Abstract
Genetic mapping of mutations in model systems has facilitated the identification of genes contributing to fundamental biological processes including human diseases. However, this approach has historically required the prior characterization of informative markers. Here we report a fast and cost-effective method for genetic mapping using next-generation sequencing that combines single nucleotide polymorphism discovery, mutation localization, and potential identification of causal sequence variants. In contrast to prior approaches, we have developed a hidden Markov model to narrowly define the mutation area by inferring recombination breakpoints of chromosomes in the mutant pool. In addition, we created an interactive online software resource to facilitate automated analysis of sequencing data and demonstrate its utility in the zebrafish and mouse models. Our novel methodology and online tools will make next-generation sequencing an easily applicable resource for mutation mapping in all model systems.
There can be little argument that genetic mapping has made a substantial contribution to our understanding of biology. For many years these studies used phenotypically defined markers, such as those used by Morgan in Drosophila and Haldane in mice (Morgan 1911; Haldane et al. 1915). The modern era of genetic analysis was heralded by the recognition that variation in genomic DNA sequence itself could be used as a facile assay for mapping (Botstein et al. 1980). This was initially accomplished using analysis of restriction fragment-length polymorphisms, which were later replaced by microsatellites and subsequently by single nucleotide polymorphisms (SNPs). Despite the remarkable technological advances, these approaches hold in common with those of Morgan and Haldane the utilization of pre-specified markers. Next-generation sequencing (NGS) technology enables simultaneous discovery of very dense sets of informative markers and actual gene mapping in the same experiment. Here, we present a strategy and computational tools to map genes in model organisms using sequencing of pooled samples. The approach can be applied to any model organism with a characterized genome and also to both spontaneous and induced mutants. We demonstrate the utility of the strategy and efficiency of the computational approach by mapping spontaneous and ethylnitrosourea (ENU)-induced developmental mutants in zebrafish and mouse.
Large-scale forward mutagenesis screens in zebrafish have been used with success to investigate fundamental developmental processes. While the recent completion of the zebrafish genome has greatly aided in the identification of genes, mapping analyses continue to rely on the use of traditional microsatellite markers. However, the utilization of SNPs for mapping of zebrafish mutants was proposed almost a decade ago (Stickney et al. 2002), large numbers of SNPs have been identified (Guryev et al. 2006; Bradley et al. 2007), and the application of NGS for SNP discovery and mutation mapping has been demonstrated (Baird et al. 2008), indicating the feasibility of modern genomic approaches.
The reasons SNP analysis has not been routinely used in zebrafish genetics may include the fact that most zebrafish strains used for genetic studies are not inbred and show abundant intrastrain variability in addition to profuse interstrain haplotype sharing (Guryev et al. 2006). Furthermore, no well-characterized panel of SNPs amenable to automated analysis has been developed in a manner similar to that which we developed for mice (Moran et al. 2006). Also, it would be desirable to develop methodology that can take advantage of the fact that large numbers of mutant zebrafish progeny can be generated and analyzed as pools (bulk segregant analysis), and while hybridization-based assays can be used to infer allele distributions in a population (Bradley et al. 2007), this is technically challenging and imprecise.
Fortuitously, the rapid progression in NGS technology makes low-pass whole-genome sequencing a practical method for genetic mapping and positional cloning. Powerful informatics tools enable SNP identification with great efficiency and reliability, such that a priori knowledge of SNP variation, while useful, is not required. Also, because individual reads can be counted, the underlying distribution of alleles in a pool can be enumerated and the sites of recombination breakpoints assessed. While these could be specifically identified by indexing (“bar-coding”) DNA from individual genomes, our method makes this additional step unnecessary. Finally, this approach has the obvious utility of potentially identifying the causal mutation directly, with a likelihood that is proportional to the depth of sequence coverage.
