Abstract
Adenosine-to-inosine (A-to-I) editing modifies RNA transcripts from their genomic blueprint. A prerequisite for this process is a double-stranded RNA (dsRNA) structure. Such dsRNAs are formed as part of the microRNA (miRNA) maturation process, and it is therefore expected that miRNAs are affected by A-to-I editing. Editing of miRNAs has the potential to add another layer of complexity to gene regulation pathways, especially if editing occurs within the miRNA–mRNA recognition site. Thus, it is of interest to study the extent of this phenomenon. Current reports in the literature disagree on its extent; while some reports claim that it may be widespread, others deem the reported events as rare. Utilizing a next-generation sequencing (NGS) approach supplemented by an extensive bioinformatic analysis, we were able to systematically identify A-to-I editing events in mature miRNAs derived from human brain tissues. Our algorithm successfully identified many of the known editing sites in mature miRNAs and revealed 17 novel human sites, 12 of which are in the recognition sites of the miRNAs. We confirmed most of the editing events using in vitro ADAR overexpression assays. The editing efficiency of most sites identified is very low. Similar results are obtained for publicly available data sets of mouse brain-regions tissues. Thus, we find that A-to-I editing does alter several miRNAs, but it is not widespread.
A-to-I editing is catalyzed by enzymes of the adenosine deaminase that act on the RNA (ADAR) family, and post-transcriptionally changes adenosine to inosine, the latter being treated by cell machinery similar to guanosine. This modification results in protein recoding of tens of genes (Nishikura 2010), shown in some cases to translate to modified biophysiological properties with critical implication on brain function (Sommer et al. 1991). However, only a minute fraction of A-to-I modifications results in protein recoding, whereas most targets of ADARs reside within the noncoding parts of the transcriptome (Li et al. 2009; Nishikura 2010).
The primary transcript of a miRNA gene (pri-miRNA) folds into a hairpin structure and undergoes cleavage by the nuclear enzyme Drosha, giving a 60- to 70-nucleotide (nt) precursor miRNA (pre-miRNA). The pre-miRNA is exported from the nucleus by Exportin-5 and processed by the cytoplasmtic enzyme Dicer into 19- to 25-nt imperfect double-stranded RNA (dsRNA) (Bartel 2004). One or both strands of the duplex may serve as the functional mature miRNA. By binding to partially complementary targets located in the 3′ untranslated region of specific mRNAs, mature miRNAs block the translation or guide the degradation of target mRNAs. Bases 2–8 at the 5′ end of the mature miRNA were found to be critical for the target recognition (Bartel 2004). By regulating protein expression, miRNAs are involved in many cellular and physiological processes, including numerous pathological conditions (Chang et al. 2008). As ADARs binds to dsRNA, they may act on the double-strand formation of pri-miRNA (Yang et al. 2006). A-to-I editing of pri-miRNA can affect the processing of the pri-miRNA to pre-miRNA or the processing of the pre-miRNA to mature miRNA (Yang et al. 2006).
Pri-miRNA editing events may lead to the expression of edited mature miRNAs. If the alterations are in the recognition site, known as the “seed” region, a change in the target genes is expected. A striking example is mouse miR-376, in which editing in the recognition site alters the target specificity of the miRNA and profoundly affects cellular processes (Kawahara et al. 2007). A-to-I editing events that change target specificity of miRNAs clearly add another layer of complexity to gene regulation pathways. It is therefore of interest to study the extent of this phenomenon. Initial reports, based on low-throughput experiments, estimated that it may be widespread (Kawahara et al. 2008). Proper utilization of next-generation sequencing (NGS) has the potential to unravel the full extent of A-to-I editing in miRNAs. Unfortunately, so far NGS-based experiments resulted in puzzling results; many types of DNA-to-reads mismatches, including adenosine-to-guanosine (A-to-G) (which may be caused by A-to-I editing), were detected (Landgraf et al. 2007; Morin et al. 2008; Ebhardt et al. 2009; Martí et al. 2010; Pantano et al. 2010; Joyce et al. 2011). These findings could mean that miRNAs undergo many kinds of biological modifications, including A-to-I editing. However, one must also consider technical explanations for the variety of mismatches observed. Indeed, de Hoon et al. (2010) have reported technical problems in the analysis of NGS data sets that may result in false detection of miRNA modification events. They have concluded that the editing events in mature miRNAs are rare (de Hoon et al. 2010). Here we set out to comprehensively identify A-to-I editing in mature miRNAs using NGS followed by bioinformatics analysis. In contrast to previous reports, we find a clear A-to-I signal in mature miRNAs.
This work focuses on human brain tissue for two main reasons: The first is the obvious importance of unraveling the scope of mature miRNAs editing in this tissue, and second is the strong line of evidence pointing toward high A-to-I editing in human brain compared with other organisms and tissues (Eisenberg et al. 2005; Paz-Yaacov et al. 2010).
