Abstract
Background
Transcription of plastid-encoded genes requires two different DNA-dependent RNA polymerases, a nuclear-encoded polymerase (NEP) and plastid-encoded polymerase (PEP). Recent studies identified two related pfkB-type carbohydrate kinases, named FRUCTOKINASE-LIKE PROTEIN (FLN1 and FLN2), as components of the thylakoid bound PEP complex in both Arabidopsis thaliana and Sinapis alba (mustard). Additional work demonstrated that RNAi-mediated reduction in FLN expression specifically diminished transcription of PEP-dependent genes.
Results
Here, we report the characterization of Arabidopsis FLN knockout alleles to examine the contribution of each gene in plant growth, chloroplast development, and in mediating PEP-dependent transcription. We show that fln plants have severe phenotypes with fln1 resulting in an albino phenotype that is seedling lethal without a source of exogenous carbon. In contrast, fln2 plants display chlorosis prior to leaf expansion, but exhibit slow greening, remain autotrophic, can grow to maturity, and set viable seed. fln1 fln2 double mutant analysis reveals haplo-insufficiency, and fln1 fln2 plants have a similar, but more severe phenotype than either single mutant. Normal plastid development in both light and dark requires the FLNs, but surprisingly skotomorphogenesis is unaffected in fln seedlings. Seedlings genetically fln1-1 with dexamethasone-inducible FLN1-HA expression at germination are phenotypically indistinguishable from wild-type. Induction of FLN-HA after 24 hours of germination cannot rescue the mutant phenotype, indicating that the effects of loss of FLN are not always reversible. Examination of chloroplast gene expression in fln1-1 and fln2-1 by qRT-PCR reveals that transcripts of PEP-dependent genes were specifically reduced compared to NEP-dependent genes in both single mutants.
Conclusions
Our results demonstrate that each FLN protein contributes to wild type growth, and acting additively are absolutely essential for plant growth and development.
Keywords: pfkB-type carbohydrate kinase, FLN, Chloroplast transcription, pTAC complex, Arabidopsis thaliana
Background
The plastid, an essential plant organelle, is the sole site of multiple biosynthetic pathways, such as de novo fatty acid and amino acid synthesis. After differentiation into the photosynthetically competent chloroplast, membrane-localized light harvesting complexes convert light energy into chemical energy and soluble enzymes reduce CO2 into various carbohydrates. Plastid genomes do not encode the complete repertoire of proteins required for these reactions; therefore, nuclear-encoded proteins are present in almost all plastid-localized multi-protein complexes. This requires import of many proteins into the plastid, their targeting to the correct sub-organellar locale, and in most cases, assembly with plastid-encoded subunits. Expression of nuclear-encoded and chloroplast-encoded genes must be coordinated to produce the appropriate subunit stoichiometries and sufficient protein complexes to meet the metabolic needs of the plant [1].
Transcription of plastid-encoded genes is achieved by two different types of DNA-dependent RNA polymerases in land plants [2]. The plastid-encoded RNA polymerase (PEP) resembles bacterial RNA polymerase in sequence identity, requirement for multiple subunits, and use of sigma factors conferring promoter specificity. Plastids contain a second unrelated type, single-subunit nuclear-encoded RNA polymerase (NEP) [3]. Deletions of individual PEP subunit-encoding genes affect the transcription of a specific subset of plastid-encoded genes, indicating the two polymerases transcribe unique sets of plastid genes, although some genes are transcribed by both PEP and NEP [4-6]. PEP can be found in two different complexes, a stromally localized soluble and a membrane attached form [2], the latter referred to as TAC for transcriptionally active chromosome [7]. Interestingly, NEP transcribes the plastid-encoded PEP core polymerase subunits, suggesting a hierarchy of transcription.
Biochemical isolation combined with mass spectrometry of the TAC complex from multiple species [8-12] revealed some additional components, with up to approximately 50 proteins (pTACs) identified in addition to the core PEP polymerase subunits [13]. Of note was the presence of two related proteins in the pkfB-type carbohydrate kinase family, initially reported by Suzuki et al. [12] and confirmed by others [9,14]. These proteins were later named FRUCTOKINASE-LIKE PROTEIN1 (FLN1, At3g54090), and FLN2 (At1g69200) [15]. FLN1 interacts with thioredoxin z and contains 2 vicinal cysteines required for this interaction [15]. The role of the FLN proteins in the PEP complex is unknown and adds yet another dimension to plastid gene transcriptional regulation.
Carbohydrate kinase-like proteins in other organisms have been reported to serve regulatory roles as opposed to a direct catalytic function, while others appear to have both functions. The budding yeast galactokinase-like protein Gal3p lacks catalytic activity but binds both ATP and galactose. The Gal3p-galactose-ATP complex binds to the transcriptional inhibitor Gal80p, reducing Gal80p’s inhibitory effect on the GAL promoter [16]. Similarly, a yeast hexokinase 2 isozyme, Hxk2p, an active kinase, has a nuclear signaling function that does not require catalytic activity [17,18]. In ArabidopsisHEXOKINASE1 (HXK1) is an active kinase, but similarly is active as a signaling protein even when its catalytic activity is eliminated by mutation [19].
Sugar kinases are grouped into three evolutionarily distinct families unrelated in amino acid sequence and structure – the hexokinase, ribokinase, and galactokinase families [20]. FLN proteins belong to the pfkB-type of carbohydrate kinase, a large group within the ribokinase family, which includes a variety of known and predicted carbohydrate, phospho-carbohydrate and pyrimidine kinases, but remains a poorly characterized group. Members include the two Arabidopsis adenosine kinases (ADK) shown to play an important role in adenylate and methyl recycling [21]. Plants with reduced ADK expression have growth defects [22] and reduced root gravitropism [23], defects linked to altered cytokinin levels [24]. In addition to the FLNs, another chloroplast-localized pfkB-type kinase, NARA5, has been characterized and is important for chloroplast development and plastid gene expression [25].
For the two closely related pfkB-type proteins, FLN1 and FLN2, previous work using inducible RNAi in Arabidopsis implied that these two proteins function equivalently in causing chlorosis of emerging leaves [15]. Curiously, using VIGS-mediated expression of RNAi in N. benthamiana, the same study showed that reducing tobacco FLN1 induced chlorosis, while reducing FLN2 had very little impact. RNAi’s effects can be variable and often incomplete. Furthermore, FLN mRNA was only transiently down-regulated in these experiments. We therefore sought to analyze the effect of FLN loss-of-function in plants genetically null for each individual FLN. We analyzed the phenotypes of individual fln1 and fln2 null plants, as well as fln1 fln2 double mutant seedlings. Our results provide new evidence regarding FLN function in Arabidopsis. In contrast to the previous results, where in Arabidopsis each protein contributes equally, analysis of null alleles reveals a larger role for FLN1. Additionally, we demonstrate for the first time, that together, FLN1 and FLN2 are essential for growth and development. We also show that initial growth in the dark is unaffected in fln, supporting the role of FLNs in development of photosynthetic competency. Similar to the previous study, transcription of PEP-dependent chloroplast genes is strongly and specifically affected in each stable fln mutant. Finally, using inducible expression, we demonstrate that FLN1 is required early during germination in the light, and expression after cotyledon emergence cannot rescue the mutant phenotype, suggesting a developmentally regulated role for FLNs.
Results
FLN1 and FLN2 loss-of-function mutants have severe chlorotic phenotypes
We took a reverse genetics approach and analyzed the available T-DNA insertion alleles to identify the role of each FLN in plant growth and development. We analyzed one FLN1 and two FLN2 insertion alleles (Figure 1A). Progeny from plants heterozygous for each T-DNA allele were assessed for segregation of any observable phenotypes after germination of seed and subsequent growth on GM plates. Two phenotypes were observed; each line segregated 3:1 for a green:pale/chlorotic phenotype (χ2 goodness-of-fit test for 3:1 segregation: fln1-1, χ2 = 3.5, p-value = 0.060; fln2-1 χ2 = 0.086, p-value = 0.76; fln2-2, χ2 = 0.030, p-value = 0.86. PCR-based genotyping revealed that the pale/chlorotic seedlings are homozygous for their respective T-DNA insertion.
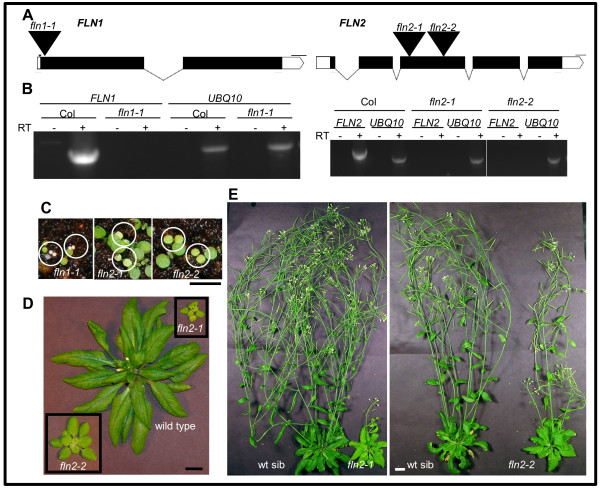
Figure 1.
