Abstract
Eukaryotic protein-coding genes are transcribed by RNA polymerase II (RNAPII) through a cycle composed of three main phases: initiation, elongation, and termination. Recent studies using chromatin immunoprecipitation coupled to high-throughput sequencing suggest that the density of RNAPII molecules is higher at the 3′-end relative to the gene body. Here we show that this view is biased due to averaging density profiles for “metagene” analysis. Indeed, the majority of genes exhibit little, if any, detectable accumulation of polymerases during transcription termination. Compared with genes with no enrichment, genes that accumulate RNAPII at the 3′-end are shorter, more frequently contain the canonical polyadenylation [poly(A)] signal AATAAA and G-rich motifs in the downstream sequence element, and have higher levels of expression. In 1% to 4% of actively transcribing genes, the RNAPII enriched at the 3′-end is phosphorylated on Ser5, and we provide evidence suggesting that these genes have their promoter and terminator regions juxtaposed. We also found a striking correlation between RNAPII accumulation and nucleosome organization, suggesting that the presence of nucleosomes after the poly(A) site induces pausing of polymerases, leading to their accumulation. Yet we further observe that nucleosome occupancy at the 3′-end of genes is dynamic and correlates with RNAPII density. Taken together, our results provide novel insight to transcription termination, a fundamental process that remains one of the least understood stages of the transcription cycle.
Eukaryotic protein-coding genes are transcribed by a molecular engine powered by RNA polymerase II (RNAPII). Transcription is a repetitive, cyclic process composed of three main phases: initiation, elongation, and termination (Fuda et al. 2009). The transcription cycle starts with RNAPII gaining access to the promoter, unwinding DNA and initiating RNA synthesis. RNAPII must then get a stable grip on both the template DNA and the growing RNA chain and proceed elongating through the entire body of the gene. Finally, the RNA is released and RNAPII can reinitiate to start a new round of transcription. In mammals, termination by RNAPII can occur anywhere from a few base pairs to several kilobases downstream from the annotated 3′-end of the gene, which corresponds to the polyadenylation [poly(A)] site (Richard and Manley 2009). Termination is tightly coupled to cleavage and polyadenylation of the nascent transcript by the 3′-end processing machinery that recognizes specific sequence elements on the pre-mRNA termed poly(A) signals (Proudfoot 2011). Mammalian poly(A) signals include a conserved hexanucleotide, AAUAAA, located ∼10–30 nt upstream of the poly(A) site, and a less-conserved GU-rich region (or downstream sequence element [DSE]) located ∼30 nt downstream from the poly(A) site (Proudfoot 2011). According to the current model, at least two distinct mechanisms contribute to transcription termination (Richard and Manley 2009). One involves conformational changes of the RNAPII complex caused by dissociation of elongation factors and/or association of termination factors as the polymerase transcribes through the poly(A) site; the other results from a “torpedo” effect on RNAPII induced by rapid exonuclease degradation of the 5′-uncapped RNA produced after cleavage at the poly(A) site.
Transcription termination plays a vital role in cells because it controls gene expression and ensures genomic partitioning (Kuehner et al. 2011). However, termination remains one of the least understood stages of the transcription cycle. Recent studies using chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) revealed higher average polymerase density downstream from the polyadenylation [poly(A)] site compared with the transcribed region (Rahl et al. 2010). A 3′ enrichment of nascent transcripts was also observed using global run-on sequencing (GRO-seq) (Core et al. 2008). Here we show that this view is biased due to averaging density profiles for “metagene” analysis. Using genome-wide occupancy data from mouse and human cells, we found that most active genes have RNAPII evenly distributed before and after the poly(A) site, and only 7% to 14% contain a 3′ enrichment. Unexpectedly, we found that 1% to 4% of actively transcribing genes contain an enrichment of RNAPII phosphorylated on Ser5 at the 3′-end, and we provide evidence suggesting that these genes are in a looped conformation, bringing together their promoter and terminator regions. Looped genes identified by accumulation of Ser5P RNAPII at the 3′ end, or through chromatin interaction analysis by paired-end-tags sequencing (ChIA-PET) (Li et al. 2012), are characterized by having the highest expression levels. We further identified features that distinguish genes with and without enrichment of RNAPII at the 3′ end, and we developed a kinetic model that explains our observations. In summary, we show that accumulation of RNAPII at the 3′-end of genes is a dynamic process that depends on the transcription rate and correlates with nucleosome organization.
Results
Not all genes accumulate RNAPII at the 3′-end
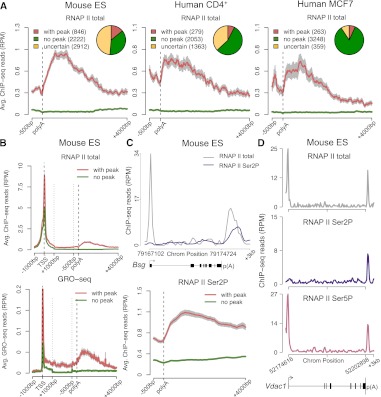
To determine whether accumulation of polymerases after the poly(A) site is a general feature of transcription termination, we systematically interrogated RNAPII ChIP-seq data for the presence of 3′ peaks. We analyzed data from mouse embryonic stem cells (Rahl et al. 2010), mouse CD4+/CD8+ T cells (Koch et al. 2011), human CD4+ T cells (Barski et al. 2007), and human MCF7 cells (Welboren et al. 2009) obtained with antibodies that recognize the largest subunit of RNAPII independently of the phosphorylation status of its C-terminal domain (CTD). Based on previous reports indicating that transcription can proceed significantly further past the poly(A) site (Gromak et al. 2006; Glover-Cutter et al. 2008), we searched for peaks in a region from the annotated gene 3′-end to 2.3 Kb downstream. Our analysis was restricted to actively transcribed genes identified by having either H3K4me3- and H3K79me2-modified chromatin at the 5′ region (Supplemental Fig. 1A,B) or medium to high expression levels according to RNA-seq or microarray data (Supplemental Table 1). To exclude the possibility that an enrichment of RNAPII at the 3′-end of a gene results from transcription of a neighboring gene, we discarded all genes for which there is another annotated gene in either strand within a region of 2.3 Kb flanking the poly(A) site (Supplemental Fig. 1C). We used three distinct peak calling tools (MACS) (Zhang et al. 2008), QuEST (Valouev et al. 2008), and SISSRs (Jothi et al. 2008), and we only considered peaks that were consistently detected by at least two of these methods (Supplemental Fig. 1D). A gene was considered devoid of a 3′ peak when no peak was detected by any of the three methods. The results show that the proportion of genes with 3′ peaks is higher in embryonic stem cells than in differentiated cells. We further detect that in differentiated cells, the majority of genes is devoid of 3′ peaks (Fig. 1A). Comparison of total RNAPII ChIP-seq data (Rahl et al. 2010) and GRO-seq data (Min et al. 2011) for mouse embryonic stem cells confirms that in genes with a 3′ peak of RNAPII occupancy, the density of nascent transcripts increases after the poly(A) site, whereas in genes devoid of peak the density of nascent transcripts is similar along the gene body and downstream from the poly(A) site (Fig. 1B). We conclude that accumulation of polymerases at the 3′-end of genes is not a global characteristic of transcription termination, but rather occurs in only a subset of all actively transcribed genes.
