Abstract
Adenosine deaminases that act on RNA (ADARs) are RNA editing enzymes that convert adenosine to inosine in double-stranded RNA (dsRNA). To evaluate effects of ADARs on small RNAs that derive from dsRNA precursors, we performed deep-sequencing, comparing small RNAs from wild-type and ADAR mutant Caenorhabditis elegans. While editing in small RNAs was rare, at least 40% of microRNAs had altered levels in at least one ADAR mutant strain, and miRNAs with significantly altered levels had mRNA targets with correspondingly affected levels. About 40% of siRNAs derived from endogenous genes (endo-siRNAs) also had altered levels in at least one mutant strain, including 63% of Dicer-dependent endo-siRNAs. The 26G class of endo-siRNAs was significantly affected by ADARs, and many altered 26G loci had intronic reads and histone modifications associated with transcriptional silencing. Our data indicate that ADARs, through both direct and indirect mechanisms, are important for maintaining wild-type levels of many small RNAs in C. elegans.
Adenosine deaminases that act on RNA (ADARs) are RNA editing enzymes that convert adenosine to inosine in double-stranded RNA (dsRNA) (Bass 2002; Zinshteyn and Nishikura 2009). Inosine prefers to pair with cytidine, so by changing AU base pairs to IU mismatches, ADARs alter base-pairing specificity, in some cases altering codon meaning. All ADARs have a conserved catalytic domain and at least one dsRNA binding motif.
ADARs exist in all studied animals, with two genes in Caenorhabditis elegans: adr-1 and adr-2 (Tonkin et al. 2002). Existing data indicate ADR-2 is an active editing enzyme, while essential residues in the catalytic domain are missing from ADR-1; C. elegans strains deficient for ADR-2 lack editing at known endogenous sites, and extracts of these animals lack deaminase activity (Tonkin et al. 2002). In contrast, in ADR-1 deficient strains, some endogenous sites show wild-type editing levels while others show decreased or increased levels, and extracts from these animals have detectable deaminase activity. Thus, through unclear mechanisms, ADR-2 edits some sites without contributions from ADR-1, but ADR-1 enhances or inhibits editing at other sites.
ADARs are challenging to study in vertebrates, as null mutations are lethal (Higuchi et al. 2000; Wang et al. 2000). However, ADAR mutant C. elegans are viable but show chemotaxis defects, a reduced lifespan, and cosuppression of transgenes in somatic cells (Knight and Bass 2002; Tonkin et al. 2002; Sebastiani et al. 2009). While many ADAR editing sites have been identified in various animals, the only site clearly correlated with a phenotype is in mouse glutamate receptor subunit B mRNA, where codon editing results in a Q to R amino acid change required for viability (Higuchi et al. 2000). Other phenotypes, in a variety of species, correlate with the absence of ADARs, but the editing events or molecular mechanisms responsible for these phenotypes are unknown.
All three phenotypes of C. elegans ADAR mutants are rescued by additional mutations in RNA interference factors (Knight and Bass 2002; Tonkin et al. 2002; Sebastiani et al. 2009). This suggests ADARs intersect with dsRNA-mediated silencing pathways, such as those involving microRNAs (miRNAs) or small interfering RNAs that derive from endogenous genes (endo-siRNAs) (Nishikura 2006; Ohman 2007; Hundley and Bass 2010).
miRNAs derive from dsRNA precursors that ADARs could bind and edit (Kim et al. 2009). For example, the primary miRNA transcript (pri-miRNA) is processed by Drosha in the nucleus, where ADARs typically localize, to yield a 50–70 nt stem–loop called a precursor miRNA (pre-miRNA). Pre-miRNAs are exported to the cytoplasm and processed by Dicer into 21–23 nt dsRNAs. While endo-siRNA biogenesis in C. elegans is not well characterized, some endo-siRNAs are Dicer-dependent, indicating that they arise from dsRNA precursors (Welker et al. 2010). A third class of small RNAs called piwiRNAs (piRNAs), whose biogenesis is also unclear, are not thought to involve a dsRNA intermediate and thus should not be affected by ADARs.
Published studies point to multiple ways ADARs can affect small RNA biogenesis. Pre-editing of certain mammalian miRNA precursors alters in vitro processing by recombinant Drosha or Dicer, although it is unknown if ADARs affect miRNA maturation in vivo (Kawahara et al. 2007a, 2008). ADARs also antagonize siRNA production from dsRNA derived from a transgene in both C. elegans and D. melanogaster (Knight and Bass 2002; Heale et al. 2009). In the latter study, ADARs' editing activity was not required for its effects on siRNA production, implying that its dsRNA-binding activity is sufficient to prevent siRNA processing. A recent study in C. elegans also found that ADARs antagonized production of siRNAs derived from transposons and inverted repeats (Wu et al. 2011). However, it remains unknown if ADAR editing and/or dsRNA binding affect siRNAs from protein coding genes. Finally, there is one example of ADAR editing that redirects a miRNA to bind and repress new mRNAs, and it is unclear if this is a common occurrence (Kawahara et al. 2007b).
At present, a global analysis to determine how ADARs affect miRNAs and other small RNAs has not been performed in any organism. Toward this end, we performed deep-sequencing of small RNAs from wild-type and ADAR mutant C. elegans. While editing within small RNAs was rare, many small RNAs had altered levels in the absence of ADARs. Our studies indicate that ADARs regulate small RNA processing and are essential for maintenance of wild-type levels of small RNAs in C. elegans.
Results
cDNA libraries of small RNAs (18–30 nt) from wild-type (WT), adr-1(tm668), adr-2(ok735), and adr-1;adr-2 double mutant strains—homozygous alleles hereafter designated (−/−)—were prepared from mixed-stage C. elegans and sequenced on an Illumina instrument, using a method that only captures RNAs with a 5′ monophosphate. This procedure enriches for small RNAs that derive from dsRNA precursors, such as miRNAs and primary (1°) endo-siRNAs. However, it excludes the abundant triphosphorylated secondary (2°) endo-siRNAs found in C. elegans that are not thought to derive from dsRNA (Sijen et al. 2007).
ADARs affect the levels of many mature miRNAs
We first compared the normalized number of sequencing reads aligning to individual miRNA loci. All three mutant strains had many miRNA loci with increased and/or decreased reads compared to WT. Overall, the program, Defined Region Scan Sequences (Nix et al. 2008), determined that 110 of 120 (92%) known miRNA loci annotated in a previous study (Warf et al. 2011) had a statistically significant change in the number of reads in at least one of three mutant strains compared to WT (P-value ≤ 0.05) (Supplemental Table S1).
