQuant Biology Software Solutions). The data represent average values from three independent blots with standard deviation. BimEL values were normalized with the loading marker, Grb-2. Statistical significance between two measurements was determined with the two-tailed Student’s t-test. (C) Flowcytometry-based evaluation of mitochondrial potential and cell death using JC-1/7-AAD test by Guava EasyCyte MitoPotential Kit. Exponentially growing LN-229 cells, in 10%FBS, were treated with 25 and 50 μM fenofibrate for 48 h. Data represent average % of cells characterized by healthy mitochondria (first quarter); partially compromised mitochondrial potential (second quarter) and severely compromised mitochondrial potential (third quarter). The percentage of 7-AAD positive cells (cells with compromised membrane potential) is indicated in the lower part of Panel C.
Since all Bim isoforms promote mitochondrial-dependent apoptosis, with the shortest form, BimS, being the most potent,28 we asked whether the observed activation of Bim is accompanied by changes in mitochondrial membrane potential, using again JC-1/7-AAD assay. The results in Figure 4C demonstrate that in control condition (FBS), 82 ± 7.3% of cells were not affected, and 13.7 ± 2.1% of cells were depolarized, among which 3.6% of the cells were partially depolarized and 10.1% severely depolarized. Following the treatment with 25 ∝M fenofibrate for 48 h, the population of unaffected cells slightly decreased, and the overall content of depolarized cells increased from 13.7 ± 2.1% to 20.3 ± 3.2%. In the presence of 50 ∝M fenofibrate, the values were 48.8 ± 6.7% for unaffected cells and 54.9 ± 8% for depolarized cells, confirming the expected mitochondrial failure in the presence of fenofibrate-induced Bim expression. Despite of this severe loss of mitochondrial membrane potential in the presence of 50 ∝M fenofibrate, the percentage of dead cells was still low at 48 h time point (Fig. 4C, lower panel).
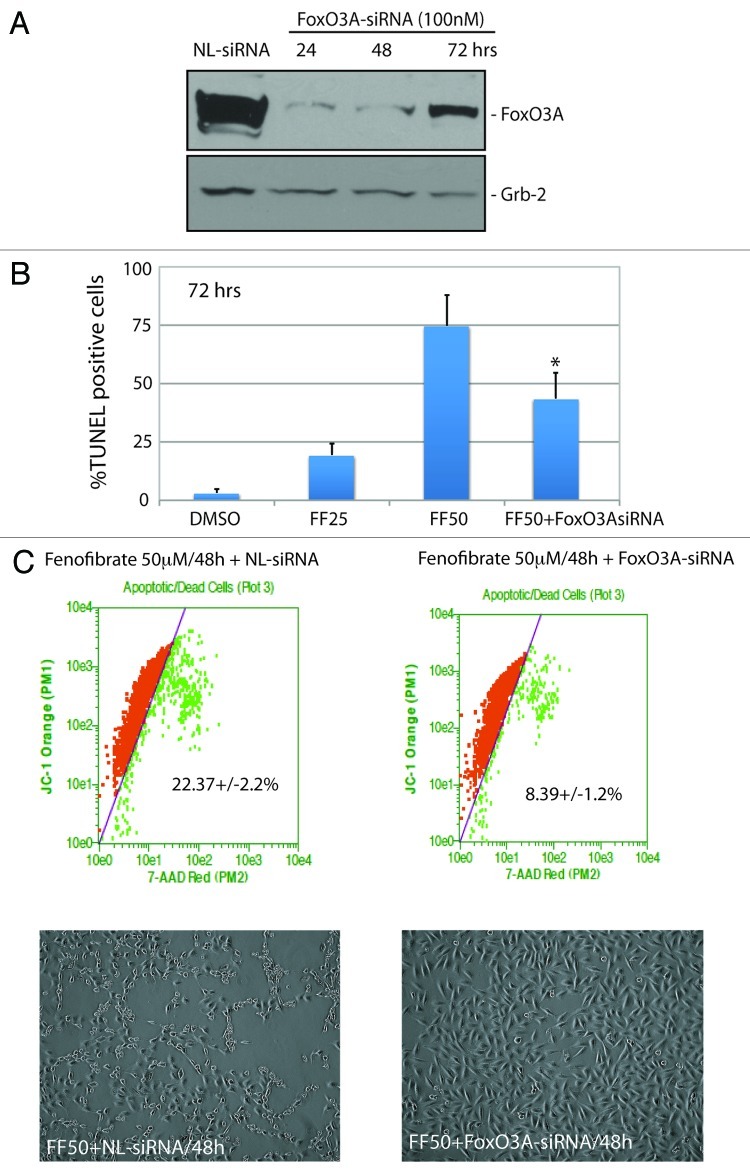
To determine if the activation of FoxO3A-Bim axis is indeed directly involved in the observed fenofibrate action against glioblastoma, we have tested FoxO3A siRNA. The results in Figure 5A demonstrate over 80% reduction in FoxO3A protein levels at 24 and 48 h following cell transfection with 100 nM FoxO3A-specific siRNA. Note, however, that the effectiveness of this treatment was partially compromised at 72 h time point. We have used this siRNA in LN-229 cells exposed to 50 ∝M fenofibrate. As shown in Figure 5B, the percentage of TUNEL-positive cells was reduced from 74.8 ± 13.2% to 43.6 ± 11% in the presence of FoxO3A siRNA, and this partial rescue of the cells was statistically significant. Considering, however, that our siRNA strategy against FoxO3A was much less effective at 72 h time point (Fig. 5A), we have also tested the cells at 48 h. The results in Figure 5C show that the siRNA-induced inhibition of FoxO3A represents a very effective protection against fenofibrate-induced cell death, demonstrated here by the 7-AAD-based cell death assay (upper panel) and by cell morphology (lower panel).