Figure 1.
Genome-wide identification of genes that accumulate RNAPII at the 3′ end. (A) Profiles of total RNAPII (Pol II) density on actively transcribed genes were analyzed using three peak calling methods (see Supplemental Fig. 1). The diagrams show numbers of genes with a consistent 3′ peak (detected by at least two methods), genes consistently devoid of 3′ peak (no peak detected by any of the three methods), and uncertain genes (genes with a 3′ peak detected by a single method) in murine ES cells, human CD4+ T, and MCF7 cells. Graphs depict average RNAPII profiles at the 3′-end of genes with (red) and without (green) peak. The 3′-end is unscaled. (B) Comparison of RNAPII ChIP-seq and Gro-seq average profiles across genes with (red) and without (green) peak in mouse ES cells. The 5′ end (TSS denotes transcription start site) and the 3′-end are unscaled. The remainder of the gene is rescaled to 2 kb. (C) Total RNAPII (gray) and Ser2P (purple) profile for a representative gene (Bsg) and average RNAPII Ser2P profiles at the 3′-end of genes with (red) and without (green) a 3′ peak of total RNAPII. Error bars (gray) represent standard error of the mean. (D) Total RNAPII (gray), Ser2P (purple), and Ser5P (pink) profile for a representative gene (Vdac1) with a peak of RNAPII Ser5P at the 3′-end. (RPM) Reads per million.
A subset of genes accumulate Ser5P RNAPII at the 3′-end
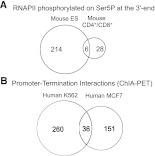
The CTD of the large subunit of RNAPII is differentially phosphorylated on serine residues throughout the transcription cycle (Buratowski 2009; Fuda et al. 2009). To determine the phosphorylation status of polymerases stalled at the 3′-end of genes, we analyzed ChIP-seq data obtained with antibodies specific for the CTD phosphorylated on either serine 5 (Ser5) or serine 2 (Ser2) residues (Schones et al. 2008; Rahl et al. 2010; Koch et al. 2011; Supplemental Table 2). As expected, the Ser5P signal was high in the promoter-proximal region of actively transcribed genes and dropped in the gene body, whereas the Ser2P signal was detected throughout the gene downstream from the promoter region (Fig. 1C,D). We further observed that genes containing a 3′ peak of total RNAPII are enriched for Ser2P in the region downstream from the poly(A) site, contrasting to genes with no 3′ peak (Fig. 1C), in agreement with the view that phosphorylation of Ser2 residues persists during termination (Buratowski 2009). Unexpectedly, we also found that some genes are enriched in RNAPII phosphorylated on Ser5P at the 3′-end (Fig. 1D; Supplemental Table 3). Increased levels of Ser5P RNAPII in the termination region were previously reported in two long yeast genes that exist in a looped conformation, bringing together their promoter and terminator regions (O'Sullivan et al. 2004). Gene looping, defined as the juxtaposition of both ends of a gene in a transcription-dependent manner has now been shown to occur in several yeast, viral, and mammalian genes (Hampsey et al. 2011). If gene loops form by physical interaction between components of the transcription initiation complex and the 3′-end processing/termination complexes (Hampsey et al. 2011), proteins associated with transcription initiation should occupy the 3′-end of looped genes. To address this view, we systematically analyzed ChIP-seq data obtained with antibodies to the TATA-box binding protein (TBP) and negative elongation factor subunit (WHSC2, also known as NELFA) (Rahl et al. 2010). TBP is a general transcription factor that binds to the promoter (Juven-Gershon et al. 2008), and WHSC2 is recruited to RNAPII concomitantly with initiation just downstream from the transcription start site (Lee et al. 2008). Consistent with our interpretation, the average signal for TBP and WHSC2 at the 3′ region is significantly higher in genes that contain a 3′ peak of Ser5P RNAPII compared with genes with a 3′ peak of total RNAPII, but devoid of Ser5P (Supplemental Fig. 2A,B). Another characteristic of gene loops is that their formation is transcription dependent (O'Sullivan et al. 2004). We therefore selected the genes that contain a 3′ peak of Ser5P RNAPII and compared the average profiles of polymerases phosphorylated on Ser5P in control mouse ES cells and cells treated with flavopiridol, an inhibitor of P-TEFb activity (Chao and Price 2001). P-TEFb is the main kinase responsible for Ser2 phosphorylation of the CTD and its activity is required for transition of RNAPII from transcription initiation to productive elongation (Fuda et al. 2009). Consistent with previous reports (Rahl et al. 2010), the levels of Ser5P at the 5′ end were largely unaffected within 60 min of flavopiridol treatment (Supplemental Fig. 2C). In contrast, the 3′ peak of Ser5P RNAPII was no longer detected in treated cells (Supplemental Fig. 2C). Taken together these results suggest that genes containing a 3′ peak of Ser5P RNAPII have their promoter and terminator regions juxtaposed. To determine the genome-wide incidence of gene looping, we searched for 3′ peaks in Ser5P RNAPII profiles from mouse ES cells (Rahl et al. 2010), mouse CD4+/CD8+ T cells (Koch et al. 2011), and human CD4+ T cells (Schones et al. 2008). The results show that 4%, 1%, and 3% of genes are enriched in Ser5P at the 3′ end, respectively (Supplemental Table 3). We then sought to validate these estimates with an independent approach to detect gene looping. For this, we analyzed recently reported maps of promoter-centered long-range chromatin interactions (Li et al. 2012). Li and colleagues used ChIA-PET (Fullwood et al. 2009) to identify genome-wide chromatin interactions associated with RNA polymerase II in human cell lines (Li et al. 2012). In the reported ChIA-PET data sets for K562 and MCF7 cells, we searched for intragenic interactions between promoter (from TSS to 2.3 Kb upstream) and termination region [from poly(A) site to 2.3 Kb downstream]. After removing all genes for which there is another annotated gene in either strand within the termination region, we found evidence for gene looping in 6% and 4% of active genes in K562 and MCF7 cells, respectively (Supplemental Table 4). Thus, both methods detect a similar proportion of active genes in a looped configuration. However, this may be an underestimate, because a significant number of genes are removed from our analysis due to the presence of another gene downstream from the poly(A) site. Furthermore, we cannot discard the possibility that many more genes form loops transiently and asynchronously in a cell population, but only the highly expressed genes are detected because these are actively transcribed by most cells in the population. Our results further show that, similarly to genes enriched in Ser5P at the 3′-end (Fig. 2A), the majority (>80%) of genes detected in a looped conformation by ChIA-PET are cell-type dependent (Fig. 2B), arguing against gene looping being an intrinsic property of a specific subset of genes.