Results
Sequencing of miRNAs from human brain tissue
Sequencing of mature miRNAs has the potential to unravel many kinds of sequence modifications, including A-to-I editing, as inosine (I) in the RNA leads to a guanosine (G) in the sequencing output. Mature miRNAs from human brain tissue (FirstChoice human brain reference RNA, Ambion) were sequenced using one lane of Illumina GAIIx instrument (Methods). The following considerations were taken into account in the sequencing data analysis: (1) The 3′ end of mature miRNAs undergoes large-scale RNA modifications in the form of adenylation and uridylation (Burroughs et al. 2010). Therefore, the last two bases of the sequencing read, which correspond to the 3′ end of the mature miRNA, were trimmed (Chiang et al. 2010). (2) As demonstrated by de Hoon et al. (2010), cross-mapping of sequencing reads can create many false alignments that may be interpreted as sequence alterations. Therefore, we have aligned all the trimmed reads against the human genome using Bowtie (Langmead et al. 2009), demanding unique best hits with up to one mismatch (Methods). Only reads that were aligned to the genomic locations of known miRNAs were further used. (3) Sequencing error hinders the discovery of editing sites. Thus, we filtered mismatches with low-quality score. Then, the sequencing error rate was estimated, and only sites with significant modification, as determined by binomial cumulative distribution, were recorded (Methods). (4) Known single nucleotide polymorphisms (SNPs) sites were removed using the dbSNP data set (Sherry et al. 2001). The detection procedure is illustrated in Figure 1 (also see Methods).
Figure 1.
A schematic representation of the procedure for identifying editing sites in mature miRNAs using the brain sample data (for details, see Methods). In each one of the four steps (A–D), the total number of mismatches of any type is given in absolute numbers (bar chart) and in relative proportions (pie chart). (A) If the untrimmed reads are aligned against the known miRNA sequences, a strong signal of adenylation and uridylation is observed. (B) After trimming the 3′ end of the reads, A-to-G modifications become dominant. (C) If the trimmed reads are aligned against the genome and not directly against the miRNA sequences, the relative proportion of the A-to-G mismatches is enhanced as a result of less cross-mapping. (D) After possible sequencing errors are removed by using binomial statistics, only A-to-G modifications are observed.
A clear A-to-I editing signal in human brain miRNAs
Overall, 19 statistically significant modification sites were detected in 18 different miRNAs expressed in this sample (Table 1). Remarkably, all of them were A-to-G modifications, suggesting that they represent A-to-I editing rather than random sequencing or alignment errors (Methods; Fig. 1; Levanon et al. 2004). Another indication for the observed modifications being editing sites comes from analysis of the sequence and structural motifs. As expected for true A-to-I editing sites (Kleinberger and Eisenberg 2010), we find enrichment of uridine (U) and depletion of G in the upstream nucleotide position (12 U and zero G out of 19 sites). The nucleotide opposing the editing site is typically cytidine (C) or U (17/19), and G is overrepresented in the downstream nucleotide (eight of 19). These sequence motifs are in accordance with the expected properties of true A-to-I editing sites (Fig. 2; Supplemental Tables 1, 2).