Figure 5. Evaluation of FoxO3A involvement in fenofibrate-induced apoptosis. (A) Western blot analysis showing FoxO3A protein levels in LN-229 cells incubated with 100 nM of OFF-TARGET siRNA against nuclear lamins (NL: 100 nM), and with 100 nM of ON-TARGRT plus SMARTpool siRNA against human FoxO3A mRNA (Thermo Scientific) at 24, 48 and 72 h time points. Grb-2 was used as a loading control. (B) Detection of apoptosis by TUNEL assay in LN-229 cells exposed to 25 and 50 μM fenofibrate (FF25 and FF50) for 72 h. To determine if fenofibrate-induced apoptosis is FoxO3A-dependent, FF50-treated cells were additionally transfected with 100 nM siRNA against FoxO3A. The TUNEL assay (Roche) was adopted for the flowcytometric analysis using GUAVA easyCyte 8HT, and according to the manufacturer recommendations (Guava/Express software). The data are presented as an average % of TUNEL positive cells at 72 h with standard deviation. *Indicates a value statistically different from FF50 (p ≤ 0.05). Statistical significance between two measurements was determined with the two-tailed Student’s t-test. (C) Flowcytometry-based evaluation of cell death using 7-AAD red fluorescent dye from Guava EasyCyte MitoPotential Kit. Exponentially growing LN-229 cells, in 10%FBS, were treated with 50 μM fenofibrate for 48 h in the presence of NL-siRNA (100 nM) or FoxO3A siRNA (100 nM). Note a significant decrease in cell death when the 50 μM fenofibrate treatment is accompanied by FoxO3A siRNA. Lower panel: Representative phase contrast images of LN-229 monolayer cultures taken before evaluation of cell death (original magnification × 20).
Discussion
Our present study demonstrates two distinct responses of Glial tumor cells treated either with 25 ∝M or 50 ∝M fenofibrate. At 25 ∝M, fenofibrate inhibited monolayer and clonogenic growth of the tumor, which involved reversible G1 arrest accompanied by only a marginal level of cell death. In contrast, the cells treated with 50 ∝M fenofibrate undergo massive apoptosis, which started after 72 h following the treatment. Interestingly, this delayed apoptotic effect was not reproduced by other agonists of PPARα; however, it was partially reversed in the presence of PPARα siRNA. In addition, we have observed a direct link between fenofibrate-treatment and FoxO3A nuclear accumulation, which was followed by FoxO3A phosphorylation on serine residue 413, increased transcription from the FoxO-responsive elements and elevated expression and accumulation of FoxO-dependent apoptotic protein, Bim. Since siRNA-mediated inhibition of FoxO3A attenuated fenofibrate-induced apoptosis, we concluded that a specific transcriptional action induced by FoxO3A is responsible for the observed apoptotic action of fenofibrate against glioblastoma.
Glial neoplasms represent almost 50% of adult primary brain tumors, with glioblastoma multiforme (GBM) being the most malignant and practically incurable CNS tumor.29,30 Currently, the only treatments that prolong, to some extent, survival of GBM patients are invasive surgery and aggressive radiotherapy, followed by chemotherapy (temolozomid);31,32 treatment with antibodies and inhibitors (imatinib, gefitinib, avastin);33 or anti-growth factor therapy (for instance antisense strategies against IGF-I or TGFβ34,35), which may prolong survival up to 18–24 mo.
