Abstract
5-methyl-cytosines at CpG sites frequently mutate into thymines, accounting for a large proportion of spontaneous point mutations. The repair system would leave substantial numbers of errors in neighboring regions if the synthesis of erased gaps around deaminated 5-methyl-cytosines is error-prone. Indeed, we identified an unexpected genome-wide role of the CpG methylation state as a major determinant of proximal natural genetic variation. Specifically, 507 Mbp (∼18%) of the human genome was within 10 bp of a CpG site; in these regions, the single nucleotide polymorphism (SNP) rate significantly increased by ∼50% (P < 10−566 by a two-proportion z-test) if the neighboring CpG sites are methylated. To reconfirm this finding in another vertebrate, we compared six single-base resolution methylomes in two inbred medaka (Oryzias latipes) strains with sufficient genetic divergence (3.4%). We found that the SNP rate also increased by ∼50% (P < 10−2170), and the substitution rates in all dinucleotides increased simultaneously (P < 10−441) around methylated CpG sites. In the hypomethylated regions, the “CGCG” motif was significantly enriched (P < 10−680) and evolutionarily conserved (P = ∼ 0.203%), and slow CpG deamination rather than fast CpG gain was seen, indicating a possible role of CGCG as a candidate cis-element for the hypomethylation state. In regions that were hypermethylated in germline-like tissues but were hypomethylated in somatic liver cells, the SNP rate was significantly smaller than that in hypomethylated regions in both tissues, suggesting a positive selective pressure during DNA methylation reprogramming. This is the first report of findings showing that the CpG methylation state is significantly correlated with the characteristics of evolutionary change in neighboring DNA.
Our understanding of the role of DNA methylation in genetic variation is limited to the observation that methylated cytosines at CpG sites mutate to thymines at very high frequencies (Lindahl and Nyberg 1972; Coulondre et al. 1978; Cooper and Krawczak 1989). This has been confirmed in genome-wide analyses (Sved and Bird 1990; Lander et al. 2001; Venter et al. 2001). Recently, precise methylation maps at single-base resolution have been created using next-generation DNA sequencing technology, confirming the overabundance of G:C→A:T transitions (Ossowski et al. 2010). In contrast, hypomethylated cytosine nucleotides seem to escape mutation; e.g., comparative genome analysis of human and primates genomes showed a lower CpG mutation rate in CpG-rich promoters that are mostly hypomethylated in the germline (Saxonov et al. 2006; Weber et al. 2007). Although the system for repairing mutations of 5-methyl cytosines to thymines is not well understood, it must be relatively ineffective because a large proportion of spontaneous point mutations are C-to-T mutations. The erroneous repair system may also leave substantial errors in neighboring regions if the synthesis of erased gaps around deaminated 5-methyl-cytosines is error-prone. The genome-wide methylation information provides an unprecedented opportunity to examine whether the CpG methylation state affects proximal genetic variation.
A related problem is the allele-specific DNA methylation (ASM), different methylation patterns in parental alleles (Chandler et al. 1987). ASM is involved in genomic imprinting and X-inactivation in females to achieve dosage compensation. Autosomal ASM has also been reported in the human and mouse genomes (Yamada et al. 2004; Heijmans et al. 2007; Zhang et al. 2009), and recent genome-wide collections of DNA methylation states have started uncovering that ASM is prevalent in the mammalian genome (Kerkel et al. 2008; Hellman and Chess 2010; Schalkwyk et al. 2010). Genome-wide studies indicate that ASM is associated with cis-acting polymorphisms (local genotypes), such as mutations in CpG sites (Schilling et al. 2009; Shoemaker et al. 2010), suggesting Mendelian inheritance patterns of ASM. A number of genomic regions of intermediate DNA methylation level are found in the human genome (Deng et al. 2009; Lister et al. 2009) and are thought to be largely a consequence of ASM and to have implications for complex disease genetics (Meaburn et al. 2010). However, little is known about fundamental cis-elements for inducing DNA methylation. Sequence motifs that are significantly enriched and evolutionarily conserved in hypomethylated regions could be candidate cis-elements; however, it is highly nontrivial to identify conserved sequence motifs due to the quite low incidence of genetic variation (∼0.1%) in the human and mouse genomes. We approach this problem through the study of correlation between CpG methylation state and proximal genetic variation in the medaka genome.