Table 1.
The statistically significant A-to-G modifications sites detected in mature miRNAs using the pooled human brain sample and the human frontal lobe samples
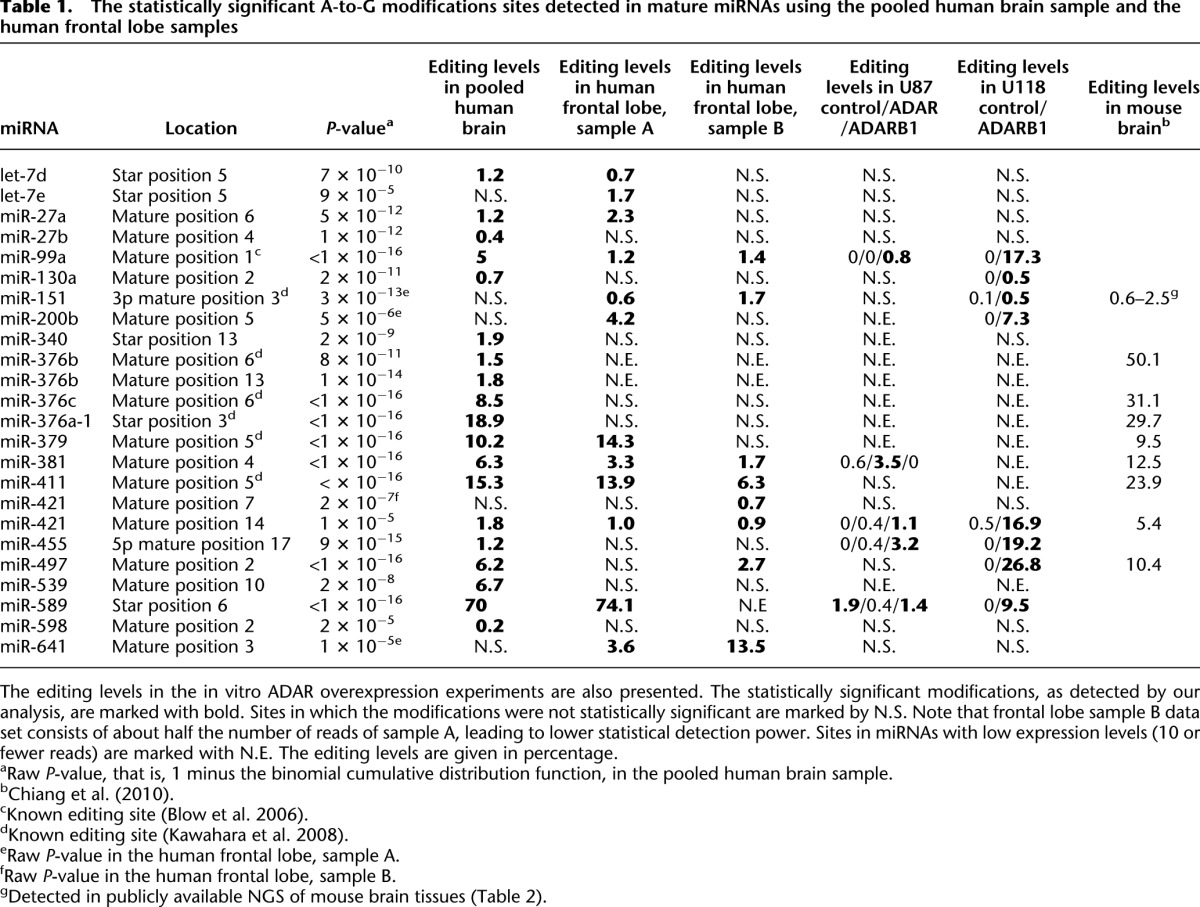
Figure 2.
Sequence preference in the bases flanking the A-to-G editing sites detected in the human brain samples (A) and the mouse brain samples (C), in sequence Logo format (Crooks et al. 2004). (B,D) Sequence preference in the bases opposing the A-to-G editing sites for the human brain samples and the mouse brain samples, respectively.
About half of the detected modifications are supported by the literature, again pointing to the validity of the list (Table 1). Moreover, out of the 12 experimentally validated and documented editing sites in mature human miRNAs that were expressed in our sample, six were identified (Methods; Supplemental Table 3), indicating a satisfactory (but not perfect) detection power. It is also of interest to compare our results with a recent report of editing in mature miRNAs from the mouse brain (Chiang et al. 2010). Editing in eight out of the 19 sites is conserved between the mouse and human, including sites with relatively low editing levels (Table 1). Surprisingly, the editing levels of human and mouse miRNAs are comparable, and in some cases, the editing levels are in fact higher in the mouse. This finding contrasts an earlier NGS-based report claiming that editing in mature miRNAs is less common in mice than in humans (Landgraf et al. 2007). This disagreement might be attributed to the many sequence modifications (other than A-to-G) observed in the previous report (Landgraf et al. 2007), suggesting a higher false-positive rate.
In vitro overexpression experiments validate the in vivo data
The clear editing signal observed makes it reasonable to assume that the novel A-to-G modification sites identified are due to A-to-I editing. Nevertheless, we sought for direct validation of the possibly novel editing sites. We note that direct Sanger sequencing is inefficient for this task, due to editing levels being too low to be detected (<2%) or miRNA in question being weakly expressed. A primer extension assay might be able to differentiate changes in the relative ratios of edited and nonedited microRNA species. However, we have taken a more global approach and tested the effect of ADAR and ADARB1 (also known as ADAR1 and ADAR2, respectively) overexpression on the full miRNA editing profile in human brain–originated U87 glioblastoma cell-line. Knockdown experiments were not considered, due to the low editing levels in this cell-line compared with the levels in brain tissues (Paz et al. 2007). We have used Illumina's HiSeq2000 apparatus to perform NGS of miRNAs from the control U87 cell-line and compared the results to the ADAR and ADARB1 overexpression (Methods). We hypothesized that some of the miRNAs that were detected as edited in the brain tissue sequencing will also be detected as edited in the cell-line overexpression experiments, if they are indeed expressed in this cell-line. Overall, out of 18 miRNAs detected as being edited in the brain tissue 12 miRNAs were expressed in the U87 cell-line. Importantly, a third were indeed validated as ADAR or ADARB1 editing targets in the cell-line overexpression experiments (Methods; Table 1), most of which were found to be ADARB1-dependent.
