Abstract
Malignant gliomas, including glioblastoma multiforme, constitute the most common and aggressive primary brain tumors in adults. The transcription factor signal transducer and activator of transcription 3 (STAT3) plays an essential role in glioblastoma pathogenesis downstream of the major oncogenic protein epidermal growth factor receptor variant III (EGFRvIII). However, the critical gene targets of STAT3 that mediate EGFRvIII-induced glial transformation have remained unknown. Here, we identify inducible nitric oxide synthase (iNOS) as a novel target gene of STAT3 in EGFRvIII-expressing mouse astrocytes. Endogenous STAT3 occupies the endogenous iNOS promoter and stimulates iNOS transcription in EGFRvIII-expressing astrocytes. STAT3 does not appear to control iNOS transcription in astrocytes deficient in the major glioblastoma tumor suppressor protein phosphatase and tensin homolog (PTEN), suggesting that STAT3 regulates iNOS transcription specifically in EGFRvIII-expressing astrocytes. Importantly, inhibition of iNOS by distinct approaches, including knockdown by RNA interference, reduces cell population growth and invasiveness of EGFRvIII-expressing astrocytes. In addition, upon iNOS knockdown or administration of a small-molecule inhibitor of iNOS, EGFRvIII-expressing astrocytes form smaller tumors in vivo. These findings suggest that inhibition of iNOS may have potential therapeutic value for EGFRvIII-activated brain tumors.
Introduction
Malignant gliomas are the most common primary tumor of the adult brain and are some of the most aggressive human cancers (Holland, 2001; Konopka and Bonni, 2003; Furnari et al., 2007). Glioblastoma tumor cells have genetic and phenotypic characteristics of astrocytes or neural stem cells, both of which may represent the cells of origin of glioblastoma (Holland, 2001; Bachoo et al., 2002; Uhrbom et al., 2002; Bajenaru et al., 2003; Louis, 2006). An emerging theme from these studies is that deregulation of developmental signaling pathways that normally drive the differentiation of neural stem cells into astrocytes may contribute to the pathogenesis of glioblastoma (de la Iglesia et al., 2009).
The transcription factor signal transducer and activator of transcription 3 (STAT3) drives astrocyte differentiation in the developing brain (Bonni et al., 1997; Rajan and McKay, 1998). We have found previously that STAT3 plays a dual role in glial cell transformation depending on the mutational profile of the tumor (de la Iglesia et al., 2008a). Using a mouse genetics approach, we found that STAT3 functions in an oncogenic manner in astrocytes upon expression of a truncated, constitutively active form of the epidermal growth factor receptor (EGFRvIII), which represents a major oncogenic stimulus in glioblastoma pathogenesis. In contrast, in the background of loss of the major tumor suppressor phosphatase and tensin homolog (PTEN), STAT3 functions in a tumor suppressive capacity, consistent with the function of STAT3 as a driver of astrocyte differentiation during brain development (de la Iglesia et al., 2008a).
How does STAT3 exert opposing functions in glial transformation in the distinct backgrounds of EGFRvIII-expression and PTEN loss? STAT3 suppresses the malignant behavior of PTEN-deficient glioblastoma cells by repressing transcription of the chemokine IL8 (de la Iglesia et al., 2008b). However, the major question of how STAT3 promotes glial transformation in the context of EGFRvIII-expression remained to be addressed.
In this study, we identify inducible nitric oxide synthase (iNOS) as a critical downstream target of STAT3 in astrocyte transformation in the background of EGFRvIII-expression. STAT3 specifically regulates iNOS transcription in EGFRvIII-expressing astrocytes but not PTEN-deficient astrocytes. We identify a STAT3 binding site within the promoter of the iNOS gene that is required for STAT3-dependent transcription and demonstrate that STAT3 directly regulates iNOS transcription in astrocytes. Inhibition of iNOS using pharmacological agents reveals a critical role for iNOS in STAT3-dependent proliferation of EGFRvIII-expressing astrocytes. In addition, a small-molecule NO donor largely reverses the deficit in population growth upon Stat3 knock-out in EGFRvIII-expressing astrocytes. Consistent with these findings, genetic knockdown of iNOS by RNA interference (RNAi) in EGFRvIII-expressing astrocytes reduces their population growth and invasiveness. Importantly, iNOS knockdown or administration of a small-molecule inhibitor of iNOS impairs the malignant transformation of EGFRvIII-expressing astrocytes in vivo. In contrast to the effect of iNOS inhibition on EGFRvIII-expressing astrocytes, inhibition of iNOS in PTEN-deficient astrocytes has little or no effect on invasiveness and proliferation. Together, these findings define iNOS as an important target of STAT3 that induces glial transformation specifically in EGFRvIII-expressing astrocytes. Our findings also suggest that inhibition of STAT3 and iNOS might represent a potential therapeutic avenue in the treatment of EGFRvIII-positive glioblastoma.
Materials and Methods
Cell culture.
EGFRvIII-expressing, control murine stem cell virus (MSCV), and PTEN knockdown (PTENi) astrocytes were generated by infection of SV40 large T-immortalized Stat3loxP/loxP astrocytes with the EGFRvIII or control MSCV retrovirus or with a pSUPER-Puro retrovirus encoding short hairpin RNAs (shRNAs) targeting PTEN as described previously (de la Iglesia et al., 2008a). The Stat3 gene flanked by loxP sites was excised in vitro using adenovirus encoding the recombinase Cre (University of Iowa) to generate Stat3−/− astrocytes. Primary astrocytes were cultured from postnatal day 2 B6 mice as described previously (Di Giorgio et al., 2007; Nagai et al., 2007).
Plasmids.
The pBABE–STAT3C construct was generated by standard subcloning techniques from RcCMV/STAT3C (Bromberg et al., 1999) into the pBABE vector. pBABE–STAT3D was generated by site-directed mutagenesis to introduce point mutations that disrupt DNA binding (Horvath et al., 1995; Nakajima et al., 1996).
Virus production and infection.
Lentiviruses were based on the pLKO.1 vector carrying the blasticidin resistance gene and obtained from the RNAi Consortium at the Broad Institute. Cloning of recombinant lentiviruses coding for shRNAs directed against iNOS was performed by annealing and insertion of complementary oligonucleotides into stuffed-pLKO.1 plasmid using AgeI and EcoRI sites. Oligonucleotides were generated as follows: iNOSi1, forward, 5′-C CGG TCC ATG CAA AGA ACG TGT TTA CTC GAG TAA ACA CGT TCT TTG CAT GGA TTTTTG-3′; iNOSi1, reverse, 5′-AATTCAAAAA TCC ATG CAA AGA ACG TGT TTA CTC GAG TAA ACA CGT TCT TTG CAT GGA-3′; iNOSi2, forward, 5′-C CGG GAG CAG GTG GAA GAC TAT TTC CTC GAG GAA ATA GTC TTC CAC CTG CTC TTTTTG-3′; iNOSi2, reverse, 5′-AATTCAAAAA GAG CAG GTG GAA GAC TAT TTC CTC GAG GAA ATA GTC TTC CAC CTG CTC-3′. Hairpin structures containing the stem sequences and loops are indicated by underlining and bold, respectively. Correct insertion of the desired oligonucleotides was confirmed by sequencing. pLKO.1–TRC026 containing the null-T sequence was used as the control.
Recombinant lentiviruses were made by transfecting human embryonic kidney 293T (HEK293T) cells with pCMV–dR8.91 (containing gag, pol, rev), pMD2.G (vesicular stomatitis virus glycoprotein), and the transfer plasmid (pLKO.1–TRC026, pLKO.1–iNOSi1, or pLKO.1–iNOSi2) via FuGENE 6 (Roche). Viral supernatants were collected and flash frozen.
EGFRvIII;Stat3loxP/loxP and MSCV;Stat3loxP/loxP astrocytes were infected in 3 cm dishes by incubating for 24 h with supernatant containing lentivirus. Cells were expanded into 10 cm plates and selected with blasticidin at a concentration of 5 μg/ml. Selection of uninfected astrocytes under the same conditions confirmed that blasticidin completely killed all cells at this concentration (data not shown). Blasticidin-resistant control and iNOS knockdown astrocytes were expanded and frozen into cell stocks for use in biochemical and functional assays.
RT-PCR analyses.
RNA was prepared from cells using Trizol extraction and purification. For gel-based RT-PCR analyses, purified RNA was quantified and reverse transcribed and amplified using the appropriate primers and SuperScript One-Step RT-PCR with Platinum Taq system (Invitrogen) according to the protocol of the manufacturer. Amplified cDNA products were resolved using agarose gel electrophoresis and visualized using ethidium bromide. Primer sequences were as follows: iNOS, forward, 5′-GTG GTG ACA AGC ACA TTT GG-3′; iNOS, reverse, 5′-GGC TGG ACT TTT CAC TCT GC-3′; Hyaluronan synthase, forward, 5′-AGT ATA CCT CGC GCT CCA GA-3′; Hyaluronan synthase, reverse, AGC AGC AGT AGA GCC CAG AG-3′; Bcl-XL, forward, 5′-TGG TGG TCG ACT TTC TCT CC-3′; Bcl-XL, reverse, 5′-TGC AAT CCG ACT CAC CAA TA-3′; matrix metalloproteinase 3 (MMP3), forward, 5′-CAG GTG TGG TGT TCC TGA TG-3′; MMP3, reverse, 5′-GCC TTG GCT GAG TGG TAG AG-3′; Pim1, forward, 5′-CAT GGA AGT GGT CCT GTT GA-3′; Pim1, reverse, GAC CCG AAG TCG ATG AGT TT-3′; VEGF, forward, 5′-CTA CAG ATG TGG GGG TTG CT-3′; VEGF, reverse, 5′-CAC AGC GGC ATA CTT CTT CA-3′; telomerase reverse transcription (TERT), forward, 5′-AGG GTA AGC TGG TGG AGG TT-3′; TERT, reverse, 5′-TGC TGA GGA AGG TTT TTG CT-3′; Survivin, forward, 5′-ATC GCC ACC TTC AAG AAC TG-3′; Survivin, reverse, 5′-CAG GGG AGT GCT TTC TAT GC-3′; Myc, forward, 5′-GCC CAG TGA GGA TAT CTG GA-3′; Myc, reverse, 5′-GAA TCG GAC GAG GTA CAG GA-3′; GAPDH, forward, 5′-ACC ACA GTC CAT GCC ATC AC-3′; GAPDH, reverse, 5′-TCC ACC ACC CTG TTG CTG TA-3′.