Results
We examined the relationship between DNA methylation and genetic variation in the human genome using publicly available data. Specifically, we examined the methylome data from human germline sperm cells (Molaro et al. 2011) and the reference single nucleotide polymorphisms (refSNPs) of the CEU population collected by the HapMap Project (The International HapMap Consortium 2003). Although substantial data are available on the human genome, the incidence of genetic variation in the human genome is quite low (∼0.1%), which is not sufficient to perform a detailed analysis on the dinucleotide patterns of genetic variations associated with hyper- and hypomethylated regions. For this purpose, the medaka (Oryzias latipes) model system provides an ideal resource in vertebrates because two medaka inbred strains, Hd-rR and HNI, have a sufficiently high incidence of genetic variation (∼3.4%) (Kasahara et al. 2007). The Hd-rR and HNI medaka inbred strains originated in the southern and northern parts of Japan, respectively, which were separated by a watershed, and the strains diverged ∼18 million years ago (Setiamarga et al. 2009). Another merit of the medaka is its abundance of germline cells in the testes and blastulae (half-day embryos, in which some of the cells remain totipotent [Hong et al. 1998]), which provides information on the primary relationships between the methylation pattern and genomic variation during the course of evolution. Thus, we newly generated six methylomes of medaka genomes at single-base resolution using genomic DNA from three libraries (liver, blastulae, and testes) of the two strains, treating it with bisulphite, and subjecting it to Illumina sequencing. In total, 1.8 billion reads were produced, and 44 billion nucleotides were uniquely mapped to the genome, with an average depth of ninefold for each cell line of each strain (Supplemental Table S1). An average of ∼78% of each medaka genome was covered by at least one read (Supplemental Table S1). The bisulphite conversion procedure was highly effective, as validated by a control experiment using yeast (Saccharomyces cerevisiae S288C) (Supplemental Table S2). In yeast, cytosine nucleotides are not methylated, and indeed, none of yeast cytosine nucleotides was uncovered to be hypermethylated (methylation level ≥ 0.8) (Supplemental Fig. S1). Data for the entire methylome of medaka at single base resolution can be accessed at http://utgenome.org/medaka/methylome/.
Substantial cytosines at non-CpG sites are methylated in human ES and Arabidopsis (Cokus et al. 2008; Lister et al. 2008, 2009); however, methylation at non-CpG sites was very rare in human sperm (Molaro et al. 2011) and in all of the three medaka tissues (Supplemental Fig. S2); i.e., few non-CpG cytosines (∼0.02%) were observed to be hypermethylated. To examine whether cytosine methylation state, unmethylated or methylated, affects proximal genetic variation, both unmethylated and methylated cytosines should be sufficiently available. In CpG sites, the ratio of unmethylated cytosines:methylated cytosines was ∼10%:∼90%, showing the abundance of unmethylated cytosines. In non-CpG sites, however, the ratio was 99.98%:0.02%, indicating the lack of methylated cytosines. Thus, we focused on methylation patterns of CpG sites in subsequent analyses. The distribution of methylation levels was found to be bimodal, and the vast majority of CpG sites were highly methylated in human sperm (Molaro et al. 2011) and in each medaka library (Supplemental Fig. S2). CpG islands may be appropriate regions in which to measure the effect of methylation pattern on the incidence of genetic variation; however, they occupy a relatively small proportion of the human genome (∼1%) as well as the medaka genome (∼3%) and exist mostly in CpG-rich regions. Therefore, regions with fewer CpG sites are overlooked. For defining more comprehensive characteristics of CpG sites in a wider range of genome sequences, we, instead, focused on neighboring genome sequences (the 10 bases upstream and downstream) of all the CpG sites, which we hereafter refer to as “CpG site blocks” (see Methods; Supplemental Table S3). Overlapping CpG site blocks were merged, and the combined length of all the CpG site blocks was 507 Mbp (∼18%) in the human genome and ∼188 Mbp in the medaka genome, accounting for ∼37% of the ∼500 Mbp that were aligned between the two medaka genomes. We then noticed that one-third of CpG site blocks were covered by <5 reads and were likely to lack accuracy. Thus, we imposed the stringent requirement that each CpG site had a read coverage of ≥5 (see Methods). CpG site blocks that met this condition accounted for ∼323 Mbp (∼11%) in the human genome and ∼139 Mbp (∼28%) in the medaka genome, which was sufficient for estimating features of the entire genomes (Supplemental Table S3).