Metabolic responses of Glial tumor cells to fenofibrate have never been tested. Although anticancer properties of fenofibrate are thought to depend, at least partially, on PPARα transcriptional activity, which supports the expression of genes involved in fatty-acid β-oxidation and inhibits glycolysis,6,7 the metabolic consequence(s) of this action in Glial tumor cells are not well defined. According to Otto Warburg’s discovery, who observed, more than six decades ago, a distinctive dependency of tumor cell metabolism from glycolysis,36,37 fenofibrate-induced PPARα activity should trigger a severe energy deficit in glycolysis/glutaminolysis dependent glioblastoma cells.38 PPARα, which is a transcriptional activator of fatty acid β-oxidation machinery, can switch energy metabolism toward fatty acid degradation and decrease glucose uptake by repressing insulin-dependent glucose transporter GLUT4.39,40 In addition, the increased rate of fatty acid and ketone oxidation forces the decline in glucose utilization through the inhibition of glycolytic enzymes.41,42 Note, however, that the human glioblastoma cells tested here are much less sensitive to other potent agonists of PPARα and are only partially rescued from the fenofibrate treatment by PPARα siRNA (Fig. 2).
In parallel to these metabolic events, fenofibrate triggers several cellular responses that seem to be PPARα-independent. First, we have observed highly encouraging in vivo effects of fenofibrate oral administration against melanoma lung metastases, in which the anticancer effect was associated with the inhibition of constitutively active Akt in these cells.2,14 In addition, we observed fenofibrate-induced attenuation of Akt activity in medulloblastoma12 and in glioblastoma13 cell lines. Interestingly, some of the most common mutations found in GBMs include phosphatase and tensin homolog, PTEN gene and overexpression of EGF receptor,43,44 which both may result in a constitutive activation of the PI-3K-Akt pathway and subsequent cytosolic retention of FoxO3A.45 Indeed, Akt and SGK have been shown to phosphorylate FoxO3A on the conserved serine and threonine residues (S52, S256 and Thr752) inhibiting its activity.21,46-48 In addition, other protein kinases, GSK1 and IKK can trigger inhibition of DNA binding and nuclear exclusion of FoxO3A, leading to its cytosolic retention.49,50 In contrast, stressful conditions are known to activate FoxO3A via a direct serine phosphorylation mediated by stress-responsive protein kinases, including MST1, AMPK and JNK, which phosphorylate FoxO3A at multiple serine and threonine residues.21 In response to energy deficit, for instance, an increasing AMP/ATP ratio triggers AMPK-mediated phosphorylation of FoxO3A transactivation domain, including serine residues S399, S413, S555, S588 and S626.51 Similarly, inhibition of the PI3K/AKT pathway results in nuclear accumulation of FoxO3A mainly via disruption of the FoxO3A-14-3-3 complex and subsequent inhibition of FoxO3A nuclear export.52 Subsequent activation of FoxO3A transcriptional activity may either lead to cell cycle arrest by cyclin-dependent kinase inhibitor (CKI) p27Kip1 activation,53 downregulation cyclin D,54 and by protection from oxidative stress, via MnSOD expression,19 or to apoptotic cell death triggered by FoxO3A-dependent genes Bim and PUMA.23,24,27 This wide spectrum of regulation of the target genes by FoxO3A may suggest a multi-factorial mechanism in its regulatory activity.25
Several studies suggest that activation of FoxO3A may affect tumor growth and survival, including prostate and breast cancer cell lines and acute myeloid leukemia, in which either growth factors withdrawal or hypomethylating agents caused FoxO3A-dependent upregulation of pro-apoptotic PUMA and BIM.21,24,26 In addition, survival analyses showed that low expression of FoxO3A is associated with a poor prognosis of patients with ovarian cancer.55 These findings may implicate the FoxO3A in the suppression of tumor growth and induction of cell death,26 especially when the PI3K/AKT pathway is compromised. In this respect, fenofibrate-induced inhibition of the IGF-IR12,13 could also contribute to the enhanced FoxO3A-Bim axis, since the IGF-IR-Akt pathway is a well-known FoxO inhibitor.56-59
We have previously reported that short exposure (up to 48 h) of glioblastoma cells to fenofibrate resulted in a significant decline in intracellular ATP, accompanied by the accumulation of reactive oxygen species (ROS) and ROS-dependent impairment of glioblastoma cell motility.13 In a different experimental setting, TNFα-treated PC12 neuron-like cells underwent FoxO3A-dependent apoptosis only when this pro-apoptotic cytokine was used in the presence of high levels of intracellular ROS, induced by high glucose concentration.58 Also, in this PC12 study, nuclear retention of FoxO3A was accompanied by elevated Bim expression, which happened in the absence of the expected activation of FoxO-dependent antioxidant protein MnSOD.58 Therefore, in glioblastoma cells treated with 50 ∝M fenofibrate, at least two independent stimuli, ROS and AMPK, should be considered as direct modulators of FoxO3A-mediated expression of Bim.