For quantitative RT-PCR analyses, cDNA was prepared using oligo-dT primers and SuperScript III First-Strand cDNA synthesis system (Invitrogen) according to the protocol of the manufacturer. Quantitative PCR was performed using the Lightcycler 480 SYBR Green 1 Master Kit on a Lightcycler 480 thermocycler (Roche). For all quantitative PCR experiments, gene expression was normalized to GAPDH levels. Specific amplification of target genes was confirmed by agarose gel electrophoresis and calculation of melting temperature of the amplified product. Primer sequences were as follows: STAT3, forward, 5′-ACC CTT AGG GAG CAG AGA TGT G-3′; STAT3, reverse, 5′-GTT CTT GGG GTT ATT GGT CAG C-3′; iNOS, forward, 5′-GGA TTG TCC TAC ACC ACA CCA A-3′; iNOS, reverse, 5′-ATC TCT GCC TAT CCG TCT CGT C-3′; GAPDH, forward, 5′-TGC TGG TGC TGA GTA TGT CG-3′; GAPDH, reverse, 5′-GCA TGT CAG ATC CAC AAC GG-3′.
Antibodies.
Rabbit iNOS antibody for immunocytochemical analyses and mouse actin antibody (Santa Cruz Biotechnology), rabbit iNOS antibody for immunoblotting analyses (BD Biosciences), polyclonal rabbit Ki67 antibody (Vector Laboratories), mouse EGFR, rabbit STAT3, and rabbit cleaved caspase 3 (CC3) antibodies (Cell Signaling Technology), mouse phospho-Tyr (clone 4G10) antibody (Millipore), mouse α-tubulin antibody (Covance), and mouse Flag antibody (Sigma) were purchased. Mouse GFP antibody was obtained through the NeuroMab antibody consortium in association with Antibodies Incorporated.
Immunocytochemistry.
For visualization of iNOS and GFP proteins, astrocytes were fixed in methanol for 10 min at −20°C and subjected to microwaving and immunocytochemical analyses with the indicated antibodies. For visualization of Ki67 and CC3, astrocytes were fixed in 4% paraformaldehyde for 20 min at room temperature and processed in a similar manner. For BrdU analyses, astrocytes were treated with BrdU for 2 h and then processed as per the protocol of the manufacturer with BrdU Labeling and Detection Kit I (Roche). Primary mouse astrocytes were incubated with BrdU for 4 h before fixing and staining.
Immunoblotting analyses.
Immunoblotting analyses were performed as described previously (Puram et al., 2011).
Luciferase assays.
The upstream 2 and 0.3 kb regions of the mouse iNOS transcriptional start site were cloned into the pGL2basic (Promega) vector to generate the iNOS–luciferase reporter genes (2 kb iNOS–pGL2b and 0.3 kb iNOS–pGL2b) using standard cloning techniques and the following primer sequences: 2 kb iNOS–pGL2b, forward, 5′-CCG CTC GAG GCC AAG CAC TCC AAT GTA AAA-3′; 0.3 kb iNOS–pGL2b, forward, 5′-CCG CTC GAG AAG CCA GCC TCC CTC CCT-3′; 2 kb and 0.3 kb iNOS–pGL2b, reverse, 5′-CCC AAG CTT CCA AGG TGG CTG AGA AGT TT-3′. Astrocytes were plated at a density of 100,000 cells per well in a 24-well format. After incubation overnight, astrocytes were transfected with the reporter plasmid and constructs of interest using FuGENE transfection reagent. Twenty-four hours after transfection, astrocytes were subjected to dual-luciferase assays (Promega). In all experiments, astrocytes were transfected with a Renilla firefly reporter to control for transfection efficiency.
Proliferation assays.
For manual counting experiments, EGFRvIII;Stat3loxP/loxP astrocytes were plated at a density of 20,000 cells per 6 cm plate. The small molecule 1400W or vehicle (water) was added to cells 12 h after plating. At each time point analyzed, cells were washed, trypsinized, and counted in triplicate for each condition via hemocytometer.
For high-throughput proliferation assays, astrocytes (250 cells), primary mouse astrocytes (1000 cells), or U87 human glioblastoma cells (250 cells) in 50 μl of media were added to each well of a 96-well plate and incubated for 24 h at 37°C. Pharmacological agents or their corresponding vehicles were then added as follows: 1400W (500 μm), S-methylisothiourea sulfate (S-MIU) (25 μm), c-PTIO [2-(4-carboxyphenyl-4,5-dihydro-4,4,5,5-tetramethyl-1H-imidazol-1-yloxy-3-oxide, potassium salt] (5 μm), and (S)-nitroso-N-acetylpenicillamine (SNAP) (1.25 μm). Cells were lysed 48, 96, and 144 h after initial plating with Cell TiterGlo reagent (Promega), agitated for 2 min, and then incubated at 25°C for 10 min. Wells were then read via a SpectraMax luminometer (Molecular Devices). For all experiments, cells were also lysed 2 h after plating to ensure that equal numbers were initially plated across conditions. Wells containing media only were used to determine background signal.
Chromatin immunoprecipitation.
EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were washed with 1× PBS and crosslinked with 1% formaldehyde in 1× PBS for 10 min. A solution containing 0.125 m glycine in 1× PBS was used for quenching for 5 min at room temperature. Cells were harvested, and the pellet was dissolved in chromatin immunoprecipitation (ChIP) lysis buffer (40 mm Tris-HCl, pH 8.0, 1.0% Triton X-100, 4 mm EDTA, and 300 mm NaCl) containing protease inhibitors. Chromatin was fragmented by sonication in a water bath sonicator at 4°C to an average length of 500 bp. The lysates were spun at 14,000 rpm for 15 min, and the supernatant was diluted to 1:1 in ChIP dilution buffer containing 40 mm Tris-HCl, pH 8.0, and 4 mm EDTA plus protease inhibitors. Immunoprecipitation was done using a ChIP-grade STAT3 antibody (Cell Signaling Technology) against the endogenous STAT3 protein. Antibody–protein–DNA complexes were collected, washed, and eluted, and crosslinks were reversed according to the instructions of the manufacturer (Millipore) as described previously (Gillespie et al., 2009). The following primer sequences were used: STAT3, forward, 5′-GAG CTA ACT TGC ACA CCC AAC-3′; STAT3, reverse, 5′-GTG GGG CCA GAG TCT CAG T-3′; iNOS, forward, GAA CAG ACA GAA AGC CAG AGA GC; iNOS, reverse, GAC ACT CCT AGT CTG TGT GCT TGA.
Matrigel invasion assays.
Matrigel was diluted and coated onto invasion chambers (BD Biosciences) with an 8 μm pore size membrane overnight. Polymerized gel was rehydrated with serum-free media, and astrocytes (2.5 × 104) in 500 μl of media were added to each of the inserts and incubated for 22 h at 37°C. Astrocytes on the lower surface of the membrane that had migrated through the matrigel were fixed, stained with crystal violet, and counted. Noninvasive NIH3T3 cells were used as a negative control. Equivalent numbers of NIH3T3 failed to invade the matrigel. The effect of iNOS knockdown or pharmacological inhibition on invasiveness was not secondary to a change in cell proliferation, because the invasive potential of these cells was measured at a time (22 h after plating) before a significant decrease in cell number on inhibition of iNOS.
Mouse injections.
Control vector transfected and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes (1 × 106) were resuspended in serum-free, antibiotic-free media and injected subcutaneously into 4- to 6-week-old male severe combined immunodeficiency (SCID) mice. For experiments with administration of 1400W, EGFRvIII;Stat3loxP/loxP astrocytes were injected subcutaneously, and 100 μl of vehicle or 1400W (500 μm) was injected locally three times per week. Four weeks after initial injection of cells, mice were killed, and the tumors were removed, measured, and fixed for histological analyses.
Histology.
Tumors were fixed in 4% paraformaldehyde, embedded in Paraplast, and serially sectioned at 7 μm. Sections were deparaffinized through xylenes and graded ethanol and subsequently stained with hematoxylin–eosin for histological evaluation. Representative areas are shown in the figures. For immunohistochemistry, antigen retrieval was performed via microwaving for 20 min in 10 mm sodium citrate, pH 6.0. Proliferating cells were stained with a rabbit polyclonal antibody to Ki67 (Vector Laboratories), and the immunolabeling was visualized with the Vectastain Elite ABC Reagent and DAB Substrate Kit (Vector Laboratories). Sections were counterstained with hematoxylin.
Statistical analyses.
All analyses were completed from a minimum of three independent experiments. Statistical analyses were performed with GraphPad Prism 4.0. All histograms are presented as mean + SEM unless otherwise noted. The Student's t test was used for comparisons in experiments with two sample groups. In experiments with more than two sample groups, ANOVA was performed followed by Bonferroni's post hoc test. For nonparametric analyses of more than two sample groups, Kruskal–Wallis test was performed, followed by Dunn's post hoc test.