As ADARB1 overexpression seems to have a larger effect on miRNA editing compared with ADAR in the U87 cell-line (Table 1), we have repeated the ADARB1 overexpression experiment. This time we used a different human brain–originated cell-line, the U118 glioblastoma, and employed a stable ADARB1 transfection assay (Methods). Illumina's HiSeq2000 apparatus was again used to perform NGS of miRNAs from control and ADARB1 overexpressing U118 cell-lines (Methods). Strikingly, all the detected editing sites in the U87 were also detected in the U118, demonstrating the robustness of our detection procedure. Three and 13 additional mature miRNAs editing sites were identified in the U87 and the U118 experiments, respectively, bringing the total number of detected editing events in mature miRNAs to over 30 (Methods; Supplemental Table 5). Note, however, that the sites detected only in the cell-line overexpression samples should be treated with caution as they may not be edited in human physiological conditions. The validation process, for four novel human editing sites, is illustrated in Figure 3.
Figure 3.
Novel human miRNA editing sites detected in the pooled brain and frontal lobe samples and validated using in vitro overexpression experiments in U87 and U118 cell-lines. The editing levels in four miRNAs are shown: (A) miR-455 5p mature position 17, confirmed by ADARB1 overexpression in both U87 and U118 cell-lines; (B) miR-421 mature position 14, confirmed by ADARB1 overexpression in both U87 and U118 cell-lines; (C) miR-381 mature position 4, confirmed by ADAR overexpression in the U87 cell-line; and (D) miR-497 mature position 2, confirmed by ADARB1 overexpression in the U118 cell-line. If the editing site is detected in the mouse brain data of Chiang et al. (2010), the editing levels in this tissue are also presented. The number of sequencing reads supporting the editing site is indicated on the bar. (E,F) The predicted change in mRNA targets as a result of editing in the binding site of miR-381 and miR-497, respectively.
A-to-I editing in miRNAs from the frontal lobe of individuals
As the human brain tissue described above was a pool of different brain regions from multiple donors, it is of interest to examine the profile of miRNA editing in a specific brain region of a single individual. Therefore, Illumina's HiSeq2000 apparatus was used to perform NGS of miRNAs from the frontal lobe of two individuals (Methods). Again, a clear A-to-I editing signal was observed (Methods). Fourteen statistically significant A-to-G modification sites were identified in these two samples, nine of which were also identified in the pooled human brain tissue (Table 1). Out of the five additional sites observed, two were confirmed as editing sites in the ADAR overexpression samples (Table 1). Taken together, the total number of editing sites found in this work for the human brain under physiological conditions adds up to 24. Editing levels in the frontal lobe were consistent between the two individuals and between them and the pooled brain tissue (Table 1). The consistency across individual samples supports the notion that miRNA editing could be utilized functionally (Greenberger et al. 2010). Obviously, more experiments are needed to validate this point.
Possible functional significance of the edited miRNAs
Many of the detected A-to-G modifications were at the recognition site of the mature miRNAs (Table 1). As demonstrated by Kawahara et al. (2007), an editing event that changes the binding specificity of the miRNA may have large impact on cellular processes. We used TargetScan (Lewis et al. 2005) to estimate the effect of the recognition site editing on the miRNA binding specificity (Methods; Supplemental Table 6). The overlap between the putative mRNA targets before and after the editing is very small (∼3% overlap on average) (see two examples in Fig. 3), meaning that editing significantly changes the binding specificity of the miRNAs. Therefore, it is possible that editing events create, de facto, “new miRNAs” in the sense that they have new sets of mRNA targets. It is tempting to speculate that the new mRNA targets have a common functional role. Indeed, functional analysis revealed that some molecular functions are overrepresented in the new mRNA targets (Methods). Most notably, the predicted set of targets for the edited versions of both miR-381 and miR-589 exhibits overrepresentation of neuronal functions (Methods). This finding may hint that the editing in these two miRNAs affects brain physiology, as previously demonstrated for miR-376 (Kawahara et al. 2007).