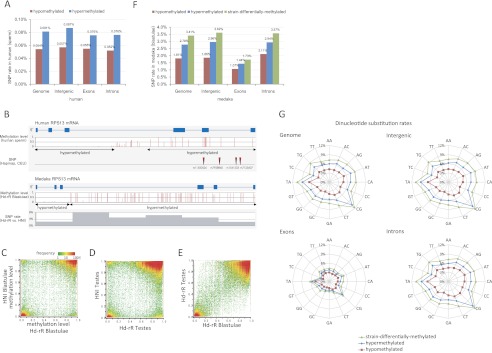
We compared the incidence of genetic variations in hypomethylated regions (methylation level ≤ 0.2) with that in hypermethylated regions (methylation level ≥ 0.8). Because single-nucleotide mutations by deamination of methylated cytosines in CpG dinucleotides are dominant, to examine single-nucleotide substitution rates in this analysis, we excluded SNPs in CpG dinucleotides and SNPs in TpG/CpA dinucleotides which might make TpG/CpA be CpG. Figure 1A shows that, in human sperm, the incidence rate in hypomethylated regions in the entire genome, 0.054%, was significantly lower than that in hypermethylated ones, 0.081% (P < 10−566 by a two-proportion z-test) (Supplemental Table S4). This significant increase of incidence rate in hypermethylated regions was also detected in intergenic regions (P < 10−305), in exons (P < 10−29), and in introns (P < 10−151) (Supplemental Table S4). We also observed a similar tendency in the medaka system. For example, Figure 1B illustrates a pair of the homologous regions of the human and medaka genomes where the RPS13 gene is encoded. We saw four SNPs in the hypermethylated human region and a higher SNP rate in the hypermethylated medaka region. We will check this characteristic precisely in the entire medaka genome in what follows.
Figure 1.
Methylation patterns and substitution rates in the inbred medaka strains, Hd-rR and HNI. (A) SNP (single-nucleotide polymorphism) rates in hyper- and hypomethylated CpG blocks in the reference human genome (hg19). The difference in SNP rates was significant in the entire genome (P < 10−566 by a two-proportion z-test) (Supplemental Table S4), in intergenic regions (P < 10−305), in exons (P < 10−29), and in introns (P < 10−151). (B) Methylation level and SNP distribution in the homologous regions of the human and medaka genomes where gene RPS13 is coded. (C,D) Comparisons of the methylation patterns in Hd-rR and HNI. The vertical and horizontal axes indicate methylation level. The heat map uses logarithmic coordinates and presents the number of corresponding CpG site blocks. Conserved hypermethylated and hypomethylated patterns between the two strains were dominant, except for a small number of hot spots observed in the differentially methylated regions (differences in methylation level ≥ 0.5). (E) Comparison of the methylation patterns in blastulae and testes in Hd-rR. (F) SNP rates in hypo-, hyper-, and strain-differentially methylated regions in medaka blastulae grouped by the entire genome, intergenic regions, exons, and introns. The differences between SNP rates of hypo- and hypermethylated regions were remarkable: P < 10−2170 (genome), P < 10−2170 (intergenic regions), P < 10−113 (exons), and P < 10−589 (introns) according to a two-proportion z-test (Supplemental Table S4). Furthermore, the differences between SNP rates of strain-differentially and hypermethylated regions were also significant (Supplemental Table S4). (G) Dinucleotide substitution rates in the whole medaka genome, intergenic regions, exons, and introns in CpG site blocks with various methylation states. Color key presents mutation rates: blue for hypermethylated (methylation level ≥ 0.8 in both strains); red for hypomethylated (methylation level ≤ 0.2 in both strains); and green for strain-differentially methylated (difference in methylation level between the two strains ≥ 0.5) in blastulae. The axes in each radar chart represent substitution rates of individual dinucleotides. Each dinucleotide shows the same substitution rate as its reverse complementary dinucleotide. Significant differences between substitution rates in hypo- and hypermethylated regions were observed for all dinucleotides, and the P-values, according to a two-proportion z-test, were P < 10−441 (genome), P < 10−263 (intergenic regions), P < 10−15 (exons), and P < 10−69 (introns) (Supplemental Table S5).