In addition, FoxO3A acetylation can modulate its transcriptional activity. Although our preliminary experiments did not show any detectable changes in FoxO3A acetylation in LN-229 cells treated with fenofibrate (not shown), there is a possibility that fenofibrate-induced mitochondrial crisis results not only in ATP deficit-induced AMPK activation but also may trigger NAD+ release from the stressed mitochondria.60 Consequently, NAD+-dependent de-acetylase, SIRT-1, could be activated and modulate FoxO3A transcriptional activity. This, however, is expected to induce FoxO3A survival mode rather that apoptosis.20,61 Further experiments are required to determine intracellular NAD+ levels and to answer why fenofibrate-treated glioblastoma cells are unable to de-acetylate FoxO3A, which seems to be a necessary step for FoxO3A-mediated survival pathways.20,61 In summary, our results demonstrate that 50 ∝M fenofibrate triggers FoxO3A nuclear retention and its specific serine phosphorylation on the residue S413, which, in the absence of FoxO3A de-acetylation, seems to favor its transcriptional activity toward the expression of pro-apoptotic protein, Bim.
Materials and Methods
Cell culture.
The tumor cells used in this study include human glioblastoma cell line, LN-229 (ATCC #CRL-2611), which has been shown in our previous studies to express PPARα, IGF-IR and wild type PTEN, and two primary human brain tumor cells isolated from patients diagnosed with grade III and grade IV Astrocytic tumors. The cells were maintained as monolayer cultures in DMEM supplemented with 50 U/ml penicillin, 50 ng/ml streptomycin and 10% fetal bovine serum (FBS) at 37°C in a 7% CO2 atmosphere. Exponentially growing cells were treated either with fenofibrate or with two other PPAR agonists, gemfibrozil and WY14643, applied at concentrations ranging from 10 to 100 ∝M. In some experiments, the cells were transfected, using oligofectamin, with ON-TARGETplus SMARTpool siRNA against human PPARα or FoxO3A; or with OFF-TARGET siRNA against nuclear lamin (Thermo Scientific) at 100 and 200 nM as indicated.
Clonogenic growth.
Exponentially growing cultures of LN-229 cells were washed with fresh serum-free medium, trypsinized and transferred to 35 mm culture dishes at 1 x 103 cells/35 mm dish. Clonogenic growth was evaluated 2 weeks after the continuous cell growth in the media containing 10% FBS ± fenofibrate used at 10, 25 and 50 ∝M concentration. The resulting clones were fixed and stained with 0.25% Cristal Violet in methanol as described in our previous work.12
Cell cycle distribution and cell viability.
Flow cytometry (GUAVA easyCyte8HT) was used to detect and quantify these cellular parameters. Briefly, the aliquots of 1 x 106 cells/ml were fixed in 70% ethanol at -20°C overnight. For cell cycle distribution, the cells were centrifuged at 1,600 rpm and the resulting pellets suspended in 1 ml of freshly prepared propidium iodide/RNaseA solution. Cell cycle was evaluated using specialized software CellCycle included in GuavaSoft 1.1. In some experiments, DNA replication was evaluated by BrdU pulse labeling using a DNA replication Assay (GuavaSoft 1.1). Cell death and cell viability were evaluated by two independent assays: TUNEL assay (Roche), which detects DNA damage associated with apoptosis and cell membrane integrity by using 7-amino-actinomycin D (7-AAD), according to the manufacturer’s recommendations (Guava/Express plus and Guava/ViaCount).
Mitochondrial membrane potential.
We used flow cytometry-based MitoPotential Kit according to manufacturer protocol (Guava EasyCyte). Loss of the mitochondrial inner transmembrane potential (ΔΨm) was measured by a cationic dye JC-1, which gives either green or orange fluorescence depending upon mitochondrial membrane depolarization. Following cell exposure to fenofibrate, the cells were harvested by trypsinization, loaded with JC-1 reagent for 30 min and immediately analyzed by Guava EasyCyte using Mito-Potential software (Millipore). The cell treatment with ionophore valinomycin, which fully depolarizes mitochondria, was utilized as a positive control.62 In some cases, JC-1 loaded cells were also labeled with 7-Aminoactinomycin D (7-AAD), which allowes detection and quantification of dead cells in the same experiment.
Luciferase assay.