A-to-I editing signal in publicly available mouse brain miRNAs
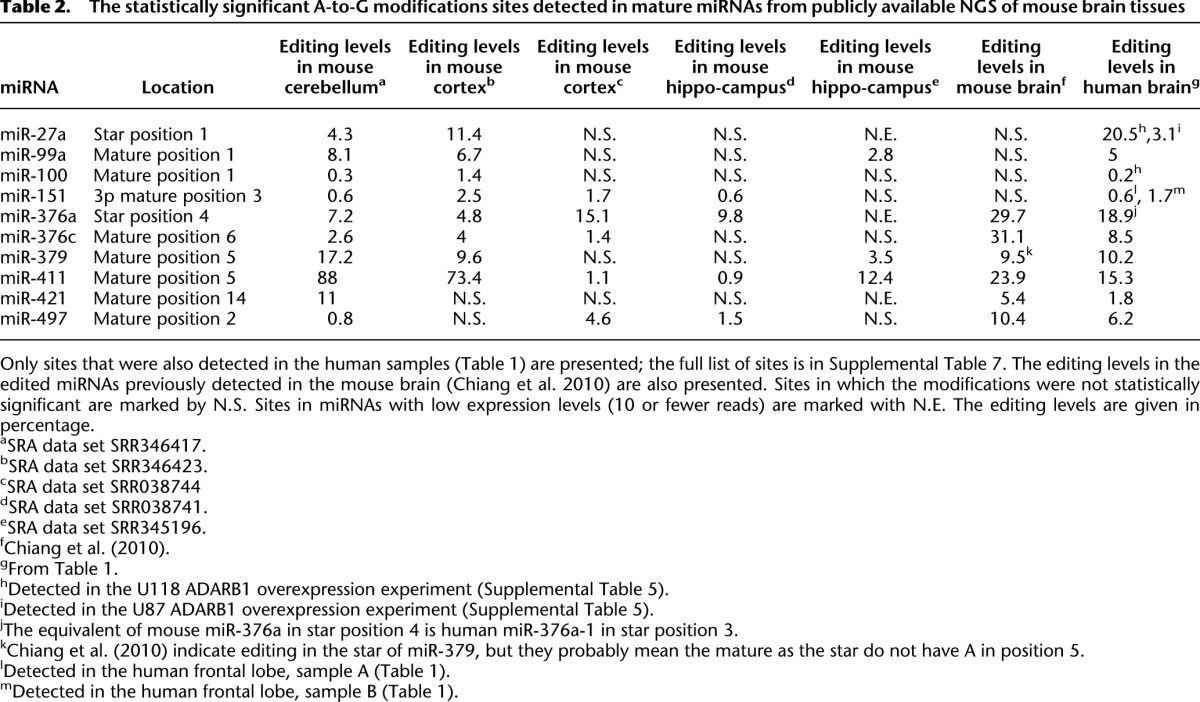
The editing detection procedure described above can be readily used on any NGS data of mature miRNA. We used five publicly available data sets to search for miRNA editing events in the mouse cortex, cerebellum, and hippocampus (Methods; Table 2). In agreement with the human brain data, we see a clear A-to-I editing signal, as 98.8% of the sequence modifications detected are A-to-G mismatches (Supplemental Fig. 5). Many of the detected sites are known mouse editing sites (16/32) (Supplemental Table 7), and many were identified in our human brain experiments above (10/32) (Table 2). In addition, 16 novel editing sites were detected (Methods; Supplemental Table 7). Different tissues typically show similar editing sites. However, in a few cases the levels of editing vary significantly between the different tissues and even between different samples of the same tissue. For example, the editing level of position 5 in the mature sequence of miR-411 varies between 1% and 90% (Table 2).
Table 2.
The statistically significant A-to-G modifications sites detected in mature miRNAs from publicly available NGS of mouse brain tissues
Discussion
Here, we generated and analyzed data of mature miRNAs from human brain tissues and observed a clear A-to-I editing signal, revealing previously characterized editing sites as well as novel editing sites. We have also examined a number of mouse brain region samples. Reassuringly, all data sets yielded similar results, providing overlapping lists of predicted editing sites. We further validated our approach using an in vitro experimental setup in which NGS data were generated from ADAR overexpression in U87 and U118 cell-lines.
Interestingly, many of the detected editing sites have relatively low editing levels (<5%) (Table 1). It is thus possible that many of these sites have no biological significance, but the finding that some of them are conserved in the mouse brain with the same level of editing hints otherwise. One possibility is that editing sites might be edited at a basal low level in generic (or pooled) tissues, being strongly edited only in specific tissues or under specific conditions. Indeed, our data show a number of miRNAs exhibiting high levels of editing in one specific brain tissue sample (e.g., miR-1251 in the mouse cerebellum) (Supplemental Table 7).
In addition, we note that low miRNA editing level does not necessarily mean that the editing has no biological significance. Editing of a miRNA has two effects. One is the reduction in the amount of the wild-type miRNA, and the other is the introduction of a new version of miRNA. A low level of editing in the recognition site of an abundantly expressed miRNA might have virtually no biological effect on the function of the original miRNA, due to the small reduction in the wild-type abundance. However, even if a small fraction of the transcripts of an abundant miRNA is edited to become a “new miRNA,” with a new set of targets, this moderately abundant miRNA might be functionally important. For example, the edited version of miR-381, representing only 6% of the wild-type miRNA count, is in the top quartile of all expressed miRNAs (Supplemental Table 5).
