Abstract
Background
The rates of multidrug-resistant (MDR), extensively drug-resistant (XDR) and pandrug-resistant (PDR) isolates among Enterobacteriaceae isolates, particularly Klebsiella pneumoniae, have risen substantially worldwide.
Methodology/Principal Findings
To better understand the molecular mechanisms of drug resistance in K. pneumoniae, we analyzed the drug resistance determinants for K. pneumoniae isolates collected from the 306 Hospital, a tertiary-care hospital in Beijing, China, for the period of September 1, 2010-October 31, 2011. Drug susceptibility testing, PCR amplification and sequencing of the drug resistance determinants were performed. Conjugation experiments were conducted to examine the natural ability of drug resistance to disseminate among Enterobacteriaceae strains using a sodium azide-resistant Escherichia coli J53 strain as a recipient. Among the 223 consecutive non-repetitive K. pneumoniae isolates included in this study, 101 (45.3%) were extended-spectrum beta-lactamases (ESBLs) positive. The rates of MDR, XDR, and PDR isolates were 61.4% (n = 137), 22.0% (n = 49), and 1.8% (n = 4), respectively. Among the tested drug resistance-associated genes, the following ones were detected at relatively high rates bla CTX-M-10 (80, 35.9%), aacC2 (73, 32.7%), dhfr (62, 27.8%), qnrS (58, 26.0%), aacA4 (57, 25.6%), aadA1 (56, 25.1%). Results from conjugation experiments indicate that many of the drug resistance genes were transmissible.
Conclusions/Significance
Our data give a “snapshot” of the complex genetic background responsible for drug resistance in K. pneumoniae in China and demonstrate that a high degree of awareness and monitoring of those drug resistance determinants are urgently needed in order to better control the emergence and transmission of drug-resistant K. pneumoniae isolates in hospital settings.
Introduction
The emergence and rapid spread of drug-resistant Klebsiella pneumoniae isolates is becoming a serious antibiotic management problem and causing a great concern worldwide [1]–[6]. For example, by late 2009, the number of unique protein sequences for beta-lactamases exceeded 890 (http://www.lahey.org/Studies) [7]. There is an increasing recognition of isolates producing newer beta-lactamases including the extended-spectrum beta-lactamase (ESBL), carbapenem-hydrolyzing enzymes (e.g., K. penumoniae carbapenemase [KPC] types and the metallo-beta-lactamases [MBLs]) [8]–[18]. Since the production of newer beta-lactamases is frequently accompanied by broad-spectrum resistance, the ESBL positivity together with the existence of newer beta-lactamases should be monitored closely as the emergence of those highly drug-resistant K. pneumoniae strains will pose a serious impact on the remaining therapeutic options [19]–[22]. In a study based on the Tigecycline Evaluation and Surveillance Trial (TEST) global surveillance database, the rate of ESBL production was highest among the K. pneumoniae isolates collected in Latin America, followed by Asia/Pacific Rim, Europe, and North America (44.0%, 22.4%, 13.3% and 7.5%, respectively) [23]. Thus the potential of drug resistant K. pneumoniae to be a global health problem is great and more intensive surveillance and more in-depth investigation into the molecular mechanisms of drug resistance in K. pneumoniae isolates are necessary in order to provide information for the development of effective molecular diagnostic methods and novel drugs against K. pneumoniae infection.
In the face of increasing resistance among multidrug-resistant (MDR) gram-negative organisms for which no adequate therapeutic options exist, a joint initiative by the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) recently created a standardized international definitions for MDR, extensively drug-resistant (XDR) and pandrug-resistant (PDR) with an aim to enhance the comparability of data and promote better comprehension of the problem of highly drug-resistant bacteria [24]. MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e. bacterial isolates remain susceptible to only one or two categories) and PDR was defined as non-susceptibility to all agents in all antimicrobial categories [24]. Though there are already many reports of drug-resistant K. pneumoniae worldwide, the extent of MDR, XDR and PDR K. pneumoniae isolates among patients is largely unknown. We thus in this study sought to determine the prevalence of MDR, XDR and PDR strains and to analyze the drug resistance determinants for K. pneumoniae isolates collected from patients being treated in the 306 Hospital, a tertiary-care hospital in Beijing, China, for the period of September 1, 2010-October 31, 2011 with an aim to better understand the current situation as well as the genetic background of the drug resistant K. pneumoniae isolates from hospital settings.
Methods
Ethics Statement
All of the investigation protocols in this study were approved by the institutional ethics committee of the 306 Hospital, Beijing, China. Written consent was given by the patients for their information to be stored in the hospital database and used for research. Permission for using the information in the medical records of the patients for research purposes was obtained from the 306 Hospital. The Institute ethics committee of the 306 Hospital reviewed that relevant ethical issues in this study were well considered.
Study Population, Bacterial Isolate Identification, and Drug Susceptibility Testing
This is a prospective surveillance study. Consecutive K. pneumoniae isolates were collected from unique patients being treated in the 306 Hospital in Beijing, China (which is a 1,100-bed tertiary-care hospital serving approximately 25,000 in-patients per year) for the period of September 1, 2010-October 31, 2011. In the case of duplicate patient samples, the first collected isolate was chosen. All strains were cultured in Luria–Bertani (LB) medium. The K. pneumoniae strains were confirmed by phenotypic tests and 16 S rDNA sequencing. Drug susceptibility testing (DST) for the K. pneumoniae strains was performed using the bioMérieux VITEK-2 AST-GN13 system following manufacturer’s instructions. The following 18 drugs were tested: ampicillin (AMP), piperacillin/tazobactam (TZP), ampicillin/sulbactam (SAM), cefazolin (CFZ), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), cefotetan (CTT), ertapenem (ETP), imipenem (IMP), aztreonam (ATM), ciprofloxacin (CIP), levofloxacin (LVX), gentamicin (GM), tobramycin (TOB), amikacin (AMK), trimethoprim-sulfamethoxazole (SXT), furadantin (FD). The ESBLs were detected by the bioMérieux VITEK-2 AST-GN13 test (which is claimed to be a confirmatory ESBL test). In some cases, the ESBL positivity was further confirmed by the double disk diffusion method [25]. Escherichia coli strains ATCC 25922 and ATCC 35218, K. pneumoniae strain ATCC 700603 and Pseudomonas aeruginosa strain ATCC 27853 were used as quality control strains for the DST. Clinical records of patients from whom the K. pneumoniae isolates were obtained were reviewed retrospectively.
PCR Amplification and Sequencing
Genomic DNA was extracted using DNeasy Tissue kit (Qiagen; Valencia, CA, USA). Drug resistance-associated genes were detected by PCR and sequencing using 37 pairs of primers listed in Table 1. Direct sequencing of positive amplicons was conducted. The primers were synthesized by the Beijing Genomics Institute (BGI, China). PCR was performed in a 50-µL reaction mixture consisting of 5 µL of 10×PCR buffer, 2.5 units of Taq DNA polymerase (Takara), 0.2 mM of dNTPs, 0.4 µM each of the primer, and 1 µL chromosomal DNA. All reaction mixtures were subjected to 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. PCR products were purified and sequenced bi-directionally with the same primers used for PCR by the Beijing Genomics Institute (BGI, China). DNA sequences were annotated using the BLAST program at http://www.ncbi.nlm.nih.gov. Mutations in the gyrA and parC genes were identified by comparing the DNA sequences with gyrA and parC sequences of the K. pneumoniae (GenBank accession numbers DQ673325 and NC009648 for gyrA and parC, respectively).