Characterization offlninsertion lines.(A) Representation of the FLN1 (At3g54090) and FLN2 (At1g69200) genes. T-DNA insertions are indicated with large, black triangles (not to scale) with allele designations above them. Exons, UTRs, introns, and deletions are indicated by black boxes, white boxes, lines, and a small triangle, respectively. Scale bars represent 100 bp. Red arrows indicate positions of primers used for RT-PCR. Graphics were generated with Exon-Intron Graphic Maker (http://wormweb.org/exonintron). (B) RT-PCR analysis of fln1 and fln2 alleles. 7-day old light grown seedlings were used for RNA extraction. The UBQ10 reactions indicate equivalent RNA inputs for Col-0 and mutant lines, and minus (−) RT reactions are samples treated equivalently except reverse transcriptase was not included in the RT reaction. Experiments are representative of 3 biological replicates. (C) 7-day old phenotype of fln mutants. Seeds segregating for fln alleles grown under a 16 h photoperiod at 18o C. Homozygous individuals are circled. Scale bar represent 0.5 cm. (D) 40-day old fln2 plants. Seedlings from (C) were transferred to individual pots for continued growth. (E) 70-day old fln2 plants. Plant from (D) after 30 more days of growth. For (D) and (E) bars represent 1 cm.
We confirmed the T-DNA insertion position for each allele with PCR and DNA sequencing across the T-DNA-gene junction (Additional file 1: Figure S1). For fln1-1 and fln2-2 only one insertion was detected. PCRs with fln2-1 T-DNA primers generated a product with either gene-specific primer, suggesting that this allele has two inverted T-DNA insertions (Additional file 1: Figure S1).
To determine if fln1 and fln2 T-DNA insertion lines synthesize authentic mRNA, RNA was extracted from pooled pale/chlorotic seedlings and used for subsequent RT-PCR experiments (Figure 1B). Using primers specific to FLN1, no product was detectable from fln1-1 cDNA, whereas a product was visible from Col-0 cDNA (Figure 1B,left panel). Similarly, a product was generated with FLN2-specific primers only from Col-0, not from cDNA synthesized from fln2-1 or fln2-2 RNA (Figure 1B, right panel). A visible PCR product in all samples using UBQ10 primers indicated the presence of roughly equivalent input of cDNA template. No product was made when reverse-transcriptase (RT) was omitted from the cDNA synthesis reaction (−RT lanes) indicating that cDNA, not genomic DNA, was the source of template in + RT reactions. These results indicate that the one FLN1 and the two FLN2 insertion lines are all likely to be null alleles.
Because the phenotypes originally observed resulted from seedling growth on media supplemented with B-vitamins and 1% sucrose, we sought to test whether the same pale/chlorotic phenotype recapitulated in seedlings grown on soil and to examine if the mutants could grow and develop autotrophically. Seeds from FLN/fln parents were sown directly on soil and grown under continuous light (116 μmol/sec-1m-2). The similar phenotypes observed on GM media were also observed in seedlings grown on soil (Figure 1C). fln1-1 seedlings have small yellow cotyledons that never green, they do not produce true leaves and die within two weeks after germination. fln2 mutants grow similarly on soil and on GM plates. They have pale green cotyledons with the cotyledons of fln2-2 slightly larger than those of fln2-1 (Figure 1C). The rosettes of the fln2 alleles are smaller than wild-type, and true leaves slowly green (Figure 1D). fln2-2 plants have slightly larger rosettes and are greener than fln2-1, likely due to the mixed ecotype background of fln2-2. Eventually, all fln2 mutants bolt and make seeds. Adult fln2-1 plants have fewer inflorescences than do their wild-type siblings, with chlorotic stems, cauline leaves and sepals (Figure 1E). fln2-2 plants, on the other hand, produce a taller inflorescence compared to fln2-1.
Inducible expression of FLN1-HA complements fln1-1
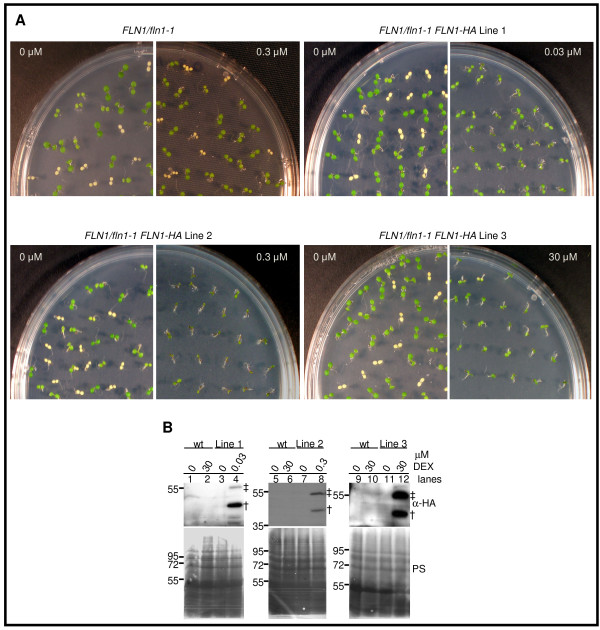
The similar phenotypes from the two independent insertion alleles of FLN2 strongly indicate that the mutant phenotype results from loss of FLN2 expression. However, only one insertion allele of FLN1 is available. To demonstrate that absence of FLN1 is responsible for the observed phenotype, FLN1/fln1-1 plants were transformed with expression constructs for constitutive expression of an HA-tagged form of FLN1. All of these transformants exhibited phenotypes consistent with transgene silencing (data not shown), preventing analysis of complementation. To overcome this problem, FLN1/fln1-1 plants were transformed with a dexamethasone (DEX)-inducible [26,27] version of the same FLN1-HA open reading frame. Progeny from three independent FLN1/fln1-1 FLN1-HA lines were assessed for phenotypic changes and FLN1-HA induction after growth on DEX-containing media (Figure 2 and Table 1). For all three lines, two anti-HA immunoreactive species could be detected in extracts from seedlings grown on DEX-containing media (Figure 2B, lanes 4, 8, 12), but not in seedlings from the same lines grown on solvent control (Figure 2B, lanes 3, 7, 11) despite loading similar amounts of total protein (Figure 2B, lower panels). This indicates that FLN1-HA expression is DEX-dependent. No anti-HA immunoreactivity could be detected in protein extracts from wild-type, non-transformed control seedlings (green progeny of FLN1/fln1-1 only) grown either on solvent control (Figure 2B, lanes 1, 5, 9) or on 30 μM DEX (Figure 2B, lanes 2, 6, 10) demonstrating that the protein induced by DEX in the FLN1/fln1-1 FLN1-HA lines requires the FLN1-HA transgene rather than DEX treatment alone. We presume the 55 kDa species (double-cross, Figure 2B) to be full-length FLN1-HA prior to import into the chloroplast, and the faster migrating species (single-cross, Figure 2B) to be FLN1-HA after import and transit peptide cleavage.
Figure 2.
Complementation offln1-1with dexamethasone-inducibleFLN1-HA.(A) Complementation of fln1-1 phenotype. BC1 F2 seeds from FLN1/fln1-1 or T3FLN1/fln1-1 FLN1-HA seeds were plated on the indicated concentration of dexamethasone and grown for 7 days at 20o C under constant light. 0 μM is DMSO solvent control. Plates shown are one representative experiment of at least three independent experiments. (B) FLN1-HA induction by dexamethasone in FLN1/fln1-1 FLN1-HA lines. Proteins were extracted from seedlings after 7 days growth, under constant light at 20o C, on GM plates supplemented with indicated concentration of dexamethasone (DEX) and immunoblotted with anti-HA antibodies. Ponceau S stained (PS) membrane is shown as a loading control. Wild-type (wt) plants were green seedlings from a FLN1/fln1-1 parent without the FLN1-HA transgene. Seedlings processed for the “0” treatment for these complementation lines were green siblings (mixed genotype) that segregated with the fln1-1 phenotype in the absence of DEX. Seedlings for other treatments consisted of a pool of all seedlings (all possible genotypes) on the plate (as fln1-1 phenotype was complemented on DEX). The presumed FLN1-HA precursor band is denoted by a double-cross, and the mature, processed FLN-HA by a single-cross. Size markers, in kDa, are to the left of blots. Lines 1, 2, 3 refer to three independent FLN/fln1-1 FLN1-HA lines. Seedlings were F3 seedlings from a FLN/fln1-1 FLN1-HA parental genotype.
Table 1.