Some of the editing sites detected in mature miRNAs lie outside the recognition site. The biological significance of these is yet to be explored. It was suggested that these editing events may affect the loading of the mature miRNAs to the RNA-induced silencing complex (RISC) complex, thereby altering the effectiveness of the mature miRNAs (Nishikura 2010).
Altogether we identified 24 editing sites in human brain miRNAs (pooled total brain or frontal lobe) and 32 sites in the mouse brain. It should be pointed out that this number is rather small considering the dsRNA structure of the pri-miRNAs. It was previously suggested that many of the pri-miRNA editing events suppress miRNA processing steps (Nishikura 2010). Therefore, it is possible that many pri-miRNAs do get edited but are then discarded and do not result in mature miRNAs. However, there are also reported cases of pri-miRNA editing events that enhance miRNA processing steps (Nishikura 2010). Therefore other explanations should be looked for, including the possibility that the pre-miRNA export from the nucleus is somehow suppressed by the editing (Nishikura 2010). These possible mechanisms may protect numerous genes from being down-regulated by large number of edited mature miRNAs.
The procedure described here allows for a reliable detection and quantification of A-to-I editing events using NGS data of mature miRNAs in varying experimental set-ups, including comparison of the editing profile between normal and pathological conditions. Such experiments will increase our knowledge about how important is the regulation of miRNA editing and whether its loss can be linked to certain pathologies.
Methods
Human brain miRNA sequencing
The RNA sample used was FirstChoice Human Brain Reference RNA (Ambion AM6050), which is pooled from multiple donors and several brain regions. All steps of the protocol for miRNA capture and library construction were conducted according to the method described by Alon et al. (2011). The mature miRNA library was sequenced on one lane of Illumina GAIIx instrument following the manufacturer's protocol. The total number of reads was ∼10 million. All the reads were filtered, demanding that the quality of each read will not be below some threshold value (chosen to be 20) in more than three positions. In addition, sequences identified as 5′ or 3′ adaptors were removed. After adaptors trimming, reads with length longer (>28 bases) or shorter (<15 bases) than the typical length of a mature miRNA were also removed. Approximately 5 million reads passed this filtering process.
Aligning the reads against known miRNAs
As the 3′ end of mature miRNAs undergoes extensive modifications (Burroughs et al. 2010), the last two bases of the read were trimmed (Chiang et al. 2010). The filtered and trimmed reads were aligned using Bowtie (Langmead et al. 2009) against the human genome (UCSC hg19/GRCh37), allowing up to one mismatch. We demanded unique best hits (i.e., reads that cannot be aligned to other locations in the genome with the same number of mismatches). This step solves, by and large, the cross-mapping problem that was reported by de Hoon et al. (2010). The importance of 3′ end trimming, as well as aligning the reads against the genome and not directly against the known miRNA sequences, is illustrated in Figure 1, A through C. If we align all the untrimmed reads against the known miRNA sequences, demanding unique best hits and up to one mismatch, a considerable amount of adenylation and uridylation events is clearly observed (Fig. 1A; Burroughs et al. 2010). When the same procedure is performed with trimmed reads (the last two bases of the reads were removed), A-to-G modifications become dominant (Fig. 1B). Finally, when the trimmed reads are uniquely aligned against the genome and not directly to the miRNAs sequences, the cross-mapping effect is suppressed and the A-to-G signal is enhanced (Fig. 1C).
Overall, ∼1.4 million reads were successfully aligned to the human genome. Only ∼1.1 million reads that were aligned against genomic regions of known pre-miRNAs, as defined in release 17 of miRBase (Kozomara and Griffiths-Jones 2011), were further used. Reads with a perfect match to regions of mature or star miRNAs (according to the definitions of miRBase) were counted. In addition, reads that had only one mismatch to regions of mature or star miRNAs but with a high-quality score (phred score) of 30 and above at this position were also counted. Finally, for each position in the mature and star miRNAs, the counts (number of supporting reads) of each sequenced nucleotide were calculated.
Estimating the sequencing error rate
The sequencing error rate was estimated using the phred score supplied by the Illumina's sequencer. As only mismatches with phred score of 30 were allowed (see above), the expected base call error rate cannot exceed 0.1% in each position. We used the aligned read data to validate that this estimate is indeed compatible with the actual sequencing error in the retained reads. By merging the data from all the mature and star miRNAs, we calculated the rate of any type of mismatch from the 12 possible mismatches. In this calculation, the location of the mismatch should also be taken into account as biological modifications appear in the 3′ of the miRNA (Burroughs et al. 2010) and sequencing quality is lower toward the end of the read (Dohm et al. 2008). Therefore, the calculation was performed separately in each position along the mature/star miRNA. The total mismatch rate (summing all the 12 types of mismatches) was 0.1%–2.6% in the different positions. These numbers are higher than the expected estimation from the phred score (<0.1%). However, the rate relevant for the statistical analysis to follow (see Identifying Editing Sites in miRNAs below) is the maximal mismatch rate of any single type of mismatch, not the sum over all mismatches. Indeed, by excluding four exceptions, no single mismatch type had rates >0.1% at any position. The four exceptions were all A-to-G modifications at positions 1, 4, 5, and 6, and they were caused by extensive A-to-G modifications in miR-99a, miR-381, miR-411, and miR-376c, respectively (Supplemental Table 5). By excluding these four A-to-G mismatches, the total mismatch rate is lowered to 0.1%–0.29%, which is compatible with the rate expected according to the phred score.