Table 1. Primers used for PCR and sequencing of drug resistance-associated genes from K. pneumoniae isolates.
| Target genes | Primer sequence (5′ to 3′) | Ampliconsize (bp) | Source ofreference | |
| Forward | Reverse | |||
| bla CTX-M-1 | GGT TAA AAA ATC ACT GCG TC | TTA CAA ACC GTC GGT GAC GA | 876 | [2] |
| bla CTX-M-2 | ATG ATG ACT CAG AGC ATT CG | TTA TTG CAT CAG AAA CCG TG | 876 | [29] |
| bla CTX-M-3 | GTT GTT GTT ATT TCG TAT CTT CC | CGA TAA ACA AAA ACG GAA TG | 934 | [10] |
| bla CTX-M-8 | ATG ATG AGA CAT CGC GTT AAG | CGG TGA CGA TTT TCG CGG CAG | 864 | [61] |
| bla CTX-M-9 | GTG ACA AAG AGA GTG CAA CGG | ATG ATT CTC GCC GCT GAA GCC | 850 | [31] |
| bla CTX-M-10 | GCA GCA CCA GTA AAG TGA TGG | GCG ATA TCG TTG GTG GTA CC | 873 | [30] |
| bla CTX-M-14 | ACA ATG ACG CTG GCA GAA CTG | TTA CAG CCC TTC GGC GAT GA | 512 | [64] |
| bla CTX-M-25 | CAC ACG AAT TGA ATG TTC AG | TCA CTC CAC ATG GTG AGT | 924 | [61] |
| bla SHV-group | TTT ATC GGC CYT CAC TCA AGG | GCT GCG GGC CGG ATA ACG | 896 | [32] |
| bla TEM | KAC AAT AAC CCT GRT AAA TGC | AGT ATA TAT GAG TAA ACT TGG | 899 | [32] |
| bla KPC | ATG TCA CTG TAT CGC CGT CT | TTT TCA GAG CCT TAC TGC CC | 882 | [8] |
| bla NDM | GGT TTG GCG ATC TGG TTT TC | CGG AAT GGC TCA TCA CGA TC | 621 | [51] |
| bla IMP | GGA ATA GAG TGG CTT AAT TCT C | CCA AAC CAC TAC GTT ATC | 624 | [27] |
| bla VIM | GGT CTC ATT GTC CGT GAT GGT GAT GAG | CTC GAT GAG AGT CCT TCT AGA G | 271 | [27] |
| bla OXA-48 | TTG GTG GCA TCG ATT ATC GG | GAG CAC TTC TTT TGT GAT GGC | 743 | [47] |
| bla CMY | TGG CCA GAA CTG ACA GGC AAA | TTT CTC CTG AAC GTG GCT GGC | 462 | [65] |
| bla DHA | AAC TTT CAC AGG TGT GCT GGG T | CCG TAC GCA TAC TGG CTT TGC | 405 | [65] |
| bla FOX | AAC ATG GGG TAT CAG GGA GAT G | CAA AGC GCG TAA CCG GAT TGG | 190 | [65] |
| dhfr | GCC AAT CGG GTT ATT GGC AA | TGG GAA GAA GGC GTC ACC CTC | 357 | [15] |
| qnrA | ATT TCT CAC GCC AGG ATT TG | GAT CGG CAA AGG TTA GGT CA | 627 | [62] |
| qnrB | GAT CGT GAA AGC CAG AAA GG | ACG ATG CCT GGT AGT TGT CC | 469 | [62] |
| qnrC | GGG TTG TAC ATT TAT TGA ATC G | CAC CTA CCC ATT TAT TTT CA | 307 | [34] |
| qnrD | CGA GAT CAA TTTA CGG GGA ATA | AAC AAG CTG AAG CGC CTG | 533 | [33] |
| qnrS | ACG ACA TTC GTC AAC TGC AA | TAA ATT GGC ACC CTG TAG GC | 417 | [28] |
| aac(6′)-Ib-cr | TTG CGA TGC TCT ATG AGT GGC TA | CTC GAA TGC CTG GCG TGT TT | 482 | [35] |
| qepA | AAC TGC TTG AGC CCG TAG AT | GTC TAC GCC ATG GAC CTC AC | 596 | [34] |
| gyrA | CGA CCT TGC GAG AGA AAT | GTT CCA TCA GCC CTT CAA | 626 | [54] |
| parC | TAC GTC ATC ATG GAC AGG | GCC ACT TCA CGC AGG TTG | 460 | [37] |
| aacA4 | ATG ACT GAG CAT GAC CTT GCG | TTA GGC ATC ACT GCG TGT TCG | 540 | [38] |
| aacC1 | ATG GGC ATC ATT CGC ACA TGT AGG | TTA GGT GGC GGT ACT TGG GTC | 873 | [38] |
| aacC2 | ATG CAT ACG CGG AAG GCA ATA AC | CTA ACC GGA AGG CTC GCA AG | 861 | [38] |
| aadA1 | ATG AGG GAA GCG GTG ATC G | TTA TTT GCC GAC TAC CTT GGT G | 792 | [38] |
| aadB | ATG GAC ACA ACG CAG GTC GC | TTA GGC CGC ATA TCG CGA CC | 534 | [38] |
| aphA6 | ATG GAA TTG CCC AAT ATT ATT C | TCA ATT CAA TTC ATC AAG TTT TA | 781 | [38] |
| armA | AGG TTG TTT CCA TTT CTG AG | TCT CTT CAT TCC CTT CTC C | 591 | [57] |
| rmtB | CCC AAA CAG ACC GTA GAG GC | CTC AAA CTC GGC GGG CAA GC | 585 | [57] |
| Integron I | GGC ATC CAA GCA CAA G | AAG CAG ACT TGA CCT GA | Variable | [63] |
Conjugation Experiments, Plasmid Analysis, and MLST Analysis
Transfer of resistance genes by conjugation experiments were carried out in LB broth using clinical isolates as donors and the E. coli J53AzR as the recipient as described previously [26]. Cultures of donor and recipient cells in logarithmic phase (0.5 mL each) were added to 4 mL of fresh LB broth and incubated overnight without shaking. Transconjugants were selected on LB plates containing 100 µg/mL sodium azide for counterselection and 100 µg/mL ampicillin to select for plasmid-encoded resistance. The drugs tested were purchased from Sigma Chemical Co. Plasmid DNA from the K. pneumoniae donor strains and E. coli transconjugants were prepared using the Plasmid Maxprep Kit (Vigorous Biotechnology, Beijing, China) and were separated on 0.7% agarose gels. Genotyping was determined by MLST analysis. MLST with seven genes (gapA, infB, mdh, pgi, phoE, rpoB and tonB) was performed on isolates according to the protocol described on the K. pneumoniae MLST website (www.pasteur.fr/mlst). Sequence types (STs) were assigned by using the MLST database (www.pasteur.fr/mlst/Kpneumoniae.html).
Statistical Analysis
SPSS software (version 15.0) was used for data analysis. Categorical variables were compared with the chi-square test or Fisher’s exact test. A p value of <0.05 was considered to be statistically significant.
Results
Demographic and Clinical Characteristics of the Patients
From September 1, 2010 to October 31, 2011, a total of 223 non-repetitive patients at the 306 Hospital who had K. pneumoniae isolates were subjected to DST using 18 antibiotics. Among which, 137 (61.4%) were MDR isolates, 49 (22.0%) were XDR isolates, 4 (1.8%) were PDR isolates, and 33 (14.8%) were other types of isolates. The proportion of the male and female were 73.5% (n = 164) and 26.5% (n = 59), respectively. Sixty-eight (30.5%) of the patients were Beijing residents and the rest were from other provinces of China (non-Beijing residents). The median (±SD) age of the patients was 74.0±20.3 years (range 1.0–98.0 years). The majority of the patients were from medical ward (97, 43.5%) and intensive care unit (75, 33.6%). The main source of the specimens was sputum (168, 75.3%). The proportion of the ESBL positive cases was 45.3% (n = 101). The proportion of XDR (42, 41.6%) and PDR (4, 4.0%) cases was significantly higher among patients whose isolates were ESBL positive as compared with those whose isolates were ESBL negative. In addition, the proportion of MDR cases (83, 68.0%) and other types of cases (32, 26.2%) was significantly higher in patients with ESBL-negative isolates than that observed for XDR (7, 5.7%) and PDR cases (0). The detailed information on relevant demographic and clinical characteristics of the study population is summarized in Table 2.