Goodness-of-fit test forfln1-1segregation in 7-day old T3populations fromFLN1/fln1-1 FLN1-HAcomplementation lines grown on the indicated dexamethasone concentration
| parental genotype | [DEX] (μM) | green | white | variegated | χ2value | p-value |
|---|---|---|---|---|---|---|
|
FLN1/fln1-1 |
0 |
64 |
26 |
0 |
0.72 |
0.39 |
| 0.3 |
88 |
26 |
0 |
0.29 |
0.58 |
|
| 30 |
90 |
22 |
0 |
1.71 |
0.19 |
|
|
FLN1/fln1-1 FLN1-HA Line 1 |
0 |
90 |
27 |
0 |
0.23 |
0.63 |
| 0.03 |
117 |
0 |
0 |
39.00 |
<0.0001 |
|
|
FLN1/fln1-1 FLN1-HA Line 2 |
0 |
85 |
27 |
0 |
0.05 |
0.82 |
| 0.3 |
96 |
0 |
0 |
32.00 |
<0.0001 |
|
| FLN1/fln1-1 FLN1-HA Line 3 | 0 |
84 |
35 |
0 |
1.23 |
0.26 |
| 3.0 |
95 |
0 |
16 |
6.63 |
0.01 |
|
| 30 | 118 | 0 | 6 | 21.73 | <0.0001 |
Complementation of the fln1-1 phenotype by FLN1-HA was assessed by observing the segregation ratio of green to white seedlings in progeny from FLN1/fln1-1 FLN1-HA parents after germination on DEX media. In FLN1/fln1-1 progeny lacking the FLN1-HA transgene, DEX treatment did not change the 3:1 segregation ratio of green to white seedlings, nor affect the phenotype (Figure 2A, Table 1). However, growth on DEX media abolished segregation of any observable fln1-1 seedlings in selfed progeny from the three independent FLN1/fln1-1 FLN1-HA parents. This result, combined with the tight induction of FLN1-HA by DEX in these same lines (Figure 2B), demonstrates that fln1-1 can be rescued by DEX-induced expression of FLN1-HA. These lines exhibited complementation on different concentrations of DEX ranging from 0.03 to 30 μM. Lines 1 and 2 fully complement the fln1-1 phenotype when grown on 0.03 and 0.3 μM DEX, respectively, although line 2 seedlings are smaller (Figure 2A, Table 1). Line 3 partially complements this phenotype when grown on both 3.0 and 30 μM DEX. No white fln1-1 seedlings were observed either concentration, but several seedlings exhibiting white-green variegated cotyledons were observed at the highest DEX dose used (Figure 2A, Table 1).
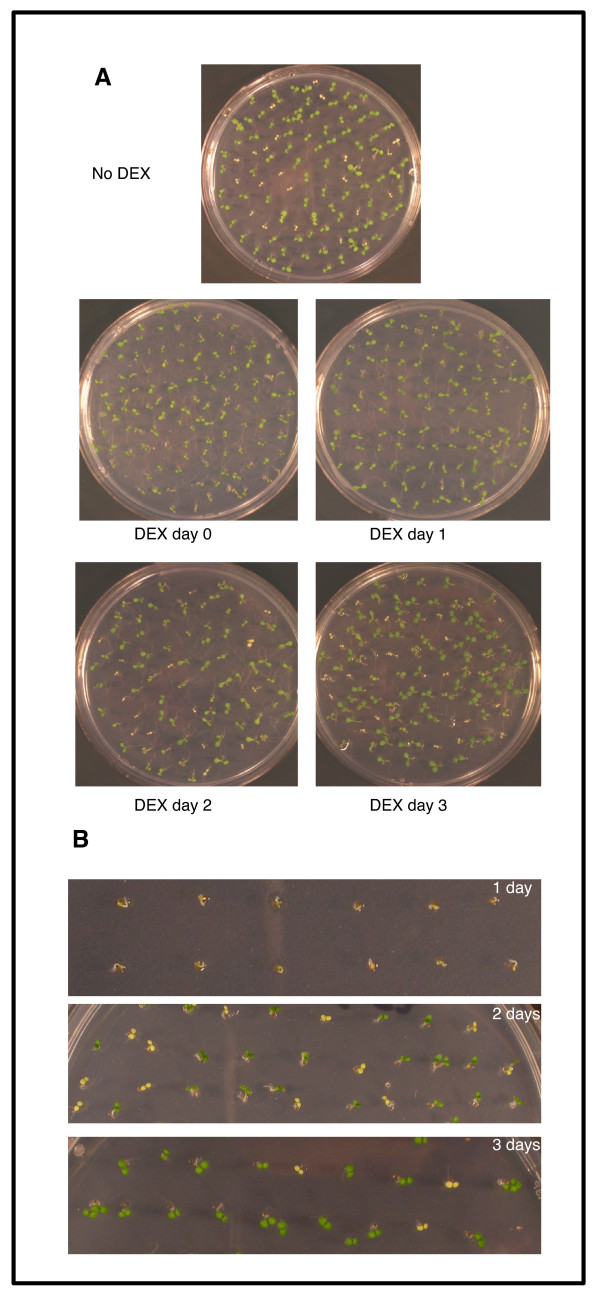
The ability of FLN1-HA to rescue the mutant phenotype at various times after germination was investigated to assess how early FLN1 expression is required for wild type growth. Seeds from FLN1/fln1-1 FLN1-HA lines were plated, stratified for 2 days at 4 °C and then grown at room temperature (RT). DEX was added to the plates when first moved to RT (0 day), or 1, 2 or 3 days later. The number of pale seedlings was scored after 7 days (Figure 3). DEX addition immediately after stratification (0 day) and after 1 day rescued the mutant phenotype, as no or only a few pale seedlings were observed (Figure 3A, Table 2). However, DEX addition after 2 or 3 days of growth failed to rescue all three lines because pale seedlings were observed at the expected ratio (Figure 3A, Table 2).
Figure 3.
Time-dependency offln1complementation with DEX treatment.(A) Seeds from FLN1/fln1-1 FLN1-HA line 2 were plated, stratified for 2 days at 4 °C, then incubated at RT. DEX was added at the indicated time after incubation at RT and photographed at 7 days. (B) Photographs of seedlings at 1, 2 and 3 days after germination immediately prior to DEX addition, showing the extent of seedling growth at time of DEX application. The experiment in (A) was performed 3 times with 3 independent lines and results were similar for all lines and replicas. Representative pictures are shown.
Table 2.
Dexamethasone induction of FLN1-HA rescues the mutant phenotype only when applied by 24 hours after plating
| parental genotype | time after dex (days) | green | white | χ2for 3:1 green:white | p-value |
|---|---|---|---|---|---|
|
FLN1/fln1-1 FLN1-HA Line 2 |
0 |
108 |
0 |
36 |
<0.0001 |
| 1 |
206 |
0 |
68.667 |
<0.0001 |
|
| 2 |
156 |
41 |
1.843 |
0.1746 |
|
| 3 |
173 |
41 |
3.894 |
0.0485 |
|
| FLN1/fln1-1 FLN1-HA Line 3 | 0 |
112 |
7 |
23.2 |
<0.0001 |
| 1 |
203 |
19 |
32.0 |
<0.0001 |
|
| 2 |
169 |
55 |
0.024 |
0.8774 |
|
| 3 | 163 | 56 | 0.038 | 0.8453 |
To determine the developmental stage at which FLN1-HA expression is able to restore fln1 seedlings to wild type, the development of the seedlings was noted immediately prior to DEX addition (Figure 3B). After 1 day, radicle emergence was observed, but cotyledon expansion had not yet occurred and identification of fln seedlings was not possible (Figure 3B, top). After 2 days, wild-type cotyledons were reflexed, fully green and almost completely expanded, and the pale fln cotyledons were evident (Figure 3B, middle). These results indicate that FLN1-HA expression must occur very early in seedling development in order to rescue the mutant phenotype.
Loss of fln1 or fln2 specifically disrupts PEP-dependent transcription
To explore the role of each FLN in chloroplast transcription, chloroplast-encoded mRNA levels were determined in fln1-1 and fln2-1. Previously, the effect of reduced FLN expression by inducible RNA-interference was investigated in Arabidopsis and tobacco leaves [15], but had not been determined in stable fln null plants. Here, we examined levels of mRNA in our fln null alleles for the three classes of chloroplast-encoded genes: PEP-dependent (Class I, Figure 4A), NEP-dependent (Class III, Figure 4B), both PEP- and NEP- transcribed C(Class II, Figure 4) and for nuclear genes encoding plastid-localized proteins (Figure 4D). RNA from 7-day old fln seedlings and their phenotypic wild-type siblings (a combination of FLN and FLN/fln genotypes) were subjected to quantitative real-time RT-PCR (qRT-PCR) analysis (Figure 4).
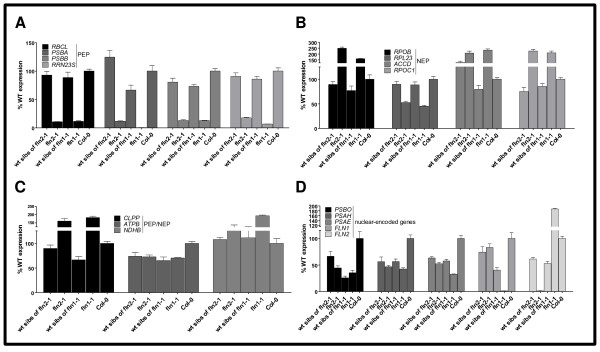
Figure 4.
Expression of chloroplast encoded genes inflnseedlings. qRT-PCR analysis of the indicated gene transcript levels was performed in 7-day-old seedlings grown as in Figure 1B. Four classes of genes were examined, PEP-dependent (A), NEP-dependent (B) PEP- and NEP-transcribed (C), and nuclear-encoded (D). Data shown are the mean ± standard error of the mean (SE) of a representative experiment (n = 3). Expression levels are presented as the percentage relative to wild type.