We further analyzed affected miRNA loci using more stringent criteria and focused on 49 loci, comprising those with reads affected ≥2-fold in at least one strain (25 loci) and those with reads affected ≥1.2-fold in multiple strains (24 loci) (Supplemental Table S1). Of these 49 loci, 44 loci (90%) had increased reads in at least one mutant strain (Fig. 1A) and only 12 loci (24%) had decreased reads in at least one mutant strain (Fig. 1B; Supplemental Fig. S1A,B). Loci were binned according to the magnitude of their differences in read numbers (Fig. 1A,B), revealing that most loci had <2.0-fold difference. However, some loci had larger differences, which were mostly in adr-1(−/−) and adr-1(−/−);adr-2(−/−) strains (Fig. 1A,B). To validate changes in miRNA levels determined by sequencing, Northern blots were probed for six miRNAs (Fig. 1C). Changes in miRNA levels determined by Northern analyses correlated well with sequencing data.
Figure 1.
ADARs affect miRNA and mRNA target levels, but few miRNAs are edited. (A,B) Number of loci with increased (A) or decreased (B) reads in ADAR mutants compared to WT were plotted on the y-axis. miRNA loci were binned (x-axis) according to their fold-change in a mutant strain compared to WT. All miRNA loci altered ≥2-fold were affected in at least one mutant strain; loci altered ≥1.2-fold but <2-fold were affected in at least two mutant strains. (C) Representative Northern blot of mirVana enriched RNA (2 μg) from indicated strains, probed for miRNAs, with U6 as a loading control. Fold-change determined by sequencing (illum) and Northern blot (north) were tabulated, comparing each mutant to WT. Blanks indicate sequencing did not predict a significant difference. (Error) Standard deviation (STD), adr-1(−/−);adr-2(−/−) abbreviated adr-1;2. (D) Fold-change of pri-miRNA in ADAR mutants compared to WT, as determined by qRT-PCR, was compared to change in miRNA. Levels of miR-800 and miR-1830 were determined by Northern blot, miR-253 by sequencing; the latter did not show a significant change in single mutants. (E) Fold-change of miR-800 (see Fig. 1C, Northern blot) compared to fold-change of predicted mRNA targets, as determined by microarray analyses (adr-1[−/−];adr-2[−/−] vs. WT worms) (Supplemental Table S2). (Error bars) Standard error of the mean (SEM) for mRNAs, STD for miR-800. (F) qRT-PCR validation of miR-800 mRNA target levels in adr-1(−/−);adr-2(−/−) worms compared to WT. (Error bars) SEM for microarray, STD for qRT-PCR. (G) Structure of pri-mir-800, with miRNA sequence underlined and bold and miRNA* sequence italicized. Percent editing at indicated adenosine (double arrowhead) is tabulated for each strain. (Single arrowheads) Drosha and Dicer cleavage sites.
We hypothesized that ADARs altered mature miRNA levels by affecting processing of pri-miRNAs, which are processed in the nucleus where ADARs typically localize. Thus, we used quantitative RT-PCR (qRT-PCR) to assay levels of three pri-miRNAs that give rise to miRNAs whose levels were strongly affected in ADAR mutants (Fig. 1D). miR-800 and miR-1830 levels were significantly increased in adr-1(−/−) and adr-1(−/−);adr-2(−/−) strains compared to WT and mildly increased in adr-2(−/−) strains (Fig. 1C,D). Correspondingly, we found substantially decreased levels of pri-mir-800 and pri-mir-1830 in adr-1(−/−) and adr-1(−/−);adr-2(−/−) worms compared to WT, and a moderate decrease in adr-2(−/−) worms (Fig. 1D).
While few miRNAs had decreased levels in ADAR mutants compared to WT, miR-253 had one of the most decreased levels, which was in the double mutant (Supplemental Table S1). Correlating with this decrease, pri-mir-253 levels were increased in adr-1(−/−);adr-2(−/−) worms compared to WT (Fig. 1D). We did not observe bands on Northern blots corresponding to a pre-miRNA for any tested miRNA. This was not surprising, as pre-miRNAs are short-lived and of low abundance.
Thus, ADAR mutant strains most often had decreased pri-miRNA levels and increased miRNA levels, suggesting that ADARs typically inhibit pri-miRNA processing in WT C. elegans. While Drosha processing of pri-miRNAs is inhibited by editing in vitro (Kawahara et al. 2008), we observed that more miRNAs were affected by ADR-1, which has binding, but not editing, activity. Our data are more consistent with a model whereby ADAR binding sequesters pri-miRNAs from Drosha.
A few miRNAs had decreased levels in ADAR mutant strains, implying ADARs sometimes increase processing efficiency. ADR-2 editing might alter pri-miRNA structure so that it is processed more efficiently, as observed for some human pri-miRNAs in vitro (Kawahara et al. 2008); however, we found no evidence of editing within pri-mir-253.
We note that ADARs could indirectly alter miRNA levels by affecting proteins involved in miRNA maturation. However, we did not observe altered mRNA expression of Drosha (DRSH-1), Pasha (PASH-1), Dicer (DCR-1), ALG-1, or RDE-4 in microarrays comparing adr-1(−/−);adr-2(−/−) and WT worms (Supplemental Table S2). Of course, while ADAR mutant C. elegans do not have apparent morphological or developmental defects, we cannot completely rule out indirect effects on processing machinery.
miRNAs with altered levels have predicted mRNA targets with affected levels
Current models indicate miRNAs promote degradation of target mRNAs (Kim et al. 2009), and we anticipated that changes in miRNA levels in ADAR mutants could lead to corresponding changes in mRNA levels. Thus, we performed microarray analyses, comparing adr-1(−/−);adr-2(−/−) and WT strains. We observed 746 genes with a ≥2.0-fold change in expression, and 3317 genes with a ≥1.2-fold change in expression (P-values ≤ 0.05) (Supplemental Table S2).
The miRNA most strongly affected in adr-1(−/−);adr-2(−/−) worms was miR-800, with levels increased eightfold compared to WT (Fig. 1C). miR-800 is expressed in L4-stage worms (Kato et al. 2009), but little else is known about this miRNA. The mRNA target prediction program TargetScan predicts 31 mRNA targets for miR-800, six of which were affected on the microarray in a significant manner (P-value ≤ 0.05). Consistent with increased miR-800 levels, all six mRNAs showed decreased levels (Fig. 1E; Supplemental Table S2), which was statistically significant (χ2 test; P-value, 0.01) when compared to the overall changes in gene expression in the microarray, where 53% of affected genes were increased and 47% decreased. Changes in mRNA levels were further validated using qRT-PCR (Fig. 1F).
While miRNA target prediction algorithms are constantly improving, their accuracy is largely unknown. It seems very possible that we observed changes in only six of 31 predicted miR-800 targets because many predicted targets are not real targets. Accepting this caveat, as a further test for correlative effects between miRNAs and their predicted targets, we analyzed the 27 miRNA loci with increased reads in adr-1(−/−);adr-2(−/−) worms compared to WT, as a group. According to TargetScan, 26 of these miRNA loci had predicted mRNA targets, and together, these 26 miRNAs had 1826 predicted mRNA targets, of which 473 were affected on the microarray in a significant manner (P-value ≤ 0.05); 163 targets (34%) had increased levels, while 310 targets (66%) had decreased levels (Supplemental Table S2; Supplemental Fig. S2A). Thus, consistent with the increased levels of the 26 miRNAs, a statistically significant number of mRNA targets have decreased levels when compared to the overall changes in genes expression on the microarray (P-value < 1 × 10−10). This correlation was most significant for the 13 miRNAs whose levels were affected ≥1.9-fold (P-value < 1 × 10−10) but still significant for the 13 miRNAs affected ≤1.9-fold (P-value, 0.01).