Table 2. Demographic and clinical characteristics of the patients.
| Characteristics | Total n = 223 (%) | Patients infected with MDR isolates n = 137 (%) | Patients infected with XDR isolates n = 49 (%) | Patients infected with PDR isolates n = 4 (%) | Patients infected with other types of isolates n = 33 (%) | P value |
| Gender | 0.991 | |||||
| Male | 164 (73.5) | 101 (73.7) | 35 (71.4) | 4 (100) | 24 (72.7) | |
| Female | 59 (26.5) | 36 (26.3) | 14 (28.6) | 0 | 9 (27.3) | |
| Age group, years | 0.400 | |||||
| <18 | 7 (3.1) | 6 (4.4) | 0 | 0 | 1 (3.0) | |
| 18–64 | 64 (28.7) | 39 (28.5) | 11 (22.4) | 2 (50.0) | 12 (36.4) | |
| >64 | 152 (68.2) | 92 (67.2) | 38 (77.6) | 2 (50.0) | 20 (60.6) | |
| Residence situation | 0.862 | |||||
| Beijing resident | 68 (30.5) | 40 (29.2) | 17 (34.7) | 0 | 11 (33.3) | |
| Non-Beijing resident | 155 (69.5) | 97 (70.8) | 32 (65.3) | 4 (100.0) | 22 (66.7) | |
| Hospital location | 0.199 | |||||
| Emergency room | 9 (4.0) | 7 (5.1) | 2 (4.1) | 0 | 0 | |
| Intensive care unit | 75 (33.6) | 51 (37.2) | 17 (34.7) | 0 | 7 (21.2) | |
| Medical ward | 97 (43.5) | 59 (43.1) | 18 (36.7) | 2 (50.0) | 18 (54.5) | |
| Surgical ward | 42 (18.8) | 20 (14.6) | 12 (24.5) | 2 (50.0) | 8 (24.2) | |
| Sources of specimens | 0.187 | |||||
| Sputum | 168 (75.3) | 108 (78.8) | 34 (69.4) | 3 (75.0) | 23 (69.7) | |
| Urine | 14 (6.3) | 7 (5.1) | 7 (14.3) | 0 | 0 | |
| Throat or nose swabs | 21 (9.4) | 11 (8.0) | 2 (4.1) | 0 | 8 (24.2) | |
| Catheters | 3 (1.3) | 2 (1.5) | 1 (2.0) | 0 | 0 | |
| Blood | 10 (4.5) | 4 (2.9) | 3 (6.1) | 1(25.0) | 2 (6.1) | |
| Puncture fluid | 1 (0.4) | 1 (0.7) | 0 | 0 | 0 | |
| Drainage fluid | 1 (0.4) | 1 (0.7) | 0 | 0 | 0 | |
| Pleural effusion | 2 (0.9) | 1 (0.7) | 1 (2.0) | 0 | 0 | |
| Plus | 1 (0.4) | 1 (0.7) | 0 | 0 | 0 | |
| Bile | 2 (0.9) | 1 (0.7) | 1 (2.0) | 0 | 0 | |
| ESBL | <0.001 | |||||
| Positive | 101 (45.3) | 54 (39.4) | 42 (85.7) | 4(100.0) | 1 (3.0) | |
| Negative | 122 (54.7) | 83 (60.6) | 7 (14.3) | 0 | 32 (97.0) |
Drug Susceptibility Patterns of the K. pneumoniae Isolates
DST was conducted for 223 K. pneumoniae isolates and the detailed information on resistance rates to all tested drugs are listed in Table 3. The highest resistance rate was observed for AMP, reaching 99.6% (n = 222), followed by resistance to FD (190, 85.2%), CTT (153, 68.6%), SXT (118, 52.9%), SAM (115, 51.6%), CFZ (114, 51.1%), CRO (110, 49.3%), CAZ (110, 49.3%), ATM (109, 48.9%), and TOB (109, 48.9%). The two carbapenems tested including ETP and IMP exhibited relatively lower resistance rates (7.2% and 5.8%, respectively). Notably, the rates of resistance to most drugs were much higher among ESBL positive isolates than ESBL negative isolates.
Table 3. Drug resistance rates of K. pneumoniae isolates.
| Antimicrobial category | Drugsa | Range (µg/mL) | MIC50 (µg/mL) | MIC90 (µg/mL) | ESBL positive n = 101 (%) | ESBL negative n = 122 (%) | Total n = 223 (%) |
| Penicillins (99.6%, 222/223) | AMP | ≤2–≧32 | ≥32 | ≥32 | 101 (100) | 121 (99.2) | 222 (99.6) |
| Antipseudomonal penicillins + beta-lactamase inhibitors (22.4%, 50/223) | TZP | ≤4–≧128 | ≤4 | ≥128 | 29 (28.7) | 21 (17.2) | 50 (22.4) |
| Penicillins + beta-lactamase inhibitors (51.6%, 115/223) | SAM | ≤2–≧32 | 16 | ≥32 | 88 (87.1) | 27 (22.1) | 115 (51.6) |
| 1st and 2nd generation cephalosporins (51.1%, 114/223) | CFZ | ≤4–≧64 | 32 | ≥64 | 96 (95.0) | 18 (14.8) | 114 (51.1) |
| 3rd and 4th generation cephalosporins (49.3%, 110/223) | CRO | ≤1–≧64 | ≤1 | ≥64 | 95 (94.1) | 15 (12.3) | 110 (49.3) |
| CAZ | ≤1–≧64 | ≤1 | ≥64 | 95 (94.1) | 15 (12.3) | 110 (49.3) | |
| FEP | ≤1–≧64 | ≤1 | ≥64 | 96 (95.0) | 12 (9.8) | 108 (48.4) | |
| Cephamycins (68.6%, 153/223) | CTT | ≤4–≧64 | ≤4 | ≤4 | 43 (42.6) | 110 (90.2) | 153 (68.6) |
| Carbapenems (7.2%, 16/223) | ETP | ≤0.5–≧8 | ≤0.5 | ≤0.5 | 5 (5.0) | 11 (9.0) | 16 (7.2) |
| IMP | ≤1–≧16 | ≤1 | ≤1 | 3 (3.0) | 10 (8.2) | 13 (5.8) | |
| Monobactams (48.9%, 109/223) | ATM | ≤1–≧64 | ≤1 | ≥64 | 96 (95.0) | 13 (10.7) | 109 (48.9) |
| Fluoroquinolones (40.4%, 90/223) | CIP | ≤0.25–≧4 | 1 | ≥4 | 63 (62.4) | 27 (22.1) | 90 (40.4) |
| LVX | ≤0.25–≧8 | 1 | ≥8 | 63 (62.4) | 27 (22.1) | 90 (40.4) | |
| Aminoglycosides (49.3%, 110/223) | GM | ≤1–≧16 | ≤1 | ≥16 | 70 (69.3) | 22 (18.0) | 92 (41.3) |
| TOB | ≤1–≧16 | ≤1 | ≥16 | 82 (81.2) | 27 (22.1) | 109 (48.9) | |
| AMK | ≤2–≧64 | ≤2 | 16 | 39 (38.6) | 15 (12.3) | 54 (24.2) | |
| Folate pathway inhibitors (52.9%, 118/223) | SXT | ≤20–≧320 | ≥320 | ≥320 | 90 (89.1) | 28 (23.0) | 118 (52.9) |
| Nitrofurantoin (85.2%, 190/223) | FD | ≤16–≧512 | 64 | ≥512 | 89 (88.1) | 101 (82.8) | 190 (85.2) |
Abbreviation of drugs: AMP, Ampicillin; TZP, Piperacillin/Tazobactam; SAM, Ampicillin/Sulbactam; CFZ, Cefazolin; CRO, Ceftriaxone; CAZ, Ceftazidime; FEP, Cefepime; CTT, Cefotetan; ETP, Ertapenem; IMP, Imipenem; ATM, Aztreonam; CIP, Ciprofloxacin; LVX, Levofloxacin; GM, Gentamycin; TOB, Tobramycin; AMK, Amikacin; SXT, Trimethoprim-Sulfamethoxazole; FD, Furadantin.
