Biochemistry students of my generation had the RNA story relatively straight: ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and messenger RNAs (mRNAs) appeared to do everything expected of RNA in the cell at that time. In the last two decades or so, the situation has changed dramatically. In addition to rRNAs and tRNAs, hundreds of other non-protein-coding RNAs have emerged, having a diverse range of functions, from structural through regulatory to catalytic (1, 2). Particularly exciting recent examples of regulatory RNAs include Xist and roX RNAs, involved in chromosome dosage compensation in mammals and Drosophila, respectively (3), and tmRNA, which directs tagging of incomplete proteins for degradation in bacteria (4). By its nature—a potential to base-pair—the recognition of sequences in other nucleic acids is an RNA molecule's major asset. Indeed, several spliceosomal small nuclear (sn) RNAs (e.g., U1, U2, and U6) base-pair with short sequences in pre-mRNAs to delineate regions to be spliced out (5, 6). Other RNAs, which identify their targets by base-pairing, include editing guide RNAs in the mitochondria of kinetoplastid protozoans (7), and lin-4 and let-7, the 21-nucleotide (nt) long mini-RNAs, which regulate timing of early developmental decisions in Caenorhabditis elegans by hybridizing to the 3′ untranslated regions of their target mRNAs (8); let-7 functions also in other bilaterian animals, from molluscs to mammals (9).
The class of noncoding RNAs that—at least in terms of numbers—is currently dominating the field is small nucleolar (sno) RNAs, which act as guides to direct pseudouridylation and 2′-O-ribose methylation in rRNA (10–14). In vertebrates, each of these modifications is found at about 100 sites per ribosome. Since a single guide snoRNA specifies one, at most two, modifications (Fig. 1A), the number of these RNAs may approach 200. Guide snoRNAs also operate in other territories; e.g., pseudouridylation-guide-like RNA and associated proteins form part of the mammalian telomerase (refs. 15 and 16 and references therein). Biogenesis of guide snoRNAs in different organisms is equally interesting (Fig. 1B). In vertebrates, all guides are encoded in introns of genes transcribed by RNA polymerase II (pol II), but some of the host genes do not code for proteins, and it is their introns and not exons that are evolutionarily conserved (10, 11, 14). The paper by Cavaillé et al. (17), appearing in this issue of PNAS, adds still another dimension to the guide snoRNA story. These authors characterize a collection of brain-specific snoRNAs whose expression is paternally imprinted. Interestingly, genes encoding most of the identified RNAs map within a region implicated in the neurogenetic disease Prader–Willi syndrome (PWS). One of the imprinted snoRNA loci [MBII-85/HBII-85; nomenclature of Cavaillé et al. (17)] is also described by De los Santos et al. (18).
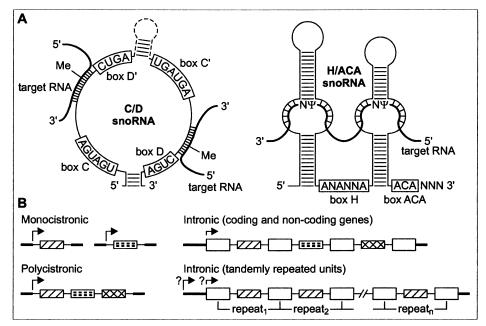
Figure 1.
Short guide to the structure (A) and biogenesis (B) of guide snoRNAs. (A) Schematic structure of the C/D-box and H/ACA-box snoRNAs, with conserved sequence elements and base-paired target RNAs indicated. Most snoRNAs contain one rather than two functional modification domains, although H/ACA-box snoRNAs always have a conserved hairpin-hinge-hairpin-tail secondary structure. The pseudouridylation pocket forms two short duplexes (3–10 bp) with the target RNA, leaving the uridine residue to be isomerized unpaired. snoRNAs of each class are associated with a set of specific proteins which are not shown. (B) Different strategies of snoRNA expression. In the yeast Saccharomyces cerevisiae, most of the snoRNA genes are transcribed as either mono- or polycistronic units, and only a few are encoded in introns. In all established cases, vertebrate guide snoRNAs originate from introns of either protein-coding or noncoding RNA polymerase II-transcribed genes. Sequences corresponding to mature snoRNAs are shown as filled-in boxes, and exons, as open boxes. Arrows indicate transcription initiation sites. Transcription start sites for the tandemly repeated brain-expressed snoRNA genes are not known.