Notably, miRNAs whose levels were not affected by ADARs had predicted mRNA targets that were not significantly different from the overall gene expression changes on the microarray (53% increased, 47% decreased). Specifically, we analyzed 10 miRNAs not affected in ADAR mutant worms that cumulatively had 91 predicted mRNA targets affected on the microarray. We observed 42 (46%) with increased levels and 49 (54%) with decreased levels (Supplemental Fig. S2B), which was not significantly different from the overall changes in gene expression (P-value, 0.98).
These data suggest ADARs can affect mRNA levels, in a statistically significant manner, by altering miRNA levels. However, the effect was only observed for miRNA whose levels were increased in the absence of ADARs.
Editing in miRNAs is exceptionally rare
Inosine is recognized as guanosine during reverse transcription and edited adenosines appear as A-to-G mismatches when cDNA sequences are aligned to the genome. In C. elegans, only animals deficient for ADR-2 completely lack editing (Tonkin et al. 2002), so A-to-G mismatches due to editing should be present in reads from WT and adr-1(−/−) strains but absent in reads from adr-2(−/−) and adr-1(−/−);adr-2(−/−) strains. Thus, we validated editing sites by their absence in the latter strains, while reads not meeting these criteria were discarded as sequencing errors.
Using an editing-sensitive version of GNUMAP (Clement et al. 2010), we identified an editing site in only one of the 120 known miRNAs, a site in the seed sequence of miR-800 (Fig. 1G; Supplemental Table S3). pri-mir-800 is extensively base-paired, but the edited adenosine is mismatched, like some mammalian editing sites (Bass 2002). Editing occurred in 23% of WT reads and 10% of adr-1(−/−) reads but was absent from adr-2(−/−) and adr-1(−/−);adr-2(−/−) reads. The observed editing site was in the seed sequence of miR-800 and, in theory, could allow a new set of mRNA targets to be silenced. Algorithms that allow prediction of targets for a custom miRNA sequence (i.e., containing inosine) and include the important parameter of conservation (Lewis et al. 2005) are not currently available for C. elegans, but future analyses in this regard will be of interest.
ADARs typically localize in the nucleus, suggesting that editing of miR-800 occurs in the pri-miRNA. We were unable to confirm this, as attempts to amplify the edited region of the pri-miRNA with RT-PCR were unsuccessful, likely due to the very low abundance and highly stable structure of pri-mir-800 and further complicated because it is encoded in an intergenic region, with an unknown primary sequence. Nonetheless, because ADR-2 affects pri-mir-800 levels, it seems likely that ADR-2 edits the pri-miRNA sequence.
We asked what unique features of miR-800 might cause it to be the only edited mature miRNA. We found that miR-800 was one of only four miRNAs with a mismatched adenosine directly adjacent to ≥15 consecutive base pairs, with the others being miR-50, miR-1830, and miR-2219. Interestingly, miR-50, miR-800, and miR-1830 all had increased levels in the absence of ADARs, and miR-800 and miR-1830 were the two miRNAs most strongly affected in the absence of ADARs. Possibly these miRNAs have a high-affinity binding site for C. elegans ADARs.
miR-1830 has three mismatched adenosines, none of which were predicted to be edited by GNUMAP. However, due to its structural similarities with miR-800, we re-examined sequencing reads from miR-1830 to determine if there was a low percent editing that was not statistically significant enough to be identified by GNUMAP. We, indeed, found that two of the three mismatched adenosines in pri-mir-1830 had a low percentage of A-to-G mismatches in reads from WT and/or adr-1(−/−) worms (≤0.2%), which were absent in reads from adr-2(−/−) or adr-1(−/−);adr-2(−/−) worms (Supplemental Fig. S1C). These data imply that both miR-800 and miR-1830 are bound and edited by ADR-2, albeit miR-1830 appears to be edited at a low frequency. The unpaired adenosine within miR-50 did not appear to be edited; however, it was in the miRNA* sequence and thus had very minimal sequencing coverage; greater coverage would be needed to ascertain if it was also edited at a low frequency.
To verify that other edited miRNA reads were not discarded when aligned to the genome, we looked for known editing sites in small RNAs from the rncs-1 locus. This locus encodes an ∼300-bp stem–loop that is both edited and processed into small RNAs (Hellwig and Bass 2008; Welker et al. 2010). With our criteria, we identified seven edited sites in reads from rncs-1 (Supplemental Table S3), suggesting GNUMAP was able to identify edited reads. As a further test, we created a synthetic data set with 15 randomly selected miRNAs with artificial editing sites (∼10% editing per site). The data set contained ∼100× coverage of known miRNAs, with a 1% sequencing error rate, and was aligned to the genome with GNUMAP under default settings or with the editing option enabled. Under default settings, we identified only 42% of the artificial editing sites, with a 6% false positive rate. However, with the editing option enabled, we identified 92% of the editing sites, with a 13% false positive rate. Analyses of the artificial data set and rncs-1 locus imply that GNUMAP can successfully identify most editing sites, supporting our conclusion that editing is rare in C. elegans miRNAs.
Levels of many endo-siRNAs are decreased in ADAR mutant worms
Effects of ADARs on endo-siRNAs are unknown. To address this, we first compiled a list of all annotated loci that produce endo-siRNAs (6994 loci) (Supplemental Table S4). Similar to previous studies (Ruby et al. 2006), for all of the strains in our study, the total number of antisense reads aligning to these loci was much greater than the total number of sense reads (Supplemental Fig. S3A).
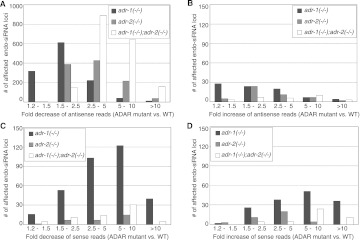
Using our list of annotated endo-siRNA loci, we identified those loci showing ≥2-fold difference in reads for any single ADAR mutant strain compared to WT, or ≥1.2-fold difference in reads in at least two strains compared to WT (P-value ≤ 0.05). This analysis revealed 2915 loci (42%) with altered reads in at least one of three ADAR mutant strains (Fig. 2; Supplemental Table S4). The majority of affected loci had a decreased number of antisense reads in mutant strains compared to WT (Fig. 2A), with 2573 loci having decreased reads in at least one mutant strain, of which 1332 were affected in at least two mutant strains (Supplemental Fig. S3B). Antisense endo-siRNA loci with the largest difference in reads (≥2.5-fold) were mostly in the double-mutant strain, indicating endo-siRNA levels are cumulatively affected by both C. elegans ADARs.
Figure 2.