Figure 2.
Cell-type specificity of gene looping. Venn diagrams depicting number of genes enriched in RNAPII phosphorylated on Ser5P at the 3′-end (A) and number of genes with intragenic interaction between promoter and termination region identified through ChIA-PET (B).
Genes with 3′ peaks of RNAPII are highly expressed
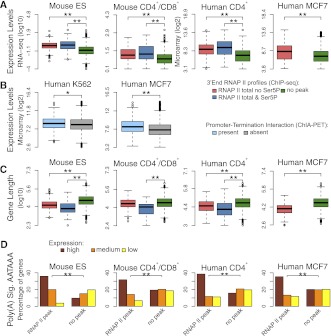
Having established that active genes can be classified into distinct groups based on RNAPII profiles at the 3′-end, we next sought to identify features characteristic of each class that could be used to predict the transcription termination pattern of any particular gene. For each gene class, we analyzed expression level, gene length, number and length of introns, and number and length of exons. We found that genes with 3′ peaks of RNAPII are significantly more expressed than genes with no peak, and genes with 3′ peaks of Ser5P RNAPII are the most highly expressed group (Fig. 3A). Similarly, genes that show chromatin interaction between the promoter and the termination region by ChIA-PET are more highly expressed than genes without such interaction (Fig. 3B). This is in agreement with the view that gene looping might be required specifically for high levels of gene expression (Ansari and Hampsey 2005). According to a proposed model, a loop forms following a pioneer round of transcription and promotes subsequent reinitiation events by “hand-off” of RNAPII from terminator to promoter (Hampsey et al. 2011).
Figure 3.
Genes with and without 3′ peaks of RNAPII have distinctive features. (A) Comparison of expression levels in genes that are devoid of any peak of total RNAPII at the 3′-end (green); genes with a 3′ peak of total RNAPII that is not phosphorylated on Ser5 (red); and genes with a 3′ peak of total RNAPII that is phosphorylated on Ser5 (blue). Gene expression levels were estimated according to RNA-seq (Mouse ES and CD4+/CD8+ cells) and microarray data (human CD4+ T and MCF7 cells). (B) Comparison of expression levels in genes with and without interactions between the promoter and termination region identified by ChIA-PET. Gene expression levels were estimated according to microarray data. (C) Comparison of gene length in the indicated categories. (*) P-value <0.05; (**) P-value <0.0005 by two-sided Mann-Whitney test. (D) Frequency of poly(A) signal AATAAA in genes that either contain a 3′ peak of total RNAPII or are devoid of peak. Genes were split into three equally sized groups according to expression level (high, medium, and low). (**) P-value <0.0005 by χ2 test. In boxplots, values inside each box correspond to the middle 50% of the data and the line within the box represents the median; the ends of the vertical lines at the top and bottom of each box indicate the maximum and minimum limits of the distribution; values behind the lines (circles) are suspected outliers.
We further observed that genes with 3′ peaks of RNAPII are significantly shorter than genes with no peak (Fig. 3C). Genes with 3′ peaks of RNAPII tend to have shorter introns, as well as shorter and fewer exons than genes with no peak (Supplemental Fig. 3A). Although a correlation between expression levels and gene length has been previously reported (Castillo-Davis et al. 2002), we detected significant differences between gene groups when considering only genes with equal expression levels or gene length (Supplemental Fig. 4). Indeed, the presence or absence of 3′ peaks correlates with both level of gene expression and gene length (Supplemental Fig. 5). We also found that the canonical poly(A) signal hexamer AATAAA (Hu et al. 2005) is more frequently present within 40 nt upstream of the poly(A) site in genes with 3′ peaks of RNAPII than in genes with no peak (Fig. 3D). Notably, the majority of the “no peak” genes that did contain a canonical poly(A) signal showed lower expression when compared with the genes with RNAPII peak. Inversely, genes with no 3′ peak contain more frequently variant forms of poly(A) signal hexamers that are presumably weaker than the canonical AATAAA (Supplemental Fig. 3B; Hu et al. 2005). In addition to the poly(A) signal hexamer, a downstream sequence element (DSE) present just past the poly(A) site plays an important role in 3′-end processing of pre-mRNA and transcription termination (Proudfoot 2011). We therefore interrogated a region of 60 nt past the poly(A) site for the presence of DSE-associated tetramers as previously described (Salisbury et al. 2006). The results show that G/TG-rich motifs (including GGGG, GGGA, GGAG, GAGG, AGGG, TCTG, CTGT, TGTC, and GTCT) are more frequently present in genes with 3′ peaks of RNAPII than in genes with no peak (Supplemental Fig. 6).