In medaka, the methylation state of each CpG site is not necessarily conserved between the two strains (Fig. 1C,D; Supplemental Table S3A), which is supported by 4.46 × 10−4 methylation polymorphisms per CG site per generation in Arabidopsis thaliana (Becker et al. 2011; Schmitz et al. 2011). In general, however, hypomethylated and hypermethylated regions were highly conserved between the two inbred strains (Fig. 1C,D) and were also highly correlated (R2 = 0.73) between testes and blastulae (Fig. 1E). Thus, hereafter, we used hypo- and hypermethylated regions that were conserved between the two strains as the representative regions. Analysis of SNPs between Hd-rR and HNI (Sasaki et al. 2009) around CpG blocks revealed that the SNP rate of 1.81% in hypomethylated regions was significantly lower than the 2.78% rate in hypermethylated regions (P < 10−2170 by a two-proportion z-test) (Fig. 1F; Supplemental Table S4). In addition to the entire genome, these characteristics were also significant in intergenic regions (P < 10−2170), in introns (P < 10−589), and in exons (P < 10−113) in spite of inherently strong purifying selection in exons (Fig. 1F; Supplemental Table S4). Furthermore, genome-wide regions that were differentially methylated in the two strains exhibited a SNP rate of 3.41%, which is significantly higher than the rate of 2.78% in hypermethylated regions (P < 10−442) (Supplemental Table S4). This indicates that genomic regions with a relaxed selective pressure display a wider divergence in cytosine methylation.
The high divergence between the two medaka strains provided a detailed analysis of mutations of dinucleotides. Figure 1G shows that the peak substitution rates occurred in CG and TA dinucleotides. The CG substitutions (∼11% in hypermethylated regions) were predominantly transitions, which can be explained by the deamination of methylated cytosines that dominates point substitutions in vertebrates (Lindahl and Nyberg 1972; Cooper and Krawczak 1989), whereas in hypomethylated regions, the CG substitution rate was moderate. Another commonly mutated dinucleotide, TA, is universally underrepresented because of its high mutation rate (Ohno 1988; Burge et al. 1992). Our novel finding is that substitution rates for all dinucleotides in CpG site blocks change simultaneously when neighboring CG dinucleotides are hypermethylated, hypomethylated, or differentially methylated. These characteristics were significant for any dinucleotide in the entire genome (P < 10−441 by a two proportion z-test), in intergenic regions (P < 10−263), in introns (P < 10−69), and in exons (P < 10−15), implying that methylation patterns have a comprehensive impact on genetic variation (see the P-values for all dinucleotides in Supplemental Table S5).