Many endo-siRNA loci show altered reads in ADAR mutant strains. Plots show number of loci (y-axis) with decreased (A) or increased (B) antisense reads in indicated strains compared to WT, or decreased (C) or increased (D) sense reads in indicated strains compared to WT. Loci were binned (x-axis) according to observed fold-change. Loci altered ≥2-fold were affected in at least one mutant strain; loci altered ≥1.2-fold but <2-fold were affected in a least two mutant strains.
Few endo-siRNA loci showed effects other than decreased levels of antisense reads, which was unexpected, as previous studies suggested endo-siRNA levels might increase in the absence of ADARs (Knight and Bass 2002; Heale et al. 2009; Wu et al. 2011). Specifically, we observed only 140 loci with increased antisense reads in at least one mutant strain (Fig. 2B). Similarly, for sense reads, only 210 loci had increased reads, and 403 loci decreased reads, in at least one mutant strain (Fig. 2C,D).
In some analyses of Figure 2, more loci were affected in adr-1(−/−) animals than other strains. This could indicate greater effects of ADR-1, but typically, a comparable number of loci were not affected in the double-mutant strain. It seems more likely that we detected more loci with altered reads in adr-1(−/−) mutant strains because of the fourfold increase in reads in this data set compared to the adr-2(−/−) or adr-1(−/−);adr-2(−/−) data sets, as the adr-1(−/−) data set was sequenced at a later date after improvements to the Illumina sequencer. The greater coverage likely increased the sensitivity of some of our methods, which was specifically helpful in analyses of loci that give rise to sense siRNAs, the least abundant small RNA we analyzed. However, in our analysis of fold-changes at various loci (Fig. 2, x-axis), we are hesitant in making conclusions based on comparison of the adr-1(−/−) strain with other strains. Regardless, in our analysis of antisense reads (Fig. 2A), loci with the greatest fold-changes were observed in the adr-1(−/−);adr-2(−/−) strain, indicating a cumulative effect from the loss of both ADARs.
In summary, the most dramatic difference at endo-siRNA loci in ADAR mutant strains was that ∼40% of annotated loci had decreased antisense reads, with most loci affected ≥2.5-fold occurring in the adr-1(−/−);adr-2(−/−) strain, consistent with a cumulative effect from the loss of both ADARs. These data indicate that ADARs' predominant effect on siRNAs from endogenous genes differs from previously observed effects on siRNAs, where loss of ADARs increases siRNA levels derived from transgenes (Knight and Bass 2002; Heale et al. 2009) or transposons and inverted repeats (Wu et al. 2011).
ADARs affect many endo-siRNA loci that are Dicer-dependent
We observed 42% of endo-siRNA loci were affected in at least one ADAR mutant strain, and because ADARs target dsRNA, this implies that the biogenesis of endo-siRNAs from these loci involves a dsRNA intermediate. As Dicer is a key enzyme in processing cellular dsRNA, we asked if these loci were also Dicer-dependent. The severe germline defects associated with null alleles of C. elegans Dicer (Knight and Bass 2001) precludes analyses of small RNAs, so we used data generated in a previous comparison of a homozygous Dicer deletion strain rescued with a WT or helicase mutant transgene (dcr-1[−/−]K39A) (Welker et al. 2010). We reanalyzed this data set and found that, of the 6994 annotated endo-siRNA loci, there were 1189 loci (17%) with levels that depend on Dicer's helicase domain. The remaining 5805 loci could involve dsRNA precursors processed by Dicer but independently of its helicase domain, or they could be processed independently of Dicer. Of the 1189 Dicer-dependent endo-siRNA loci, 63% were also affected in ADAR mutant strains (Supplemental Table S4), which is a statistically significant overlap (χ2 test; P-value < 1 × 10−10). These data are consistent with the idea that these loci have a dsRNA intermediate.
While many loci affected by Dicer were also affected by ADARs, only 739 loci (27%) of the 2321 loci affected in either adr-1(−/−) and/or adr-2(−/−) strains were dependent on Dicer's helicase domain (Fig. 3). Therefore, according to the previously published data set (Welker et al. 2010), most endo-siRNA loci affected by ADARs were independent of Dicer's helicase domain (1582 of 2231 loci) (Fig. 3). However, it is also possible that not all Dicer helicase-dependent loci were identified in the previous study, since expression of the Dicer transgene was at least partially silenced in the germline. Finally, the Dicer helicase-independent loci may not require Dicer at all, and another protein, such as DRH-3 (Dicer Related Helicase 3) (Gu et al. 2009), might process these loci. However, we did not see a significant enrichment for these 1582 loci among known DRH-3-regulated loci (Supplemental Table S4).
Figure 3.
Overview of endo-siRNA loci affected by ADARs. (Pie charts) Number and percent of loci affected by different ADARs or Dicer's helicase domain. Loci affected in both adr-1(−/−) and adr-2(−/−) are abbreviated ADR-1 + 2.
Delineation of two categories of endo-siRNA loci affected by both ADAR and Dicer
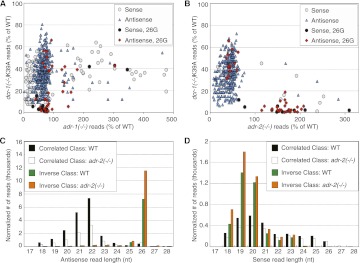
In an effort to understand how ADARs might affect endo-siRNA biogenesis, we further analyzed the 739 Dicer-dependent endo-siRNA loci that were also affected by ADARs (Welker et al. 2010). We delineated these 739 loci by plotting the change in read number, as percent of WT reads, for the dcr-1(−/−)K39A strain compared to an ADAR mutant strain (Fig. 4A,B). We observed two distinct clusters of loci. The largest cluster (Correlated class) consisted of loci with decreased reads in both the dcr-1(−/−)K39A and ADAR mutant strains and is observed on the left edge of the plots. Data points to the right of this cluster (Inverse class) were loci with decreased reads in the dcr-1(−/−)K39A strain and increased reads in ADAR mutants.
Figure 4.
Many endo-siRNA loci have levels affected by both Dicer and ADARs. (A,B) Plots show change in read numbers (as % of WT) observed in the dcr-1(−/−)K39A strain compared to adr-1(−/−) (A) or adr-2(−/−) (B) strains. Points are differentiated based on the strand affected in the ADAR mutant strain (sense or antisense) and whether the affected locus gives rise to 26G endo-siRNAs. Of the 1173 loci with decreased reads in the dcr-1(−/−)K39A strain compared to WT, 299 had altered sense reads, 791 had altered antisense reads, and 99 had changes in both. (C,D) Distributions of antisense and sense reads from the Inverse and Correlated classes, differentiating reads based on length.
To reveal further patterns, we used different symbols to designate loci that showed altered sense or antisense reads, as well as loci that give rise to 26G endo-siRNAs, a type of small RNA enriched among loci affected by Dicer's helicase domain (Welker et al. 2010). For both ADAR mutant strains, Correlated loci reiterated the predominant effect of ADARs, which was a decrease in antisense reads. Most Inverse loci affected by ADR-1 showed moderate changes in the dcr-1(−/−)K39A strain, but notably, Inverse loci affected by ADR-2 were typically those most strongly decreased in the dcr-1(−/−)K39A strain. The double mutant had intermediate effects (Supplemental Fig. S4A).
