The cycling of carbon between organic and inorganic forms controls levels of O2 in the global environment. Through its relationships to CO2 and CH4, both greenhouse gases, the carbon cycle also influences climate. For such reasons, geoscientists are broadly interested in understanding processes within the carbon cycle, both now and in ancient times. Microbial catalysis is pervasive, so organic geochemists—who once focused largely on petroleum—are taking a great interest in recognizing and interpreting molecular fossils, or “biomarkers,” that reflect the presence and activities of specific microorganisms. As they search, they occasionally come across products that surprise everyone. The report by Stefan Schouten et al. in this issue of PNAS is a recent and particularly interesting example (1). By using a new technique applicable to lipids containing as many as 86 carbon atoms, they found molecules that expand the range of known microbial products. Moreover, they have discovered some carbon skeletons in which archaeal and bacterial structural elements are combined.
This work is intriguing because biomarkers are in some cases very specific to a particular microorganism. A few species of haptophyte algae, for example, produce methyl ketones with 37 carbon atoms and 2, 3, or 4 double bonds (2). These alkenones can comprise up to 5% by weight of the biomass, and the number of double bonds varies systematically with the temperature of growth (3). Fortunately for geochemists, the alkenones are relatively resistant to biodegradation. Reaching the sea floor in pelletal debris, they are incorporated into sediments. When a core is raised from the sea floor and its lipids extracted and analyzed, the abundance of alkenones at each depth within the core indicates the extent to which conditions favored haptophyte algae, and the degree of unsaturation provides a quantitative estimate of sea-surface temperature. Such data usefully supplement other lines of paleoclimatic evidence. Less specific but still useful biomarkers exist for many eukaryotic microbes.
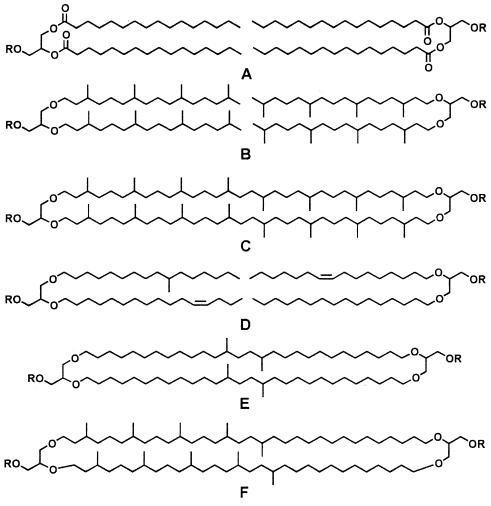
The last acts within the carbon cycle—those in which carbon is either locked in a sedimentary prison for a geological term or retained as a participant in the surface mêlée—determine the chemical balances that control our environment. The conditions for this part of the drama are commonly anaerobic. The actors, whose roles we would like to understand in detail, are prokaryotes from the bacterial and archaeal domains. The bacteria, in particular, seem almost to have designed their lipids to provide anonymity. Apart from the hopanoids (4), bacterial products that are both distinctive and preservable are rare. By default, attention falls on details of the common lipids, the fatty acids and related products. Pertinent examples are shown in Fig. 1.
Figure 1.
Representative structures of microbial membrane lipids. R, polar head group, typically bearing both positive and negative charges.
The structures shown are all based on glycerol. To it long-chain aliphatic moieties are attached by ester or ether links (respectively, structure A and structures B–F). For decades, such a generalized display would have included only structure A, a ubiquitous product of both eukaryotes and bacteria. That example shows, at the left and right, two fatty acyl diglycerides. In the functional natural product, a polar “head group” would be attached to the third OH group on each glycerol. A represents the basic motif of a “lipid bilayer” membrane. With their polar hydrophilic surfaces and nonpolar hydrophobic interiors, such membranes, pierced by proteins that regulate the transmission of specific reactants, functionally define the boundaries of cells. Such diglycerides provide useful information about the composition of microbial communities only when they happen to include fatty acids more distinctive than the common-as-dirt linear C16 species shown in structure A.