FoxO transcriptional activity was evaluated in exponentially growing LN-229 in the presence of 0, 25 and 50 ∝M fenofibrate by utilizing the reporter construct in which a region from Fas-ligand promoter, containing three canonical forkhead responsive elements (FHREs), was inserted 5' of the basal promoter controlling the expression of luciferase.58 Fenofibrate-induced activation of FoxO elements was measured by dual-Firefly/Renilla luciferase reporter system (Promega), using a Synergy 2 micro-plate reader (BioTech) and calculated using Gene 5 software (BioTech).
Western blotting.
Total protein extracts were obtained by lysing the cells with 400 ∝l of buffer A [50 mM HEPES; pH 7.5; 150 mM NaCl; 1.5 mM MgCl2; 1 mM EGTA; 10% glycerol; 1% Triton X-100; 1 mM phenylmethylsulfonyl fluoride (PMSF); 0.2 mM Na-orthovanadate and 10 mg/ml aprotinin] on ice for 5 min. Protein extracts (50 ∝g) were separated on a 4–15% gradient SDS-PAGE (BioRad) and transferred using Trans-Blot Turbo system (BioRad). Subcellular fractionation was applied to separate cytoplasmic and nuclear proteins using the Nuclear/Cytoplasmic Fractionation Kit (Thermo Scientific) with some modifications.63 The blots were analyzed by following primary antibodies: anti-PPARα rabbit polyclonal antibody (Abcam); anti-Bim rabbit polyclonal antibody (Cell Signaling); anti-FoxO3A rabbit polyclonal antibody (Cell Signaling); anti-FoxO3A (pS413) rabbit polyclonal (Cell Signaling). Anti-Grb-2 mouse monoclonal antibody (BD Bioscience) was used to monitor loading conditions, and purities of nuclear and cytosolic fractions were determined by anti-Lamin A/C rabbit polyclonal and anti-GAPDH mouse monoclonal antibody (Fitzgerald), respectively.
Immuno-cytofluorescence.
The cells were fixed and permeabilized with the buffer containing 0.02% Triton X-100 and 4% formaldehyde in PBS, followed by washing (3x in PBS) and blocking in 5% BSA for 30 min. Subcellular distribution of FoxO3A was evaluated by utilizing anti-FoxO3A rabbit polyclonal antibody (Cell Signaling) followed by FITC-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Inc.,), and the nuclei were labeled with the DNA binding blue fluorescent dye 4',6-diamidino-2-phenylindole (DAPI). We utilized dual color co-localization analysis (DAPI blue fluorescence and Foxo3A-associated green fluorescence) to quantify FoxO3A nuclear content during the course of the fenofibrate treatment. The images were visualized with the Nikon Eclipse E400 upright fluorescence microscope equipped with Nikon Plan Fluor 100x/1.3 Oil objective, EXI aqua camera (Qimaging), motorized Z-axis, and SlideBook5 acquisition/deconvolution software (Intelligent Imaging Innovations, Inc.). A series of three-dimensional images of each individual picture were deconvoluted to one two-dimensional picture and resolved by adjusting the signal cut-off to near maximal intensity to increase resolution. Nuclear content of FoxO3A was calculated from the entire volume of the nucleus, DAPI labeled, by the Mask Operation procedure included in SlideBook5 software.
Real time RTPCR detection of Bim and MnSOD mRNAs:
The experimental protocol for this methodology has been described in our previous publication 58. Briefly, total RNA was extracted from 1.5x106 LN-229 cells with RNAqueous kit (Ambion) according to the manufacturer protocol. Total RNA (3 μg) was denatured and reverse transcribed using oligo (dT)15 primers and M-MLV reverse transcriptase (Invitrogen). PCR reactions were performed with the LightCycler 480 and SYBRGreen I Master (Roche) using the following primers for human:
Bim.
(forward primer) 5'-AGA TCC CCG CTT TTC ATC TT-3' (reverse primer) 5'-TCT TGG GCG ATC CAT ATC TC-3'
MnSOD.
(forward primer) 5'-TCC ACT GCA AGG AAC AAC AG-3' (reverse primer) 5'-TCT TGC TGG GAT CAT TGA GG-3'.
Acknowledgments
This work was supported by a grant from NIH awarded to KR (RO1CA095518) and from Foundation for Polish Science, POMOST, awarded to M.G.