Results
STAT3 specifically stimulates iNOS transcription in EGFRvIII-expressing astrocytes
The identification of dual oncogenic and tumor suppressive functions for STAT3 in genetic studies of EGFRvIII-expressing and PTEN-deficient mouse astrocytes, respectively (de la Iglesia et al., 2008a), raises the major question of how STAT3 regulates tumorigenesis in these distinct genetic contexts. We reasoned that as a transcription factor STAT3 might regulate specific targets within the context of EGFRvIII expression and PTEN loss. Previously, we identified IL8 as a direct, repressed gene target of STAT3. Inhibition of STAT3 in PTEN-deficient glioblastoma cells leads to derepression of IL8 and the consequent promotion of proliferation and invasiveness of these cells (de la Iglesia et al., 2008b). To identify potential targets of STAT3 that operate downstream of EGFRvIII in glial transformation, we characterized the expression of a panel of STAT3-regulated gene targets, previously reported in non-neural cells (Kiuchi et al., 1999; Shirogane et al., 1999; Niu et al., 2002; Yu et al., 2002; Aoki et al., 2003; Selander et al., 2004; Yu and Jove, 2004; Lo et al., 2005; Chau et al., 2007; Lassmann et al., 2007; Ho et al., 2010; Ohara et al., 2010; van der Poel et al., 2010).
Using RT-PCR analyses, we measured the mRNA levels of these candidate targets in astrocytes harboring a floxed Stat3 allele (Stat3loxP/loxP) or in astrocytes in which the Stat3 gene was knocked out (Stat3−/−) using the recombinase Cre, in the context of EGFRvIII expression or PTEN knockdown (PTENi) (Fig. 1A). Remarkably, among the panel of STAT3-regulated genes, only iNOS was specifically downregulated in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII;Stat3loxP/loxP astrocytes (Fig. 1A). In contrast, iNOS mRNA levels were unchanged in PTEN-deficient Stat3 knock-out astrocytes compared with control PTEN-deficient Stat3 floxed astrocytes (Fig. 1A). The expression of other STAT3 targets was similar in astrocytes among the different genotypes, suggesting that iNOS may represent a specific target of STAT3 in the context of EGFRvIII expression in astrocytes.
Figure 1.
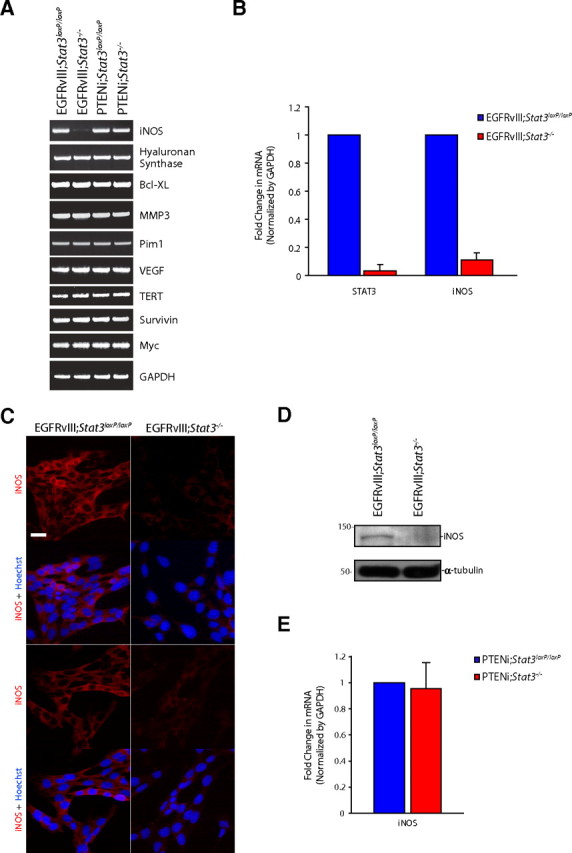
STAT3 regulates iNOS expression in EGFRvIII-expressing astrocytes. A, RNA was isolated from mouse EGFRvIII;Stat3loxP/loxP, EGFRvIII;Stat3−/−, PTENi;Stat3loxP/loxP, and PTENi;Stat3−/− astrocytes and subjected to RT-PCR with primers specific for the indicated genes. GAPDH served as control. iNOS mRNA levels were specifically reduced in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII;Stat3loxP/loxP astrocytes. B, RNA isolated from mouse EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes was subjected to quantitative RT-PCR analyses using primers specific for STAT3 and iNOS. mRNA levels were normalized to GAPDH. STAT3 and iNOS mRNA levels were significantly reduced in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.0005, n = 3). C, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were subjected to immunocytochemistry using the rabbit iNOS antibody. Representative images are shown. The expression of iNOS was substantially reduced in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII;Stat3loxP/loxP astrocytes. Scale bar, 20 μm. D, Lysates of EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were immunoblotted with the iNOS or α-tubulin antibody. The levels of iNOS protein were substantially reduced in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII;Stat3loxP/loxP astrocytes. E, RNA isolated from mouse PTENi;Stat3loxP/loxP and PTENi;Stat3−/− astrocytes was subjected to quantitative RT-PCR analyses using primers specific for iNOS. mRNA levels were normalized to GAPDH. iNOS mRNA levels were not significantly different between PTENi;Stat3loxP/loxP astrocytes compared with PTENi;Stat3−/− astrocytes.
To further characterize the role of STAT3 in the regulation of iNOS expression in EGFRvIII-expressing astrocytes, we used real-time RT-PCR analyses to quantitatively assess iNOS mRNA levels in astrocytes. We confirmed that STAT3 knock-out cells had little or no detectable STAT3 mRNA compared with floxed cells (98% reduction of STAT3 in EGFRvIII;Stat3−/− astrocytes compared with EGFRvIII; Stat3loxP/loxP astrocytes) (Fig. 1B). Importantly, iNOS mRNA levels were reduced by 90% in EGFRvIII-expressing Stat3 knock-out astrocytes compared with the control floxed cells (Fig. 1B). Consistent with these results, immunocytochemical and immunoblotting analyses revealed that the levels of iNOS protein were substantially reduced upon STAT3 knock-out in EGFRvIII-expressing astrocytes (Fig. 1C,D). These data suggest that STAT3 plays a crucial role in the regulation of iNOS gene expression in EGFRvIII-expressing astrocytes. In other experiments, we confirmed that iNOS mRNA levels were unaltered after deletion of Stat3 in the background of PTEN loss (Fig. 1E), indicating that STAT3 specifically regulates iNOS gene expression in the context of EGFRvIII expression but not PTEN deficiency. These data suggest that STAT3 may have unique transcriptional targets depending on the genetic background of the tumor.
STAT3 directly regulates iNOS transcription in astrocytes
To determine the mechanism by which STAT3 promotes iNOS gene expression, we first analyzed the activity of the iNOS promoter in transient expression assays in EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes. We found that the expression of a luciferase reporter gene that is controlled by 2 kb or 300 nt of the 5′ regulatory sequences of the iNOS gene was high in Stat3loxP/loxP astrocytes but was significantly reduced in EGFRvIII;Stat3−/− astrocytes (Fig. 2A). These data suggest that STAT3 regulates iNOS transcription via the iNOS promoter containing 300 nt upstream of the iNOS transcriptional start site.
Figure 2.
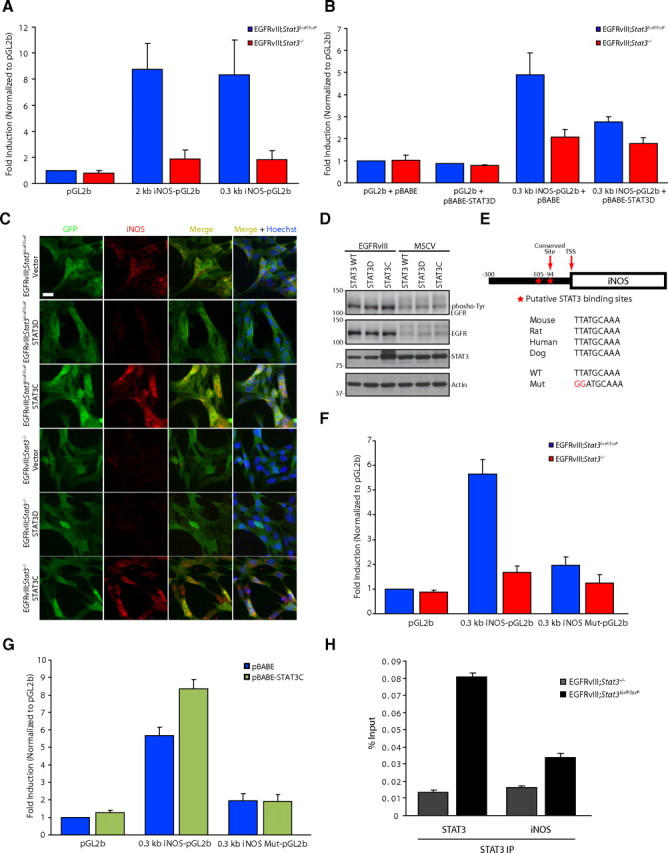
STAT3 directly regulates iNOS transcription in astrocytes. A, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes transfected with a luciferase reporter plasmid driven by a promoter containing the 2 or 0.3 kb region upstream of the iNOS transcriptional start site or the control pGL2-basic reporter plasmid together with a Renilla expression plasmid were subjected to dual luciferase assay 48 h after transfection. Expression of the luciferase reporter from both the 2 and 0.3 kb regions of the iNOS promoter was significantly increased in EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes (Kruskal–Wallis test, p < 0.01, n = 4). B, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes transfected with the 0.3 kb iNOS–luciferase reporter plasmid together with dominant-negative Stat3 (STAT3D) or the control vector and the Renilla expression plasmid were subjected to dual luciferase assay 48 h after transfection. Expression of the iNOS–luciferase reporter was significantly increased in EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes in the background of control vector. Expression of STAT3D significantly reduced the expression of the iNOS–luciferase reporter in EGFRvIII;Stat3loxP/loxP astrocytes (Kruskal–Wallis test, p < 0.005, n = 4). C, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes transfected with vector control, STAT3D, or STAT3C were subjected to immunocytochemistry using the GFP and iNOS antibodies. Endogenous iNOS expression was substantially reduced in EGFRvIII;Stat3loxP/loxP astrocytes upon expression of STAT3D and substantially increased in EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes upon expression of STAT3C. Scale bar, 20 μm. D, Lysates of EGFRvIII-expressing and control MSCV-infected astrocytes transfected with wild-type Stat3 (STAT3 WT), STAT3D, or STAT3C were immunoblotted with the phospho-Tyr (4G10), EGFR, STAT3, or actin antibody. Expression of STAT3D or STAT3C had little or no effect on the tyrosine phosphorylation of EGFRvIII. E, Top, STAT3 binding site in iNOS 94 bp upstream of the transcriptional start site (TSS) is conserved. Bottom, Mutations introduced into the iNOS promoter to disrupt STAT3 binding. F, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes transfected with the 0.3 kb iNOS–luciferase or 0.3 kb iNOS–luciferase mutant reporter plasmid together with the Renilla expression plasmid were subjected to dual luciferase assay 48 h after transfection. Expression of the iNOS–luciferase reporter was significantly increased in EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes (Kruskal–Wallis test, p < 0.005, n = 5). Mutation of the STAT3 binding site significantly reduced iNOS-promoter-mediated expression in EGFRvIII;Stat3loxP/loxP astrocytes (Kruskal–Wallis test, p < 0.005, n = 5). G, EGFRvIII;Stat3loxP/loxP astrocytes transfected with the 0.3 kb iNOS–luciferase or 0.3 kb iNOS–luciferase mutant reporter plasmid together with STAT3C or the control vector and the Renilla expression plasmid were subjected to dual-luciferase assay 48 h after transfection. Expression of STAT3C significantly increased expression of the iNOS–luciferase reporter (Kruskal–Wallis test, p < 0.0005, n = 5) but had little or no effect on the expression of the iNOS–luciferase mutant reporter. H, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were subjected to ChIP analyses using the STAT3 antibody. The STAT3 gene served as positive control. Endogenous STAT3 was significantly enriched at the endogenous iNOS promoter in EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes (ANOVA, p < 0.001, n = 3).