We observed that 24 of the 42 ADR-2 Inverse loci were also annotated 26G loci (Fig. 4B; Supplemental Table S4), which was a statistically significant enrichment (χ2 test, P-value < 1.0 × 10−10); additionally, 26G loci were depleted from the ADR-2 Correlated loci (17 of 363 loci; P-value, 0.02). Correspondingly, the majority of ADR-2 Inverse antisense reads were 26 nt in length, while most ADR-2 Correlated antisense reads were ∼20–23 nt in length (Fig. 4C). We also observed thousands of 18- to 20-nt ADR-2 Inverse sense reads but only a few hundred ADR-2 Correlated 18- to 20-nt sense reads, which were 18–26 nt in length (Fig. 4D).
Given that there was an increase in both sense and antisense reads for 13 of the 42 ADR-2 Inverse loci (Supplemental Table S4), we asked if reads from these individual loci could base-pair. On average, 52% of sense reads and 15% of antisense reads within individual ADR-2 Inverse 26G loci were capable of forming structures (Supplemental Fig. S5), while <1% of reads from ADR-2 Inverse non-26G loci could form structures. This was a significant enrichment for 26G loci with reads that could form structures when compared to non-26G loci (χ2 test; P-value < 1.0 × 10−10).
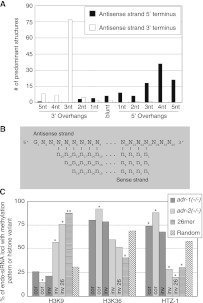
Most structures from 26G loci had antisense reads with 3-nt 3′ overhangs at the 3′ terminus of the antisense read (Fig. 5A,B; Supplemental Fig. S5), consistent with previous data (Ruby et al. 2006). Such overhangs contrast with the 2-nt 3′ overhangs made when Dicer processes pre-miRNAs (Warf et al. 2011) but are consistent with Dicer processing long dsRNA (Welker et al. 2011). A χ2 test verified 3-nt 3′ overhangs were enriched in Inverse loci structures compared to Dicer cleavage sites in miRNAs (P-value < 1.0 × 10−10), insinuating Dicer has altered functionality when processing 26G precursors. In contrast to the 3′ terminus, the 26G structures were more heterogeneous at the 5′ terminus of the antisense reads, where 3- to 5-nt 5′ overhangs were all abundant, due almost entirely to variability at the 3′ termini of sense reads. This indicates that a different and less exact mechanism defines this terminus.
Figure 5.
ADAR-affected 26G loci have small RNA reads that can base-pair in specific structures and have histone modification patterns associated with transcriptional silencing. (A) Bar height indicates number of termini with a given terminus from putative Inverse 26G structures, relative to the antisense strand. If multiple reads could form different structures at a given location, the structure with the predominate read was analyzed. (B) Consensus structure of predominant structures from 26G loci. Nearly all reads had a 5′ guanosine, while other positions varied. (C) Histone modification and histone variant patterns for various groups of endo-siRNA loci. χ2 tests compare patterns from each group to the random data set, with P-values < 0.05 (*) or < 1 × 10−10 (**). (cor) Correlated loci, (inv) Inverse loci, (inv 26) Inverse 26G loci.
ADARs target AU base pairs to create IU mismatches, and it has been proposed that such mismatches inhibit processing of dsRNA by Dicer (Bass 2000), allowing ADARs to regulate gene silencing. This model is supported by the observation that ADARs inhibit biogenesis of siRNAs derived from transgenes, in both C. elegans and D. melanogaster (Knight and Bass 2002; Heale et al. 2009), and those derived from C. elegans transposons and inverted repeats (Wu et al. 2011). Similarly, ADAR editing inhibits the production of siRNAs in vitro in Drosophila extracts (Scadden and Smith 2001). The Inverse endo-siRNA loci affected by ADR-2, the active C. elegans deaminase, may represent the first examples of endogenous protein coding genes where ADAR editing antagonizes the production of endo-siRNAs.
Some 26G endo-siRNAs are enriched for specific histone methylation patterns and may be processed cotranscriptionally
Only a small percentage of C. elegans endo-siRNAs align to introns (Welker et al. 2010), suggesting that the spliced mRNA in the cytoplasm is the template for their synthesis. While our data indicate ADARs affect many endo-siRNA loci, ADARs typically localize in the nucleus. To address this conundrum, we determined where sequencing reads aligned within Dicer-dependent loci affected by ADR-2. We observed that 88% of ADR-2 Inverse loci, but only 22% of ADR-2 Correlated loci, had ≥2 sequencing reads aligning within an intron (Supplemental Fig. S4B,C), which was a significant enrichment (χ2 test; P-value < 1 × 10−10). These data imply that at least a subset of adr-2-affected Inverse loci, including many that give rise to 26G endo-siRNA, are produced in the nucleus prior to splicing.
Given the evidence for nuclear localization, we wondered if the 26G endo-siRNAs affected by ADR-2 were involved in RNA-Induced Transcriptional Silencing (RITS). In RITS, small RNAs induce degradation of nascent mRNAs in the nucleus and lead to transcriptional silencing at that locus, mainly through increased levels of H3K9 histone methylation (Buhler and Moazed 2007). RITS is well documented in organisms such as fission yeast but poorly understood in C. elegans. Some proteins are implicated in a RITS-like mechanism in worms (Robert et al. 2005; She et al. 2009; Guang et al. 2010; Burkhart et al. 2011), but it is unclear if the ERI/Dicer complex, which makes 26G RNAs (Gent et al. 2010; Vasale et al. 2010), is involved. Supporting this possibility are studies showing that ERI-1, an endonuclease found in the C. elegans ERI/Dicer complex, is required for H3K9 methylation of at least four endo-siRNA loci (Burkhart et al. 2011), as well as proper regulation of H3K9 methylation of two transgenes in fission yeast (Iida et al. 2006).
To determine if 26G loci and the ERI/Dicer complex might be involved in RITS, we analyzed data from a recent study of histone methylation patterns across the C. elegans genome (Liu et al. 2011). We observed 18 of 25 ADR-2 Inverse loci had an enrichment of H3K9 methylation compared to 100 random genes (Fig. 5C), which was statistically significant (χ2 test; P-value, 4.0 × 10−5). Correspondingly, these loci were significantly depleted of patterns associated with high levels of transcription, such as H3K36 methylation (P-value, 0.1) and occupancy of the histone 2 variant HTZ-1 (P-value, 6.8 × 10−4). Furthermore, when only ADR-2 Inverse 26G loci were considered, we observed an even greater enrichment for patterns of silencing. In contrast, ADR-2 Correlated loci and ADR-1 Inverse and Correlated loci all had methylation patterns associated with much higher levels of transcription. This may suggest that, of Dicer-dependent loci affected by ADARs, only ADR-2 Inverse 26G loci are transcriptionally silenced.