Drug Resistance Determinants of the K. pneumoniae Isolates
PCR and sequencing analysis were conducted for 223 K. pneumoniae isolates to analyze ESBL genes as well as drug resistance determinants conferring resistance to carbapenems, folate pathway inhibitors, fluoroquinolones, and aminoglycosides. The detailed information on the percentage of drug resistance-associated genes detected in K. pneumoniae isolates were summarized in Table 4 and Table 5. The percentage of isolates with 8 or more drug resistance-associated genes was 24.2% (n = 54). Among the beta-lactamase genes, the most frequently detected ones include: bla CTX-M-10 (80, 35.9%), bla SHV-1 (55, 24.7%), bla SHV-11 (47, 21.1%), bla CTX-M-1 (37, 16.6%), bla CTX-M-14 (37, 16.6%), bla CTX-M-15 (34, 15.2%) and bla CTX-M-3 (32, 14.3%). Except for bla NDM, all the other 4 examined carbapenemase genes including bla KPC-2 (3, 1.3%), bla IMP (1, 0.4%), bla VIM (1, 0.4%), and bla OXA-48 (5, 2.2%) were detected in this study. The prevalence of AmpC beta-lactamases including bla CMY-2, bla DHA-1 and bla FOX were 3.1%, 4.0% and 0, respectively in this study. Among the 7 plasmid-encoded fluoroquinolone resistance-associated genes including qnrA, qnrB, qnrC, qnrD, qnrS, aac(6′)-Ib-cr, and qepA, the highest rates were observed for qnrS (58, 26.0%) and aac(6′)-Ib-cr (53, 23.8%). In addition, gyrA gene mutations including T247A (Ser83Ile) (21, 9.4%), C248T (Ser83Phe) (15, 6.7%), and A260C (Asp87Ala) (16, 7.2%) were identified. No mutations were detected in parC gene. Among the aminoglycosides resistance-associated genes, the highest rates were observed for aacC2 (73, 32.7%), aacA4 (57, 25.6%), and aadA1 (56, 25.1%). The prevalence of the plasmid-encoded 16 S rRNA methylases armA and rmtB were detected to be 5.8% and 3.6%, respectively. Class 1 integrons were detected in 47.5% (n = 106) of the isolates. In order to evaluate the correlation between phenotypic and genotypic drug resistance profiles, we also calculated the proportion of antibiotic resistance-associated genes among the phenotypic resistant isolates as well as the phenotypic susceptible isolates. We found that while some of the previously reported resistance-associated genes were indeed detected at relatively higher rates among corresponding phenotypic resistant isolates, some others were detected in very low proportion of the phenotypic resistant isolates. In addition, some of the resistance-associated genes were also detected in a sizable proportion of the phenotypic susceptible isolates.
Table 4. Percentage of beta-lactamase antibiotics resistance-associated genes detected in K. pneumoniae isolates.
| Target antimicrobial category | Resistance-associated genes | Resistance-associated genes detected in phenotypic resistant isolates, n/Na (%) | Resistance-associated genes detected in phenotypic susceptible isolates, n/Nb (%) | Resistance-associated genes detected in all isolates, n/Nc (%) |
| Antipseudomonal penicillins + beta-lactamase inhibitors, penicillins + beta-lactamase inhibitors, 1st and 2nd generation cephalosporins, 3rd and 4th generation cephalosporins, cephamycins (n = 204) | bla CTX-M-1 | 35/204 (17.2) | 2/19 (10.5) | 37/223 (16.6) |
| bla CTX-M-2 | 1/204 (0.5) | 1/19 (5.3) | 2/223 (0.9) | |
| bla CTX-M-3 | 31/204 (15.2) | 1/19 (5.3) | 32/223 (14.3) | |
| bla CTX-M-8 | 20/204 (9.8) | 2/19 (10.5) | 22/223 (9.9) | |
| bla CTX-M-9 | 18/204 (8.8) | 2/19 (10.5) | 20/223 (9.0) | |
| bla CTX-M-14 | 37/204 (18.1) | 0 | 37/223 (16.6) | |
| bla CTX-M-15 | 34/204 (16.7) | 0 | 34/223 (15.2) | |
| bla CTX-M-10 | 76/204 (37.3) | 4/19 (21.1) | 80/223 (35.9) | |
| bla CTX-M-25 | 2/204 (1.0) | 1/19 (5.3) | 3/223 (1.3) | |
| bla CTX-M-55 | 3/204 (1.5) | 0 | 3/223 (1.3) | |
| bla SHV-1 | 55/204 (27.0) | 0 | 55/223 (24.7) | |
| bla SHV-2 | 2/204 (1.0) | 0 | 2/223 (0.9) | |
| bla SHV-11 | 44/204 (21.6) | 3/19 (15.8) | 47/223 (21.1) | |
| bla SHV-85 | 15/204 (7.4) | 0 | 15/223 (6.7) | |
| bla TEM-1 | 19/204 (9.3) | 0 | 19/223 (8.5) | |
| bla TEM-186 | 3/204 (1.5) | 0 | 3/223 (1.3) | |
| bla CMY-2 | 7/204 (3.4) | 0 | 7/223 (3.1) | |
| bla DHA-1 | 9/204 (4.4) | 0 | 9/223 (4.0) | |
| bla FOX | 0 | 0 | 0 | |
| Carbapenems (n = 16) | bla KPC-2 | 3/16 (18.8) | 0 | 3/223 (1.3) |
| bla NDM-1 | 0 | 0 | 0 | |
| bla IMP | 1/16 (6.3) | 0 | 1/223 (0.4) | |
| bla VIM | 1/16 (6.3) | 0 | 1/223 (0.4) | |
| bla OXA-48 | 5/16 (31.3) | 0 | 5/223 (2.2) |
n/N: No. of designated drug resistance-associated genes/No. of isolates resistant to the corresponding drugs.
n/N: No. of designated drug resistance-associated genes/No. of isolates susceptible to the corresponding drugs.
n/N: No. of designated drug resistance-associated genes/No. of all isolates.
Table 5. Percentage of non-beta-lactamase antibiotics resistance-associated genes detected in K. pneumoniae isolates.
| Target antimicrobial category | Resistance-associated genes | Resistance-associated genes detected in phenotypic resistant isolates, n/Na (%) | Resistance-associated genes detected in phenotypic susceptible isolates, n/Nb (%) | Resistance-associated genes detected in all isolates, n/Nc (%) |
| Folate pathway inhibitors (n = 118) | Dhfr | 52/118(44.1) | 10/105 (9.5) | 62/223 (27.8) |
| Fluoroquinolones (n = 90) | qnrA | 1/90 (1.1) | 1/133 (0.8) | 2/223 (0.9) |
| qnrB | 33/90 (36.7) | 6/133 (4.5) | 39/223 (17.5) | |
| qnrC | 1/90 (1.1) | 1/133 (0.8) | 2/223 (0.9) | |
| qnrD | 23/90 (25.6) | 12/133 (9.0) | 35/223 (15.7) | |
| qnrS | 22/90 (24.4) | 36/133 (27.1) | 58/223 (26.0) | |
| aac(6′)-Ib-cr | 45/90 (50.0) | 8/133 (6.0) | 53/223 (23.8) | |
| qepA | 1/90 (1.1) | 1/133 (0.8) | 2/223 (0.9) | |
| gyrA mutations | ||||
| T247A(Ser83Ile) | 18/90 (20.0) | 3/133 (2.3) | 21/223 (9.4) | |
| C248T (Ser83Phe) | 12/90 (13.3) | 3/133 (2.3) | 15/223 (6.7) | |
| A260C(Asp87Ala) | 13/90 (14.4) | 3/133 (2.3) | 16/223 (7.2) | |
| parC mutations | None | None | None | |
| Aminoglycosides (n = 110) | aacA4 | 50/110 (45.5) | 7/113 (6.2) | 57/223 (25.6) |
| aacC1 | 3/110 (2.7) | 0 | 3/223 (1.3) | |
| aacC2 | 66/110 (60.0) | 7/113 (6.2) | 73/223 (32.7) | |
| aadA1 | 49/110 (44.5) | 7/113 (6.2) | 56/223 (25.1) | |
| aadB | 4/110 (3.6) | 0 | 4/223 (1.8) | |
| aphA6 | 1/110 (0.9) | 0 | 1/223 (0.4) | |
| armA | 13/110 (11.8) | 0 | 13/223 (5.8) | |
| rmtB | 8/110 (7.3) | 0 | 8/223 (3.6) | |
n/N: No. of designated drug resistance-associated genes/No. of isolates resistant to the corresponding drugs.
n/N: No. of designated drug resistance-associated genes/No. of isolates susceptible to the corresponding drugs.
n/N: No. of designated drug resistance-associated genes/No. of all isolates.