The snoRNAs fall into two major classes, each characterized by specific conserved sequence elements (“boxes”) and a set of associated proteins (Fig. 1A). The C/D box snoRNAs, associated with fibrillarin, guide a site-specific 2′-O-methylation, and the H/ACA box snoRNAs, associated with protein GAR1, target specific conversions of uridine to pseudouridine. Guiding of 2′-O-methylation involves base-pairing of the 10- to 21-nt-long sequence positioned upstream of box D (or D′) to the target RNA, with the nucleotide positioned 5 base pairs (bp) upstream of the D/D′ box being selected for methylation. In the H/ACA snoRNAs, one or both of the two RNA hairpins are interrupted by an internal loop, the pseudouridylation pocket, which contains two short (3- to 10-nt) sequences complementary to nucleotides flanking the site of isomerization (refs. 10–14; Fig. 1A).
When searching for small RNAs expressed in mouse brain, Cavaillé et al. (17) identified three novel C/D box snoRNAs (MBII-13, MBII-52, and MBII-85) and one H/ACA box snoRNA, MBI-36. In mouse or rat, all four RNAs are present exclusively in the brain, with the three C/D box RNAs accumulating at similar levels in all brain areas except the choroid plexus, and the H/ACA RNA MBI-36 having the reverse distribution, being found mainly in the choroid plexus. In humans, all mouse RNA orthologs are likewise either exclusively (HBII-52 and HBI-36) or predominantly (HBII-13 and -85) expressed in brain.
The most interesting aspect of the work is that the genes encoding all three human C/D box snoRNAs map to chromosome 15q11–q13, more precisely to the ≈1.5-Mb region linked to PWS; they also map to the syntenic chromosome 7C region in mouse (17, 18). Genes in the PWS region are parentally imprinted, with only alleles inherited from the father being expressed (19–23). The loss of imprinted gene expression, most frequently caused by paternal deletions or maternal uniparental disomy, results in severe developmental and neurobehavioral problems. PWS occurs in about 1 in 15,000 births. Its clinical features include infant failure to thrive with hypotonia, hyperphagia leading to severe obesity, hypogonadism, and mild to medium mental retardation with learning and other disabilities. Among the paternally controlled genes in the PWS region are some that encode proteins—e.g., the dicistronic gene SNURF-SNRPN, coding for the brain-expressed protein SmN, a counterpart of the common U-snRNP Sm proteins B and B′. Other regions produce RNAs without apparent protein-coding potential, and some of the noncoding transcripts are antisense to translatable mRNAs, a situation frequently encountered in different parentally imprinted regions (20, 21, 23).
Consistent with their chromosomal localization, the C/D box snoRNA genes are paternally imprinted and are not expressed in PWS patients or mouse models of the disease (17, 18). Detailed analysis of the imprinted snoRNA loci revealed their very unusual organization. snoRNA sequences of one type (MBII-52/HBII-52) are embedded in ≈2-kb DNA units which, in humans, are tandemly repeated 47 times (17); snoRNAs appear to be processed from the excised introns by the exonucleolytic mechanism (24). The MBII-85/HBII-85 snoRNAs are likewise present in multiple cotranscribed repeats, but they are less uniformly spaced and their mode of processing is not certain (17, 18). Sequences flanking the snoRNA regions are not phylogenetically conserved and have no protein-coding potential, making the host genes of these brain-expressed snoRNAs similar to the noncoding hosts of other snoRNAs (Fig. 1B).
The organization and expression of other genes in the PWS region is also very complex. In addition to snoRNA loci, many other DNA repeats are present; it is conceivable that DNA repeats contribute to the establishment of imprinted states. Expression of all genes in the PWS region, and also maternally imprinted genes distal to it and affected in Angelman syndrome (AS, another neurogenetic disease), is controlled by the large bipartite imprinting center (IC). The IC region partially overlaps the upstream portion of SNURF-SNRPN, making functional analysis of individual genes difficult. Current models propose that loss of expression of multiple paternal-specific genes contributes to the PWS phenotype. At least two of the described snoRNA loci map to the regions implicated in the disease. It is likely, however, that other paternally imprinted genes will be found in the many still-uncharacterized regions of 15q11–q13. PWS and AS phenotypes strongly argue for a function of imprinting in brain development. Furthermore, the expression data for 15q11–q13, and also other imprinted loci, indicate that noncoding RNAs may play an important role, either in imprinting itself or in transmission of imprinted information (19–23).