Identifying editing sites in miRNAs
All the locations in each mature or star miRNA were screened for mismatches that were overrepresented considering the expected sequencing error rate. This was done by applying the binomial cumulative distribution on the counts of each sequenced nucleotide. The expression filter used was very permissive, taking into account any miRNA with more than five reads. By using a Benjamini-Hochberg false-discovery rate of 5% (Benjamini and Hochberg 1995), we found 19 statistically significant locations (Table 1). Remarkably, almost all the A-to-G mismatches that were recorded before the binomial statistical test were indeed clustered to these 19 locations, whereas all the other types of modifications were filtered out (Fig. 1C,D).
Note that the statistical significance threshold for detection strongly depends on the number of reads available for this miRNA. Thus, it is possible to have sites with the same level of editing in two samples, where in one sample the modification is statistically significant and in the other it is not.
The sequence properties of the detected A-to-G modification sites are illustrated by the example of miR-455 (Supplemental Fig. 1) and summarized in Supplemental Table 2. Sequence preference in the bases flanking the A-to-G editing sites detected in the human brain samples as well as sequence preference in the bases opposing the A-to-G editing sites were created using the WebLogo tool http://weblogo.berkeley.edu/ (Fig. 2; Crooks et al. 2004).
We note that some low-abundance isomirs display 5′ sequence modifications similar to the biological modifications reported at the 3′ of mature miRNAs (Burroughs et al. 2010). We identified 42 isomers that start one or two bases upstream of a different isomer and display sequence modifications in the 5′ end in the form of adenylation and uridylation. In all these events, the abundance of the modified isomer was at least two orders of magnitude lower than the unmodified isomer. Eleven out of these 42 events were at position 1 of the mature miRNA according to the miRBase definition (Kozomara and Griffiths-Jones 2011), and the rest were at upstream positions. Detailed characterization of the 3′ modifications in mature miRNAs is still in process (Burroughs et al. 2010; Wyman et al. 2011), and it is possible that some low abundant 5′ modifications also exist in mature miRNAs. Therefore, we have discarded all these events from our analysis.
Comparison to known editing sites in mature human miRNAs
Experimentally validated editing sites in human miRNAs (Blow et al. 2006; Kawahara et al. 2008) were examined in order to see how many of them were detected by our method (Supplemental Table 3). Only editing in mature and star miRNAs was included in this comparison as our brain samples sequencing data only include mature and star miRNAs.
In vitro overexpression experiments
The overexpression of ADAR and ADARB1 in the U87 and the U118 human glioblastoma cell-lines was performed according to the method described by Paz et al. (2007) and Cenci et al. (2008), respectively (Supplemental Methods). MiRNA capturing and library construction, for the U87 and U118 cell-lines samples, were conducted using Illumina's TruSeq Small RNA Sample Prep Kit. The miRNAs were sequenced using Illumina HiSeq2000 instrument (Supplemental Methods). The sequencing data were analyzed as described above for the human brain miRNA sequencing data (Supplemental Methods).
Human frontal lobe miRNA sequencing and analyzing
Normal frontal white matter samples, obtained from pediatric patients undergoing focal brain resection for head injury (e.g., brain contusion), were used. The two frontal lobe samples (A and B) were obtained from a 3-yr-old boy and a 3-mo-old girl, respectively. MiRNAs were sequenced using Illumina HiSeq2000 instrument, and the sequencing data were analyzed as described above (Supplemental Methods).
Analyzing publicly available mouse brain miRNAs sequencing data
Five data sets were downloaded from the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra): SRR346417, SRR346423, SRR038744, SRR038741, and SRR345196, which contain mature miRNA NGS reads from mouse cerebellum, cortex (first sample), cortex (second sample), hippocampus (first sample), and hippocampus (second sample), respectively. These data sets were analyzed as described above (Supplemental Methods).