Characteristics of Carbapenem-resistant K. pneumoniae Isolates
Among the 223 isolates, 16 were detected to be carbapenem-resistant and were used for further characterization. Most of the patients from whom the isolates obtained were male 87.5% (14/16) and aged patients (all were 56 years old or above). Among the 14 patients whose treatment outcome information was available, 6 died. Although carbapenemase genes were detected only in 7 of the 16 isolates, the majority of them exhibited resistance to a high number of drugs and contained a variety of corresponding drug resistance-associated genes. The proportion of MDR, XDR and PDR were 12.5% (n = 2), 62.5% (n = 10), 25.0% (n = 4), respectively. The more detailed characteristics of those carbapenem-resistant isolates are shown in Table S1.
Transmissibility of Drug Resistance of ESBL Positive MDR, XDR and PDR K. pneumoniae Isolates
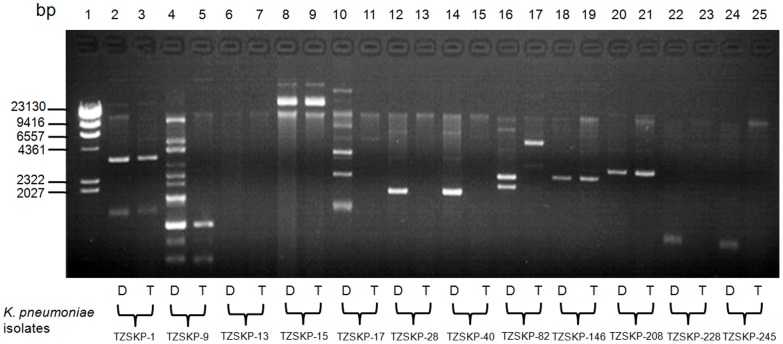
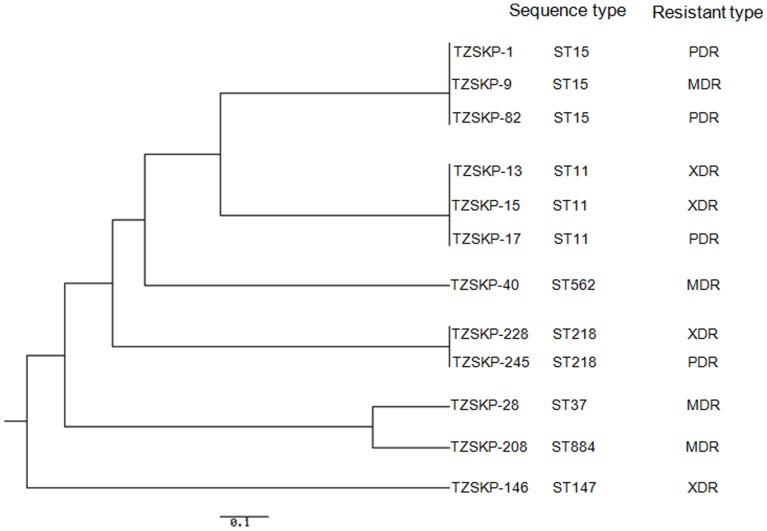
Twelve K. pneumoniae isolates including 4 MDR, 4 XDR, and 4 PDR isolates were selected to test the natural transmissibility of antibiotic resistance by conjugation experiments. As shown in Table 6, the resistance to various drugs and the corresponding resistance-associated genes were transferred in all tested isolates, though to different extent. The most frequently transferred genes include aac(6′)-Ib-cr (6/7, 85.7%), bla CTX-M-14 (4/5, 80.0%), bla CTX-M-9 (2/3, 66.7%), aacA4 (5/8, 62.5%) and bla SHV-11 (4/8, 50.0%). Five transconjugants contained plasmids with the same size as those in their respective donors (TZSKP-1, 9, 15, 146, and 208) (Figure 1). In addition, the transconjugant for TZSKP-82 harbored new plasmid with different size than that in the donor strain. The phylogenetic tree based on the MLST analysis results for the isolates is shown in Figure 2. Seven different STs were identified for those 12 isolates. Three isolates (TZSKP-1, 9, and 82) belonged to ST15, three isolates (TZSKP-13, 15, and 17) belonged to ST11, two isolates (TZSKP-228 and 245) belonged to ST218, and the rest of the isolates had unique STs. The epidemiological links were further determined for the patients from whom the clustered isolates were obtained.
Table 6. Transmissibility of drug resistance of ESBL positive MDR, XDR and PDR K. pneumoniae isolates by conjugation.
| K. pneumoniae isolates | Resistance profile of K. pneumoniae isolatesa,b | Resistance-associated genes detected in K. pneumoniae isolatesb |
| MDR isolates | ||
| TZSKP-9 | AMP,SAM,CFZ,CRO,CAZ,FEP,ATM,CIP,LVX,SXT,FD | bla CTX-M-14,dhfr |
| TZSKP-28 | AMP,SAM,CFZ,CRO,CAZ,FEP,ATM,GM,TOB,SXT,FD | bla SHV-11 |
| TZSKP-40 | AMP,SAM,CFZ,CRO,CAZ,FEP,ATM,GM,TOB,SXT,FD | bla CTX-M-14,bla CTX-M-10,bla SHV-11,bla TEM-1,qnrS,aacC2 |
| TZSKP-208 | AMP,SAM,CFZ,CRO,CAZ,FEP,ATM,SXT,FD | bla CTX-M-9,bla CTX-M-10,bla SHV-11,qnrS |
| XDR isolates | ||
| TZSKP-13 | AMP,SAM,CFZ,CRO,CAZ,FEP,ATM,CIP,LVX,TOB,AMK,SXT,FD | bla CTX-M-14,dhfr,qnrB,aacA4 |
| TZSKP-15 | AMP,SAM,CFZ,CRO,CAZ,FEP,CTT,ETP,IMP,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | bla CTX-M-1,bla CTX-M-14,bla CTX-M-10,bla SHV-11,bla TEM,dhfr,qnrA,qnrB,aac(6′)-Ib-cr,aacA4,aadA1 |
| TZSKP-146 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | bla CTX-M-1,bla CTX-M-3,bla CTX-M-9,bla CTX-M-10,bla SHV-11,bla TEM,dhfr,qnrD,aac(6′)-Ib-cr,aacA4,aacC2,aadA1 |
| TZSKP-228 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,CTT,ETP,IPM,ATM,TOB,AMK,SXT | bla CTX-M-3,bla CTX-M-10,bla SHV-11,bla TEM-1,bla KPC-2,bla IMP,bla VIM,bla OXA-48,dhfr,qnrD,qnrS,aac(6′)-Ib-cr,aacA4,aadB,aphA6 |
| PDR isolates | ||
| TZSKP-1 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,CTT,ETP,IMP,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | blaCTX-M-3,blaCTX-M-10,blaSHV-11,dhfr,qnrB,aac(6′)-Ib-cr,aacA4,aacC2 |
| TZSKP-17 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,CTT,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | bla CTX-M-2,bla CTX-M-14,bla CTX-M-10,bla SHV-11,bla TEM-1,bla CMY2,bla DHA1,dhfr,qnrS,aac(6′)-Ib-cr,aacA4,aacC2,aadA1 |
| TZSKP-82 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,CTT,ETP,IPM,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | bla CTX-M-3,bla CTX-M-9,bla CTX-M-10,bla OXA-48,aac(6′)-Ib-cr,aacA4,aacC2,armA |
| TZSKP-245 | AMP,TZP,SAM,CFZ,CRO,CAZ,FEP,CTT,ETP,IPM,ATM,CIP,LVX,GM,TOB,AMK,SXT,FD | bla CTX-M-10,bla TEM-1,bla KPC-2,dhfr,aac(6′)-Ib-cr,aacA4,aacC2 |
Abbreviation of drugs: AMP, Ampicillin; TZP, Piperacillin/Tazobactam; SAM, Ampicillin/Sulbactam; CFZ, Cefazolin; CRO, Ceftriaxone; CAZ, Ceftazidime; FEP, Cefepime; CTT, efotetan; ETP, Ertapenem; IMP, Imipenem; ATM, Aztreonam; CIP, Ciprofloxacin; LVX, Levofloxacin; GM, Gentamycin; TOB, Tobramycin; AMK, Amikacin; SXT, Trimethoprim-Sulfamethoxazole; FD, Furadantin.