We wondered if the methylation patterns of ADR-2 Inverse 26G loci were emblematic of all 26G loci. Indeed, we observed a highly significant enrichment of H3K9 methylation at 49 known 26G loci (Vasale et al. 2010) compared to 100 random genes (P-value < 1 × 10−10) (Fig. 5C). We also observed that, compared to the random data set, 26G loci were depleted for methylation patterns associated with high levels of transcription—for example, H3K36 methylation (P-value, 9.8 × 10−4) and occupancy of the histone 2 variant HTZ-1 (P-value, 1.7 × 10−3).
These data suggest that adr-2-affected Inverse 26G loci, and more generally, a majority of 26G endo-siRNAs, are made in the nucleus prior to splicing and mediate transcriptional silencing of the loci from which they derive. While 26G loci are known to be silenced in a manner that involves the ERI/DCR complex (e.g., Welker et al. 2010), our data indicate this silencing may occur in the nucleus cotranscriptionally.
Editing within endo-siRNAs
Double-stranded RNA incubated in D. melanogaster extract is processed into siRNAs, but dsRNA that remains unprocessed contains many inosines, while the siRNAs have few (Zamore et al. 2000; Scadden and Smith 2001). These data indicate ADAR editing inhibits processing of dsRNA, and we did not expect to find substantial editing in mature endo-siRNA reads. As anticipated, we identified only 54 editing sites within reads from 32 endo-siRNA loci, which was only 0.5% of annotated loci (Supplemental Table S3).
Of loci with editing, the F43E2.6 locus had the most abundant number of total reads. Interestingly, the predominant sense and antisense reads from this locus form a dsRNA with 2-nt 3′ overhangs. By examining all reads from this locus, we identified five edited sites within or directly adjacent to this duplex, one in antisense reads and four in sense reads (Fig. 6). All five sites were edited in WT and adr-1(−/−) reads, with little or no editing in adr-2(−/−) or adr-1(−/−);adr-2(−/−) reads. Sense and antisense F43E2.6 reads were increased in the absence of ADARs (Supplemental Table S4), suggesting ADARs inhibit maturation of a dsRNA precursor. However, the F43E2.6 locus is atypical, as there are ∼500 antisense and ∼3000 sense reads in WT worms, >97% of which align within a 30-nt window. Most other endo-siRNA loci have a few hundred total reads, which align throughout the entire locus, with many more antisense than sense reads.
Figure 6.
Reads from the F43E2.6 locus are edited. Putative dsRNA is shown with the predominant sense read (underlined, bold) pairing with the predominant antisense read (italicized). (Double arrowheads) Edited adenosines, with percent editing tabulated for each strain. (Single arrowheads) Termini of predominant reads.
The four sites with editing in sense reads were within the coding region of the gene, but all caused synonymous mutations at the third codon position and did not alter coding potential (Supplemental Fig. S6). Thus, the coding region of the F43E2.6 gene appears to be edited without affecting the encoded protein. The F43E2.6 locus has one predicted orthologous gene, TFA1 in Saccharomyces cerevisiae (http://www.wormbase.org), which is an essential subunit of the RNA polymerase II factor that assists in promoter opening (Feaver et al. 1994).
ADARs inhibit maturation of siRNAs from the long noncoding RNA rncs-1
We determined ADARs' effects on the processing of the long noncoding RNA rncs-1, which is an ∼300-bp stem–loop that is both edited and processed into small RNAs (Hellwig and Bass 2008; Welker et al. 2010). Reads from this locus were increased 4.9-fold in adr-2(−/−) animals compared to WT, but only 1.6-fold in adr-1(−/−) animals (P-values < 1 × 10−10). The double mutant showed a cumulative effect, with reads increased 6.1-fold. These data indicate that, like the 26G Inverse loci, editing, rather than RNA binding, has a larger effect on the processing of certain RNA substrates. This is in contrast to miRNAs and most endo-siRNAs, where RNA binding had a predominate effect.
Few piRNAs are edited, but many have decreased levels in ADAR mutants
We did not anticipate ADARs would affect piRNAs, as piRNAs are not thought to derive from a dsRNA precursor in C. elegans (Kim et al. 2009). However, compared to WT, total piRNA reads were decreased ∼40% in adr-1(−/−) and adr-1(−/−);adr-2(−/−) strains, and ∼15% in adr-2(−/−) worms (Supplemental Fig. S7A). Only a few piRNA loci had increased reads, mainly in the adr-1(−/−) strain, but hundreds of loci had decreased reads in mutant strains (Supplemental Table S5; Supplemental Fig. S7B,C). U21R-1 levels determined by Northern blots correlated well with sequencing data (Supplemental Fig. S8D). As with other mature small RNAs, very few mature piRNAs were edited, and only nine sites in eight mature piRNA sequences met our criteria for editing, constituting 0.08% of annotated piRNA loci (Supplemental Table S3).
Discussion
Here we report a genome-wide analysis of ADARs' effects on small RNAs from gene-silencing pathways. While editing of C. elegans small RNAs was rare, ADARs profoundly affected small RNA levels, with our most stringent criteria indicating that at least 40% of miRNAs and endo-siRNAs are altered in ADAR mutant strains. In most cases, miRNA levels were increased in mutant strains, with a corresponding decrease in pri-miRNA levels, suggesting ADARs function to inhibit pri-miRNA processing. miRNAs with the most dramatically altered levels had predicted mRNA targets with reciprocal changes, indicating ADARs are required for normal gene expression. Recently, similar results were obtained for miRNAs of mouse embryos (Vesely et al. 2012).
Previous data from candidate studies and in vitro experiments (Blow et al. 2006; Kawahara et al. 2007a, 2008) suggest a model whereby editing inhibits pri-miRNA processing. However, we observed the greatest effects in the absence of ADR-1, an inactive deaminase. Thus, our data are more consistent with a model whereby, in most cases, ADARs inhibit Drosha processing by binding and sequestering pri-miRNAs. This model agrees with a study that showed ADAR editing was not required to inhibit siRNA production from a transgene in D. melanogaster (Heale et al. 2009).
Many studies support the idea that ADAR editing inhibits processing of long dsRNA into siRNAs (Knight and Bass 2002; Heale et al. 2009; Wu et al. 2011). Our studies indicate the long, noncoding RNA rncs-1 and the adr-2 Inverse loci are regulated by ADR-2 in this manner. Further, analyses of adr-2 Inverse loci provided an intriguing association with 26G endo-siRNAs and transcriptional silencing. However, the largest class of affected endo-siRNAs showed decreased levels in ADAR mutants, contrasting with most previous models.
Do ADARs directly affect the maturation of some small RNAs and indirectly affect others?