What could be the biological role of the newly described brain-specific snoRNAs? Lack of complementarity to rRNA and their tissue-specific expression argue against the possibility that they specify rRNA modification. Significantly, recent work has shown that guide snoRNAs can also function in posttrancriptional modification of cellular RNAs other then rRNA. In vertebrates, addition of several 2′-O-methyls to the spliceosomal U6 snRNA is directed by classical snoRNAs (25, 26), and the 2′-O-methylation and pseudouridylation at two adjacent positions in U5 snRNA are both guided by a novel C/D-H/ACA-box “hybrid” RNA (T. Kiss, personal communication). Identification of “orphan” snoRNAs for which targets are unknown (27), and the demonstration that snoRNAs can, although inefficiently, modify sequences inserted into RNA polymerase (pol) II-transcribed mRNA-like molecules (28), further argue for substrate diversity. The small C/D-box-like RNAs present in Archaea direct methylation of not only rRNAs but also tRNAs (refs. 29 and 30; P. Dennis, personal communication; J.-P. Bachellerie and C. Gaspin, personal communication).
The three brain C/D box snoRNAs show no significant potential to base-pair with known cellular small RNAs but one of them, MBII-52/HBII-52, has a guide region with an 18-nt phylogenetically conserved complementarity to the serotonin receptor 5-HT2C mRNA that is expressed in brain (17). Interestingly, a potential acceptor of the methyl group, an A residue, happens to be the subject of the physiologically relevant A-to-I editing in this mRNA (31, 32). Because 2′-O-methylation of adenosine in a model substrate is known to decrease its deamination by 200-fold (33), regulation of mRNA editing by snoRNAs is an attractive possibility, particularly in brain. The A-to-I editing enzymes are most abundant in brain (34, 35), and quantification of the inosine content in poly(A)+ RNA suggests that hundreds of mRNAs might undergo editing in this tissue (34). If the snoRNA-directed 2′-O-methylation of mRNA indeed occurs in vivo (although 2′-O-methyls have been found only in 5′-cap-proximal nucleotides of mRNA to date; ref. 36), editing might not be the only process potentially regulated by such modification. The 2′-O-methylation of 3′ splice site and polyadenylation signal (AAUAAA) nucleotides strongly inhibits the respective pre-mRNA processing reactions in vitro (37, 38). One could also envisage effects of base-pairing of snoRNAs to mRNAs (or other RNAs), independently of methylation. Such associations, related to the proposed chaperonin function of snoRNAs in ribosome biogenesis (10, 14), could modify maturation patterns of target RNAs by masking the processing signals or interfering with binding of protein factors. Interestingly, the snoRNA complementarity in 5-HT2C mRNA occurs near a site used for alternative splicing (17).
How could guide snoRNAs, localized predominantly in the nucleolus, target nucleoplasmic substrates such as snRNAs or mRNAs? One possibility is that they meet within Cajal (coiled) bodies (CBs). Nearly all components of C/D box and H/ACA box snoRNAs, and also U snRNPs, are present in CBs of mammalian cells (refs. 16 and 39, and references therein). Alternatively, some guide snoRNPs, in particular those encoded by multicopy genes expressed in brain, can be present in the nucleoplasm, their most-likely site of assembly (40), at concentrations high enough to interact with their targets. Finally, some small RNAs (including U6 snRNA) and mRNAs are known to transit the nucleolus en route to their final destinations (41–43). It will be interesting to look at the morphology of the active multiarray snoRNA loci in neuronal nuclei, to find out whether they are associated with CBs (39) or some other structures functioning as the small nucleolar ribonucleoprotein (snoRNP) maturation “factory.”
The four RNAs discussed above are the first tissue-specific snoRNAs to be described; four additional brain-localized C/D box RNAs have already been identified in rodents (J. Cavaillé, P. Vitali, Z. Basyuk, and J. P. Bachellerie; J. Brosius and A. Hüttenhofer; personal communications). How many more will be found? Do tissues other than brain also have their own sets of snoRNAs? What is the biological role of the newly discovered RNAs and the relevance of their tandemly repeated gene organization? Given the unusual diversity of RNA functions already documented, no doubt many more noncoding RNAs will be found to operate in cells. RNomics approaches, involving either cDNA bank screens similar to the screen used by Cavaillé et al. (17) and others (44), or analyses of “intergenic gaps,” like that carried out in yeast (45), may be rewarding in identifying additional players in RNA games.
Acknowledgments
I thank Helen and Mike Rothnie for their help with the figure and the text, and all of the colleagues who shared with me their unpublished results. Friedrich Miescher Institute is part of the Novartis Research Foundation.
Footnotes
See companion article on page 14311.