Expression of PEP-dependent genes rbcL, psbA, psbB, and rrn23S was strongly reduced in both fln1-1 and fln2-1 seedlings compared to Col-0 and to their respective phenotypically wild-type siblings (Figure 4A). Transcripts for NEP-dependent genes rpoB, accD, and rpoC1, on the other hand, were slightly elevated in fln seedlings, except for rpl23 transcripts with only 50% of wild type expression in the mutants. clpP, atpB, and ndhB are transcribed by both PEP and NEP. Both clpP and ndhB mRNAs were slightly up-regulated in the mutants, while atpB mRNA was similar in both fln mutants to their respective wild type siblings (Figure 4C).
Expectedly, mRNAs for FLN1 and FLN2 were absent in cDNA prepared from their respective mutants. Interestingly, FLN2 mRNA is two-fold higher in fln1-1 than Col-0, but the opposite is not true, FLN1 mRNA is not elevated in fln2-1 (Figure 4D). For other nuclear-encoded chloroplast-localized proteins, fln and wild-type siblings had similar transcript levels, approximately a 50% reduction compared to Col-0 (Figure 4D). This reduction likely results from the mixed genotypes of the phenotypically wild type sibling seedlings consisting of FLN/FLN and FLN/fln1-1 genotypes. In summary, only PEP-dependent transcripts were consistently and severely down-regulated in fln seedlings, indicating that FLNs are positive regulators of TAC complex function.
FLN proteins are not required for plant cell elongation in the dark
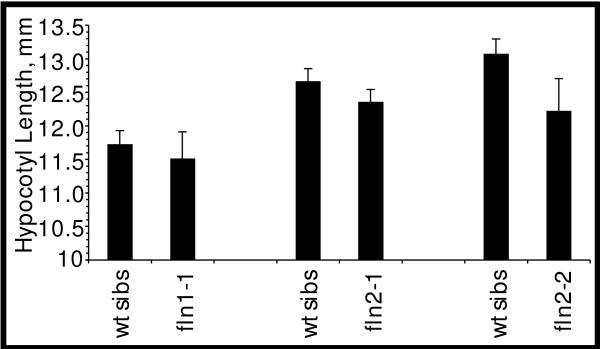
Given the strong dependence on FLN expression for wild-type growth of light-grown seedlings, we sought to ascertain if FLN proteins are required for skotomorphogenesis. We examined hypocotyl elongation in dark-grown 7 day-old seedlings from FLN/fln parents. All seedlings had elongated hypocotyls, an apical hook and unexpanded cotyledons (data not shown). We measured hypocotyl length of etiolated seedlings to assess if cell expansion in the dark was affected (Figure 5). fln hypocotyl lengths were not significantly shorter than those of their corresponding wild-type siblings as assessed by a Student’s t-test (p-values for comparison to wild-type siblings: fln1-1, 0.607; fln2-1, 0.202; and fln2-2, 0.0821; α = 0.05).
Figure 5.
Hypocotyl length of 7 day old dark-grownflnmutants. Progeny from FLN/fln parents were grown in the dark for 7 days at 20o C. Measurements of fln hypocotyls are shown next to their respective phenotypically wild-type siblings and combined for two different growth experiments. No fln mutant was significantly different from wild type siblings using a Student’s T-test (α=0.05). Bars are se. (35≥ n ≥27).
Dark-grown seedlings were then exposed to light. Cotyledons of fln1-1 mutants never greened, but did unhook in the light. fln2 seedlings unhooked and turned pale green, eventually resembling cotyledons of fln2 mutants grown under continuous light. Thus, prior expansion in the dark did not change the phenotype of fln cotyledons in the light. We therefore conclude that neither FLN1 nor FLN2 are required for skotomorphogenesis.
FLN proteins are required for normal plastid development
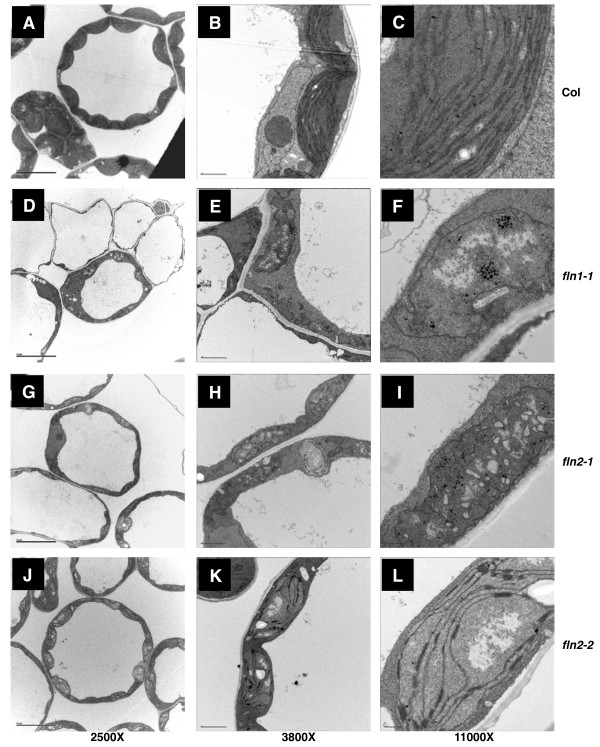
To investigate plastid development in fln1 and fln2 seedlings, we examined chloroplast ultrastructure in cotyledons of 7 day-old light-grown seedlings grown on GM by transmission electron microscopy (TEM) (Figure 6). Columbia (Col-0) cells contained a population of morphologically homogenous chloroplasts (Figure 6A). Wild-type chloroplasts are typified by stacks of thylakoid grana membrane connected via stromal thylakoid membranes with a few starch granules and plastoglobuli interspersed throughout the stroma (Figure 6C). Consistent with fln1 mutants having a more severe morphological phenotype (Figure 1), cells of fln1-1 were extremely vacuolated and contained chloroplasts with more extreme defects (Figure 6D, E) than those of fln2 cells (Figure 6G, J). Almost all fln1 chloroplasts lack internal membrane structures with a large number of densely staining plastoglobuli. (Figure 6F).
Figure 6.
Chloroplast ultrastructure inflnmutants. Cotyledons from 7-day old seedlings were fixed and sectioned for TEM imaging. (A) to (C) Col-0, (D) to (F)fln1-1, (G) to (I)fln2-1, (J) to (L)fln2-2. Scale bars for first, second, and third columns are 10, 2, and 0.5 μm, respectively.
In contrast, fln2 cells contained a population of chloroplasts with a range of defects (Figure 6G, J). In both fln2 alleles, chloroplasts have highly disrupted internal membrane structures compared to wild-type Col-0 (Figure 6H, K). The majority of fln2-1 and fln2-2 chloroplasts were missing thylakoid membranes, highly vacuolated, containing many more plastoglobuli than wild-type, and filled with unidentifiable suborganellar structures (Figure 6I). Other chloroplasts contained some stromal thylakoid membrane and a few grana stacks (Figure 6L). In summary, fln2-1 chloroplasts were either lacking any typical membrane structures or contained only a few thylakoid membranes and grana stacks. fln2-2 chloroplasts, on the other hand, had some contiguous stromal thylakoid membrane forming a reticulum with reduced granal stacks (Figure 6L).
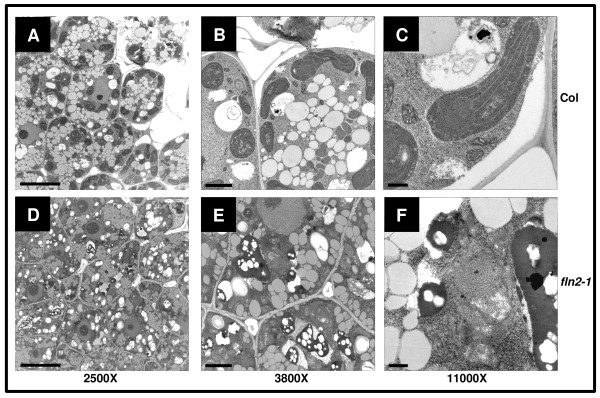
While mature chloroplasts of fln mutants fail to develop properly and mutants have severe growth defects in the light, cell elongation in the dark is largely unaffected (Figure 5). These observations lead to us investigate whether etioplast development was affected in fln mutants. Only fln2 could be analyzed because fln1 homozygous seed cannot be obtained [fln1-1 is seedling lethal (Figure 1)], and fln1-1 segregants cannot be phenotypically distinguished from FLN1 and FLN1/fln1-1 siblings in the dark (Figure 5). fln 2–1 etioplasts were examined from cotyledons of 3-day old dark-grown seedlings by TEM (Figure 7). For both Col-0 and fln2-1 dark-grown cells, the cytoplasm was dense with multiple small vacuoles and oil bodies (Figure 7A, D). fln2-1 cells also contained subcellular components that did not fix during the preparation of the samples, leaving holes in the sections (Figure 7E). Etioplasts in Col-0 could easily be identified and stained densely (Figure 7B), but fln2-1 etioplasts stained less densely and were difficult to discern from other cellular contents (Figure 7E). Col-0 etioplasts contained obvious prolamellar bodies with stromal strands of thylakoid membrane extending from them, as classically observed (Figure 7C). fln2-1 etioplasts did contain prolamellar bodies, but were atypical and lacked any obvious associated stromal thylakoids (Figure 7f). Altogether, these data indicate that FLN2 is not only required for chloroplast development in the light, but also for development of etioplasts in the dark.