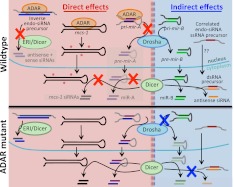
To explain our data, we propose a model (Fig. 7) whereby ADARs have direct (red X) and indirect (blue X) effects on small RNA biogenesis. In its essence, it is a competition model, based on the fact that dsRNA binding proteins (dsRBPs) are not sequence-specific and can bind each other's substrates. Direct effects involve ADAR sequestering a dsRNA away from another dsRBP, for example, a pri-miRNA from Drosha, or Inverse endo-siRNAs from the ERI/Dicer complex. Many of the effects we observed correlated with the loss of ADR-1, indicating that binding plays a large role. However, editing could also inhibit processing, as suggested by observations with rncs-1 and the fact that Inverse endo-siRNA loci are affected by the loss of ADR-2, the active deaminase. Direct effects lead to increased small RNA levels in the absence of ADARs, and as ADARs are mainly nuclear, direct effects may be limited to pathways with nuclear dsRNAs.
Figure 7.
Model of ADARs' effects on the biogenesis of C. elegans small RNAs. Direct (red X) and indirect (blue X) effects are indicated. In WT cells (top), ADAR binding to certain dsRNAs competes with binding by other dsRBPs, in some cases inhibiting processing (e.g., pri-miRNAs, Inverse endo-siRNA precursors), while ADAR editing (red *) inhibits processing of other RNAs (e.g., rncs-1). Indirect effects arise in mutant strains when dsRNAs normally bound or edited by ADARs (e.g., miRNAs, rncs-1) are released to compete with dsRNA in other pathways.
Indirect effects are only observed in ADAR mutants, when dsRNAs are liberated from ADAR binding/editing, allowing them to compete with pathways not typically affected by ADARs. Specifically, increased levels of free pri-mir-800 or pri-mir-1830 in the nucleus could titrate Drosha from processing pri-mir-253, and increased levels of pre-mir-800, pre-mir-1830, or unedited rncs-1 in the cytoplasm could titrate a protein, such as Dicer, from processing Correlated endo-siRNA precursors.
Insights into 26G endo-siRNA biogenesis
The ERI/Dicer complex makes 26G endo-siRNAs (Gent et al. 2010; Vasale et al. 2010), but their precursors are uncharacterized. Our data have interesting implications regarding 26G biogenesis and function. 26G loci have increased reads in ADAR mutants, suggesting that ADARs directly bind 26G precursors and inhibit their processing. ADARs are mostly nuclear (Hundley et al. 2008), implying that 26G precursors are also processed in the nucleus and prior to splicing, as ∼80% of ADR-2 Inverse 26G loci have reads aligning within introns. Splicing is usually cotranscriptional (Bentley 2005), insinuating that 26G precursors are also processed cotranscriptionally, consistent with the function we propose for them in RITS. Further evidence that 26G loci are involved in a RITS-like process is their enrichment for H3K9 histone methylation.
We also observed that 26G loci were enriched for reads that can base-pair. Paired structures showed a 3-nt 3′ overhang, as expected of products cleaved by Dicer from a long dsRNA precursor (Welker et al. 2011) but have variable 3- to 5-nt 5′ overhangs, which is inconsistent with Dicer processing. We propose the 3′ to 5′ ERI-1 nuclease (Kennedy et al. 2004), of the ERI/Dicer complex, might imprecisely process sense strands, resulting in their variable 3′ termini.
Possible mechanisms for phenotypes of ADAR mutant C. elegans
There are three known phenotypes in ADAR mutant worms: chemotaxis defects, cosuppression of transgenes in somatic cells, and a reduced lifespan (Knight and Bass 2002; Tonkin et al. 2002; Sebastiani et al. 2009). Interestingly, all of these phenotypes are rescued by additional mutations in RDE-1 and/or RDE-4, which are involved in processing of siRNAs, but the mechanism(s) giving rise to these phenotypes is unclear. One hypothesis is that ADARs prevent processing of specific RNAs (e.g., endo-siRNAs), resulting in a phenotype when these RNAs are processed in ADAR mutant strains (Knight and Bass 2002). In the additional absence of RDE-1 and/or RDE-4, these RNAs are again no longer made, rescuing the defects. In our data set, only Inverse endo-siRNA loci and the noncoding RNA rncs-1 fit this model, and it is unknown if increased levels of any of these specific siRNAs give rise to a phenotype.
With our microarray data set, it was also possible to identify candidate genes whose altered expression in adr-1(−/−);adr-2(−/−) worms could lead to observed phenotypes. Specifically, mutations in the abnormal dauer formation proteins (DAFs) alter C. elegans lifespan (Shmookler Reis et al. 2009). Eight DAF mRNAs had altered expression in adr-1(−/−);adr-2(−/−) worms, which might lead to a reduced lifespan in ADAR mutant worms (Supplemental Table S2). Additionally, we also observed increased mRNA levels for eight of 12 respiratory proteins (MTCEs), which could also lead to oxidative damage and reduce a worm's lifespan. It is unclear if ADARs directly affect DAF or MTCE genes. If the altered expression of any of these genes does give rise to a phenotype, it is likely that ADARs regulate them through a small RNA pathway, as all known phenotypes were rescued in previous studies by additional mutations to RDE-1 and/or RDE-4.
Methods
Sequencing of small RNAs
Five to ten worms were used to seed 6-cm plates containing OP50 as food. Worms were grown at 20°C and harvested after 4–5 d, at which point worms of each developmental stage were present (mixed-stage) in reproducible ratios. Total RNA was extracted from the worms using TRIzol, 10 μg was run on a 15% denaturing gel, and ∼18- to 30-nt RNAs were isolated. cDNA libraries were made only from 5′ monophosphate RNAs, using the Illumina Small RNA Prep Kit v1.0 or v1.5, and were sequenced on an Illumina Genome Analyzer IIx. The program Novoalign (http://www.novocraft.com) was used to trim 3′ adaptors and align sequences to the genome (r-value, 0.2; q-value, 5). The USeq component Novalign Parser (Nix et al. 2008) was used to remove poorly aligned sequences with a posterior probability >0.1.
Reads aligning to the genome, in millions, were: 6.9 (WT), 5.8 (adr-2[−/−]), and 6.8 (adr-1[−/−];adr-2[−/−]). At a later date, we sequenced and obtained reads, in millions: 11.1 (WT) and 25.6 (adr-1[−/−]). Data from the first WT data set are shown for normalized read levels, while the second WT data set was used for comparison with the adr-1(−/−) data set.
Analysis of aligned reads
Coordinates were compiled from previous studies of miRNAs (Warf et al. 2011), piRNAs (Ruby et al. 2006; Batista et al. 2008; Kato et al. 2009), and endo-siRNAs (references in Supplemental Table S4). The total number of reads within each data set was normalized when comparing data sets. We used the USeq component Defined Region Scan Sequences (Nix et al. 2008) to determine significant differences in read levels between strains, which uses a standard binomial model, with a Benjamini-Hochberg correction. We reanalyzed data from the dcr-1(−/−)K39A strain (Welker et al. 2010), using our updated endo-siRNA list, to compare with loci affected in ADAR mutants.
Target mRNAs of miRNAs were predicted using release 5.1 of TargetScan (http://www.targetscan.org/worm_12). When analyzing potential structures from loci, we evaluated all antisense and sense reads with >10 pairing nt, with ≥2 sense and ≥2 antisense reads pairing. The most abundant read was used if multiple reads aligned.