Glossary
Abbreviations:
- PPARα
peroxisome proliferator activated receptor alpha
- FoxO3A
forkhead-box O 3A transcription factor
- siRNA
small interfering RNA
- IGF-IR
receptor for insulin-like growth factor I
- Bim
pro-apoptotic BH3-only member of Bcl-2 family
- CNS
central nervous system
- FBS
fetal bovine serum
- ROS
reactive oxygen species
- TNF
tumor necrosis factor
- MnSOD
Mn-dependent superoxide dismutase
- NAD+
nicotinamide adenine dinucleotide
- DAPI
4',6-diamidino-2-phenylindole
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick end labeling
- AMPK
AMP-activated protein kinase
- PTEN
phosphatase and tensin homolog
- IKK
IκB-kinase [kinase which phosphorylated the inhibitor (IκB) of nuclear factor kappa B (NFκB)]
- PI3K
phosphatidylinositol 3-kinases
- SGK
serum-and glucocorticoid-induced protein kinase
- MST1
macrophage stimulating 1 (hepatocyte growth factor-like)
- JNK
c-Jun N-terminal kinase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21015
References
- 1.Egerod FL, Nielsen HS, Iversen L, Thorup I, Storgaard T, Oleksiewicz MB. Biomarkers for early effects of carcinogenic dual-acting PPAR agonists in rat urinary bladder urothelium in vivo. Biomarkers. 2005;10:295–309. doi: 10.1080/13547500500218682. [DOI] [PubMed] [Google Scholar]
- 2.Grabacka M, Plonka PM, Urbanska K, Reiss K. Peroxisome proliferator-activated receptor alpha activation decreases metastatic potential of melanoma cells in vitro via down-regulation of Akt. Clin Cancer Res. 2006;12:3028–36. doi: 10.1158/1078-0432.CCR-05-2556. [DOI] [PubMed] [Google Scholar]
- 3.Panigrahy D, Kaipainen A, Huang S, Butterfield CE, Barnés CM, Fannon M, et al. PPARalpha agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc Natl Acad Sci USA. 2008;105:985–90. doi: 10.1073/pnas.0711281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saidi SA, Holland CM, Charnock-Jones DS, Smith SK. In vitro and in vivo effects of the PPAR-alpha agonists fenofibrate and retinoic acid in endometrial cancer. Mol Cancer. 2006;5:13. doi: 10.1186/1476-4598-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorin Y, Kim NH, Feliers D, Bhandari B, Choudhury GG, Abboud HE. Angiotensin II activates Akt/protein kinase B by an arachidonic acid/redox-dependent pathway and independent of phosphoinositide 3-kinase. FASEB J. 2001;15:1909–20. doi: 10.1096/fj..01-0165com. [DOI] [PubMed] [Google Scholar]
- 6.Grabacka M, Reiss K. Anticancer Properties of PPARalpha-Effects on Cellular Metabolism and Inflammation. PPAR Res. 2008;2008:930705. doi: 10.1155/2008/930705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabacka M, Pierzchalska M, Reiss K. Peroxisome Proliferator Activated Receptor α Ligands As Anti-Cancer Drugs Targeting Mitochondrial Metabolism. Curr Pharm Biotechnol. 2010 doi: 10.2174/1389201011314030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardette V, Bongard V, Dallongeville J, Arveiler D, Bingham A, Ruidavets JB, et al. Ten-year all-cause mortality in presumably healthy subjects on lipid-lowering drugs (from the Prospective Epidemiological Study of Myocardial Infarction [PRIME] prospective cohort) Am J Cardiol. 2009;103:381–6. doi: 10.1016/j.amjcard.2008.09.092. [DOI] [PubMed] [Google Scholar]
- 9.Shigeto T, Yokoyama Y, Xin B, Mizunuma H. Peroxisome proliferator-activated receptor alpha and gamma ligands inhibit the growth of human ovarian cancer. Oncol Rep. 2007;18:833–40. [PubMed] [Google Scholar]
- 10.Yokoyama Y, Xin B, Shigeto T, Mizunuma H. Combination of ciglitazone, a peroxisome proliferator-activated receptor gamma ligand, and cisplatin enhances the inhibition of growth of human ovarian cancers. J Cancer Res Clin Oncol. 2011;137:1219–28. doi: 10.1007/s00432-011-0993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strakova N, Ehrmann J, Bartos J, Malikova J, Dolezel J, Kolar Z. Peroxisome proliferator-activated receptors (PPAR) agonists affect cell viability, apoptosis and expression of cell cycle related proteins in cell lines of glial brain tumors. Neoplasma. 2005;52:126–36. [PubMed] [Google Scholar]
- 12.Urbanska K, Pannizzo P, Grabacka M, Croul S, Del Valle L, Khalili K, et al. Activation of PPARalpha inhibits IGF-I-mediated growth and survival responses in medulloblastoma cell lines. Int J Cancer. 2008;123:1015–24. doi: 10.1002/ijc.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drukala J, Urbanska K, Wilk A, Grabacka M, Wybieralska E, Del Valle L, et al. ROS accumulation and IGF-IR inhibition contribute to fenofibrate/PPARalpha -mediated inhibition of glioma cell motility in vitro. Mol Cancer. 2010;9:159. doi: 10.1186/1476-4598-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grabacka M, Placha W, Plonka PM, Pajak S, Urbanska K, Laidler P, et al. Inhibition of melanoma metastases by fenofibrate. Arch Dermatol Res. 2004;296:54–8. doi: 10.1007/s00403-004-0479-y. [DOI] [PubMed] [Google Scholar]