Figure 7.
Etioplast ultrastructure infln2-1mutants. Cotyledons from 3-day old dark grown seedlings grown at 20° C were fixed and sectioned for TEM imaging. (A) to (C) Col-0, (D) to (F)fln2-1. Scale bars for first, second, and third columns are 10, 2, and 0.5 μm, respectively.
FLNs function additively in plant growth and development
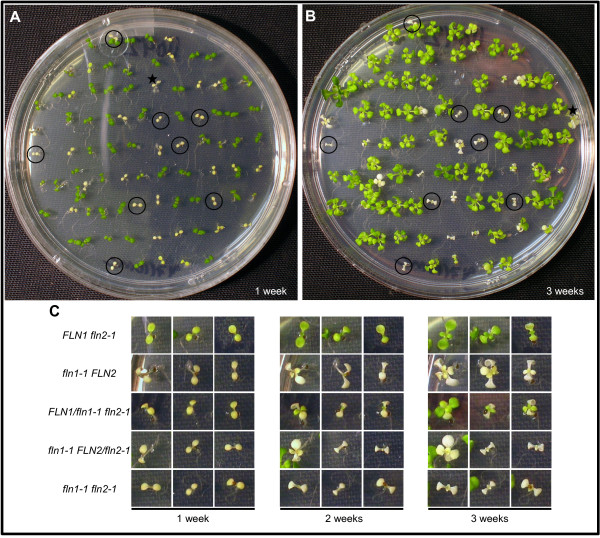
To determine the effect on seedling growth when both FLN proteins are absent, we crossed fln2-1 homozygotes to FLN1/fln1-1 heterozygotes to obtain fln1 fln2 double homozygous mutants. Seeds from FLN1/fln1-1 FLN2/fln2-1 plants were plated and growth monitored over a three-week period. One week-old pale/chlorotic seedlings were marked and photographed (Figure 8A, B). The same plate was incubated for two additional weeks, after which these plants were sacrificed for fln1-1 and fln2-1 genotyping. Double fln1 fln2 mutants were identified (circled, Figure 8A, B), and recovered with the expected 1/16 frequency in a large F2 population (χ2 = 1.142, p-value = 0.2853, α = 0.05).
Figure 8.
fln1 fln2double mutant analysis. F2 progeny from FLN1/fln1-1 FLN2/fln2-1 parents are shown. (A) and (B) F2 seeds were plated on GM and grown for three weeks with images taken at 1 week (A), 2 weeks, and 3 weeks (B). Pale and white seedlings were sacrificed for genotyping. Individuals identified as fln1 fln2 are circled. One individual accidentally moved on the plate during the growth period and is denoted with a star. (C) Left, middle and right sets of pictures, magnification of seedlings from the plate shown in (A), after 2 weeks growth, and from the plate in (B), respectively. Three representative seedlings are shown for each indicated genotype, with their phenotypes shown at different ages.
Also among the one week-old pale seedlings were single mutant siblings and siblings heterozygous at one locus and homozygous at the other (Figure 8C). Three different seedlings of each genotype, representative of the phenotypes observed, are shown at different ages. Single fln2-1 seedlings have pale cotyledons, and a few green true leaves by three weeks of growth; unlike on soil, other leaves remain pale (Figure 8C, first row). As observed on soil (Figure 1), single mutant fln1-1 cotyledons are white and totally void of visible chlorophyll. Unlike on soil, fln1-1 seedlings grown on media supplemented with 1% sucrose produce true leaves. These leaves are white and never green (Figure 8C, second row).
Analysis of FLN1/fln1-1 fln2-1 and fln1-1 FLN2/fln2-2 plants revealed haplo-insufficiency for FLN function in these genotypes. FLN1/fln1-1 fln2-1 seedlings are more similar in appearance to the more severe single fln1-1 rather than to single fln2-1 after one week of growth, and the cotyledons never green. True leaves for this genotypic class emerge pale green, but are less expanded than those of fln2-1. By three weeks, some FLN1/fln1-1 fln2-1 plants resemble fln2-1, while the majority has a more severe phenotype (Figure 8C, third row). fln1-1 FLN2/fln2-1 seedlings appear like fln1-1 after seven days of growth, but by 14 days a fraction of this genotypic class begins to have shriveled cotyledons, while others continued to look like fln1-1 (Figure 8C, fourth row). Even after three weeks, the more severe individuals never expand their true leaves, and resemble the fln1-1 fln2-1 double mutant (Figure 8C, bottom row).
Double mutant fln1-1 fln2-1 plants are indistinguishable from their fln1-1 siblings after 7 days of growth, but are clearly different after three weeks (Figure 8c, fifth row). All double mutants produce one set of very small, white true leaves visible by two weeks, but these never expand. The double mutant and a subset of fln1-1 FLN2/fln2-1 seedlings cannot be distinguished from each other phenotypically, and together demonstrate that seedlings lacking all or with insufficient FLN activity are seedling lethal even when supplemented with sucrose. This result suggests that FLN function, in some capacity, is required for development even when sucrose is present. This genetic analysis also suggests that FLN1 and FLN2 probably do not have functionally specificity, but rather work additively to regulate gene expression.
Discussion
Two proteins with predicted domains related to phosphofructokinases in the pfkB family are found in plastid TAC complexes from multiple plant species, but their roles remain enigmatic. E. coli possesses two phosphofructokinases, and PfkB/Pfk-2 was originally discovered as a suppressor of a PfkA/Pfk-1 mutation [28,29]. While both PfkA and PfkB are catalytically active, they are not similar in sequence and have different allosteric regulation [30-32]. Thus, kinases with sequence similarity to PfkB became known as pfkB-type kinases. Other PfkB-type kinases phosphorylate a variety of substrates including fructose, fructose-6-phosphate, ribose, adenosine, phosphomethylpyrimidine, and tagatose-6-phosphate [33], but many remain poorly characterized. To unravel the roles of pfkB-type kinases in plant growth and development we have taken a reverse genetics approach to assess the phenotypes of pfkB-type kinase loss-of-function mutants, beginning with genes encoding two closely-related FRUCTOKINASE-LIKE PROTEINs, FLN1 and FLN2, identified as components of the PEP TAC complex [9,12,14]. Using semi-quantitative and qRT-PCR, we demonstrate that these fln insertion alleles do not produce detectable mRNA and are null alleles, and through complementation and characterization of multiple alleles, indicate that the observed phenotypes result from loss of fln expression. These null alleles were used to separately assess FLN’s individual and collective contribution to plant growth and development.
The work initially describing FLNs, addressed their roles using RNAi in adult plants [14,15]. Silencing of either FLN1 or FLN2 caused similar chlorosis in young and developing Arabidopsis rosette leaves [15]. In contrast, our FLN loss-of-function analysis revealed unequal roles for FLN1 and FLN2. Accordingly, we observed that chloroplasts of fln1 plants are more severely affected than those of fln2. The differences between the two experiments might be because the differential contributions of FLN1 and FLN2 are greater during early seedling growth than in rosette leaves. The effect of reduced FLN expression was also assessed previously in tobacco. RNAi of only FLN1 in tobacco resulted in chlorosis, consistent with our proposed greater role for FLN1. Chloroplast ultrastructure was not examined in the Arabidopsis RNAi lines; however, the authors did examine chloroplasts of FLN1 RNAi in tobacco cells [15]. Chloroplasts in FLN1 RNAi tobacco were very similar to those in Arabidopsis fln1 plants reported here, indicating that loss of this protein in both tobacco and Arabidopsis results in similar plastid defects.
FLN’s localization to TAC complexes suggests that PEP loss of function mutants should have similar phenotypes. The PEP holoenzyme is similar in composition to eubacterial RNA polymerases and comprised of four different subunits α2ββ′β′′ encoded by the plastid genes rpoArpoBrpoC1, and rpoC2, respectively. PEP-dependent transcription additionally requires nuclear-encoded sigma factors to ensure the correct transcription start site [reviewed in [34]]. Mutations in rpo genes in tobacco result in albinism and severe defects in plastid development [6,35]. rpo- plants can grow only when supplemented with sucrose; however, these plants resemble wild-type plants morphologically with the exception of pigmentation loss, and can develop normally, flower and set seed [35]. Here we show that fln1 plants also require sucrose for growth, but are not identical to rpo- plants. fln1 grows and flowers on sucrose, but plants are extremely small and infertile (data not shown). fln2 plants, on the other hand, remain autotrophic with more normal chloroplasts than found in either fln1 or rpo- both of which are lacking thylakoid membranes and are highly vacuolated [6,35]. The fln1-1 fln2-1 double mutant is seedling lethal even when grown on sucrose and only one set of two true leaves emerges without subsequent expansion before dying. This result, in light of the viability of rpo- plants grown on sucrose, suggests that the FLNs may serve another function in addition to their role in PEP complexes or that loss of FLNs is more toxic than loss of core PEP subunits. A challenge to this interpretation is that no rpo-Arabidopsis have been described. Those plants may be more severe than the tobacco mutants and more closely resemble the fln double mutant. A FLN substrate remains elusive [15], therefore, a full understanding of their role both inside and out of the TAC complex awaits the identification of a substrate. One approach to dissect a function independent of the TAC complex would be to assess phenotypes and chloroplast-encoded transcript levels in fln nulls expressing FLNs that do not localize to TAC complexes. However, such mutations have not been identified to date.