Functional analyses based on the association of Gene Ontology (GO) terms revealed that genes with and without 3′ peaks of RNAPII tend to be enriched for different functions (Supplemental Fig. 7A; Supplemental Table 5). We further detect a similar pattern of GO-term enrichment for genes with 3′ peaks and highly expressed genes (Supplemental Fig. 7B), in agreement with the observation that genes with 3′ peaks are highly expressed.
Formation of 3′ peaks depends on the transcription rate
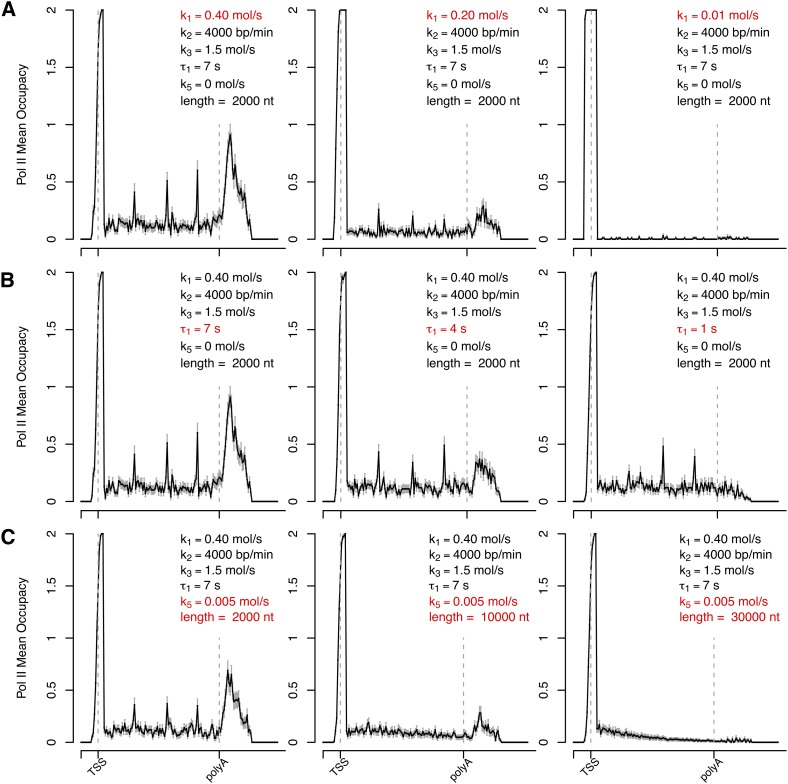
Taken together, the results described so far suggest that accumulation of RNAPII at the 3′-end of genes may be determined by the combinatorial contribution of expression level, gene length, and strength of the poly(A) signal. To rigorously test this hypothesis we developed a stochastic computational model that allows the observed profiles of RNAPII to be derived. We reasoned that the distribution of RNAPII along any gene can be modeled assuming that polymerases bind to random positions of the promoter region. In our model, an initiating polymerase is cleared from the promoter-proximal region at variable rate (k1) to start elongation, which proceeds at constant rate (k2). Because we systematically detect discrete intragenic accumulations of RNAPII, particularly in genes with a 3′ peak (Supplemental Fig. 8), we consider that polymerases slow down or pause at certain positions within the gene body. Therefore, we introduced in the model pause sites at a regular distance of 500 nt along the gene body, where polymerases are randomly selected to stop with a certain probability (k3). To model transcription termination, we considered two possible scenarios. One assumes a pause site downstream from the poly(A) site (Fig. 4); the other assumes that the speed of the polymerase becomes variable after the poly(A) site (Supplemental Fig. 9). The model further considers a processivity rate (k5) that reflects the number of nucleotides transcribed by an elongating polymerase before it falls off the DNA template. With the exception of elongation rate, for which we used a value estimated experimentally (Darzacq et al. 2007), all other values were arbitrarily chosen based on the sole criterion that the simulated average distribution of polymerases was similar to the experimental data. Although a peak forms at the 3′-end when the elongation rate of the polymerase is reduced after the poly(A) site (k4; Supplemental Fig. 9), the peak has an asymmetric shape that is not observed in experimental profiles (Supplemental Fig. 8). A more symmetric peak is detected when transcription termination is modeled as a pause with mean duration τ1 (Fig. 4), suggesting that the mechanism for 3′ enrichment is more likely a pause than a slowing down of the polymerase. However, the modeled 3′ peak in Figure 4 does not entirely reproduce the qualitative features observed in experimental profiles (which tend to have broader and less-defined peaks), and therefore we cannot conclude on the exact mechanism leading to RNAPII accumulation at the 3′-end of genes.
Figure 4.
Prediction of RNAPII distribution by stochastic computational modeling. The values of predicted RNAPII density along a hypothetical gene were estimated using the following parameters: promoter-proximal clearance rate (k1), elongation rate throughout the gene body (k2), pausing rate at intragenic sites positioned at a regular distance of 500 nt along the gene body (k3), pausing time downstream from the poly(A) site (τ1), and premature termination rate (k5). (Dashed lines) The positions of TSS and poly(A) site. (A–C) depict a parameter sensitivity analysis: the changes of predicted RNAPII average density profile with variation of either rate of productive transcription (A, reflective of k1); pausing time past the poly(A) site (B, reflective of τ1); and gene length with premature termination (C, reflective of k5).