If iNOS is a bona fide transcriptional target of STAT3, then transcription from the iNOS promoter should depend on the ability of STAT3 to bind to DNA. Previous studies have described a dominant interfering mutant of STAT3 that contains mutations in the DNA binding domain of STAT3 (STAT3D) (Horvath et al., 1995; Nakajima et al., 1996). Because STAT3 forms dimers, expression of STAT3D inhibits the binding of endogenous STAT3 to responsive genes. We found that expression of STAT3D significantly reduced iNOS promoter-mediated transcription in EGFRvIII;Stat3loxP/loxP astrocytes but not in EGFRvIII;Stat3−/− astrocytes (Fig. 2B), suggesting that STAT3 regulates iNOS transcription in a DNA binding-dependent manner. Consistent with these results, expression of STAT3D reduced the levels of endogenous iNOS protein in EGFRvIII;Stat3loxP/loxP but not EGFRvIII;Stat3−/− astrocytes (Fig. 2C). In control experiments, expression of STAT3D had little or no effect on the level of tyrosine phosphorylation of EGFRvIII in astrocytes (Fig. 2D), suggesting that STAT3D does not disrupt upstream signaling by EGFRvIII. Together, these data suggest that STAT3 operates in a DNA binding-dependent manner to stimulate iNOS transcription and expression in EGFRvIII-expressing astrocytes.
Analysis of the 300 bp upstream of the mouse iNOS promoter revealed two potential sites that fulfill criteria for a STAT3 binding site (Seidel et al., 1995; Schaefer et al., 2000), but only one of these sites, 94 bp upstream of the iNOS transcriptional start site, is conserved (Fig. 2E). Strikingly, mutation of the conserved putative STAT3-binding site primarily abrogated the ability of STAT3 to induce iNOS promoter-mediated transcription in EGFRvIII;Stat3loxP/loxP astrocytes (Fig. 2F,G). In other analyses, we determined whether a constitutively active form of STAT3 (STAT3C) might exert a gain-of-function effect on iNOS promoter-mediated transcription (Bromberg et al., 1999). We found that expression of STAT3C augmented the expression of the iNOS–luciferase reporter gene (Fig. 2G). In contrast, expression of STAT3C had little or no effect on the expression of an iNOS–luciferase reporter gene containing a mutation in the STAT3 binding site in astrocytes (Fig. 2G). Consistent with these results, expression of STAT3C increased the levels of endogenous iNOS protein in EGFRvIII;Stat3loxP/loxP astrocytes and restored iNOS protein expression in EGFRvIII;Stat3−/− astrocytes (Fig. 2C). In control experiments, expression of STAT3C had little or no effect on EGFRvIII activation as monitored by tyrosine phosphorylation (Fig. 2D). Together, these data suggest that STAT3 stimulates iNOS transcription via a STAT3 binding site located 94 bp upstream of the transcriptional start site of the iNOS gene.
To determine whether endogenous STAT3 occupies the promoter of the iNOS gene in EGFRvIII-expressing astrocytes, we performed ChIP analyses. In control experiments, we confirmed that STAT3 occupies the endogenous STAT3 promoter (Fig. 2H). We also found significant enrichment of endogenous STAT3 at the endogenous iNOS promoter in EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes (Fig. 2H). Thus, endogenous STAT3 occupies the endogenous iNOS promoter in EGFRvIII-expressing astrocytes. Collectively, our data suggest that STAT3 directly activates iNOS transcription in astrocytes.
iNOS supports the proliferation of EGFRvIII-expressing astrocytes
The identification of iNOS as a direct target gene of STAT3 in EGFRvIII-expressing astrocytes led us to explore the question of whether iNOS might mediate the proliferation of astrocytes in response to the oncogenic stimulus of EGFRvIII expression. To address this question, we first used a pharmacological approach targeting distinct aspects of the biosynthetic pathway of NO, which is regulated by iNOS (Fig. 3A). The small molecule 1400W is a potent and specific inhibitor of iNOS but not neuronal NOS (nNOS) or endothelial NOS (eNOS) (Fig. 3A) (Garvey et al., 1994; Jafarian-Tehrani et al., 2005). Exposure of EGFRvIII;Stat3loxP/loxP astrocytes to 1400W significantly reduced the population growth of these cells, with increasing efficacy over time (Fig. 3B).
Figure 3.
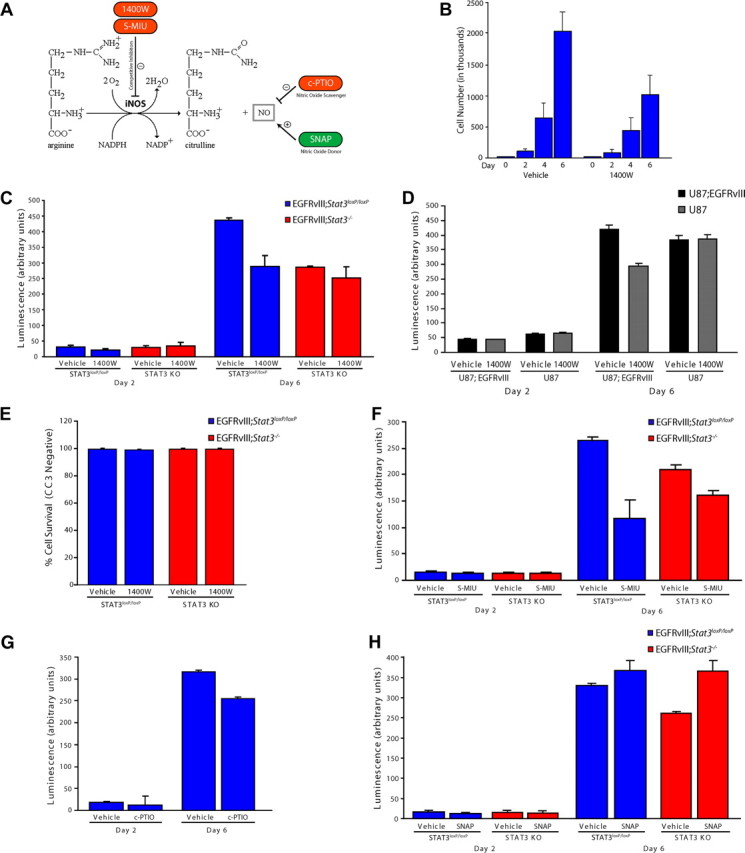
iNOS is required for the proliferation of EGFRvIII-expressing astrocytes. A, Schematic of NOS catalyzed by iNOS. Pharmacological agents used for analyses influence NO levels by distinct mechanisms. B, Population growth of EGFRvIII;Stat3loxP/loxP astrocytes exposed to vehicle or the iNOS inhibitor 1400W. Pharmacological inhibition of iNOS significantly reduced EGFRvIII-expressing astrocyte population growth (representative experiment of 3 independent experiments performed in triplicate; ANOVA, p < 0.0001, n = 3). C, Population growth of EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes was assessed using an ATP-based assay of cell proliferation (Cell-Titer Glo). Pharmacological inhibition of iNOS with 1400W significantly reduced the population growth of EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.0001, n = 5), with little or no effect on EGFRvIII;Stat3−/− astrocytes. D, Population growth of control and EGFRvIII-expressing U87 human glioblastoma cells was assessed as in C. Pharmacological inhibition of iNOS with 1400W significantly reduced the population growth of EGFRvIII-expressing U87 cells but had little or no effect on control U87 cells (representative experiment of 3 independent experiments performed in triplicate; ANOVA, p < 0.001, n = 3). E, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were treated with the iNOS inhibitor 1400W and subjected to immunocytochemistry using the CC3 antibody. Exposure to 1400W had little or no effect on the percentage of cells that were CC3 negative in both EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes. F, Population growth of EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes was assessed as in C. Pharmacological inhibition of iNOS with S-MIU significantly reduced the population growth of EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.0001, n = 3), with a more modest effect on EGFRvIII;Stat3−/− astrocytes. G, Population growth of EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes was assessed as in C. The NO scavenger c-PTIO significantly reduced the population growth of EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.0001, n = 3). H, Population growth of EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes was assessed as in C. The NO donor SNAP modestly increased the population growth of EGFRvIII;Stat3loxP/loxP astrocytes and significantly increased the population growth of EGFRvIII;Stat3−/− astrocytes (ANOVA, p < 0.0001, n = 3). KO, Knock-out.