In this work, we also sought to assess the role of FLN proteins during skotomorphogenesis. We evaluated etioplast structure in fln2 dark-grown seedlings. We were unfortunately unable to examine etioplasts in fln1 seedlings because seedling lethality prevented collection of a population of homozygous seed and fln1-1 seedlings are indistinguishable from wild-type siblings when grown in the dark. However, based on the mild defect in fln2 etioplasts and the normal skotomorphogenesis of both fln1 and fln2 plants, we conclude that FLNs have a more prominent role in plastid development in the light than mediating growth in the dark.
FLNs appear to be regulated by the thioredoxin system that modulates enzyme activities in response to illumination, which for FLNs, requires a specific thioredoxin, TRX-z [15]. trx-z null plants are viable and develop morphologically normal leaves abeit without chlorophyll when supplied with sucrose [15]. In contrast, complete loss of FLN expression in a fln1 fln2 background results in a seedling lethal phenotype, even when supplied with exogenous sucrose. The differences in the growth abilities between trx-z and fln1 fln2 mutants on sucrose synthetic media suggests that there is some FLN activity in trx-z plants and hence, redox regulation by TRX-z is not strictly required for FLN activity.
Consistent with their location in TAC complexes [15] and previous RNA analyses with RNAi plants [15], single fln mutants dramatically affect PEP-dependent chloroplast RNA accumulation, while most of the NEP and NEP/PEP-dependent mRNAs slightly accumulate or are not affected. These findings are in line with previous observations that disruption of PEP-dependent transcription results in an elevation of class III and some class II chloroplast genes (reviewed in [36]). One exception in our experiments was rpl23, a NEP-dependent gene whose mRNA was reduced in fln seedlings. A recent study showed that rpl23 transcript level is reduced when PEP is inhibited during germination [37], suggesting that this gene can be transcribed by both NEP and PEP during this phase of seedling establishment. Arsova et. al 2010 only examined NEP-dependent transcripts in mature rosette leaves, and did not include rpl23. This study adds new insight into the requirement of FLNs for PEP activity during early seedling growth.
PEP-dependent transcription is maximal during greening, although a role early in germination is proposed [37,38]. A recent report suggests that PEP is present and active during imbibition; however, PEP transcribes only genes encoding ribosomal proteins during this stage of development as revealed by using tagetin, a PEP-specific inhibitor. PEP begins to transcribe photosynthesis related genes within 24 hours of imbibition, prior to completion of germination [37]. The ability of FLN1-HA to complement the fln1 phenotype when expressed after seed stratification and after one day of germination suggests that FLN activity is not required early, but rather as the seedlings are developing photosynthetic competency. These plants do, however, express FLN2, so it is possible that FLN2 provides sufficient activity early, but is insufficient during rapid chloroplast maturation. The plastids in fln1 cotyledons do not resemble chloroplasts (Figure 6), and are missing most internal membranes. Addition of FLN1-HA to these plastids by DEX-induction cannot rescue the pale phenotype, implying that the effects of reduced FLN are not reversible. In other words, once photomorphogenesis has proceeded past a specific point, addition of FLN1 cannot restore normal greening. These observations are consistent with the inability of grossly mis-developed/damaged chloroplasts to reorganize and repair themselves, after development from either etioplasts or pro-plastids.
PEP core subunits are found in soluble PEP complexes and in the insoluble TAC complexes associated with plastid DNA [9]. TACs are estimated to contain 40–60 different proteins in addition to PEP subunits [9]. Although mutant analysis has not been reported for all components of TAC, plants with mutations in PEP and pTAC encoding genes have diverse pleiotropic phenotypes, but all share, to some degree, loss of pigmentation and abnormal plastid development. To our knowledge, the following TAC components have been assessed by loss of function analysis: pTAC1/WHIRLY1 [39], pTAC2 [9], pTAC4/VESICLE-INDUCING PROTEIN IN PLASTIDS [40], pTAC6 [9], pTAC11/WHIRLY3 [39], pTAC12/HEMERA [9,41], IRON SUPEROXIDE DISMUTASE 1 and 3 [42], DNA GYRASE [43,44], THIOREDOXIN Z [14,15], URIDINE DIPHOSPHATE-N-ACETYLMERAMIC ACID LIGASE E [45], pTAC14 [46], GENOMES UNCOUPLED 1 [47,48], PLASTID REDOX INSENSITIVE 2 [49] and FRUCTOKINASE-LIKE PROTEIN 1 and 2 [[11,15] and this work]. With the exception of WHIRLY, DNA GYRASE, GENOMES UNCOUPLED 1, and PLASTID REDOX INSENSITIVE 2, all mutants listed above require an exogenous carbon source for viability. WHIRLY mutants have a low penetrance of chlorotic variegation, and these mutants are fully autotrophic. DNA GYRASE mutants, similar to fln double mutants, only produce two true leaves that fail to expand, with no further growth even when supplemented with sucrose.
While the exact mechanism of FLN action remains to be elucidated, our genetic analysis points to an overlapping role of FLN1 and FLN2 in terms of plastid development and plastid-specific gene expression. Phylogenetic analysis of plant pfkB-type kinases revealed that the FLNs clade with enzymes shown to have fructokinase activity and are more distantly related to known adenosine kinases [15]. Attempts to identify a FLN substrate were unsuccessful using recombinant FLN and a variety of sugars [15]. FLNs could serve both an enzymatic function not yet identified and/or additionally act as a metabolic sensor in PEP complexes to coordinate gene expression with metabolic status of the plastid. Proteins with dual function in metabolism and signaling have been characterized across all kingdoms of life [reviewed in [50]], and Arabidopsis HEXOKINASE1 has been shown to chromatin immunoprecipitate with nuclear DNA and other proteins required for glucose signaling [51].
Conclusions
Our analysis of fln loss-of-function mutants further substantiates the RNAi phenotypes reported previously for Arabidopsis and tobacco, and reveals an essential role for these proteins in plant growth and development. Transcripts of PEP-dependent chloroplast genes are severely reduced in fln seedlings by qRT-PCR analysis, indicating that FLN proteins are indispensible components of the TAC complex. Genetic analyses show that FLN1 plays a more dominant role than FLN2; single mutant analysis showed FLN1 to be required for autotrophic growth; double mutant analysis revealed that complete loss of FLN function results in seedling lethality even on an exogenous carbon source. This analysis demonstrates that seedlings are extremely sensitive to FLN levels.
Methods
Plant materials and growth conditions
All plants are Arabidopsis thaliana, ecotype Columbia-0 (Col-0) unless otherwise noted. At3g54090 T-DNA insertion line GK-443A08 (fln1-1) was obtained from The European Arabidopsis Stock Centre (http://Arabidopsis.info/) [52]. T-DNA insertion lines for At1g69200 were SALK_008812 (fln2-1) obtained from the Arabidopsis Biological Resource Center (ABRC) (http://abrc.osu.edu/) [53] and FLAG_110F06 (fln2-2, in Wassilevskija (WS-4) ecotype) obtained from INRA Arabidopsis thaliana Resource Centre for Genomics (http://www-ijpb.versailles.inra.fr/en/sgap/equipes/ variabilite/crg/index.htm) [54]. A putative third allele from ABRC [SALK_005734 (fln2-3)], was found to have the identical insertion site as SALK_008812 (Additional file 1: Figure S1) and is likely an inadvertent duplicate of the same insertion event. It exhibited the same phenotypes as fln2-2. PCR genotyping primers are listed in Additional file 2: Table S1. All plants lines were F2 progeny from a backcross (BC1) to Col-0. Growth media (GM) for plants grown in plates was prepared as previously described [55]. Seeds were surface sterilized in 30% bleach, 0.1% Triton-X 100 prior to plating. All plate-grown seedlings were grown at 20o C under constant white light at 40–50 μmol/sec-1m-2.
For hypocotyl measurements, progeny from FLN/fln were grown on solid GM in the dark for 7 days. Seedlings were then transferred to new plates, imaged, their positions noted, and hypocotyl lengths determined from this image. To identify which hypocotyls were fln, seedlings were placed under lights and allowed to grow an additional 7 days. Homozygous fln hypocotyls were identified based on white/pale phenotype of cotyledons and true leaves. Measurements were obtained using Image J 1.36 (http://rsb.info.nih.gov/ij/).