To explore the specific contribution of each rate constant in 3′ peak formation, we changed one rate at a time, keeping the rest unaltered. The results show that decreasing the rate at which a polymerase starts to elongate is sufficient to abolish formation of peaks, both after the poly(A) site and along the gene body (Fig. 4A; Supplemental Fig. 9A). This is consistent with our observation that genes with a 3′ peak of RNAPII have higher expression levels than genes devoid of peak (Fig. 3A), and that genes with a 3′ peak also have more intragenic peaks of RNAPII (Supplemental Fig. 8). The model further shows that reducing the pausing time after the poly(A) site (τ1; Fig. 4B) or increasing the speed of the polymerase after the poly(A) site (k4; Supplemental Fig. 9B) reduces the 3′ peak. This could explain why the canonical poly(A) signal hexamer, which is presumably more effective in pausing or slowing down the polymerase, is more frequently present in genes with a 3′ peak of RNAPII (Fig. 3D). The model also shows that increasing gene length alone is not sufficient to change the 3′ peak (Supplemental Fig. 9C). An additional factor was introduced in order to fit the observation that genes with 3′ peaks of RNAPII are shorter than genes with no peak (Fig. 3B). We changed k5, assuming that not all polymerases that start to elongate reach the end of the gene. Increasing k5 from zero to 0.005 does not alter the profile of polymerase density along a 2-kb gene, but is sufficient to abrogate the 3′ peak in a 30-kb gene (Fig. 4C; Supplemental Fig. 9D).
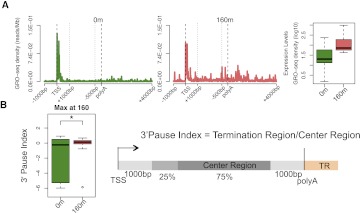
These quantitative analyses suggest that accumulation of polymerases downstream from the poly(A) site is not an intrinsic characteristic of transcription termination in certain genes, but rather a dynamic process that depends on kinetic competition between transcription rates during the different stages of the transcription cycle. If so, a given gene should undergo transitions between presence and absence of a 3′ peak. We validated this prediction by analyzing GRO-seq data from MCF7 cells treated for short periods with estrogen (Hah et al. 2011). We selected genes that show maximal up-regulation at 160 min after estrogen treatment (Fig. 5A) and we calculated the 3′ pausing index (3′PI) defined as the ratio of GRO-seq signal at the 3′-end to the average signal over gene bodies (Fig. 5B). The calculated 3′PI is typically a negative number in untreated cells, indicating no enrichment at the 3′ end. After estrogen treatment the 3′PI is significantly different and becomes positive, indicating accumulation at the 3′-end (Fig. 5B). This provides direct evidence for dynamic transitions in the relative enrichment of RNAPII at the 3′-end of genes.
Figure 5.
Accumulation of RNAPII at the 3′-end of genes is a dynamic process. (A) Average GRO-seq profiles for 31 estrogen-responsive genes that show maximal up-regulation at 160 min after treatment (Hah et al. 2011). To show the average profiles across genes, a “metagene” profile was plotted. The 5′ end [1 kp upstream of the transcription start site (TSS) to 1 kb downstream] and the 3′-end [500 bp upstream of the poly(A) site to 4 kb downstream] are unscaled. The remainder of the gene is rescaled to 2 kb. The boxplot depicts the expression levels. (B) The 3′ PI was calculated as shown in the schematic using unscaled GRO-seq tag counts from individual genes in each group. The 3′ PI values are significantly different between untreated (green) and estrogen-treated cells (red) as depicted by the boxplots.
Genes with and without 3′ peaks of RNAPII differ in nucleosome organization
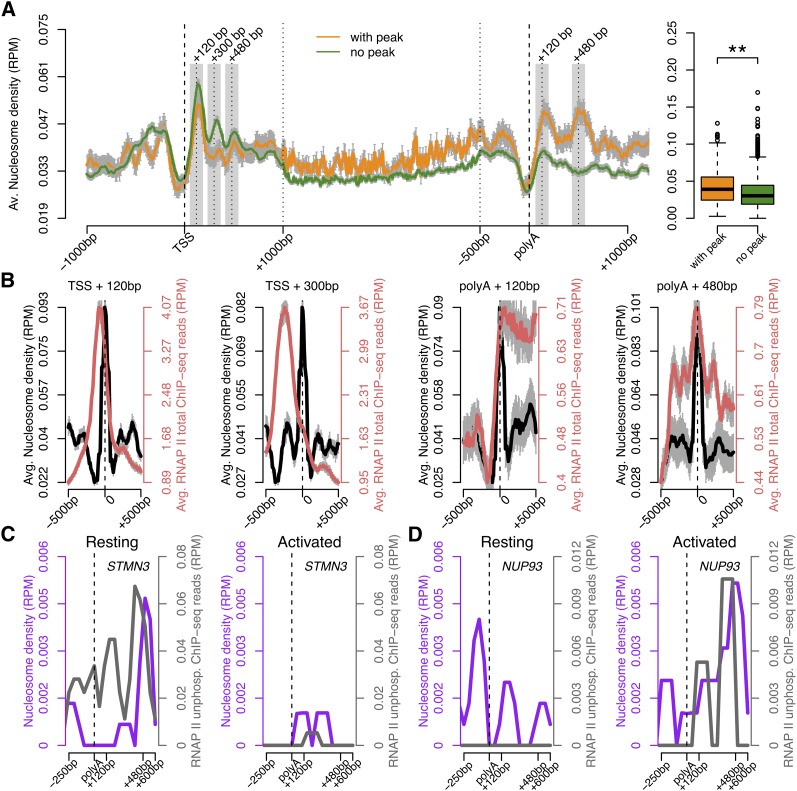
In vitro, nucleosomes induce pausing of RNAPII (Kireeva et al. 2005; Hodges et al. 2009) and in vivo RNAPII density peaks before nucleosomes (Churchman and Weissman 2011). To determine whether nucleosomes interfere with RNAPII accumulation at the 3′ end, we compared nucleosome organization in genes with and without 3′ peaks using genome-wide data from resting and activated human CD4+ T cells (Schones et al. 2008). The results show that nucleosome occupancy in the region downstream from the poly(A) site is significantly higher in genes with 3′ peaks of RNAPII than in genes devoid of peak (Fig. 6A). Furthermore, we found that in genes with 3′ peaks of RNAPII nucleosomes align at regular intervals from the poly(A) site, whereas nucleosome alignment is much less obvious in genes devoid of RNAPII 3′ peaks (Fig. 6A). In contrast, in the region surrounding the TSS, an array of highly positioned nucleosomes is equally observed in genes with and without a 3′ RNAPII peak (Fig. 6A). We then plotted the average profiles of RNAPII density relative to nucleosome position (Fig. 6B). At the 5′ end, the promoter-proximal peak of RNAPII correlates with the position of the first (TSS + 120), but not the second (TSS + 300) nucleosome. In contrast, in the 3′ region the two well-positioned nucleosomes [poly(A) + 120 bp and poly(A) +480 bp] associate with a high density of RNAPII (Fig. 6B). We next compared RNAPII and nucleosome occupancy downstream from the poly(A) site of individual genes that are either down- (Fig. 6C) or up-regulated (Fig. 6D) upon activation of CD4+ T cells. We observe a positive correlation between nucleosome occupancy and accumulation of RNAPII at the 3′ end. Compared with resting cells, the STMN3 gene is less expressed in activated cells; at the 3′-end of this gene, nucleosome occupancy decreases and enrichment of RNAPII density is no longer detected (Fig. 6C). Conversely, expression of the NUP93 gene is higher in activated cells; at the 3′-end of this gene we detect higher nucleosome occupancy and accumulation of RNAPII (Fig. 6D). Thus, the dynamic transcription-dependent transitions in RNAPII density profiles at the 3′-end of genes correlate with dynamic nucleosome reorganization downstream from the poly(A) site.