The ones found in both donor strains and transconjugants were underlined to demonstrate the difference.
Figure 1. Analysis of plasmid DNA from K. pneumoniae parental donor strains (Designated D) and derived E. coli transconjugants (Designated T) by agarose gel electrophoresis.
Lanes: 1, molecular marker (λ-Hind β digest DNA Marker, TaKaRa Code: D3403A); 2, TZSKP-1; 3, Transconjugant from TZSKP-1; 4, TZSKP-9; 5, Transconjugant from TZSKP-9; 6, TZSKP-13; 7, Transconjugant from TZSKP-13; 8, TZSKP-15; 9, Transconjugant from TZSKP-15; 10, TZSKP-17; 11, Transconjugant from TZSKP-17; 12, TZSKP-28; 13, Transconjugant from TZSKP-28; 14, TZSKP-40; 15, Transconjugant from TZSKP-40; 16, TZSKP-82; 17, Transconjugant from TZSKP-82; 18, TZSKP-146; 19, Transconjugant from TZSKP-146; 20, TZSKP-208; 21, Transconjugant from TZSKP-208; 22, TZSKP-228; 23, Transconjugant from TZSKP-228; 24, TZSKP-245; 25, Transconjugant from TZSKP-245.
Figure 2. Phylogenetic tree for the 7 housekeeping loci in K. pneumoniae constructed using the UPGMA method, displaying the clonal relationship among the K. pneumoniae isolates.
Discussion
Worldwide emergence and dissemination of ESBL and carbapenemase genes among Enterobacteriaceae, especially in K. pneumoniae isolates, poses a considerable threat to public health. The major goal of this study was to evaluate the current situation and genetic background of drug-resistant K. pneumoniae isolates from patients in hospital settings. The highest and lowest resistance rates were observed for penicillins (99.6%) and carbapenems (7.2%), respectively. The rates of MDR, XDR and PDR isolates observed in this study are alarmingly high. This could cause difficulty in treating K. pneumoniae-associated infections since fewer and fewer effective drugs are available for treating those highly drug-resistant isolates. We also found that the proportion of MDR and other types of cases was significantly higher in patients with ESBL-negative isolates than that for XDR and PDR cases. This could be partially explained by the fact that the ESBL-positive isolates are normally resistant to many drugs, leaving only a few effective drugs available for treatment, which could lead to further resistance to those drugs. Indeed, this study further reveals that resistance to most of the drugs was found to be associated with ESBL positivity. Infections due to those ESBL positive and highly resistant strains are reported to be associated with higher morbidity and mortality rates [19]–[22], thus globally coordinated surveillance of epidemiology of those resistant isolates are warranted.
Another goal of this study was to evaluate the correlation between resistance phenotypes and the genetic determinants clinical K. penumoniae isolates, so as to give a “snapshot” of the background of those resistant isolates. A striking feature of this study is the large number of antibiotic resistance-associated genes detected in the examined isolates. We also found that while some of the previously reported resistance-associated genes were indeed detected at relatively higher rates among corresponding phenotypic resistant isolates, some others were detected in very low proportion of the phenotypic resistant isolates, suggesting the existence of unknown drug resistance mechanisms such as reduced permeability of the outer membrane or up-regulated unknown efflux pumps in some clinical isolates [27], [28]. In addition, some of the resistance-associated genes were also detected in a sizable proportion of the phenotypic susceptible isolates, suggesting that individual resistance gene alone is not sufficient to cause resistance phenotype and only when some of them were accumulated can the resistance become detectable in the clinical isolates.
Rapid detection of genetic determinants associated with drug resistance in clinical K. pneumoniae isolates is crucial for appropriate antimicrobial therapy and infection control measures. We detected relatively high percentage of previously reported genes associated with resistance to beta-lactams [2], [10], [29]–[32], fluoroquinolones [28], [33]–[37], aminoglycosides [38], [39], and folate pathway inhibitors [15] in K. pneumoniae isolates. CTX-M-type beta-lactamase genes (such as bla CTX-M-14 and bla CTX-M-15) have been reported to be prevalent worldwide [40]–[42]. For example, a recent study from China reported that among the 21 K. pneumoniae isolates from 1270 specimens collected in a prospective multi-center study in eight teaching hospitals in China from June to December in 2007, 3 were detected to have bla CTX-M-14 (3, 14.3%) [40]. Another study conducted in Scotland showed that 16 of the 219 (7.3%) clinical isolates of K. pneumoniae collected in 2006 and 2007 at the Royal Infirmary of Edinburgh, Scotland had bla CTX-M-15 [41]. In the present study, the highest rate of CTX-M-type beta-lactamase genes was observed for bla CTX-M-10 (35.9%), followed by bla CTX-M-1 (16.6%), bla CTX-M-14 (16.6%), and bla CTX-M-15 (15.2%). The rates of the non-ESBL SHV-type beta-lactamase genes bla SHV-1 and bla SHV-11 genes were 24.7% and 21.1%, respectively in this study, which were relatively lower compared to that reported by some previous studies conducted in other regions. For example, according to a study conducted in Korea, the rates of the bla SHV-1 and bla SHV-11 genes among K. pneumoniae isolates collected from May to July, 2002 were 35% (50/142) and 62% (62/142), respectively [43]. Another study from Brazil reported that 55.8% (29/52) of the K. pneumoniae isolates collected in Recife, PE, Brazil during 1998 to 2005 harbored the bla SHV genes [44]. Thus, the prevalence of some beta-lactamase genes such as the bla SHV genes could be greatly variable geographically and timewise.
The more recently reported carbapenemases genes (such as bla IMP, bla VIM, bla NDM, plasmid-mediated clavulanic acid-inhibited class A beta-lactamases genes such as bla KPC, and the class D beta-lactamase gene bla OXA-48) were rarely detected or undetected in this study. Carbapenemases increasingly have been reported in Enterobacteriaceae in the past decade. KPC carbapenemases have been reported in the United States and then worldwide [6], [8]. VIM and IMP metallo-beta-lactamases also have been reported in many regions of the world, with a higher prevalence in southern Europe and Asia [1], [5], [45]–[47]. Carbapenemases of the oxacillinase-48 type (OXA-48) have been identified mostly in Mediterranean and European countries and in India [48], [49]. Although the world-alarming New Delhi metallo-beta-lactamase-1 (NDM-1) was not detected in this study, it has been detected worldwide since it was first identified in India and its variants have emerged [13], [50]–[52]. Thus resistance caused by those recently emerging beta-lactamases is still worrisome and needs continuous monitoring. The association of alterations in gyrA (gene encoding for GyrA subunit of DNA gyrase) and parC (gene encoding for ParC subunit of DNA topoisomerase IV) with fluoroquinolone resistance in K. pneumoniae is still not clear. Some studies suggested that in K. pneumoniae, DNA gyrase A is a primary target of quinolones and that ParC alterations play a complementary role in the development of higher-level fluoroquinolone resistance [53], [54], while a study reported that hypermutation in K. pneumoniae is uncommon and does not contribute to accumulation of gyrA mutations or directly to ciprofloxacin resistance [55]. We identified 3 types of gyrA mutations including the previously reported C248T (Ser83Phe) and A260C (Asp87Ala) [53] and the unreported T247A (Ser83Ile). No mutations in parC were detected in this study. The plasmid-encoded 16 S rRNA methylases armA and rmtB has emerged as a new mechanism of resistance to aminoglycosides, and the concomitant presence of armA or rmtB with bla CTX-M type beta-lactamase genes, especially the group 1 (CTX-M-3 and CTXM-15) or group 9 (CTX-M-14), among amikacin-resistant ESBL-producing K. pneumoniae isolates was reported in Taiwan and Belgium [56], [57]. In this study, both armA and rmtB were detected (5.8% and 3.6%, respectively). One isolate was found to harbor both armA and rmtB genes, and consistent with previous reports, the armA and rmtB genes were coexistent with at least one of the bla CTX-M type beta-lactamases tested in this study.