To further test pharmacological inhibitors and activators of the NO pathway, we developed a high-throughput assay for cell proliferation using an ATP-based luminescence reagent. We validated the sensitivity of this assay and optimized it for astrocytes used throughout our study (data not shown). We first confirmed that 1400W significantly reduced the luminescent-based readout of EGFRvIII;Stat3loxP/loxP astrocytes, consistent with impaired cell proliferation (Fig. 3C). Notably, exposure to 1400W reduced the population growth of EGFRvIII;Stat3loxP/loxP astrocytes to similar levels as EGFRvIII;Stat3−/− astrocytes (Fig. 3C). Exposure of EGFRvIII;Stat3−/− astrocytes to 1400W had little or no effect on population growth in these assays (Fig. 3C). In other experiments, we found that 1400W significantly reduced the population growth of human EGFRvIII-expressing U87 glioblastoma cells but had little or no effect on the population growth of U87 glioblastoma control cells (Fig. 3D). In control experiments, exposure of EGFRvIII;Stat3loxP/loxP or EGFRvIII;Stat3−/− astrocytes to the iNOS inhibitor 1400W had little or no effect on cell survival, as monitored by expression of CC3 (Fig. 3E). These data suggest a critical role for iNOS in STAT3-dependent proliferation of EGFRvIII-expressing astrocytes.
The small molecule S-MIU is another potent, competitive inhibitor that selectively inhibits iNOS but not eNOS or nNOS (Fig. 3A) (Southan et al., 1995; Iuvone et al., 1997). Like 1400W, exposure to S-MIU preferentially inhibited the population growth of EGFRvIII;Stat3loxP/loxP astrocytes compared with EGFRvIII;Stat3−/− astrocytes (Fig. 3F). Quantification of the percentage inhibition of EGFRvIII-expressing Stat3 floxed and knock-out astrocytes upon exposure to S-MIU revealed a differential effect on Stat3 floxed cells compared with knock-out astrocytes (55.8 ± 19.3% inhibition of population growth with S-MIU treatment in Stat3 floxed cells compared with 22.8 ± 1.2% in knock-out astrocytes; t test, p < 0.05). These results corroborate the conclusion that iNOS mediates STAT3-dependent proliferation of EGFRvIII-expressing astrocytes. We also treated EGFRvIII-expressing astrocytes with the small-molecule inhibitor c-PTIO, an NO scavenger that converts free NO to NO2 (Fig. 3A) (Akaike et al., 1993; Tsunoda et al., 1994; Rand and Li, 1995). Exposure of EGFRvIII;Stat3loxP/loxP astrocytes to c-PTIO reduced population growth (Fig. 3G), suggesting that free NO is essential for the proliferation of these cells. These data suggest that the iNOS-catalyzed product, NO, plays a critical role in the proliferation of EGFRvIII-expressing astrocytes.
If iNOS is the critical target gene of STAT3 that mediates the oncogenic effect of STAT3, then increasing NO levels in Stat3 knock-out astrocytes should restore cell population growth to a level comparable with Stat3 floxed astrocytes. Consistent with this prediction, exposure of EGFRvIII;Stat3−/− astrocytes to the NO donor SNAP augments cell population growth to a level similar to EGFRvIII;Stat3loxP/loxP astrocytes (Fig. 3A,H). SNAP also modestly stimulated the population growth of EGFRvIII;Stat3loxP/loxP astrocytes, suggesting that NO has a gain-of-function effect on EGFRvIII-astrocyte population growth (Fig. 3H). Collectively, these data reveal that iNOS plays a key role downstream of STAT3 in promoting proliferation of EGFRvIII-expressing astrocytes.
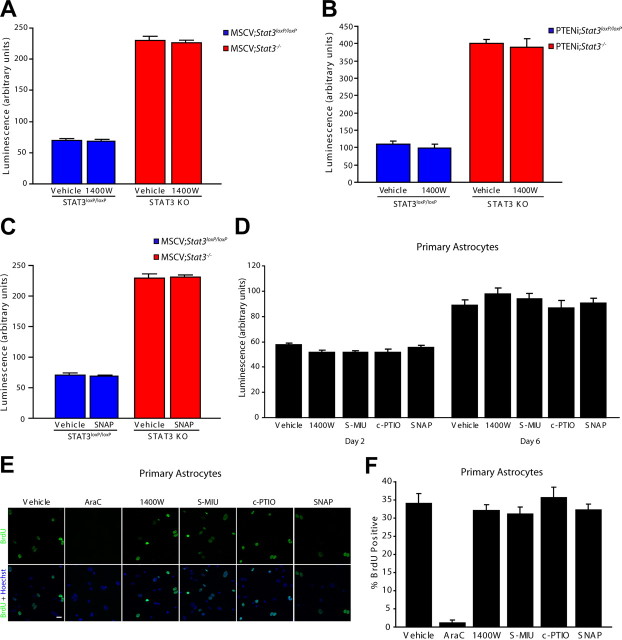
We next explored whether iNOS is also required for the proliferation of PTEN-deficient or control astrocytes. We found that pharmacological inhibition of iNOS using 1400W had little or no effect on the population growth of Stat3loxP/loxP or Stat3−/− astrocytes infected with the control MSCV retrovirus or on the population growth of Stat3loxP/loxP or Stat3−/− astrocytes that were PTEN deficient (Fig. 4A,B). Likewise, treatment of control MSCV astrocytes with the NO donor SNAP had little or no effect on population growth (Fig. 4C). In other experiments, we found that exposure of primary mouse astrocytes to pharmacological activators or inhibitors or the iNOS pathway had little or no effect on cell population growth (Fig. 4D). Likewise, pharmacological inhibition or activation of the iNOS pathway had little or no effect on BrdU incorporation in primary astrocytes (Fig. 4E,F). In control analyses, inhibition of DNA synthesis with the nucleoside analog arabinose C (AraC) blocked BrdU incorporation in primary astrocytes (Fig. 4E,F). Together, our data suggest that iNOS is specifically required for the proliferation of EGFRvIII-expressing astrocytes.
Figure 4.
iNOS is not required for the proliferation of control and PTEN-deficient astrocytes. A, Population growth of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes was assessed as in Figure 3C. Pharmacological inhibition of iNOS with 1400W had little or no effect on the population growth of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes compared with vehicle treatment (n = 3). B, Population growth of PTENi;Stat3loxP/loxP and PTENi;Stat3−/− astrocytes was assessed as in Figure 3C. Pharmacological inhibition of iNOS with 1400W had little or no effect on the population growth of PTENi;Stat3loxP/loxP and PTENi;Stat3−/− astrocytes compared with vehicle treatment (n = 3). C, Population growth of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes was assessed as in Figure 3C. The NO donor SNAP had little or no effect on the population growth of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes compared with vehicle treatment (n = 3). D, Population growth of primary mouse astrocytes was assessed as in Figure 3C. Pharmacological inhibitors and activators of the iNOS pathway had little or no effect on the population growth of primary mouse astrocytes compared with vehicle treatment (n = 3). E, Primary mouse astrocytes treated with inhibitors and activators of the iNOS pathway were subjected to immunocytochemistry using the BrdU antibody. Inhibition or activation of the iNOS pathway had little or no effect on BrdU incorporation of primary mouse astrocytes compared with vehicle treatment. In control experiments, the nucleoside analog AraC substantially reduced BrdU incorporation in primary astrocytes compared with vehicle treatment. Scale bar, 20 μm. F, Primary mouse astrocytes analyzed as in E were quantified for the percentage of BrdU-positive cells. Inhibition or activation of the iNOS pathway had little or no effect on BrdU incorporation in primary astrocytes. In control experiments, the nucleoside analog AraC significantly reduced BrdU incorporation compared with vehicle treatment (ANOVA, p < 0.0001, n = 3). KO, Knock-out.
Having identified a role for iNOS in the proliferation of EGFRvIII-expressing astrocytes using a pharmacological approach, we next used a genetic approach to determine whether expression of endogenous iNOS is required for the population growth of EGFRvIII-expressing astrocytes. We used a lentiviral-based method of RNAi to induce the knockdown of iNOS in astrocytes. We first confirmed that these shRNAs significantly reduced the levels of iNOS mRNA in EGFRvIII-expressing astrocytes (Fig. 5A). In addition, iNOS shRNAs substantially reduced the levels of exogenous iNOS protein in HEK293T cells and endogenous iNOS protein in astrocytes (Fig. 5B,C). Immunocytochemical analyses further confirmed that iNOS RNAi led to substantially reduced levels of iNOS protein in EGFRvIII-expressing astrocytes (Fig. 5D). Importantly, in assays of population growth, we found that iNOS knockdown astrocytes had reduced population growth compared with control-infected astrocytes (Fig. 5E), with little or no effect on cell survival as monitored by CC3 expression (Fig. 5F). Exposure of iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes to the NO donor SNAP augmented cell population growth to a level similar to control EGFRvIII;Stat3loxP/loxP astrocytes (Fig. 5G). In other experiments, we found that iNOS knockdown significantly reduced BrdU incorporation and the percentage of cells that were positive for the proliferation marker Ki67 in EGFRvIII-expressing astrocytes (Fig. 5H–K). In contrast, iNOS knockdown had little or no effect on population growth or BrdU incorporation in control MSCV-infected astrocytes (data not shown). Collectively, these findings provide genetic evidence that iNOS is required specifically for the proliferation of EGFRvIII-expressing astrocytes.
Figure 5.