One might be concerned that the high CG substitution rate could affect the substitution rates for dinucleotides other than CG; however, the effects are limited to dinucleotides immediately before CG dinucleotides and are considerably small due to the rarity of CG dinucleotides (2.0% in the medaka genome). In fact, the substitution rates of dinucleotides excluding the dinucleotides overlapping CGs by one base remain essentially unchanged (Supplemental Fig. S3). Another possible explanation for the slower mutation rate in evolutionarily conserved hypomethylated regions is that these regions are around transcription start site (TSS) regions that are highly conserved (Taylor et al. 2006; Sasaki et al. 2009); however, evolution in such regions remained approximately the same even after hypomethylated regions near TSSs (those lying in the 500 bp immediately upstream of and downstream from the TSSs for 23,531 predicted medaka genes) were excluded from the analysis (Supplemental Fig. S4; Supplemental Table S3A,B). The higher substitution rate in evolutionarily conserved hypermethylated regions may be influenced by transposable elements which are highly mutated and methylated in vertebrates (Goll and Bestor 2005). However, transposons in the medaka genome are extremely rare and can be ignored (Kasahara et al. 2007). Therefore, methylation-dependent factors other than the influence of gene transcription or transposons may contribute to the slow evolution of hypomethylated regions.
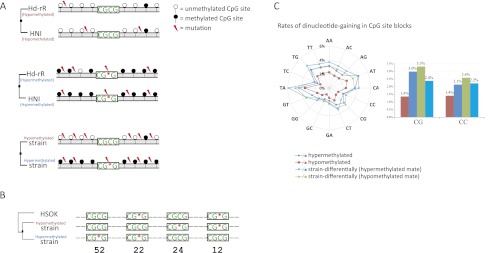
A related problem is that methylation patterns may be affected by cis-acting polymorphisms, such as mutations in CpG sites (Schilling et al. 2009; Shoemaker et al. 2010), but little is known about fundamental cis-elements. It is highly nontrivial to identify cis-elements that induce the CpG methylation state. We attempted to find sequence motifs that were enriched and evolutionarily conserved in hypomethylated regions because such motifs could be candidate cis-elements and would be informative for further studies. We, therefore, searched for potential conserved motifs that were capable of distinguishing two collections of 10,000 hypo- or hypermethylated CpG site blocks using AdaBoost, an established machine learning algorithm (see Methods). The most significant motif was “CGCG,” and the incidence of “CGCG” in 10,000 hypomethylated regions was 61.39%, compared with 21.84% in 10,000 hypermethylated regions, showing the significant relevance of “CGCG” sequence enrichment to the methylation state (P < 10−680 by a two-proportion z-test), as illustrated in Figure 2A. We then examined whether the CGCG motif is evolutionarily conserved in hypomethylated regions. For this purpose, it was necessary to predict the ancestor of Hd-rR and HNI strains. Therefore, we assembled a draft genome of an outgroup medaka strain, HSOK. Figure 2B shows that CGCG is significantly more conserved in hypomethylated regions than in hypermethylated regions (P = ∼ 0.203% by a two-proportion z-test). This evolutionarily preserved CGCG motif in hypomethylated regions may be attributable to slow CpG deamination or fast CpG-gaining. To clarify this issue, we calculated dinucleotide-gaining substitution rates by comparing the genomes of Hd-rR and HNI to their ancestor which was estimated from the outgroup HSOK genome (Fig. 2C; see Methods). The rate of CpG gain in hypomethylated regions, 1.4%, was much smaller than the 3.0% rate observed in hypermethylated ones, supporting the hypothesis that slow CpG deamination is the dominant factor for maintaining CGCG richness in hypomethylated regions (Cohen et al. 2011). Meanwhile, we observed an intriguing property in differentially methylated regions in different strains: The rates of CG/CC gain in the hypomethylated mate are higher than those in the hypermethylated mate (3.3% and 2.4% compared with 2.6% and 2.2%, respectively), suggesting that CG/CC gains are involved in the evolution of methylation among different strains.
Figure 2.
(A) Representative mutation patterns in evolutionarily conserved hypo- and hypermethylated regions and in regions that are differentially methylated in different strains, where substitution rates ascend from top to bottom. The CGCG motif is significantly conserved in hypomethylated regions (P < 10−680 by a two-proportion z-test). (B) Number of strain-differentially methylated CpG site blocks with either gain or loss of the CGCG motif. Of 1656 CGCG motif occurrences in multiple alignments of the genomes of the three strains, 52 (24, respectively) were conserved in hypomethylated (hypermethylated) regions of one of Hd-rR or HNI but were mutated in hypermethylated (hypomethylated) regions of the other strain. We then evaluated the significance of the difference between the means in the two groups, 52/1656 vs. 24/1656, to obtain a P-value of 0.203% according to a two-proportion z-test. (C) The rates of dinucleotide gain in CpG site blocks. Gain rates in the hypo- or hypermethylated CpG site blocks and each mate in strain-differentially methylated blocks are shown.