Figure 6.
Nucleosome organization correlates with RNAPII density at the 3′-end. (A) Average nucleosome occupancy across genes with (orange) and without (green) a peak of total RNAPII at the 3′-end. The y-axis shows the normalized number of sequence tags of DNA at each position. The 5′- and 3′-ends are unscaled. The remainder of the gene is rescaled to 2 kb. The boxplot represents the average nucleosome occupancy for each gene in the termination region [poly(A) site to 500 bp downstream]. (**) P-value <0.00005 by two-sided Mann-Whitney test. (B) Average nucleosome distribution (black) and total RNAPII density (red) around the position of maximum nucleosome occupancy (0) at the indicated regions. Error bars (gray), SEM. (C,D) Nucleosome (purple) and RNAPII (gray) occupancy along two genes, the expression of which is either down- (STMN3, C) or up-regulated (NUP93, D) in human CD4+ T cells activated by TCR signaling. (RPM) Reads per million.
Discussion
Our genome-wide analysis of RNAPII distribution in mouse and human cells reveals that the majority of genes exhibit little, if any, detectable accumulation of polymerases during transcription termination. The proportion of genes with a relative enrichment of RNAPII density in the region downstream from the poly(A) site ranges between 14% in embryonic stem cells to 7%–8% in differentiated cells. To quantify the accumulation of polymerases at the 3′-end we determined the 3′ pausing index defined as the ratio of RNAPII density in the region after the poly(A) site to the average density over gene bodies, and we show that it is influenced by transcriptional activity. The 3′ pausing index differs in the same set of genes, depending on their expression level, arguing against DNA sequence being the major determinant of polymerase accumulation. However, for genes with similar expression levels, the presence of the canonical poly(A) signal appears to promote polymerase accumulation, whereas the presence of variant hexamers is more frequently associated with the lack of accumulation. This suggests that the DNA sequence can also contribute to the process of RNAPII enrichment at the 3′ end. Our mathematical simulations indicate that polymerases must either pause or reduce their speed after the poly(A) site in order to accumulate; it is therefore conceivable that DNA elements such as the poly(A) signal may act as intrinsic modulators of speed as the enzyme moves toward the end of gene bodies. Another feature that distinguishes genes with and without RNAPII accumulation at the 3′-end is gene length: Genes with a 3′ peak of RNAPII are shorter than genes with no peak. The observed differences between long and short genes could be explained by RNAPII processivity (Mason and Struhl 2005). Indeed, our modeling results suggest that if all elongating polymerases reach the end of a gene, then accumulation of RNAPII past the poly(A) site occurs; however, if a few polymerases dissociate from the template before reaching the end, this is sufficient to abrogate accumulation past the poly(A) site.
We also find a striking correlation between RNAPII accumulation and nucleosome organization at the region after the poly(A) site. Most likely, the presence of nucleosomes after the poly(A) site induces pausing of polymerases, leading to their accumulation. While multiple chromatin remodeling factors and histone chaperones help the polymerase to resume elongation each time it encounters a nucleosome along the gene body (Li et al. 2007), there is evidence suggesting that these factors dissociate from the transcription complex near the poly(A) site (Mayer et al. 2010). This may increase the time a polymerase is paused behind nucleosomes located at the 3′-end of genes, leading to the observed enrichment of RNAPII density. However, our results further show that nucleosome occupancy at the 3′-end of genes is dynamic and correlates with RNAPII density. These observations should be useful for future investigations aimed at determining whether RNAPII stalled in the termination region of highly expressed genes drives a local pattern of nucleosome spacing and thereby contributes to gene regulation.
Methods
ChIP-seq, RNA-seq, GRO-seq, and ChIA-PET data sets and preprocessing
High-throughput sequencing data was obtained from previously published studies for mouse embryonic stem cells (GSE8024, GSE20485, GSE22303, GSE20851, and GSE27037); mouse CD4+/CD8+ T cells (GSE29362); human CD4+ T cells (GSE10437, SRA000206, SRA000287, and SRA000234); human MCF7 cells (GSE14664, GSE27463, GSE33664, and GSE18912) and human K562 cells (GSE33664 and GSE14083) (detailed information in Supplemental Tables 1, 2). High-throughput sequencing reads were aligned to the reference human (hg19) or mouse (mm9) genomes using BWA (Li and Durbin 2009). Reads from samples with multiple sequencing lanes were merged. Reads and alignment quality was assessed using Rsamtools (http://www.bioconductor.org/packages/2.6/bioc/html/Rsamtools.html) and ShortRead (Morgan et al. 2009) R packages. We filtered out reads with bad-quality scores, reads not uniquely mapped and PCR duplicates (identical coordinates). SAMtools (Li et al. 2009) and BEDtools (Quinlan and Hall 2010) were used for filtering steps and file formats conversion. Processing of ChIP-seq data and nucleosome data involved read extension calculated and performed using the Pyicos tool (Althammer et al. 2011). In addition, mouse embryonic stem cells ChIP-seq data was normalized and background subtracted using whole-cell extract in matched cell samples (Mikkelsen et al. 2007) through the Pyicos tool. This additional step could not be applied to human CD4+ T cells data, since no input sample was available. All data sets were analyzed as described above, with the exception of data for mouse CD4+/CD8+ T cells, GRO-seq data of human MFC7 cells, and ChIA-PET data for human K562 and MCF7 cells for which preprocessed data was obtained from original works (Hah et al. 2011; Koch et al. 2011; Li et al. 2012).