Our study further confirmed the notion that patients infected with carbapenem-resistant K. pneumoniae isolates normally have worse treatment outcome. In addition, the conjugation results suggest that certain ESBL genes (such as bla CTX-M-14), aac(6′)-Ib-cr and aacA4 were frequently co-transmitted and co-selected in MDR, XDR and PDR isolates and can be naturally transferred to susceptible E. coli strains by conjugation. Five transconjugants contained plasmids with the same size as those in their respective donors. Nevertheless, plasmids of the same sizes found in both donor and recipient isolates could not guarantee that the resistance transfer was plasmid-mediated. A subsequent DNA-DNA hybridization experiment with probes made by the respective resistance genes is warranted to show that the plasmids of the same sizes do carry the same resistance genes. On the other hand, we noticed that plasmids were not identified in some of the donor and recipient isolates. This could be explained by the existence of some other non-plasmid-mediated mechanisms involved in the occurrence and transfer of drug resistance in those isolates. For example, the drug resistance-associated genes could be carried on chromosomally located transposons and integrons. Although the isolate TZSKP-28 was detected to be ESBL-positive, only the non-ESBL bla SHV-11 gene was detected in it. After conjugation experiments, the recipient strain became multidrug resistant and again only the bla SHV-11 was detected. We also noticed that the beta-lactamase genes found in the transconjugants of the isolates TZSKP-1 and TZSKP-40 (bla SHV-11 or bla TEM-1) were not ESBLs, either. This result suggested that some other mechanisms may be involved in causing the ESBL positivity and MDR phenotypes in those isolates. As horizontal transmission event can result in the acquisition of multidrug resistance by wild-type strains, thus this could presumably contribute to the rapid increase in the prevalence of multidrug resistance among clinical bacteria. Another important aspect for infection control is to know whether there is a clonal spread among the highly drug-resistant isolates. Relatively diverse genotypes were identified for those 12 isolates used in conjugation analysis by MLST analysis. Three clusters (which belonged to ST15, ST11, and ST218, respectively) each consisted of 2 or 3 isolates were detected and the epidemiological links were not observed for the patients from whom the clustered isolates were obtained. Further investigation of the transmission patterns of a larger sample of drug-resistant K. pneumoniae isolates by MLST analysis is warranted and which is currently underway.
Emerging plasmid-encoded ESBLs and carbapenemases are increasingly reported worldwide [58]. Carbapenemase production encoded by genes located on mobile genetic elements is typically accompanied by genes encoding resistance to other drug classes, and are frequently located on the same mobile DNA elements such as integrons, which act as bacterial recombination systems that mediate the capture and expression of gene cassettes and are considered as the primary mechanism for antibiotic resistance gene acquisition among bacteria and are frequently associated with transposons and conjugative plasmids (http://integrall.bio.ua.pt 2009) [59], [60]. In this study, class 1 integrons were detected in a relatively high percentage of the isolates. Further sequencing analysis of class 1 integrons and gene cassette arrays is currently undergoing in the lab.
In summary, our results indicate that there is a high prevalence and possible transmission of MDR, XDR and PDR K. pneumoniae isolates among hospitalized patients. In addition, our data give a “snapshot” of the complex genetic background responsible for drug resistance in those highly drug-resistant K. pneumoniae isolates. Thus our study demonstrate that a high degree of awareness and monitoring of those drug resistance determinants are urgently needed in order to better control the emergence and transmission of drug-resistant K. pneumoniae isolates in hospital settings.
Supporting Information
Characteristics of carbapenem-resistant K. pneumoniae isolates.
(DOC)
Acknowledgments
We are grateful to Professor George A. Jacoby from Lahey Clinic Medical Center, for kindly sending us the E. coli J53 AzR for the conjugation experiments.
Funding Statement
This work was supported by the National Basic Research Program of China (2012CB518700), the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-J-6), the National Natural Science Foundation of China (grant No. 30700975), and the Merieux Research Grant program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sánchez-Romero I, Asensio A, Oteo J, Muñoz-Algarra M, Isidoro B, et al. (2012) Nosocomial Outbreak of VIM-1-producing Klebsiella pneumoniae of multilocus sequence type 15: Molecular basis, clinical risk factors, and outcome. Antimicrob Agents Chemother 56: 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jemima SA, Verghese S (2008) Molecular characterization of nosocomial CTX-M type β-lactamase producing Enterobacteriaceae from a tertiary care hospital in south India. Indian J Med Microbiol 4: 365–368. [DOI] [PubMed] [Google Scholar]
- 3. Ko KS, Lee MY, Song JH, Lee H, Jung DS, et al. (2008) Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated in Korean hospitals. Diagn Microbiol Infect Dis 61: 453–459. [DOI] [PubMed] [Google Scholar]
- 4. Kohlenberg A, Schwab F, Rüden H (2012) Wide dissemination of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella spp . in acute care and rehabilitation hospitals Epidemiol Infect 140: 528–534. [DOI] [PubMed] [Google Scholar]
- 5. Ptout JD (2008) Multidrug resistant enterobacteriaceae: new threat of an old problem. Expert Rev Anti Infect Ther 6: 657–669. [DOI] [PubMed] [Google Scholar]
- 6. Zhang R, Wang XD, Cai JC, Zhou HW, Lv HX, et al. (2011) Outbreak of Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae with high qnr prevalence in a Chinese hospital J Med Microbiol. 60: 977–982. [DOI] [PubMed] [Google Scholar]
- 7. Bush K, Jacoby GA (2010) Updated Functional Classification of β-Lactamases. Antimicrob Agents Chemother 54: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, et al. (2004) Emergence of Carbapenem-Resistant Klebsiella Species Possessing the Class A Carbapenem-Hydrolyzing KPC-2 and Inhibitor-Resistant TEM-30 β-Lactamases in New York City. Clin Infect Dis 39: 55–60. [DOI] [PubMed] [Google Scholar]
- 9. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, et al. (2011) Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother 55: 1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chmelnitsky I, Carmeli Y, Leavitt A, Schwaber MJ, Navon-Venezia S (2005) CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob Agents Chemother 49: 4745–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chudácková E, Bergerová T, Fajfrlík K, Cervená D, Urbásková P, et al. (2010) Carbapenem-nonsusceptible strains of Klebsiella pneumoniae producing SHV-5 and/or DHA-1 beta-lactamases in a Czech hospital. FEMS Microbiol Lett 309: 62–70. [DOI] [PubMed] [Google Scholar]
- 12. Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P (2011) Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55: 2420–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hammerum AM, Toleman MA, Hansen F, Kristensen B, Lester CH, et al. (2010) Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis 10: 829–830. [DOI] [PubMed] [Google Scholar]
- 14. Jean SS, Hsueh PR, Lee WS, Chang HT, Chou MY, et al. (2009) Nationwide surveillance of antimicrobial resistance among Enterobacteriaceae in intensive care units in Taiwan. Eur J Clin Microbiol Infect Dis 28: 215–220. [DOI] [PubMed] [Google Scholar]
- 15. Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S (2009) Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 65: 243–248. [DOI] [PubMed] [Google Scholar]
- 16. Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y (2008) Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother 52: 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nordmann P, Naas T, Poirel L (2011) Global Spread of Carbapenemase-producing Enterobacteriaceae . Emerg Infect Dis 17: 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomson KS (2010) Extended-Spectrum-β-Lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol 48: 1019–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ben-David D, Kordevani R, Keller N, Tal I, Marzel A, et al. (2012) Outcome of carbapenem resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect 18: 54–60. [DOI] [PubMed] [Google Scholar]
- 20. Daikos GL, Petrikkos P, Psichogiou M, Kosmidis C, Vryonis E, et al. (2009) Prospective observational study of the impact of VIM-1 metallo-beta-lactamase on the outcome of patients with Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother 53: 1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO (2001) Extended-spectrumβ-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin Infect Dis 32: 1162–1171. [DOI] [PubMed] [Google Scholar]
- 22. Marchaim D, Gottesman T, Schwartz O, Korem M, Maor Y, et al. (2010) National multicenter study of predictors and outcomes of bacteremia upon hospital admission caused by Enterobacteriaceae producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother 54: 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reinert RR (2007) Antimicrobial susceptibility among organisms from the Asia/Pacific Rim, Europe and Latin and North America collected as part of TEST and the in vitro activity of tigecycline. J Antimicrob Chemother 60: 1818–1829. [DOI] [PubMed] [Google Scholar]
- 24. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. [DOI] [PubMed] [Google Scholar]
- 25. Clinical and Laboratory Standards Institute (CLSI) (2011) Performance Standards for antimicrobial Susceptibility Testing; Twenty-first Informational Supplement. CLSI document M100-S21.