RT-PCR analysis of FLN transcripts in insertion lines
BC1 F2 seeds segregating for each fln allele were plated on GM and grown for 7 days. Homozygous mutants were identified by phenotype, harvested, and stored in liquid N2 for RNA extraction. RNA extraction was performed using the RNeasy plant mini kit (Qiagen). DNase digestion was carried out on the column prior to RNA elution to eliminate contaminating genomic DNA. For fln1 and fln2 experiments, 2.5 and 2.8 μg total RNA, respectively, was used in reverse-transcriptase (RT) reactions using SuperScript III RT (Invitrogen, Carlsbad, CA) and oligo(dT) primer to synthesize cDNA. cDNA was then used as a template to amplify FLN full-length coding sequences (CDS). PCR products were 1437, 1851, and 1446 bp for FLN1, FLN2, and UBIQUITIN10 (UBQ10) respectively. UBQ10 was used a control to ensure that equal RNA was used for all cDNA synthesis reactions. The FLN1 product included the entire 5’UTR as annotated and ended at the stop codon sequence, while the FLN2 was the coding sequence from start to stop codon sequence. Primers used for RT-PCR experiments are listed in Additional file 2: Table S1.
qRT-PCR analysis of chloroplast gene expression
Plants were grown and RNA extracted as described above for RT-PCR. cDNA was prepared from 2 μg total RNA using the Maxima first strand cDNA synthesis kit (Fermentas). 5 μL of a 1:400 cDNA dilution was used as template for qRT-PCR. Real time PCR was performed using a CFX384TM Real-Time System (BIO-RAD) using SYBR Green (Invitrogen). The following standard thermal profile was used for all PCR reactions: 95 °C for 3 min, 40 cycles of 95 °C for 10s and 60 °C for 1 min. Primers for psbOpsaHpsaE, and ndhB were as previously published [15]. Additional primers are listed in Additional file 2: Table S1. Expression levels were normalized to 18S rRNA content.
Transmission electron microscopy
Seedlings were grown on GM plates as described above. Light grown seedlings were 7 days old, and dark grown seedlings were 3 days old. Cotyledons were fixed in Karnovsky’s fixative [56] using a microwave [57], rinsed in 0.1 M PBS and post-fixed in buffered 1% osmium Tetroxide for 2 hours. Samples were then incubated in 0.1% tannic acid (aqueous) for 30 minutes before dehydrating [58-60] in ascending concentrations of ethyl alcohol (ETOH) for 20 minutes each step (30%-50%). They were then stained with 2% uranyl acetate in 50% ETOH [61]. Dehydration was completed with 70%, three changes of 95% and two changes of 100% ethanol for 20 minutes each [57]. Pure epoxy resin (Epon/Araldite) replaced the final ethanol step and was infiltrated overnight [62]. The samples were then carefully placed in the bottom of a flat embedding mold (Electron Microscopy Sciences, PA, USA), and the capsules filled with fresh resin and polymerized overnight in a 70o C oven. Sections were cut on a Leica Ultracut UCT Ultramicrotome using a diamond knife (Diatome, Switzerland, EMS U.S.A. distributor). The samples were viewed and images were taken using a Philips CM120 Biotwin TEM, (FEI Company, Hillsboro, OR, USA) using a Gatan MegaScan 794/20 digital camera (2 K X 2 K) (Pleasanton, CA, USA). All work was completed in the Electron Microscopy Laboratory, Department of Medical Pathology and Laboratory Medicine, School of Medicine, University of California at Davis.
Genetic Complementation of fln1-1 mutants and inducible expression of FLN1
We obtained a full-length clone of the FLN1 (At3g54090.1) coding sequence (Clone U17866) from the ABRC [63] in a pENTR/SD/D-TOPO backbone. To clone this ORF without a stop codon for making C-terminal tagged translational fusions, we used U17866 as a template using Phusion high-fidelity DNA polymerase (NEB). Primers used are listed in Additional file 2: Table S1. The PCR product was gel-purified and recombined into pDonr201 using a GATEWAY Technology BP-clonase reaction (Invitrogen). The ORF was sequenced verified then recombined into pBAV154 plant transformation vector [27] according to manufacturer’s directions. The final plant expression cassette after recombination allows for dexamethasone (DEX)-induced expression of FLN1-HA. The plant transformation vector was introduced into Agrobacterium tumefaciens, strain AGL1, which was then used to transform fln1-1 heterozygotes using the floral dip method [64]. Transformants were selected by spraying progeny with a 0.578% solution of Finale (1% glufosinate-ammonium, Bayer CropScience http://www.bayercropscience.com/); resistant T1 plants were genotyped for the fln1-1 insertion and 14 heterozygotes (independent transformation events) carried forward to the T2 generation. Because only fln1-1 heterozygotes survive on GM supplemented with sulfadiazine, T2 seeds from these 14 lines were plated on 5.25 μg/mL sulfadiazine sodium (Sigma-Aldrich, S6387) to select for fln1-1 heterozygotes. Resistant individuals were transferred to soil and sprayed with Finale to determine the number of FLN1-HA transgenes present in each line. Only lines segregating 3:1 for glufosinate resistance were carried forward. T3 seeds from these T2 families were sown on soil and again selected for glufosinate resistance to determine which T2 family individuals were fixed for the FLN1-HA transgene. T3 seeds homozygous for the FLN1-HA transgene and segregating for fln1-1 were used dexamethasone-dependent complementation experiments. Seed were plated on varying concentrations of dexamethasone and the number of seedlings with green or pale cotyledons were scored after 7 days. For the developmental time course, seed were plated, stratified for 2 days at 4 °C, then placed at room temperature (RT). Dexamethasone was added to a plate the same day or after 1 or 2 days at RT and scored for green cotyledons after 7 days.
Western blot analysis
Total protein extracts were prepared from seedlings grown for complementation experiments described above. Tissue was homogenized in a 1:1 seedling weight (mg) to buffer (μl) ratio using 5x Laemmli sample buffer (125 mM Tris pH 6.8, 20% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.2 mg/mL bromophenyl blue). After homogenization, lysates were boiled 5 min and cleared by centrifugation at 16,000xg for 20 min. Proteins were separated by SDS-PAGE on 8% gels, and transferred to PDVF Immobilon-P membrane (Millipore, Billerica, MA, USA) for immunoblotting. Membranes were blocked with BLOTTO [5% non-fat dry milk in TBS-T (50 mM Tris, 200 mM NaCl, 0.1% Tween-20), 0.165% antifoam Y emulsion (Sigma)] for 15 min, then immunoblotted with 1:1000 anti-HA-peroxidase (3 F10 rat monoclonal antibody, Roche Diagnostics, Mannheim, Germany) in BLOTTO for 2 hr. Membranes were washed in TBS-T, then developed with Amersham ECL Plus western detection system (GE Healthcare, Buckinghamshire, United Kingdom) and X-ray film following the manufacturer’s instructions.
Abbreviations
NEP, Nuclear-encoded polymerase; PEP, Plastid-encoded polymerase; FLN1 and FLN2, FRUCTOKINASE-LIKE PROTEIN; TAC, Transcriptionally active chromosome; qRT-PCR, Quantitative real-time RT-PCR; TEM, Transmission electron microscopy; ADK, Adenosine kinase.
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
JG and JCallis conceived the experiments, and JG, JMPR and JCallis performed the experiments. JChory participated in the design of the qRTPCR study. JG wrote the first draft, which was edited by JCallis and JChory. All authors read and approved the final manuscript.
Supplementary Material
Sequence of T-DNA borders in fln alleles. The T-DNA specific bands from PCRs were sequenced from the left border of the T-DNA for the three different lines. Orientation of the T-DNA is denoted by “L” for left and “R” for right borders. Boxed sequence is additional DNA not in T-DNA or at locus prior to insertion. Right border sequence at insertion site was not determined. Because fln2-1 and fln2-3 have two inverted T-DNA insertions in tandem, sequence on each side of the insertion could be determined. Unboxed sequence is present in endogenous locus.
DNA Primer sequences used for PCR. Contains a list of primer sequences, by purpose, used in this study.
Contributor Information
Jonathan Gilkerson, Email: jgilkerson@salk.edu.
Juan Manuel Perez-Ruiz, Email: jperez@salk.edu.
Joanne Chory, Email: jchory@salk.edu.
Judy Callis, Email: jcallis@ucdavis.edu.
Acknowledgements
The authors acknowledge the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy to JCallis (DE-FG02-09ER16077) and JChory (FG02-11ER15540). Additionally, the qPCR experiment was supported in part by The Howard Hughes Medical Institute (JChory). We acknowledge the UC-Davis Controlled Environment Facility (CEF) for assistance in the propagation of transgenic plants. We thank Meliza Castro and Gurjeet Bath for excellent assistance, and Patricia Kysar, UCD Electron Microscopy Laboratory, for assistance with TEM experiments. We especially thank Kentaro Inoue for helpful discussion and assistance with experiments not shown here, and Jesse Woodson for helpful discussions.