Indeed, NGS analysis of SNP variation has been used for mutation discovery in C. elegans (Sarin et al. 2008; Doitsidou et al. 2010) and mice (Arnold et al. 2011). Furthermore, Schneeberger et al. (2009) and Mokry et al. (2011) compared heterozygous and homozygous areas in Arabidopsis to identify mutation regions; this approach, however, has not been successfully applied to larger vertebrate genomes. Our aim in this report was to determine the map position by integrating SNP variant detection informatics with an algorithm that compares allele distribution between mutant and wild-type pools. This automated method indicates a maximum likelihood interval within which the mutation should reside. Our initial approach used a simple comparative allele-counting method similar to what has been previously reported (Schneeberger et al. 2009; Mokry et al. 2011). This analysis, however, can be undermined by the fact that SNP variation between strains is not randomly distributed, and large noninformative regions can be present. Our method, based on maximization of log likelihood computed by a hidden Markov model (HMM), is much more precise as it integrates allele distribution inferences over large intervals. Moreover, it is tolerant of ambiguity and errors such as sequencing artifacts and misscoring of mutant embryos.
Results
Genetic mapping experiments for recessive mutations rely on identifying regions of homozygosity in the mutant pool. Mutant pools lack diversity in the genomic region harboring the mutation, and the diversity gradually increases with distance from the mutation position due to recombination. Given the incomplete characterization of SNP variants in zebrafish, sequencing of the pool of unaffecteds is required to identify the complete spectrum of informative variants. This is in contrast to previously reported methods in other species, where only affected mutants were sequenced and compared with the reference genome (Sarin et al. 2008). This mapping process can be represented by a hidden Markov model with the architecture presented in Figure 1. Specifically, the number of recombination events from the mutation position can be considered a hidden state. What is observable, however, are sequencing reads covering informative variants in the mutant pool. The number of recombination events from the mutant position determines allele frequency in the sample. If none of the fish chromosomes recombined (HMM state 0), all informative SNPs would be homozygous in the mutant pool. If one recombination occurs (state 1), major allele frequency of an informative SNP in the pool of N chromosomes depends on whether both parents were heterozygous for this SNP or one of the parents was homozygous. If both parents were heterozygous, the major allele frequency is (N-1)/N. If one of the parents is homozygous, there is an equal chance that either the mutant pool will be homozygous for that SNP or it will have the same frequency as in the case of two heterozygous parents (depending on the parental origin of the recombination event). The same logic applies to additional recombinations, corresponding to higher HMM states. Short-read sequencing can then be modeled as a binomial sampling. Overall, the emission probability for state k (the probability to observe i reads supporting major allele out of total m reads in state k) is given by
 |
Here p(het) and p(hom) are probabilities that both parents are heterozygous or one parent is homozygous for any given SNP. In the case of the outcross of inbred lines, both heterozygous parents should always be expected. In the case of an incross, having one homozygous parent is more probable. These probabilities can be easily estimated from the data. Each individual term in the equation represents the contribution of each type of SNP event to the overall probability, i.e., reference allele is major and either both parents are heterozygous or one parent is homozygous, nonreference allele is major and either both parents are heterozygous or one parent is homozygous. Transition probabilities between the mentioned HMM states are determined by the recombination rate. Recombination rate is assumed to be constant along the genome. This assumption greatly facilitates computation and is justified by robustness of the method with respect to local variation of the recombination rate determined in simulation experiments. Starting at the mutation position, at which point there are assumed to be no recombination events, the model can successively move to states with an increasing number of recombinants. We also allow the model to leave the identity by descent (IBD) region and move to the state representing a “random” genome location or to a state corresponding to sequencing or mapping error.
Figure 1.
Schematic representation of mutation analysis by NGS and SNP mapping.
We compute the log likelihood of the HMM using the Viterbi algorithm starting from discrete locations on the chromosome, given that the starting position corresponds to the location of mutation (Nielsen and Sand 2011). The rationale for this approach, as opposed to simply estimating positions of recombination events using HMM, is that the data carry additional information about the mutation location within the completely homozygous region, so the log likelihood is not flat within this homozygous sequence region. That is, the likelihood peak should correspond to the position of the mutation.