Microarray data analysis
Microarray data was obtained from previously published studies for human CD4+ T, MCF7, and K562 cells (detailed information in Supplemental Table 1). Data analysis was done using R and several packages available from CRAN (The R Development Core Team 2011) and Bioconductor (Gentleman et al. 2004). The raw data (CEL files) were normalized and summarized with the Robust MultiArray Average method from the affy package (Gautier et al. 2004). Absence or presence of expression was statistically determined by using the mas5calls function from affy package. Differentially expressed genes for human CD4+ T cell activation were selected using linear models and empirical Bayes methods (Smyth 2004) as implemented in the limma package (Smyth 2005) verifying the P-values corresponding to moderated F-statistics and selecting as differentially expressed genes those that had adjusted P-values adjusted using the Benjamini and Hochberg (1995) method lower than 0.05.
Gene classification
Actively transcribed genes in mouse embryonic stem cells and human CD4+ T cells were identified using ChIP-seq data of chromatin modifications characteristic of transcription initiation and elongation (H3K4m3 and H3K79me2). For mouse CD4+/CD8+ T cells and human MCF7 cells, we selected genes with high and medium levels (5664 and 7895, respectively) of expression based on RNA-seq (Koch et al. 2011) and microarray data (Heintzman et al. 2009; Hou et al. 2011), respectively. We filtered out all genes for which there was another annotated gene in either strand within a region of 2.3 Kb flanking the poly(A) site according to the UCSC KnownGene genes annotations for hg19 and mm9 genome versions (Rhead et al. 2010). All transcribed regions that are annotated (including ncRNA genes) were considered. Our analysis assessed a total of 5980 genes for mouse ES cells; 3195 genes for mouse CD4+/CD8+ T cells; 3695 genes for human CD4+ T cells; 3870 genes for human MCF7 cells, and 5109 for human K562 cells. Three different peak calling tools were used to detect enrichment of RNAPII density at the 3′-end of genes: MACS (Zhang et al. 2008), QuEST (Valouev et al. 2008), and SISSRs (Jothi et al. 2008). The analyses were performed using the default tools parameters for all samples, with the exception of human CD4+ T cells data, where no input sample could be used as control; thus, FDR was not estimated (MACS, QuEST) or was estimated from a random background model based on Poisson probabilities (SISSRs). Intragenic interactions between promoter (from TSS to 2.3 Kb upstream) and termination region [from poly(A) site to 2.3 Kb downstream] were identified from ChIA-PET data (Li et al. 2012). Only interactions identified in both saturated replicates for K562 and MCF7 cells were selected.
Metagene profiles
To show the average profiles across genes with and without a 3′ peak, a “metagene” profile was plotted for each group. Genes were aligned at the first and last nucleotides of the annotated transcripts and sequencing tags were scaled as follows. The 5′ end (1 kp upstream of the transcription start site [TSS] to 1 kb downstream) and the 3′-end [500 bp upstream of the poly(A) site to 4 kb downstream] were unscaled and averaged in a 10-bp window. The remainder of the gene was scaled to 200 equally sized bins so that all genes appear to have the same length (2 kb). Individual profiles were produced using a 100-bp window. All profiles were plotted on a normalized read per million (RPM) basis. Statistical significance of difference between gene groups was assessed using a two-sided Mann-Whitney test (n indicated in the figure legends).
Gene feature analysis
Gene features (gene length, exon number, intron and exon length) and DNA sequences were obtained from UCSC for hg19 and mm9 genome versions (Rhead et al. 2010). Statistical significance of differences between groups was assessed using two-sided Mann-Whitney test for an n of 3068 (mouse ES cells); 3195 (mouse CD4+/CD8+ T cells); 2333 (human CD4+ T cells); and 3511 (human MCF7 cells). Poly(A) signal frequency was evaluated for the canonical AATAAA hexamer and its variants and for tetramers characteristic of DSE (Salisbury et al. 2006). Statistical significance of differences between poly(A) signal frequencies was assessed using χ2 test. Boxplots were produced using the boxplot() R function with the default arguments. The outliers were determined as the most extreme data points that are more than 1.5 times the interquartile range from the box.
Functional analysis
We used DAVID (Huang da et al. 2009) to test enrichment of biological processes in each gene group using all genes as the background. Analysis was restricted to GO fat subset (which filters broadest terms so that they do not overshadow the more specific terms). GO terms with Benjamini corrected P-value <0.05 for Fisher's exact test were selected. Networks of GO terms were produced using Enrichment Map (Merico et al. 2010) Cytoscape (Smoot et al. 2011) plugin using default values.
Simulation of RNAPII density profiles
We simulated the average distribution of polymerases along a hypothetical gene, assuming that all events in the transcription cycle are Poisson processes. The model assumes that an initiating polymerase is cleared from the promoter-proximal region to start elongation at a variable rate (k1), and that polymerases elongate (k2) at a constant speed of 4 kb/min (Darzacq et al. 2007). Intragenic pause sites were positioned at a regular distance of 500 nt along the gene body. At a pause site a decision is made as to whether a polymerase continues elongating or stops for a certain amount of time. Polymerases are randomly selected to stop with a specific rate (k3), a parameter that controls the probability of pausing. The duration of the pause is a random variable with an exponential distribution with mean τ2. After this time the polymerase resumes the elongation process. Polymerases that elongate past the poly(A) site are eventually released within a region of 1000 nt. The polymerase pauses inside this region at a random selected position with uniform distribution. The duration of the pause is a random variable with an exponential distribution with mean τ1. After this time, the polymerase is removed from the simulation. As an alternative to the pause at the 3′ end, we considered that the elongation speed of the polymerase becomes variable as it transcribes past the poly(A) site (k4). Polymerases are released from the DNA after a constant time period once they reach the poly(A) site. However, some elongating polymerases may not reach the poly(A) site, depending on their processivity rate (k5), which is defined as the number of nucleotides transcribed by an elongating polymerase before it falls off the DNA template. Due to the nature of the transcription process, a polymerase can never outpace a leading polymerase. Also, taking into account spatial constraints imposed by the size of polymerases, we divided the gene into segments and set a limit on the number of polymerases that can simultaneously occupy the same segment.