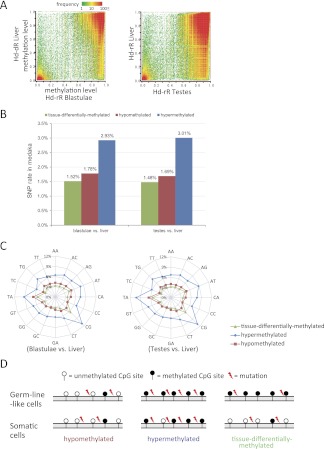
Next, we examined the methylation and genetic variations of two germline-like tissue types (blastulae and testes) and somatic liver cells that are fully differentiated and mostly homogeneous. In this analysis, we focused on tissue types in a single strain, Hd-rR. Figure 3A shows an abundance of CpG site blocks with tissue-differentially methylation states such that differences in methylation levels between each of the germline-like tissues and liver are more than 0.5. Notably, ∼99% of tissue-differentially methylated regions were hypomethylated in the liver but were hypermethylated in blastulae and testes. Remarkably, as illustrated in Figure 3B, the tissue-differentially methylated regions showed a significantly lower SNP rate than did the hypomethylated ones (P < 10−82) (Supplemental Table S6). Further analysis on dinucleotides revealed that the substitution rates of dinucleotides except for CC/GG/CG in differentially methylated regions were also significantly lower than were those in hypomethylated regions (P < 10−3) (Fig. 3C; Supplemental Table S7A), which was also seen even after excluding regions near TSSs (Supplemental Fig. S6). The high CG substitution rate in these regions could have been a consequence of deamination mutations occurring in primary germline-like cells. These results suggest that, because change in DNA methylation can affect transcription so that it can regulate reprogramming during somatic development, a positive selective pressure of genomic sequence could be involved in these differentially methylated regions (Fig. 3D).
Figure 3.
Methylation patterns and substitution rates in different tissue types. (A) Comparison of the methylation patterns in Hd-rR: blastulae vs. liver, and testes vs. liver. The vertical and horizontal axes show methylation level. The methylation patterns in HNI are presented in Supplemental Fig. S5 and are similar to those in these figures. (B) SNP rates in CpG site blocks with three methylation states: hypomethylated in both of the two tissue types, hypermethylated in both, and differentially methylated between the two tissue types. Significant differences in SNP rates were seen between tissue-differentially and hypomethylated regions (P < 10−82 by a two-proportion z-test) (Supplemental Table S6), and between hypo- and hypermethylated CpG site blocks (P < 10−2170) (Supplemental Table S6). (C) Dinucleotide substitution rates in CpG site blocks with the three methylation states. The substitution rates of all dinucleotides, except for CC/GG/CG, in tissue-differentially methylated regions were significantly lower than those in hypomethylated regions (P < 10−3 by a two-proportion z-test) (Supplemental Table S7A). (D) Representative mutation patterns in hypomethylated, hypermethylated, and somatic cell-specific hypomethylated (germline-like-specific hypermethylated) regions. Somatic cell-specific hypomethylated regions exhibited the lowest mutation rates.