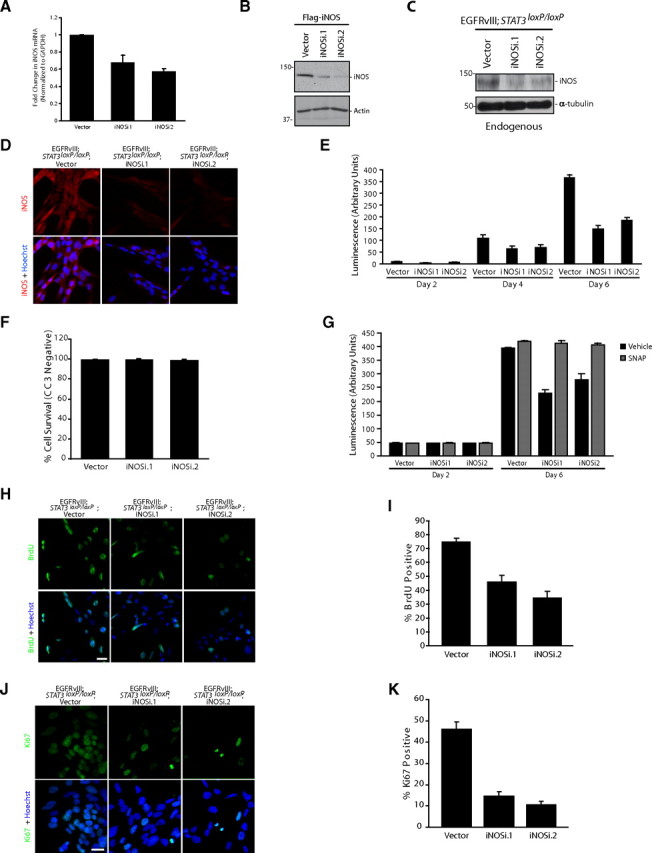
iNOS knockdown reduces the proliferation of EGFRvIII-expressing astrocytes. A, RNA isolated from mouse EGFRvIII;Stat3loxP/loxP astrocytes infected with iNOS RNAi virus or control virus was subjected to quantitative RT-PCR analyses using primers specific for iNOS. mRNA levels were normalized to GAPDH. iNOS mRNA levels were significantly reduced in EGFRvIII;Stat3loxP/loxP iNOS knockdown astrocytes compared with vector-transfected astrocytes (ANOVA, p < 0.005, n = 4). B, Lysates of HEK293T cells transfected with an expression plasmid encoding Flag–iNOS together with the iNOS RNAi or control RNAi plasmid were immunoblotted with the Flag or actin antibody. Knockdown of iNOS reduced the levels of iNOS protein in HEK293T cells. C, Lysates of control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were immunoblotted with the iNOS or α-tubulin antibody. Knockdown of iNOS substantially reduced the levels of endogenous iNOS protein in astrocytes. D, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were subjected to immunocytochemistry using the iNOS antibody. Representative images are shown. The expression of endogenous iNOS protein was substantially reduced in iNOS knockdown astrocytes compared with control astrocytes. Scale bar, 20 μm. E, Population growth of control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes was assessed as in Figure 3C. iNOS knockdown significantly reduced the population growth of EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.0001, n = 3). F, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were subjected to immunocytochemistry using the CC3 antibody. Knockdown of iNOS had little or no effect on the percentage of astrocytes that were CC3 negative (n = 3). G, Population growth of control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes was assessed as in Figure 3C. The NO donor SNAP significantly increased the population growth of iNOS knockdown EGFRvIII;Stat3−/− astrocytes (representative experiment of 3 independent experiments performed in triplicate; ANOVA, p < 0.001, n = 3). H, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were subjected to immunocytochemistry using the BrdU antibody. Incorporation of BrdU in astrocytes was substantially reduced upon iNOS knockdown. Scale bar, 20 μm. I, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes analyzed as in H were quantified for the percentage of BrdU-positive cells. Incorporation of BrdU was substantially reduced in iNOS knockdown astrocytes compared with control astrocytes (ANOVA, p < 0.0001, n = 3). J, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were subjected to immunocytochemistry using the Ki67 antibody. Immunoreactivity of Ki67 was substantially reduced in iNOS knockdown astrocytes. Scale bar, 20 μm. K, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes analyzed as in J were quantified for the percentage of Ki67-positive cells. The percentage of Ki67-positive cells was significantly reduced in iNOS knockdown astrocytes compared with control astrocytes (ANOVA, p < 0.0001, n = 3).
iNOS is required for the invasiveness of EGFRvIII-expressing astrocytes
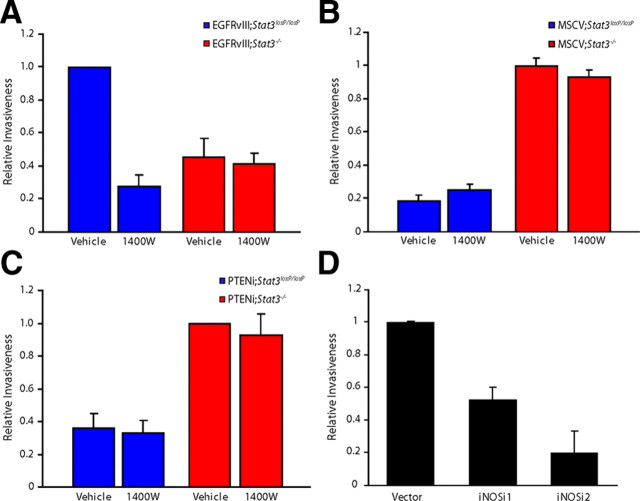
In addition to uncontrolled proliferation, glioblastoma cells are characterized by the property of invasiveness. We therefore characterized the role of iNOS in the invasiveness of EGFRvIII-expressing, PTEN-deficient, and control MSCV-infected astrocytes using a matrigel assay. Pharmacological inhibition of iNOS with 1400W significantly reduced the relative invasiveness of EGFRvIII;Stat3loxP/loxP astrocytes, with little or no effect on EGFRvIII;Stat3−/− astrocytes (Fig. 6A). In contrast, 1400W had little or no effect on the invasiveness of control MSCV-infected or PTEN-deficient astrocytes (Fig. 6B,C), suggesting that iNOS specifically promotes invasiveness of EGFRvIII-expressing astrocytes. In complementary analyses, we found that iNOS knockdown substantially reduced the relative invasiveness of EGFRvIII-expressing astrocytes (Fig. 6D), with little or no effect on the invasiveness of control MSCV-infected astrocytes (data not shown). Thus, iNOS plays a critical role in both proliferation and invasiveness of EGFRvIII-expressing astrocytes.
Figure 6.
iNOS knockdown impairs the invasiveness of EGFRvIII-expressing astrocytes. A, EGFRvIII;Stat3loxP/loxP and EGFRvIII;Stat3−/− astrocytes were seeded on top of an 8 μm pore size insert coated with matrigel in the presence of 1400W or control vehicle and allowed to invade through the matrigel matrix for 22 h. 1400W treatment significantly reduced the invasiveness of EGFRvIII;Stat3loxP/loxP astrocytes (ANOVA, p < 0.005, n = 3) but had little or no effect on EGFRvIII;Stat3−/− astrocytes. The effect of 1400W treatment on invasiveness was not secondary to a change in cell proliferation, because the invasive potential of these cells was measured at a time (22 h after plating) before a significant decrease in cell number upon iNOS inhibition. B, Invasiveness of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes exposed to 1400W was assessed as in A. Exposure to 1400W had little or no effect on the invasiveness of MSCV;Stat3loxP/loxP and MSCV;Stat3−/− astrocytes compared with vehicle treatment (n = 3). C, Invasiveness of PTENi;Stat3loxP/loxP and PTENi;Stat3−/− astrocytes exposed to 1400W was assessed as in A. Exposure to 1400W had little or no effect on the invasiveness of PTENi;Stat3loxP/loxP and PTENi;Stat3−/− astrocytes compared with vehicle treatment (n = 3). D, Invasiveness of control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes was assessed as in A. Knockdown of iNOS significantly reduced EGFRvIII;Stat3loxP/loxP astrocyte cell invasiveness (ANOVA, p < 0.005, n = 4). The effect of iNOS knockdown on invasiveness was not secondary to a change in cell proliferation, because the invasive potential of these cells was measured at a time (22 h after plating) before a significant decrease in cell number upon iNOS knockdown.
iNOS is required for the ability of EGFRvIII-expressing astrocytes to form tumors in vivo
Identification of a function for iNOS in EGFRvIII-induced astrocyte proliferation and invasiveness led us next to characterize the role of iNOS in glial transformation in vivo. We determined the ability of control and iNOS knockdown EGFRvIII-expressing astrocytes to form subcutaneous tumors in SCID mice. Control EGFRvIII-expressing astrocytes formed large solid tumors in these mice. In some cases, the tumor was highly invasive, growing into the surrounding muscle and connective tissue and even ulcerating through the skin. In contrast, iNOS knockdown EGRFvIII-expressing astrocytes produced smaller tumors and, in some cases, did not form tumors (Fig. 7A and data not shown). Examination of excised tumors using fluorescence imaging revealed that these tumors were GFP positive (Fig. 7B), confirming that tumor masses were derived from the injected EGFRvIII-expressing astrocytes. To determine the histology of these tumors, we used hematoxylin–eosin staining. Tumors formed by control EGFRvIII-expressing astrocytes had numerous mitotic figures, nuclear atypia, and hypercellularity. In contrast, tumors derived from iNOS knockdown EGFRvIII-expressing astrocytes had few mitotic figures (Fig. 7C). In addition, tumors derived from iNOS knockdown EGFRvIII-expressing astrocytes had fewer Ki67-positive cells compared with tumors formed by control EGFRvIII-expressing astrocytes (Fig. 7D). Consistent with these histological criteria, average tumor mass was significantly reduced in iNOS knockdown tumors compared with control tumors (Fig. 7E). Although control EGFRvIII-expressing astrocytes generated tumors that were on average 1.2 g, the mass of iNOS knockdown tumors was reduced by 75% overall, weighing on average 0.4 g. Thus, iNOS plays a critical role in malignant glial transformation in vivo.