- 15.Araki H, Tamada Y, Imoto S, Dunmore B, Sanders D, Humphrey S, et al. Analysis of PPARalpha-dependent and PPARalpha-independent transcript regulation following fenofibrate treatment of human endothelial cells. Angiogenesis. 2009;12:221–9. doi: 10.1007/s10456-009-9142-8. [DOI] [PubMed] [Google Scholar]
- 16.Nadanaciva S, Dykens JA, Bernal A, Capaldi RA, Will Y. Mitochondrial impairment by PPAR agonists and statins identified via immunocaptured OXPHOS complex activities and respiration. Toxicol Appl Pharmacol. 2007;223:277–87. doi: 10.1016/j.taap.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Zungu M, Felix R, Essop MF. Wy-14,643 and fenofibrate inhibit mitochondrial respiration in isolated rat cardiac mitochondria. Mitochondrion. 2006;6:315–22. doi: 10.1016/j.mito.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Wybieralska E, Szpak K, Górecki A, Bonarek P, Miękus K, Drukała J, et al. Fenofibrate attenuates contact-stimulated cell motility and gap junctional coupling in DU-145 human prostate cancer cell populations. Oncol Rep. 2011;26:447–53. doi: 10.3892/or.2011.1321. [DOI] [PubMed] [Google Scholar]
- 19.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–21. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 20.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–63. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;9:1091–6. doi: 10.4161/cc.9.6.11035. [DOI] [PubMed] [Google Scholar]
- 22.Chow KU, Boehrer S, Geduldig K, Krapohl A, Hoelzer D, Mitrou PS, et al. In vitro induction of apoptosis of neoplastic cells in low-grade non-Hodgkin’s lymphomas using combinations of established cytotoxic drugs with bendamustine. Haematologica. 2001;86:485–93. [PubMed] [Google Scholar]
- 23.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 24.Thépot S, Lainey E, Cluzeau T, Sébert M, Leroy C, Adès L, et al. Hypomethylating agents reactivate FOXO3A in acute myeloid leukemia. Cell Cycle. 2011;10:2323–30. doi: 10.4161/cc.10.14.16399. [DOI] [PubMed] [Google Scholar]
- 25.Liu JW, Chandra D, Rudd MD, Butler AP, Pallotta V, Brown D, et al. Induction of prosurvival molecules by apoptotic stimuli: involvement of FOXO3a and ROS. Oncogene. 2005;24:2020–31. doi: 10.1038/sj.onc.1208385. [DOI] [PubMed] [Google Scholar]
- 26.Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27:2312–9. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–22. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber A, Paschen SA, Heger K, Wilfling F, Frankenberg T, Bauerschmitt H, et al. BimS-induced apoptosis requires mitochondrial localization but not interaction with anti-apoptotic Bcl-2 proteins. J Cell Biol. 2007;177:625–36. doi: 10.1083/jcb.200610148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–78. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terzis AJ, Niclou SP, Rajcevic U, Danzeisen C, Bjerkvig R. Cell therapies for glioblastoma. Expert Opin Biol Ther. 2006;6:739–49. doi: 10.1517/14712598.6.8.739. [DOI] [PubMed] [Google Scholar]
- 31.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 32.Stupp R, Weber DC. The role of radio- and chemotherapy in glioblastoma. Onkologie. 2005;28:315–7. doi: 10.1159/000085575. [DOI] [PubMed] [Google Scholar]
- 33.Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, et al. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99-08. Clin Cancer Res. 2006;12:4899–907. doi: 10.1158/1078-0432.CCR-06-0773. [DOI] [PubMed] [Google Scholar]
- 34.Trojan J, Cloix JF, Ardourel MY, Chatel M, Anthony DD. Insulin-like growth factor type I biology and targeting in malignant gliomas. Neuroscience. 2007;145:795–811. doi: 10.1016/j.neuroscience.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 35.Hau P, Jachimczak P, Schlingensiepen R, Schulmeyer F, Jauch T, Steinbrecher A, et al. Inhibition of TGF-beta2 with AP 12009 in recurrent malignant gliomas: from preclinical to phase I/II studies. Oligonucleotides. 2007;17:201–12. doi: 10.1089/oli.2006.0053. [DOI] [PubMed] [Google Scholar]
- 36.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 37.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–70. [PubMed] [Google Scholar]