Discussion
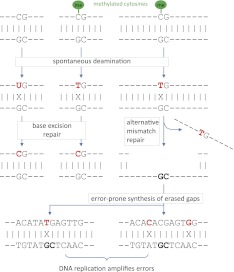
Because DNA methylation may influence genetic variation, we measured the incidence of genetic variations in methylation states in the human genome as well as in the two medaka inbred strains. We focused on germline-like tissue types, blastulae and testes, to determine the primary effects of methylation and evaluated a total of six single-base-resolution DNA methylomes in the medaka system. Our findings provide novel insights into the relationships between the dynamics of methylation states and genomic variations (Figs. 2A, 3D). Figure 4 illustrates our hypothesis for explaining the difference observed in genetic evolution between hyper- and hypomethylated region. Given that deaminated cytosine (U:G mismatch) is recognized and repaired by base excision repair (BER), deaminated 5-methyl-cytosine (T:G mismatch) would be corrected via more complicated mismatch-repair pathways (Molaro et al. 2011; Fig. 4). Previous studies revealed that two glycosylases, thymine DNA glycosylase (TDG) and methyl-CpG-binding domain protein 4 (MBD4), are associated with T:G repair, but processing by these enzymes is biased in the genome and is relatively ineffective (Neddermann et al. 1996; Yoon et al. 2003). An alternative mismatch-repair system (MMR) might gain access to T:G mismatches, which could involve DNA polymerases with lower fidelity (Loeb and Monnat 2008). Thus, we speculate that the error-prone synthesis of the erased gaps around deaminated 5-methyl-cytosines could lead to a higher mutation rate in hypermethylated regions (Fig. 4).
Figure 4.
A working model for a higher mutation rate in hypermethylated regions. Deaminated cytosine (U:G mismatch) is repaired by base excision repair (BER), but deaminated 5-methyl-cytosine (T:G mismatch) is corrected by more complicated repair pathways. An alternative mismatch repair system (MMR) might involve low-fidelity DNA polymerase, resulting in the error-prone synthesis of erased gaps.
In an effort to search for candidate cis-elements for affecting the CpG methylation state, we observed a significant correlation between the methylation state of CpG sites and the presence of CGCG in the neighborhood. This CGCG motif was evolutionarily conserved in hypomethylated regions with statistical significance. A comparative genomic analysis showed slow CpG-gaining in hypomethylated regions, supporting the hypothesis that the evolutionary conservation of CGCG was brought about by slow CpG deamination (Cohen et al. 2011). It is intriguing to ask whether or not CGCG and other candidate motifs are associated with or affect allele-specific methylation. Breeding the F1 generation of the two medaka inbred strains is a promising approach to this problem, and high-throughput short-read bisulfite sequencing is an inexpensive way of observing DNA methylome, though it suffers from a technical difficulty in distinguishing short reads of ∼100 bp from two different alleles. The high incidence of genetic variation (∼3.4%) between the two inbred strains would facilitate achieving this classification task because an average of 3.4 SNPs per 100-bp read would make it easier to align a read to its originating allele.
Overall, these genetic consequences of methylation, including the slower evolution rate of the cis-element “CGCG” found in hypomethylated regions of the medaka system, should also be explored in other species.
Methods
Construction and sequencing of medaka bisulfite-treated DNA libraries
Genomic DNA from medaka tissue cells (blastulae, testes, and liver from two medaka strains, Hd-rR and HNI) was isolated and sonicated to a desired size range (100–400 bp). The DNA fragments were treated with DNA polymerase to generate blunt ends and were ligated with double-stranded DNA adaptors containing methylated cytosines, which were designed to amplify only those DNA fragments carrying bisulfite-converted adaptor sequences at both ends. Followed by 7–10 cycles of PCR, 250 –450-bp size-fractionated DNA was sequenced using an Illumina GAIIx genome analyzer. A validation experiment was performed according to the same procedure using genomic DNA from S. cerevisiae S288C.
Computational and statistical methods
We converted all cytosines in reads and in both the Watson and Crick strands of the reference genome to thymines for primary mapping and used Smith-Waterman alignments between the original sequences of primary best hits. All possible methylation patterns were evaluated, and only uniquely mapped reads were retained for further analyses. The level of methylation of a particular cytosine was estimated by dividing the number of mapped reads reporting a cytosine (C) by the total number of reads reporting a C or T (thymine). The comparison of substitution rate and methylation level was performed among CpG site blocks, which consist of at least one CpG site and its surrounding upstream or downstream 10 bases. Overlapping CpG site blocks were merged. A two-proportion z-test was performed to test the significance of the substitution rate difference observed between hypo-/hypermethylated CpG site blocks. The AdaBoost algorithm was used to find conserved motifs in hypo-/hypermethylated CpG site blocks. Estimation of the ancestor sequence of Hd-rR and HNI was given by multiple alignments using an out-group medaka strain HSOK which was assembled using the SOAPdenovo assembler (Li et al. 2010). Details of methods can be found in Supplemental Methods.