Figure 7.
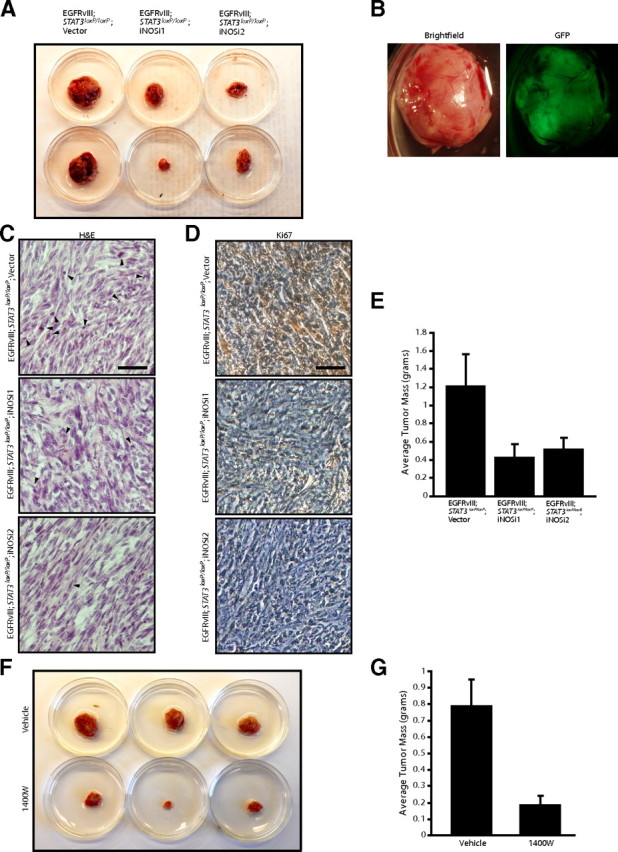
iNOS is required for the ability of EGFRvIII-expressing astrocytes to form tumors in vivo. A, Control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were injected subcutaneously into SCID mice. Four weeks after injection, tumors were removed, measured, and stained. iNOS knockdown reduced tumor size. B, Representative excised subcutaneous tumor revealed that masses removed were GFP positive and derived from injected cells. C, Tumors derived from control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were analyzed by hematoxylin and eosin (H&E) staining. Control EGFRvIII;Stat3loxP/loxP astrocytes showed histological features of neoplastic transformation, including nuclear atypia, hypercellularity, and frequent mitotic figures (arrowheads), although these features were less prominent in iNOS knockdown tumors. Scale bar, 200 μm. D, Tumors derived from control and iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes were analyzed by immunohistochemical analyses using the Ki67 antibody. Tumors derived from control EGFRvIII;Stat3loxP/loxP astrocytes had substantially greater Ki67 immunoreactivity compared with tumors derived from iNOS knockdown EGFRvIII;Stat3loxP/loxP astrocytes. Scale bar, 200 μm. E, Average tumor mass of tumors from A was measured. iNOS knockdown significantly reduced the mass of EGFRvIII;Stat3loxP/loxP tumors (ANOVA, p < 0.05, n = 33 animals). F, EGFRvIII;Stat3loxP/loxP astrocytes were injected subcutaneously into SCID mice, and the iNOS inhibitor 1400W or vehicle control was locally administered. Four weeks after initial injections, tumors were removed, measured, and stained. Treatment with the iNOS inhibitor 1400W substantially reduced tumor size. G, Average tumor mass of tumors from F was measured. Treatment with the iNOS inhibitor 1400W substantially reduced the mass of EGFRvIII;Stat3loxP/loxP tumors (t test, p < 0.005, n = 20 animals). Two animals in the 1400W-treated cohort did not have detectable tumors at the endpoint.
To determine whether inhibition of iNOS might represent a useful therapeutic strategy to reduce tumor growth and proliferation, we injected 1400W or vehicle locally at the site of subcutaneous tumor formation. EGFRvIII-expressing astrocytes treated with vehicle control formed large solid tumors that were invasive in SCID mice (Fig. 7F and data not shown). In contrast, injection of 1400W locally at the site of tumor formation substantially reduced tumor growth, leading to smaller tumors that were well circumscribed (Fig. 7F,G and data not shown). Notably, two animals in the 1400W-treated group failed to form detectable tumors in vivo. Together, our findings suggest that iNOS represents an important transcriptional target of oncogenic STAT3 and an essential regulator of the proliferation, invasiveness, and transformation of EGFRvIII-expressing astrocytes.
Discussion
In this study, we have identified iNOS as a novel gene target of STAT3 in EGFRvIII-expressing astrocytes. STAT3 specifically regulates iNOS transcription in EGFRvIII-expressing astrocytes but not PTEN-deficient astrocytes. STAT3 directly binds to the iNOS promoter and thereby stimulates iNOS transcription and expression in EGFRvIII astrocytes. Importantly, iNOS mediates EGFRvIII-induced STAT3-dependent proliferation of astrocytes. In particular, pharmacological inhibition of iNOS with distinct agents reduces the population growth of EGFRvIII-expressing astrocytes but has little or no effect on PTEN-deficient astrocytes. Consistent with these findings, genetic knockdown of iNOS limits the proliferation and invasiveness of EGFRvIII-expressing astrocytes. Knockdown of iNOS also diminishes the ability of EGFRvIII-expressing astrocytes to form glial tumors in vivo, and administration of the iNOS inhibitor 1400W at the site of tumor formation markedly reduces tumor size. Collectively, our findings define an important role for STAT3 regulation of iNOS gene expression in EGFRvIII-induced proliferation and transformation.
Our findings provide mechanistic insight into how STAT3 promotes the malignant behavior of EGFRvIII-expressing astrocytes. In the context of PTEN loss, STAT3 acts as a tumor suppressor by repressing transcription of the chemokine IL8 (de la Iglesia et al., 2008b). In the present study, we have found that STAT3 stimulates transcription of iNOS in EGFRvIII-expressing astrocytes and thereby promotes their transformation. Thus, the oncogenic switch in STAT3 function upon EGFRvIII-expression may drive a transcriptional switch in the targets that are regulated. A recent study has identified a critical role for iNOS in the transformation of glial cancer stem cells (Eyler et al., 2011), raising the possibility that cancer stem cells expressing EGFRvIII may rely on STAT3-dependent transcription of iNOS. Consistent with findings in our study, STAT3 also appears to operate downstream of EGFR signaling in breast cancer models to stimulate iNOS expression (Lo et al., 2005). Whether EGFRvIII triggers a transcriptional switch in STAT3 function outside of the brain will be an important question for future studies.
The identification of iNOS as a novel STAT3-regulated target in glial transformation suggests an important link to angiogenesis. NO has important functions in regulating vascular flow and permeability (Jordan et al., 2000; Van Buren et al., 2006). NO levels are frequently elevated in tumor cells (Swana et al., 1999; Fitzpatrick et al., 2008), and accordingly, as a NO-producing enzyme, iNOS has been linked to several human malignancies (Cobbs et al., 1995; Thomsen et al., 1995; Gallo et al., 1998; Lagares-Garcia et al., 2001; Crowell et al., 2003). Consistent with these findings, glioblastoma tumors are highly invasive, vascularized tumors (Jaeckle et al., 1996), raising the possibility that iNOS might support tumor cell nourishment and, hence, tumor cell proliferation and tumor growth in vivo.
Our findings indicate that STAT3 directly binds to the iNOS promoter and stimulates iNOS transcription downstream of EGFRvIII in astrocytes. Interestingly, in other cell types, such as endothelial cells, STAT3 represses iNOS transcription by directly inhibiting nuclear factor-κΒ (Yu et al., 2002; Yu and Kone, 2004). STAT3 does not appear to be regulated by the oncogene EGFRvIII in these cells, raising the possibility that EGFRvIII specifically triggers a switch from STAT3 repression to STAT3 activation of iNOS transcription. Alternatively, STAT3 regulation of iNOS might be distinct in different cell types and tissues. Additional research to clarify these possibilities is essential to understand whether the EGFRvIII–STAT3–iNOS signaling pathway can be generalized to diverse aspects of biology.
iNOS is an attractive target for therapeutic intervention for malignant gliomas. Recent pathological analyses of human glioma specimens have revealed that many of these tumors have elevated iNOS (Ellie et al., 1995; Hara and Okayasu, 2004), suggesting that intervention at this critical signaling node might be effective. Indeed, administration of the iNOS inhibitor 1400W at the site of injection of EGFRvIII-expressing astrocytes in SCID mice dramatically reduced tumor size and in some animals prevented tumor formation. Notably, several selective and potent iNOS inhibitors are readily available, making steps toward iNOS-based therapeutics for glioblastoma a real possibility. More generally, our study supports the emerging idea that patient-tailored treatment of glioblastoma might be necessary. Depending on the genetic background of a tumor, patients might need to receive specific therapies that target pathways active in their cancers. For example, patients with activating mutations in EGFR signaling might receive a mixture of STAT3 and iNOS inhibitors, whereas those with PTEN loss-of-function mutations might be treated with STAT3 activators and IL8 inhibitors. In the future, it will be essential to identify additional target genes that mediate the oncogenic switch in STAT3 function in astrocytes. These analyses will improve our understanding of the differences in STAT3 function in glioblastoma in distinct genetic contexts.
Footnotes
This work was supported by National Institutes of Health Grant NS064007 (A.B.), a James F. Petersen Chair of Research Grant from the National Brain Tumor Society (A.B.), and a postdoctoral fellowship from the Canadian Institutes of Health Research (A.J.-A.). We thank members of the Bonni laboratory for helpful discussions and critical reading of this manuscript.