References
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Börner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y, Onda Y, Shiina T, Nakahira Y. Plastid transcription in higher plants. Crit Rev Plant Sci. 2005;24:59–81. doi: 10.1080/07352680590910438. [DOI] [Google Scholar]
- Serino G, Maliga P. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 1998;117:1165–1170. doi: 10.1104/pp.117.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Hallick RB, Lipper C, Richards OC, Rutter WJ. Isolation of a transcriptionally active chromosome from chloroplasts of Euglena gracilis. Biochemistry. 1976;15:3039–3045. doi: 10.1021/bi00659a016. [DOI] [PubMed] [Google Scholar]
- Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins K, Barkan A, van Wijk K. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and −12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G. Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol. 1994;25:69–81. doi: 10.1007/BF00024199. [DOI] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 2011;157:1043–1055. doi: 10.1104/pp.111.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P. Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. Plant J. 2004;40:164–172. doi: 10.1111/j.1365-313X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T. The transcription machineries of plant mitochondria and chloroplasts: composition, function and regulation. J Plant Physiol. 2011;168:1345–1360. doi: 10.1016/j.jplph.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Schröter Y, Steiner S, Matthäi K, Pfannschmidt T. Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics. 2010;10:2191–2204. doi: 10.1002/pmic.200900678. [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Campbell RN, Reece RJ. Galactose metabolism in yeast-structure and regulation of the leloir pathway enzymes and the genes encoding them. Int Rev Cell Mol Biol. 2008;269:111–150. doi: 10.1016/S1937-6448(08)01003-4. [DOI] [PubMed] [Google Scholar]
- Moreno F, Ahuatzi D, Riera A, Palomino CA, Herrero P. Glucose sensing through the Hxk2-dependent signalling pathway. Biochem Soc Trans. 2005;33:265–268. doi: 10.1042/BST0330265. [DOI] [PubMed] [Google Scholar]
- Peláez R, Herrero P, Moreno F. Functional domains of yeast hexokinase 2. Biochem J. 2010;432:181–190. doi: 10.1042/BJ20100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Wang L, Allen MS, Stevens YY, Qin W, Snider J, von Schwartzenberg K. Adenosine kinase of Arabidopsis. Kinetic properties and gene expression. Plant Physiol. 2000;124:1775–1785. doi: 10.1104/pp.124.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt BA, Stevens YY, Allen MS, Snider JD, Pereira LA, Todorova MI, Summers PS, Weretilnyk EA, Martin-McCaffrey L, Wagner C. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 2002;128:812–821. doi: 10.1104/pp.010880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Harrison BR, Narayana Murthy UM, Moffatt BA, Gilroy S, Masson PH. Adenosine kinase modulates root gravitropism and cap morphogenesis in Arabidopsis. Plant Physiol. 2006;142:564–573. doi: 10.1104/pp.106.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoor S, Farrow S, Blaschke H, Lee S, Perry G, von Schwartzenberg K, Emery N, Moffatt B. Adenosine kinase contributes to cytokinin interconversion in Arabidopsis. Plant Physiol. 2011;157:659–672. doi: 10.1104/pp.111.181560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Nishimura K, Aoki T, Takase H, Tomizawa K, Ashida H, Yokota A. A phosphofructokinase B-type carbohydrate kinase family protein, NARA5, for massive expressions of plastid-encoded photosynthetic genes in Arabidopsis. Plant Physiol. 2009;151:114–128. doi: 10.1104/pp.109.139683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313X.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- Vinatzer BA, Teitzel GM, Lee MW, Jelenska J, Hotton S, Fairfax K, Jenrette J, Greenberg JT. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol. 2006;62:26–44. doi: 10.1111/j.1365-2958.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- Morrissey AT, Fraenkel DG. Suppressor of phosphofructokinase mutations of Escherichia coli. J Bacteriol. 1972;112:183–187. doi: 10.1128/jb.112.1.183-187.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel DG, Kotlarz D, Buc H. Two fructose 6-phosphate kinase activities in Escherichia coli. J Biol Chem. 1973;248:4865–4866. [PubMed] [Google Scholar]
- Daldal F. Nucleotide sequence of gene pfkB encoding the minor phosphofructokinase of Escherichia coli K-12. Gene. 1984;28:337–342. doi: 10.1016/0378-1119(84)90151-3. [DOI] [PubMed] [Google Scholar]
- Babul J. Phosphofructokinases fromEscherichia coli. Purification and characterization of the nonallosteric isozyme. J Biol Chem. 1978;253:4350–4355. [PubMed] [Google Scholar]
- Hellinga HW, Evans PR. Nucleotide sequence and high-level expression of the major Escherichia coli phosphofructokinase. Eur J Biochem. 1985;149:363–373. doi: 10.1111/j.1432-1033.1985.tb08934.x. [DOI] [PubMed] [Google Scholar]
- Park J, Gupta RS. Adenosine kinase and ribokinase–the RK family of proteins. Cell Mol Life Sci. 2008;65:2875–2896. doi: 10.1007/s00018-008-8123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess WR, Borner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- De Santis-MacIossek G, Kofer W, Bock A, Schoch S, Maier RM, Wanner G, Rüdiger W, Koop HU, Herrmann RG. Targeted disruption of the plastid RNA polymerase genes rpoA, B and C1: molecular biology, biochemistry and ultrastructure. Plant J. 1999;18:477–489. doi: 10.1046/j.1365-313X.1999.00473.x. [DOI] [PubMed] [Google Scholar]
- Gray J, Sullivan J, Wang J-H, Jerome C, MacLean D. Coordination of plastid and nuclear gene expression. Phil Trans R Soc Lond B. 2003;358:135–145. doi: 10.1098/rstb.2002.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Buhr F, Lambert E, Lerbs-Mache S. Characterization of the plastid-specific germination and seedling establishment transcriptional programme. J Exp Bot. 2012;63:925–939. doi: 10.1093/jxb/err322. [DOI] [PubMed] [Google Scholar]
- Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Machi S. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 2006;142:993–1003. doi: 10.1104/pp.106.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Véronneau-Lafortune F, Joyeux A, Lang BF, Brisson N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:14693–14698. doi: 10.1073/pnas.0901710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell. 2010;141:1230–1240. doi: 10.1016/j.cell.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell. 2008;20:3148–3162. doi: 10.1105/tpc.108.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MK, Mitchenall LA, Maxwell A. Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc Natl Acad Sci USA. 2004;101:7821–7826. doi: 10.1073/pnas.0400836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HS, Lee SS, Kim KD, Hwang I, Lim JS, Park YI, Pai HS. DNA gyrase is involved in chloroplast nucleoid partitioning. Plant Cell. 2004;16:2665–2682. doi: 10.1105/tpc.104.024281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 2008;53:924–934. doi: 10.1111/j.1365-313X.2007.03379.x. [DOI] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 2011;157:1733–1745. doi: 10.1104/pp.111.184762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim IJ, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. 10.1126/science. 1140516. [PubMed] [Google Scholar]
- Susek R, Ausubel F, Chory J. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development . Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco N, de Dios Barajas López J, Piñas Fernández A, Tellgren-Roth C, Small I, Strand A. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 2012;70:279–291. doi: 10.1111/j.1365-313X.2011.04865.x. [DOI] [PubMed] [Google Scholar]
- Huberts DH, van der Klei IJ. Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta. 2010;1803:520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiol. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Samson F, Brunaud V, Balzergue S, Dubreucq B, Lepiniec L, Pelletier G, Caboche M, Lecharny A. FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 2002;30:94–97. doi: 10.1093/nar/30.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J, Hu J, Brown J, Jones A, Sun TP, Callis J. Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics. 2009;181:945–963. doi: 10.1534/genetics.108.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovsky MJ. A formaldehyde-glutaraldehyde fixative for high osmolality for use in electron microscopy. J Cell Biol. 1965;27:137–138A. [Google Scholar]
- Russin WA, Trivett CL. In: Microwave Techniques and Protocols. Giberson RT Jr, Demaree RS, editor. Human Press, Totowa, NJ; 2001. Vacuum-Microwave Combination for Processing Plant Tissues for Electron Microscopy. [Google Scholar]
- Bozzola JJ, Russell LD. Specimen preparation for transmission electron microscopy In Electron Microscopy Principles and Techniques for Biologists. Jones and Bartlett Publishers, Boston, MA; 1992. pp. 16–47. [Google Scholar]
- Kalina M, Pease DC. The preservation of ultrastructure in saturated phosphatidyl cholines by tannic acid in model systems and type II pneumocytes. J Cell Biol. 1977;74:726–741. doi: 10.1083/jcb.74.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionescu N, Simionescu M. Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J Cell Biol. 1976;70:608–621. doi: 10.1083/jcb.70.3.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzola JJ, Russell LD. Specimen staining and contrast methods for transmission electron microscopy. In Electron Microscopy Principles and Techniques for Biologists. Jones and Bartlett Publishers, Boston, MA; 1992. pp. 48–71. [Google Scholar]
- Heumann HG. Microwave-stimulated glutaraldehyde and osmium tetroxide fixation of plant tissue: ultrastructural preservation in seconds. Histochemistry. 1992;97:341–347. doi: 10.1007/BF00270036. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of T-DNA borders in fln alleles. The T-DNA specific bands from PCRs were sequenced from the left border of the T-DNA for the three different lines. Orientation of the T-DNA is denoted by “L” for left and “R” for right borders. Boxed sequence is additional DNA not in T-DNA or at locus prior to insertion. Right border sequence at insertion site was not determined. Because fln2-1 and fln2-3 have two inverted T-DNA insertions in tandem, sequence on each side of the insertion could be determined. Unboxed sequence is present in endogenous locus.
DNA Primer sequences used for PCR. Contains a list of primer sequences, by purpose, used in this study.