Data access
All sequence data are deposited at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) (accession number SRA026693).
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (A23241058) from the MEXT, by the Global COE program (Deciphering Biosphere from Genome Big Bang) from the MEXT, and by the Innovative Cell Biology by Innovative Technology from the MEXT. Computational time was provided by the Information Technology Center, the University of Tokyo.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.140236.112.
Freely available online through the Genome Research Open Access option.
References
- Becker C, Hagmann J, Muller J, Koenig D, Stegle O, Borgwardt K, Weigel D 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249 [DOI] [PubMed] [Google Scholar]
- Burge C, Campbell AM, Karlin S 1992. Over-representation and under-representation of short oligonucleotides in DNA-sequences. Proc Natl Acad Sci 89: 1358–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler LA, Ghazi H, Jones PA, Boukamp P, Fusenig NE 1987. Allele-specific methylation of the human c-Ha-ras-1 gene. Cell 50: 711–717 [DOI] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A 2011. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell 145: 773–786 [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng SH, Zhang XY, Chen ZG, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 83: 181–188 [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780 [DOI] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH 2005. Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514 [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE 2007. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16: 547–554 [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A 2010. Extensive sequence-influenced DNA methylation polymorphism in the human genome. Epigenetics Chromatin 3: 11 doi: 10.1186/1756-8935-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Winkler C, Schartl M 1998. Production of medakafish chimeras from a stable embryonic stem cell line. Proc Natl Acad Sci 95: 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium 2003. The International HapMap Project. Nature 426: 789–796 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719 [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40: 904–908 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11: 3610–3618 [DOI] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Monnat RJ Jr 2008. DNA polymerases and human disease. Nat Rev Genet 9: 594–604 [DOI] [PubMed] [Google Scholar]
- Meaburn EL, Schalkwyk LC, Mill J 2010. Allele-specific methylation in the human genome: Implications for genetic studies of complex disease. Epigenetics 5: 578–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD 2011. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146: 1029–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J 1996. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem 271: 12767–12774 [DOI] [PubMed] [Google Scholar]
- Ohno S 1988. Universal rule for coding sequence construction: TA/CG deficiency-TG/CT excess. Proc Natl Acad Sci 85: 9630–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Lucas-Lledo JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, Ogawa M, Matsushima K, Gu SG, Kasahara M, Ahsan B, et al. 2009. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science 323: 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J 2010. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet 86: 196–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling E, El Chartouni C, Rehli M 2009. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res 19: 2028–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, Schork NJ, Ecker JR 2011. Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiamarga DH, Miya M, Yamanoue Y, Azuma Y, Inoue JG, Ishiguro NB, Mabuchi K, Nishida M 2009. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biol Lett 5: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R, Deng J, Wang W, Zhang K 2010. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res 20: 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci 87: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Kai C, Kawai J, Carninci P, Hayashizaki Y, Semple CA 2006. Heterotachy in mammalian promoter evolution. PLoS Genet 2: e30 doi: 10.1371/journal.pgen.0020030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Watanabe H, Miura F, Soejima H, Uchiyama M, Iwasaka T, Mukai T, Sakaki Y, Ito T 2004. A comprehensive analysis of allelic methylation status of CpG islands on human chromosome 21q. Genome Res 14: 247–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Iwai S, O'Connor TR, Pfeifer GP 2003. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res 31: 5399–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. 2009. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet 5: e1000438 doi: 10.1371/journal.pgen.1000438 [DOI] [PMC free article] [PubMed] [Google Scholar]