- 38.Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget. 2010;1:552–62. doi: 10.18632/oncotarget.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed W, Ziouzenkova O, Brown J, Devchand P, Francis S, Kadakia M, et al. PPARs and their metabolic modulation: new mechanisms for transcriptional regulation? J Intern Med. 2007;262:184–98. doi: 10.1111/j.1365-2796.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 40.Finck BN, Kelly DP. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol. 2002;34:1249–57. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 41.Randle PJ. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev. 1998;14:263–83. doi: 10.1002/(SICI)1099-0895(199812)14:4<263::AID-DMR233>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe RR. Metabolic interactions between glucose and fatty acids in humans. Am J Clin Nutr. 1998;67(Suppl):519S–26S. doi: 10.1093/ajcn/67.3.519S. [DOI] [PubMed] [Google Scholar]
- 43.Reis RM, Könü-Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H. Genetic profile of gliosarcomas. Am J Pathol. 2000;156:425–32. doi: 10.1016/S0002-9440(10)64746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber GL, Parat MO, Binder ZA, Gallia GL, Riggins GJ. Abrogation of PIK3CA or PIK3R1 reduces proliferation, migration, and invasion in glioblastoma multiforme cells. Oncotarget. 2011;2:833–49. doi: 10.18632/oncotarget.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J Biol Chem. 2002;277:47928–37. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 46.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 47.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–65. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skurk C, Maatz H, Kim HS, Yang J, Abid MR, Aird WC, et al. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J Biol Chem. 2004;279:1513–25. doi: 10.1074/jbc.M304736200. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Wang Z, Kong D, Li R, Sarkar SH, Sarkar FH. Regulation of Akt/FOXO3a/GSK-3beta/AR signaling network by isoflavone in prostate cancer cells. J Biol Chem. 2008;283:27707–16. doi: 10.1074/jbc.M802759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/S0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 51.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 52.Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, et al. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–6. [PubMed] [Google Scholar]
- 53.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–7. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, et al. Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol. 2002;22:7842–52. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fei M, Zhao Y, Wang Y, Lu M, Cheng C, Huang X, et al. Low expression of Foxo3a is associated with poor prognosis in ovarian cancer patients. Cancer Invest. 2009;27:52–9. doi: 10.1080/07357900802146204. [DOI] [PubMed] [Google Scholar]
- 56.Dávila D, Torres-Aleman I. Neuronal death by oxidative stress involves activation of FOXO3 through a two-arm pathway that activates stress kinases and attenuates insulin-like growth factor I signaling. Mol Biol Cell. 2008;19:2014–25. doi: 10.1091/mbc.E07-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Bruyne E, Bos TJ, Schuit F, Van Valckenborgh E, Menu E, Thorrez L, et al. IGF-1 suppresses Bim expression in multiple myeloma via epigenetic and posttranslational mechanisms. Blood. 2010;115:2430–40. doi: 10.1182/blood-2009-07-232801. [DOI] [PubMed] [Google Scholar]
- 58.Wilk A, Urbanska K, Yang S, Wang JY, Amini S, Del Valle L, et al. Insulin-like growth factor-I-forkhead box O transcription factor 3a counteracts high glucose/tumor necrosis factor-α-mediated neuronal damage: implications for human immunodeficiency virus encephalitis. J Neurosci Res. 2011;89:183–98. doi: 10.1002/jnr.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng WH, Kar S, Quirion R. Insulin-like growth factor-1-induced phosphorylation of the forkhead family transcription factor FKHRL1 is mediated by Akt kinase in PC12 cells. J Biol Chem. 2000;275:39152–8. doi: 10.1074/jbc.M002417200. [DOI] [PubMed] [Google Scholar]
- 60.Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64:166–87. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 62.Troiano L, Ferraresi R, Lugli E, Nemes E, Roat E, Nasi M, et al. Multiparametric analysis of cells with different mitochondrial membrane potential during apoptosis by polychromatic flow cytometry. Nat Protoc. 2007;2:2719–27. doi: 10.1038/nprot.2007.405. [DOI] [PubMed] [Google Scholar]
- 63.Trojanek J, Ho T, Del Valle L, Nowicki M, Wang JY, Lassak A, et al. Role of the insulin-like growth factor I/insulin receptor substrate 1 axis in Rad51 trafficking and DNA repair by homologous recombination. Mol Cell Biol. 2003;23:7510–24. doi: 10.1128/MCB.23.21.7510-7524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]