Identification of editing sites
Reads were realigned using GNUMAP, with improvements to identify editing sites (http://dna.cs.byu.edu/gnumap/index.shtml). Editing sites were identified if ≥3 reads had A-to-G mismatches in WT or adr-1(−/−) data sets, with ≤1% of reads with A-to-C or A-to-T mismatches. Sites were discarded if ≥1% of reads (with >1 read) had A-to-G mismatches in adr-2(−/−) or adr-1(−/−);adr-2(−/−) data sets (with at least 10 reads required in an ADR-2 mutant), or if the site was the last nt in a read, which has a high error rate.
Analysis of methylation patterns
To determine histone modification patterns, we analyzed data from Liu et al. (2011), visualized at modENCO (http://www.modencode.org). Endo-siRNA loci were positively scored if a modification was within any part of the locus. We scored 49 known 26G loci (Vasale et al. 2010), all ADR-1 and ADR-2 Inverse loci, and a random selection of ADR-1 (120 of 525) and ADR-2 (200 of 367) Correlated loci.
Northern blots
Total RNA (75 μg), as previously prepared, was enriched for small RNAs using the mirVana isolation kit (Ambion), and 2 μg was run on a 15% denaturing gel and transferred onto a Hybond-NX membrane (Amersham Biosciences) (see Supplemental Methods).
For Northern analysis, membranes were preincubated with ULTRAhyb-Oligo buffer (Ambion) for ∼1 h at 42°C and probed overnight at 42°C with a DNA oligonucleotide end labeled using T4 PNK (New England Biolabs). Blots were washed 4 × 20 min with 2× SSC at 42°C, autoradiographed, and then quantified using ImageQuant (Molecular Dynamics). The U6 snRNA was used for normalization of each lane, and 3–4 biological replicate experiments were performed for each tested RNA.
RT-PCR
For quantitative RT-PCR (qRT-PCR), total RNA, as previously prepared, was DNased, and then reverse transcription (RT) was performed on 5 μg of DNased RNA (see Supplemental Methods). qPCR was then performed on a LightCycler 2.0. For primary miRNAs, an RT primer specific to each pri-miRNA was designed to bind within the pre-miRNA loop and destabilize the stem–loop structure. The forward PCR primer was outside of the pre-miRNA sequence, allowing only the pri-miRNA sequence to amplify. RT was performed on gpd-3, C18C4.7, and F29F11.6 using an oligo-dT primer, with gene-specific primers used for PCR. The gpd-3 gene was used for normalization between samples. Three to five biological replicates were performed for each tested RNA species.
Gene expression microarrays
Total RNA (1 μg), as previously prepared, was used to probe a C. elegans v2 Gene Expression array (Agilent) (see Supplemental Methods). Results were analyzed using GeneSifter (Geospiza) to determine which genes in adr-1(−/−);adr-2(−/−) were changed in a significant manner compared to WT, using a t-test with a Benjamini and Hochberg correction. Four biological replicate arrays were performed.
Data access
Raw and aligned data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE28888.
Acknowledgments
We thank A. Krauchuk for help with qRT-PCR, and N. Walker, D. Nix, B. Dalley, and B. Milash for help with sequencing and microarrays. This work was supported by funds to B.L.B. from the NIH (GM044073) and to M.B.W. by an NIH Training Grant (5T32 HD07491).
Author contributions: M.B.W. designed and performed all experiments, performed data analysis, and prepared the manuscript. B.A.S. and W.E.J. performed data analysis. B.L.B. designed experiments, performed data analysis, and prepared the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.134841.111.
Note added in proof
Related studies have appeared recently for human miRNAs (Alon et al. 2012).
References
- Alon S, Mor E, Vigneault F, Church GM, Locatelli F, Galeano F, Gallo A, Shomron N, Eisenberg E 2012. Systematic identification of edited microRNAs in the human brain. Genome Res (this issue). doi: 10.1101/gr.131573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL 2000. Double-stranded RNA as a template for gene silencing. Cell 101: 235–238 [DOI] [PubMed] [Google Scholar]
- Bass BL 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL 2005. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR 2006. RNA editing of human microRNAs. Genome Biol 7: R27 doi: 10.1186/gb-2006-7-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Moazed D 2007. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S 2011. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet 7: e1002249 doi: 10.1371/journal.pgen.1002249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement NL, Snell Q, Clement MJ, Hollenhorst PC, Purwar J, Graves BJ, Cairns BR, Johnson WE 2010. The GNUMAP algorithm: Unbiased probabilistic mapping of oligonucleotides from next-generation sequencing. Bioinformatics 26: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver WJ, Henry NL, Bushnell DA, Sayre MH, Brickner JH, Gileadi O, Kornberg RD 1994. Yeast TFIIE. Cloning, expression, and homology to vertebrate proteins. J Biol Chem 269: 27549–27553 [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycom JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct Argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA 2009. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J 28: 3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwig S, Bass BL 2008. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci 105: 12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hundley HA, Bass BL 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 35: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL 2008. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14: 2050–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida T, Kawaguchi R, Nakayama J 2006. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr Biol 16: 1459–1464 [DOI] [PubMed] [Google Scholar]
- Kato M, de Lencastre A, Pincus Z, Slack FJ 2009. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54 doi: 10.1186/gb-2009-10-5-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K 2007a. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K 2007b. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Natl Rev 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2002. The role of RNA editing by ADARs in RNAi. Mol Cell 10: 809–817 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K 2006. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Natl Rev 7: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix DA, Courdy SJ, Boucher KM 2008. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics 9: 523 doi: 10.1186/1471-2105-9-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman M 2007. A-to-I editing challenger or ally to the microRNA process. Biochimie 89: 1171–1176 [DOI] [PubMed] [Google Scholar]
- Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev 19: 782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Scadden AD, Smith CW 2001. RNAi is antagonized by A→I hyper-editing. EMBO Rep 2: 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Montano M, Puca A, Solovieff N, Kojima T, Wang MC, Melista E, Meltzer M, Fischer SE, Andersen S, et al. 2009. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS ONE 4: e8210 doi: 10.1371/journal.pone.0008210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM 2009. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet 5: e1000624 doi: 10.1371/journal.pgen.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis RJ, Bharill P, Tazearslan C, Ayyadevara S 2009. Extreme-longevity mutations orchestrate silencing of multiple signaling pathways. Biochim Biophys Acta 1790: 1075–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J 21: 6025–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci 107: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF 2012. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res (this issue). doi: 10.1101/gr.133025.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Warf MB, Johnson WE, Bass BL 2011. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 17: 563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S, Bass BL 2010. Dicer's helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16: 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL 2011. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell 41: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire A 2011. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat Struct Mol Biol 18: 1094–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zinshteyn B, Nishikura K 2009. Adenosine-to-inosine RNA editing. Wiley Interdiscip Rev Syst Biol Med 1: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]