Functional analysis of the edited miRNAs
TargetScan (Lewis et al. 2005) was used in order to detect possible targets of the miRNAs, before and after the editing. We used TargetScanHuman 5.2 Custom (http://www.targetscan.org/vert_50/seedmatch.html) to identify conserved mRNA targets for positions 2–8 in the mature miRNAs. Following the method described by Kawahara et al. (2007), we approximate the set of targets for the edited version of the miRNA by treating inosine as a guanosine. Like guanosines, inosines bind strongly to cytidines and more weakly to uridines. The binding energy of inosine to cytidine is roughly the same as that of guanosine, while the binding energy of inosine to uridine is lower than that of guanosine. On the other hand, inosine weakly base-pairs to adenosine, while guanosine does not (Vendeix et al. 2009). The following TargetScan estimates should therefore be considered only as a rough estimate to the change in miRNA specificity due to the editing event. The total number of mRNA targets before and after the editing was recorded, as well as the targets overlap (Supplemental Table 6). The overlap between the putative mRNA targets before and after the editing was ∼3% on average, meaning that editing significantly changes the binding specificity of the miRNAs.
The mRNA targets that were created by the editing events (i.e., were not detected as targets before the editing) were analyzed in order to find overrepresented molecular functions in them. The DAVID bioinformatics tools were used for that purpose (Huang et al. 2009), focusing on overrepresented gene ontology (GO) categories and KEGG pathways (Ashburner et al. 2000; Kanehisa and Goto 2000). The DAVID's default human genes background was used. From the list of 18 miRNAs that were edited in their binding sites (Table 1), six had statistically significant (Benjamini-Hochberg corrected P < 0.05) overrepresentation of molecular functions in their mRNA targets (Supplemental Methods). Similar analysis was performed for the edited miRNAs detected in the mouse brain samples (Supplemental Table 10; Supplemental Methods).
Data access
The sequencing data have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRA029326, SRA044983, and SRA049814. A detailed protocol describing the editing detection scheme, including our scripts, is given at webpage www.tau.ac.il/∼elieis/miR_editing.
Acknowledgments
We thank Professor Gideon Rechavi and Dr. Nurit Paz for providing us with the human ADARs expression vectors, Professor C. Di Rocco for tissues samples, and Dr. Federica Cattonaro from IGA Technology Services for the technical support in the NGS experiments. This work was partially supported by a grant from the United States–Israel Binational Science Foundation (grant no. 2009/290), Jerusalem, Israel, to E.E. This work was also supported by the I-CORE Program of the Planning and Budgeting Committee, The Israel Science Foundation (grant no. 41/11); the Ministry of Health, Israel; the Israeli Cancer Association; and the Wolfson Family Charitable Fund to N.S. This work was also supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC, Milan, Italy) grant to A.G. and a special project 5x1000 AIRC grant to F.L. S.A. was supported by a Clore Fellowship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131573.111.
References
- Alon S, Vigneault F, Eminaga S, Christodoulou DC, Seidman JG, Church GM, Eisenberg E 2011. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res 21: 1506–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR 2006. RNA editing of human microRNAs. Genome Biol 7: R27 doi: 10.1186/gb-2006-7-4-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, et al. 2010. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res 20: 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A 2008. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 283: 7251–7260 [DOI] [PubMed] [Google Scholar]
- Chang T-C, Yu D, Lee Y-S, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, Kishima M, Lassmann T, Faulkner GJ, Mattick JS, et al. 2010. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res 20: 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP 2009. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37: 2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY 2005. Is abundant A-to-I RNA editing primate-specific? Trends Genet 21: 77–81 [DOI] [PubMed] [Google Scholar]
- Greenberger S, Levanon EY, Paz-Yaacov N, Barzilai A, Safran M, Osenberg S, Amariglio N, Rechavi G, Eisenberg E 2010. Consistent levels of A-to-I RNA editing across individuals in coding sequences and non-conserved Alu repeats. BMC Genomics 11: 608 doi: 10.1186/1471-2164-11-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W, Bowcock AM 2011. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 20: 4025–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K 2008. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger Y, Eisenberg E 2010. Large-scale analysis of structural, sequence and thermodynamic characteristics of A-to-I RNA editing sites in human Alu repeats. BMC Genomics 11: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Martí E, Pantano L, Bañez-Coronel M, Llorens F, Miñones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38: 7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu A-L, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K 2010. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 79: 321–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano L, Estivill X, Martí E 2010. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res 38: e34 doi: 10.1093/nar/gkp1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. 2007. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17: 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G 2010. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci 107: 12174–12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Köhler M, Sprengel R, Seeburg PH 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19 [DOI] [PubMed] [Google Scholar]
- Vendeix FAP, Munoz AM, Agris PF 2009. Free energy calculation of modified base-pair formation in explicit solvent: A predictive model. RNA 15: 2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M 2011. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res 21: 1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg Peter H, Shiekhattar R, Nishikura K 2006. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]