References
- 1.Gesteland R F, Cech T R, Atkins J F, editors. The RNA World. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 2.Eddy S R. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- 3.Kelley R L, Kuroda M I. Cell. 2000;103:9–12. doi: 10.1016/s0092-8674(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 4.Karzai A W, Roche E D, Sauer R T. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 5.Burge C B, Tuschl T, Sharp P A. In: The RNA World. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 6.Staley J P, Guthrie C. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 7.Estevez A M, Simpson L. Gene. 1999;240:247–260. doi: 10.1016/s0378-1119(99)00437-0. [DOI] [PubMed] [Google Scholar]
- 8.Moss E G. Curr Biol. 2000;10:R436–R439. doi: 10.1016/s0960-9822(00)00528-5. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli A E, Reinhart B J, Slack F, Martindale M Q, Kuroda M I, Maller B, Hayward D C, Ball E E, Degnan B, Müller P, et al. Nature (London) 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Bachellerie J-P, Cavaillé J, Qu L-H. In: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Garrett R A, Douthwaite S R, Liljas A, Matheson A T, Moore P B, Noller H F, editors. Washington, DC: Am. Soc. Microbiol. Press; 2000. pp. 191–203. [Google Scholar]
- 11.Filipowicz W, Pelczar P, Pogacic V, Dragon F. Acta Biochim Pol. 1999;46:377–389. [PubMed] [Google Scholar]
- 12.Lafontaine D L J, Tollervey D. Trends Biochem Sci. 1998;23:383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 13.Ofengand J, Fournier M J. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol. Press; 1998. pp. 229–253. [Google Scholar]
- 14.Weinstein L, Steitz J A. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell J R, Collins K. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 16.Pogacic V, Dragon F, Filipowicz W. Mol Cell Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan C I, Horsthemke B, Bachellerie J-P, Brosius J, Hüttenhofer A. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. . (First Published December 5, 2000; 10.1073/pnas.250426397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Los Santos T, Schweizer J, Rees C A, Francke U. Am J Hum Genet. 2000;67:1067–1082. doi: 10.1086/303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholls R D, Saitoh S, Horsthemke B. Trends Genet. 1998;14:194–200. doi: 10.1016/s0168-9525(98)01432-2. [DOI] [PubMed] [Google Scholar]
- 20.Brannan C I, Bartolomei M S. Curr Opin Genet Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 21.Tilghman S M. Cell. 1999;96:185–193. doi: 10.1016/s0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls R D. Nature Genet. 1999;23:132–134. doi: 10.1038/13758. [DOI] [PubMed] [Google Scholar]
- 23.Nicholls R D. J Clin Invest. 2000;105:413–418. doi: 10.1172/JCI9460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiss T, Filipowicz W. Genes Dev. 1995;9:1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 25.Tycowski T H, You Z H, Graham P J, Steitz J A. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 26.Ganot P, Jady B E, Bortolin M-L, Darzacq X, Kiss T. Mol Cell Biol. 1999;19:6906–6917. doi: 10.1128/mcb.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jady B E, Kiss T. Nucleic Acids Res. 2000;28:1348–1354. doi: 10.1093/nar/28.6.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavaillé J, Nicoloso M, Bachellerie J P. Nature (London) 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 29.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 30.Gaspin C, Cavaillé J, Erauso G, Bachellerie J-P. J Mol Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 31.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 32.Niswender C M, Copeland S C, Herrick-Davis K, Emeson R B, Sanders-Bush E. J Biol Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 33.Yi-Brunozzi H Y, Easterwood L M, Kamilar G M, Beal P A. Nucleic Acids Res. 1999;27:2912–2917. doi: 10.1093/nar/27.14.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C-X, Cho D-S C, Wang Q, Lai F, Carter K C, Nishikura K. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bokar J A, Rottman F M. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol. Press; 1998. pp. 183–200. [Google Scholar]
- 37.Moore M J, Sharp P A. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 38.Bardwell V J, Wickens M, Bienroth S, Keller W, Sproat B S, Lamond A I. Cell. 1991;65:125–133. doi: 10.1016/0092-8674(91)90414-t. [DOI] [PubMed] [Google Scholar]
- 39.Matera A G. Trends Cell Biol. 1999;9:302–317. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 40.Samarsky D A, Fournier M J, Singer R H, Bertrand E. EMBO J. 1998;13:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmo-Fonseca M, Mendes-Soares L, Campos I. Nat Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 42.Olson M O J, Dundr M, Szebeni A. Trends Cell Biol. 2000;10:189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- 43.Pederson T, Politz J C. J Cell Biol. 2000;148:1091–1095. doi: 10.1083/jcb.148.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 45.Olivas W M, Muhlrad D, Parker R. Nucleic Acids Res. 1997;22:4619–4625. doi: 10.1093/nar/25.22.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]