The values for k1, k3, k4, and k5 were arbitrarily chosen based on the sole criterion that the simulated average distribution of polymerases was similar to the experimental data. Supplemental Table 6 shows the parameters chosen for the simulations. Simulations run for a period of time sufficient for a polymerase to start and reach the end of the gene 10 times (i.e., 10 × GeneLength/ Elongation rate), allowing the system to reach a stationary state. The actual position of all the polymerases in the gene at that moment is recorded. The process was repeated 100 times, and the average occupancy of polymerases along the gene was computed.
Calculation of the 3′-end pause index
To quantify accumulation of RNAPII at the 3′ end, we have defined the 3′ pause index (3′PI) that compares the average GRO-seq signal at the 3′-end [1 kb downstream from the poly(A) site] to the average signal over gene bodies. For calculation of gene body signal, the first and last 1 kb of each gene was excluded to eliminate the effect of proximity to the 5′ and 3′ peaks. The remainder of the coding region was further subdivided into 5′ and 3′ segments (25% and 75%, respectively), as previously described (Larschan et al. 2011).
Acknowledgments
We thank Dr. Nick Proudfoot (University of Oxford) and Dr. Laszlo Tora (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg) for stimulating discussions and critical comments. We are also grateful to Dr. Pierre Ferrier and Dr. Jean-Christophe Andrau (Centre d'Immunologie de Marseille-Luminy) for help on the bioinformatics preprocessing of mouse CD4+ /CD8+ T cell data. This work was supported by grants from Fundação para a Ciência e Tecnologia, Portugal (PTDC-SAU-GMG-113440-2009 to M.C.-F. and PTDC-BIA-BCM-111451-2009 to S.F.deA.). S.F.deA. and A.R.G. are supported by fellowships from Fundação para a Ciência e Tecnologia (SFRH-BPD-34679-2007 and SFRH-BPD-62911-2009).
Author contributions: A.R.G, S.F.deA., and M.C.-F. conceived the project. A.R.G designed and carried out the bioinformatic and computacional analysis. J.B. conceived the framework of the simulation studies. The manuscript was prepared by A.R.G and M.C.-F., with the participation of S.F.deA. All authors reviewed the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138057.112.
References
- Althammer S, González-Vallinas J, Ballaré C, Beato M, Eyras E 2011. Pyicos: A versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 27: 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Hampsey M 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19: 2969–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA 2002. Selection for short introns in highly expressed genes. Nat Genet 31: 415–418 [DOI] [PubMed] [Google Scholar]
- Chao S, Price D 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 [DOI] [PubMed] [Google Scholar]
- Churchman L, Weissman J 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L, Waterfall J, Lis J 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N, Ardehali M, Lis J 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad B, Irizarry R 2004. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley D 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot N 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26: 3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko C, Core L, Waterfall J, Siepel A, Lis J, Kraus W 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S 2011. Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv Enzyme Regul 51: 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C 2009. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325: 626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Huang F, Carboni JM, Flatten K, Asmann YW, Ten Eyck C, Nakanishi T, Tibodeau JD, Ross DD, Gottardis MM, et al. 2011. Drug efflux by breast cancer resistance protein is a mechanism of resistance to the benzimidazole insulin-like growth factor receptor/insulin receptor inhibitor, BMS-536924. Mol Cancer Ther 10: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lutz C, Wilusz J, Tian B 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K 2008. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36: 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu J, Theisen J, Kadonaga J 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva M, Hancock B, Cremona G, Walter W, Studitsky V, Kashlev M 2005. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 18: 97–108 [DOI] [PubMed] [Google Scholar]
- Koch F, Fenouil R, Gut M, Cauchy P, Albert T, Zacarias-Cabeza J, Spicuglia S, de la Chapelle A, Heidemann M, Hintermair C, et al. 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18: 956–963 [DOI] [PubMed] [Google Scholar]
- Kuehner J, Pearson E, Moore C 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Bishop E, Kharchenko P, Core L, Lis J, Park P, Kuroda M 2011. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471: 115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Li X, Hechmer A, Eisen M, Biggin M, Venters B, Jiang C, Li J, Pugh B, Gilmour D 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol Cell Biol 28: 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P, Struhl K 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader G 2010. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 5: e13984 doi: 10.1371/journal.pone.0013984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T, Ku M, Jaffe D, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T, Koche R, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min I, Waterfall J, Core L, Munroe R, Schimenti J, Lis J 2011. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 25: 742–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R 2009. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25: 2607–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan J, Tan-Wong S, Morillon A, Lee B, Coles J, Mellor J, Proudfoot N 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A, Hall I 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Development Core Team. 2011. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rahl P, Lin C, Seila A, Flynn R, McCuine S, Burge C, Sharp P, Young R 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley J 2009. Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J, Hutchison K, Graber J 2006. A multispecies comparison of the metazoan 3′-processing downstream elements and the CstF-64 RNA recognition motif. BMC Genomics 7: 55 doi: 10.1186/1471-2164-7-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D, Cui K, Cuddapah S, Roh T, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T 2011. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: Linear models for microarray data. In Bioinformatics and computational biology solutions using R and bioconductor (ed. R Gentleman et al.), pp. 397–420. Springer, New York. [Google Scholar]
- Valouev A, Johnson D, Sundquist A, Medina C, Anton E, Batzoglou S, Myers R, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren W, van Driel M, Janssen-Megens E, van Heeringen S, Sweep F, Span P, Stunnenberg H 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer C, Eeckhoute J, Johnson D, Bernstein B, Nussbaum C, Myers R, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]