Interesting and potentially informative variety came on the scene when it was discovered that members of the other prokaryotic domain, the archaea, had glycerol-based lipids with ether linkages and isoprenoidal aliphatic substituents (5). Examples are structures B and C. Individual products become distinctive when their biosyntheses incorporate specific substitutions. Some methanogenic archaea in the order Methanosarcinales, for example, produce hydroxyarchaeol, a glycerol diether like either half of B except that the methyl-branched carbon position nearest the middle O of the glycerol bears an OH group. Finding hydroxyarchaeol in sediments near a methane seep in the Eel River Basin, offshore of northern California, and noting in addition that its natural content of 13C was particularly low, Hinrichs and coworkers at the Woods Hole Oceanographic Institution and Monterey Bay Aquarium Research Institute (mbari) were able to refine the identification of the archaeon that mediates the first step in the anaerobic oxidation of methane (6).
More recently, the Woods Hole–mbari team has found additional ether-linked lipids that are depleted in 13C and thus also produced by organisms involved in the recycling of methane. But in these (D), the alkyl chains are linear and methyl branched and thus typical of bacteria rather than archaea (7). The first report of glycerol diethers in bacteria appeared in 1983 (8). The organisms were thermophilic sulfate reducers. Subsequent findings have also been in thermophiles (9, 10). All are from deeply branching lineages on the bacterial side of the phylogenetic tree, relatively close to the archaea. Eel River bacterial ethers do indeed seem to come from sulfate reducers (which oxidize products from the methane-consuming archaea). Their placement in a deeply branching lineage may be suggested by their involvement with archaea in the recycling of methane. But they thrive at 4°C and are hardly strict thermophiles.
Molecules like C sidestep the requirement for formation of a bilayer. The left and right sides are connected by chemical bonds. The resulting single molecule is a cyclic tetraether. With formulas like C86H172O6, such compounds are not amenable to gas chromatographic separation and have been studied mostly after cleavage of the ether links. The group with which Schouten is associated at the Netherlands Institute for Sea Research (nioz) has, however, successfully used liquid chromatography with mass spectrometry for the analysis of intact tetraethers (11). The present report (1) definitively extends the roster of bacterial (as opposed to archaeal) products to include cyclic tetraethers (E) and provides new information on archaeal products. Most notably, some of the molecules combine archaeal and bacterial characteristics. In tetraethers like F, one-half of each hydrocarbon chain is isoprenoidal, whereas the other half looks like the alkyl portion of a common bacterial fatty acid.
We are discussing molecules from mud, not from organisms. But the nioz team shows that these molecules are very widely distributed and regular in their structure. They are undoubtedly microbial products and must point to a group of organisms of great phylogenetic interest. Can the producer(s) of structures like F be isolated and grown? Can the sequences of their 16S rRNAs at least be identified in gene libraries? Will they prove to be bacteria, archaea, or perhaps even something else?
Footnotes
See companion article on page 14421.
References
- 1.Schouten S, Hopmans E C, Pancost R D, Sinninghe Damsté J S. Proc Natl Acad Sci USA. 2000;97:14421–14426. doi: 10.1073/pnas.97.26.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marlowe I T, Brassell S C, Eglinton G, Green J C. Org Geochem. 1984;6:135–141. [Google Scholar]
- 3.Müller P J, Kirst G, Ruhland G, von Storch I, Rosell-Melé A. Geochim Cosmochim Acta. 1998;62:1757–1772. [Google Scholar]
- 4.Ourisson G, Poralla K, Rohmer M. Annu Rev Microbiol. 1987;41:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 5.De Rosa M, Gambacorta A, Gliozzi A. Microbiol Rev. 1986;50:70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinrichs K-U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Nature (London) 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 7.Hinrichs K-U, Summons R E, Orphan V, Sylva S P, Hayes J M. Org Geochem. 2000;31:1685–1701. [Google Scholar]
- 8.Langworthy T A, Holzer G, Zeikus J G, Tornabene T G. Syst Appl Microbiol. 1983;4:1–17. doi: 10.1016/S0723-2020(83)80029-0. [DOI] [PubMed] [Google Scholar]
- 9.DeRosa M, Gambacorta A, Huber R, Lanzotti V, Nicolaus B, Stetter K O, Trincone A. J. Chem. Soc. Chem. Commun. 1988. , 1300–1301. [Google Scholar]
- 10.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, Rachel R, Rockinger I, Fricke H, Stetter K O. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 11.Hopmans E C, Schouten S, Pancost R D, van der Meer M J T, Sinninghe Damsté J S. Rapid Comm Mass Spectrom. 2000;14:585–589. doi: 10.1002/(SICI)1097-0231(20000415)14:7<585::AID-RCM913>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]