References
- Akaike T, Yoshida M, Miyamoto Y, Sato K, Kohno M, Sasamoto K, Miyazaki K, Ueda S, Maeda H. Antagonistic action of imidazolineoxyl N-oxides against endothelium-derived relaxing factor/.NO through a radical reaction. Biochemistry. 1993;32:827–832. doi: 10.1021/bi00054a013. [DOI] [PubMed] [Google Scholar]
- Aoki Y, Feldman GM, Tosato G. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood. 2003;101:1535–1542. doi: 10.1182/blood-2002-07-2130. [DOI] [PubMed] [Google Scholar]
- Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH. Optic nerve glioma in mice requires astrocyte Nf1 gene inactivation and Nf1 brain heterozygosity. Cancer Res. 2003;63:8573–8577. [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Chau MN, El Touny LH, Jagadeesh S, Banerjee PP. Physiologically achievable concentrations of genistein enhance telomerase activity in prostate cancer cells via the activation of STAT3. Carcinogenesis. 2007;28:2282–2290. doi: 10.1093/carcin/bgm148. [DOI] [PubMed] [Google Scholar]
- Cobbs CS, Brenman JE, Aldape KD, Bredt DS, Israel MA. Expression of nitric oxide synthase in human central nervous system tumors. Cancer Res. 1995;55:727–730. [PubMed] [Google Scholar]
- Crowell JA, Steele VE, Sigman CC, Fay JR. Is inducible nitric oxide synthase a target for chemoprevention? Mol Cancer Ther. 2003;2:815–823. [PubMed] [Google Scholar]
- de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008a;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA, Mischel PS, Louis DN, Bonni A. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci. 2008b;28:5870–5878. doi: 10.1523/JNEUROSCI.5385-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia N, Puram SV, Bonni A. STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med. 2009;9:580–590. doi: 10.2174/156652409788488739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. 2007;10:608–614. doi: 10.1038/nn1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellie E, Loiseau H, Lafond F, Arsaut J, Demotes-Mainard J. Differential expression of inducible nitric oxide synthase mRNA in human brain tumours. Neuroreport. 1995;7:294–296. [PubMed] [Google Scholar]
- Eyler CE, Wu Q, Yan K, MacSwords JM, Chandler-Militello D, Misuraca KL, Lathia JD, Forrester MT, Lee J, Stamler JS, Goldman SA, Bredel M, McLendon RE, Sloan AE, Hjelmeland AB, Rich JN. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick B, Mehibel M, Cowen RL, Stratford IJ. iNOS as a therapeutic target for treatment of human tumors. Nitric Oxide. 2008;19:217–224. doi: 10.1016/j.niox.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gallo O, Masini E, Morbidelli L, Franchi A, Fini-Storchi I, Vergari WA, Ziche M. Role of nitric oxide in angiogenesis and tumor progression in head and neck cancer. J Natl Cancer Inst. 1998;90:587–596. doi: 10.1093/jnci/90.8.587. [DOI] [PubMed] [Google Scholar]
- Garvey EP, Oplinger JA, Tanoury GJ, Sherman PA, Fowler M, Marshall S, Harmon MF, Paith JE, Furfine ES. Potent and selective inhibition of human nitric oxide synthases. Inhibition by non-amino acid isothioureas. J Biol Chem. 1994;269:26669–26676. [PubMed] [Google Scholar]
- Gillespie MA, Le Grand F, Scim è A, Kuang S, von Maltzahn J, Seale V, Cuenda A, Ranish JA, Rudnicki MA. p38-γ-dependent gene silencing restricts entry into the myogenic differentiation program. J Cell Biol. 2009;187:991–1005. doi: 10.1083/jcb.200907037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara A, Okayasu I. Cyclooxygenase-2 and inducible nitric oxide synthase expression in human astrocytic gliomas: correlation with angiogenesis and prognostic significance. Acta Neuropathol. 2004;108:43–48. doi: 10.1007/s00401-004-0860-0. [DOI] [PubMed] [Google Scholar]
- Ho JN, Kang GY, Lee SS, Kim J, Bae IH, Hwang SG, Um HD. Bcl-XL and STAT3 mediate malignant actions of gamma-irradiation in lung cancer cells. Cancer Sci. 2010;101:1417–1423. doi: 10.1111/j.1349-7006.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- Horvath CM, Wen Z, Darnell JE., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- Iuvone T, Van Osselaer N, D'Acquisto F, Carnuccio R, Herman AG. Differential effect of l-NAME and S-methyl-isothiourea on leukocyte emigration in carrageenin-soaked sponge implants in rat. Br J Pharmacol. 1997;121:1637–1644. doi: 10.1038/sj.bjp.0701317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeckle KA, Cohen ME, Duffner PK. Clinical presentation and therapy of nervous system tumors. Oxford: Butterworth-Heinemann; 1996. pp. 1131–1149. [Google Scholar]
- Jafarian-Tehrani M, Louin G, Royo NC, Besson VC, Bohme GA, Plotkine M, Marchand-Verrecchia C. 1400W, a potent selective inducible NOS inhibitor, improves histopathological outcome following traumatic brain injury in rats. Nitric Oxide. 2005;12:61–69. doi: 10.1016/j.niox.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Jordan BF, Misson P, Demeure R, Baudelet C, Beghein N, Gallez B. Changes in tumor oxygenation/perfusion induced by the no donor, isosorbide dinitrate, in comparison with carbogen: monitoring by EPR and MRI. Int J Radiat Oncol Biol Phys. 2000;48:565–570. doi: 10.1016/s0360-3016(00)00694-5. [DOI] [PubMed] [Google Scholar]
- Kiuchi N, Nakajima K, Ichiba M, Fukada T, Narimatsu M, Mizuno K, Hibi M, Hirano T. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med. 1999;189:63–73. doi: 10.1084/jem.189.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bonni A. Signaling pathways regulating gliomagenesis. Curr Mol Med. 2003;3:73–84. doi: 10.2174/1566524033361609. [DOI] [PubMed] [Google Scholar]
- Lagares-Garcia JA, Moore RA, Collier B, Heggere M, Diaz F, Qian F. Nitric oxide synthase as a marker in colorectal carcinoma. Am Surg. 2001;67:709–713. [PubMed] [Google Scholar]
- Lassmann S, Schuster I, Walch A, Göbel H, Jütting U, Makowiec F, Hopt U, Werner M. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J Clin Pathol. 2007;60:173–179. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, Heller R, Ellis LM, Karras J, Bromberg J, Pardoll D, Jove R, Yu H. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- Ohara H, Ichikawa S, Matsumoto H, Akiyama M, Fujimoto N, Kobayashi T, Tajima S. Collagen-derived dipeptide, proline-hydroxyproline, stimulates cell proliferation and hyaluronic acid synthesis in cultured human dermal fibroblasts. J Dermatol. 2010;37:330–338. doi: 10.1111/j.1346-8138.2010.00827.x. [DOI] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Ikeuchi Y, Wilson-Grady JT, Merdes A, Gygi SP, Bonni A. A CaMKIIbeta signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat Neurosci. 2011;14:973–983. doi: 10.1038/nn.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, McKay RD. Multiple routes to astrocytic differentiation in the CNS. J Neurosci. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MJ, Li CG. Discrimination by the NO-trapping agent, carboxy-PTIO, between NO and the nitrergic transmitter but not between NO and EDRF. Br J Pharmacol. 1995;116:1906–1910. doi: 10.1111/j.1476-5381.1995.tb16681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer LK, Menter DG, Schaefer TS. Activation of stat3 and stat1 DNA binding and transcriptional activity in human brain tumour cell lines by gp130 cytokines. Cell Signal. 2000;12:143–151. doi: 10.1016/s0898-6568(99)00077-7. [DOI] [PubMed] [Google Scholar]
- Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci U S A. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander KS, Li L, Watson L, Merrell M, Dahmen H, Heinrich PC, Müller-Newen G, Harris KW. Inhibition of gp130 signaling in breast cancer blocks constitutive activation of Stat3 and inhibits in vivo malignancy. Cancer Res. 2004;64:6924–6933. doi: 10.1158/0008-5472.CAN-03-2516. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- Southan GJ, Szab ó C, Thiemermann C. Isothioureas: potent inhibitors of nitric oxide synthases with variable isoform selectivity. Br J Pharmacol. 1995;114:510–516. doi: 10.1111/j.1476-5381.1995.tb13256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swana HS, Smith SD, Perrotta PL, Saito N, Wheeler MA, Weiss RM. Inducible nitric oxide synthase with transitional cell carcinoma of the bladder. J Urol. 1999;161:630–634. [PubMed] [Google Scholar]
- Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda R, Okumura K, Ishizaka H, Matsunaga T, Tabuchi T, Yasue H, Akaike T, Sato K, Maeda H. Vasodilator effect of carboxy-2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl in the coronary circulation: in vivo and in vitro studies. Eur J Pharmacol. 1994;262:55–63. doi: 10.1016/0014-2999(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002;62:5551–5558. [PubMed] [Google Scholar]
- Van Buren G, 2nd, Camp ER, Yang AD, Gray MJ, Fan F, Somcio R, Ellis LM. The role of nitric oxide in mediating tumour blood flow. Expert Opin Ther Targets. 2006;10:689–701. doi: 10.1517/14728222.10.5.689. [DOI] [PubMed] [Google Scholar]
- van der Poel HG, Zevenhoven J, Bergman AM. Pim1 regulates androgen-dependent survival signaling in prostate cancer cells. Urol Int. 2010;84:212–220. doi: 10.1159/000277601. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Yu Z, Kone BC. The STAT3 DNA-binding domain mediates interaction with NF-kappaB p65 and iuducible nitric oxide synthase transrepression in mesangial cells. J Am Soc Nephrol. 2004;15:585–591. doi: 10.1097/01.asn.0000114556.19556.f9. [DOI] [PubMed] [Google Scholar]
- Yu Z, Zhang W, Kone BC. Signal transducers and activators of transcription 3 (STAT3) inhibits transcription of the inducible nitric oxide synthase gene by interacting with nuclear factor kappaB. Biochem J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]