- 26. Jacoby GA, Han P (1996) Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli . J Clin Microbiol 34: 908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD (2006) High-Level Carbapenem Resistance in a Klebsiella pneumoniae Clinical Isolate Is Due to the Combination of blaACT-1 β-Lactamase Production, Porin OmpK35/36 Insertional Inactivation, and Down-Regulation of the Phosphate Transport Porin PhoE. Antimicrob Agents Chemother 50: 3396–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robicsek A, Sahm DF, Strahilevitz J, Jacoby GA, Hooper DC (2005) Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob Agents Chemother 49: 3001–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas JM (1996) Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob Agents Chemother 40: 509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oliver A, Pérez-Díaz JC, Coque TM, Baquero F, Cantón R (2001) Nucleotide Sequence and Characterization of a Novel Cefotaxime-Hydrolyzing beta-Lactamase (CTX-M-10) Isolated in Spain. Antimicrob Agents Chemother 45: 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sabaté M, Tarragó R, Navarro F, Miró E, Vergés C, et al. (2000) Cloning and sequence of the gene rncoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob Agents Chemother 44: 1970–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schlesinger J, Navon-Venezia S, Chmelnitsky I, Hammer-Münz O, Leavitt A, et al. (2005) Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob Agents Chemother 49: 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cavaco LM, Hasman H, Xia S, Aarestrup FM (2009) qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob Agents Chemother 53: 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, et al. (2009) Prevalence of Plasmid-Mediated Quinolone Resistance Determinants over a 9-Year Period. Antimicrob Agents Chemother 53: 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC (2006) Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50: 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, et al. (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51: 3354–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zimhony O, Chmelnitsky I, Bardenstein R, Goland S, Hammer Muntz O, et al. (2006) Endocarditis caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae: emergence of resistance to ciprofloxacin and piperacillin-tazobactam during treatment despite initial susceptibility. Antimicrob Agents Chemother 50: 3179–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, et al. (2006) Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50: 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez MS, Tolmasky ME (2010) Aminoglycoside modifying enzymes. Drug Resist Updat 13: 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang XR, Chen JC, Kang Y, Jiang N, An SC, et al. (2012) Prevalence and characterization of plasmid-mediated blaESBL with their genetic environment in Escherichia coli and Klebsiella pneumoniae in patients with pneumonia. Chin Med J (Engl) 125: 894–900. [PubMed] [Google Scholar]
- 41. Younes A, Hamouda A, Dave J, Amyes SG (2011) Prevalence of transferable blaCTX-M-15 from hospital- and community-acquired Klebsiella pneumoniae isolates in Scotland. J Antimicrob Chemother 66: 313–318. [DOI] [PubMed] [Google Scholar]
- 42. Luo Y, Yang J, Zhang Y, Ye L, Wang L, et al. (2011) Prevalence of β-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents 37: 352–325. [DOI] [PubMed] [Google Scholar]
- 43. Lee YH, Cho B, Bae IK, Chang CL, Jeong SH (2006) Klebsiella pneumoniae strains carrying the chromosomal SHV-11 beta-lactamase gene produce the plasmid-mediated SHV-12 extended-spectrum beta-lactamase more frequently than those carrying the chromosomal SHV-1 beta-lactamase gene. J Antimicrob Chemother. 57: 1259–1261. [DOI] [PubMed] [Google Scholar]
- 44. Veras DL, Alves LC, Brayner FA, Guedes DR, Maciel MA, et al. (2011) Prevalence of the bla SHV gene in Klebsiella pneumoniae isolates obtained from hospital and community infections and from the microbiota of healthy individuals in Recife, Brazil. Curr Microbiol 62: 1610–1616. [DOI] [PubMed] [Google Scholar]
- 45. Giakkoupi P, Xanthaki A, Kanelopoulou M, Vlahaki A, Miriagou V, et al. (2003) VIM-1 Metallo-β-Lactamase-Producing Klebsiella pneumoniae strains in Greek hospitals. J Clin Microbiol 41: 3893–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Poulou A, Spanakis N, Pournaras S, Pitiriga V, Ranellou K, et al. (2010) Recurrent health-care-associated community-onset infections due to Klebsiella pneumoniae producing VIM-1 metallo-beta-lactamase. J Antimicrob Chemother 65: 2538–2542. [DOI] [PubMed] [Google Scholar]
- 47. Tato M, Morosini M, García L, Albertí S, Coque MT, et al. (2010) Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: consequences for routine susceptibility testing. J Clin Microbiol 48: 4089–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goren MG, Chmelnitsky I, Carmeli Y, Navon-Venezia S (2011) Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J Antimicrob Chemother 66: 672–673. [DOI] [PubMed] [Google Scholar]
- 49. Poirel L, Héritier C, Tolün V, Nordmann P (2004) Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae . Antimicrob Agents Chemother 48: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan,and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR (2010) How To Detect NDM-1 Producers. J Clin Microbiol 49: 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53: 5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, et al. (1997) Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae . Antimicrob Agents Chemother 41: 699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weigel LM, Steward CD, Tenover FC (1998) gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae . Antimicrob Agents Chemother 42: 2661–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aathithan S, French GL (2010) Hypermutability in clinical isolates of Klebsiella pneumoniae is uncommon and is unrelated to ciprofloxacin resistance. Int J Antimicrob Agents 36: 239–242. [DOI] [PubMed] [Google Scholar]
- 56. Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, et al. (2007) Emergence of ArmA and RmtB aminoglycoside resistance 16 S rRNA methylases in Belgium. J Antimicrob Chemother 59: 459–464. [DOI] [PubMed] [Google Scholar]
- 57. Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, et al. (2009) Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother 53: 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schultsz C, Geerlings S (2012) Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs 72: 1–16. [DOI] [PubMed] [Google Scholar]
- 59. Mokracka J, Koczura R, Pawlowski K, Kaznowski A (2011) Resistance patterns and integron cassette arrays of the Enterobactercloacae complex strains of human origin. Med Microbiol 60: 737–743. [DOI] [PubMed] [Google Scholar]
- 60. Moura A, Soares M, Pereira C, Leitão N, Henriques I, et al. (2009) INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 61. Chmelnitsky I, Hermesh O, Navon-Venezia S, Strahilevitz J, Carmeli Y (2009) Detection of aac(6′)-Ib-cr in KPC-producing Klebsiella pneumoniae isolates from Tel Aviv, Israel. J Antimicro Chemother 64: 718–722. [DOI] [PubMed] [Google Scholar]
- 62. Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, et al. (2006) Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12: 83–88. [DOI] [PubMed] [Google Scholar]
- 63. Bissonnette L, Roy PH (1992) Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol 174: 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee H, Yong D, Yum JH, Roh KH, Lee K, et al. (2006) Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis 56: 305–312. [DOI] [PubMed] [Google Scholar]
- 65. Pérez-Pérez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40: 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of carbapenem-resistant K. pneumoniae isolates.
(DOC)