For simplicity and computational efficiency, we initially map the mutation to a chromosome by calculating the homozygosity score, expressed as a ratio of heterozygous SNP calls between control and mutant pools multiplied by the number of informative homozygous SNP calls in the mutant pool. This score surveys the entire genome in 10-kb windows and can identify an isolated high-scoring region on a single chromosome. Then, we fine map the mutation position using the algorithm outlined above.
We tested the method in a series of simulation experiments. We varied pool sizes, defined as the number of individual mutants or unaffected siblings analyzed together, and coverage depths to assess the expected maximal likelihood interval size as well as minimum coverage required to roughly map the mutation region (Supplemental Figs. S1–S4). These simulations demonstrated that larger pools may result in narrower mapping intervals, but would require higher coverage depth. Computational experiments with 5–7× coverage for a pool size of 20 individuals resulted in the expected size of the mapping interval of ∼2–3 Mb. Increasing the pool size to 40 individuals narrowed the expected interval to ∼1 Mb (Supplemental Figs. S2–S4). The estimates obtained by simulations may be optimistic, because simulations did not take into account the reference genome quality, the density of errors due to read misalignments, and heterogeneity of SNP density along the genome. We also analyzed simulated data sets made by downsampling coverage of obtained experimental results (described below) and producing coverage as low as 1–1.5× per pool. The homozygosity score can still detect the correct region amid increased noise level on the whole genome plot (Supplemental Fig. S1). However, the HMM score suffers from the inherently low signal and a genomic coverage of at least 2–3× is recommended.
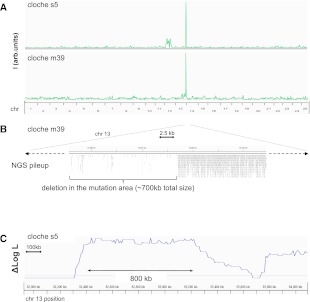
To test the feasibility and accuracy of our approach, we analyzed two distinct alleles of the same mutant phenotype, cloche (clo). This mutant lacks endothelial and hematopoietic cells (Stainier et al. 1995). One allele, clo m39, is a spontaneous deletion mutant originally described in a semi-wild population from an Indonesian fish farm (Stainier et al. 1995, 1996), while another one, clo s5, is caused by a presumed point mutation induced by ENU chemical mutagenesis. The cloche mutation has been previously mapped to the telomeric region of chromosome 13 (Liao et al. 2000). Lysocardiolipin acyltransferase (lycat) has been shown to be located in the clo m39 deletion region and described as a candidate gene responsible for the cloche phenotype (Xiong et al. 2008). For sequencing, 20 pooled mutants and 20 unaffected siblings were obtained from incrosses of clo s5 genotype, and pools of 160 individuals each for the clo m39. Sequencing each pool on a separate lane of Illumina HiSeq2000 platform using 100-bp single-end reads gave between 90 and 110 million reads, resulting in approximately six- to sevenfold genomic coverage (see Supplemental Table ST1). Homozygosity scores for both clo s5 and clo m39 revealed corresponding peaks on the telomeric region of chromosome 13 (Fig. 2A). The clo m39 peak borders an area where no sequence reads were returned for the mutant pool (Fig. 2B), corresponding to the previously described deletion area (Xiong et al. 2008). Log likelihood analysis with HMM revealed a well-defined ∼800-kb interval for both clo alleles, overlapping with the area indicated by the homozygosity score (Fig. 2C). Analysis of the clo s5 allele revealed a log likelihood peak aligned with the deletion interval determined for the clo m39 mutant. The area under the peak that spans a genomic interval of ∼800 kb contains about 22 presumptive genes, including lycat. No homozygous nonsynonymous SNPs in the lycat coding region or splice junctions were found in s5 mutants. In our analysis with 20, and even 160, pooled embryos, the region of maximal likelihood spanned several genes. This is consistent with the results of computer simulations described above.
Figure 2.
Mapping of two alleles of the cloche mutant. (A) Homozygosity scoring of the clo s5 and clo m39 alleles reveals corresponding overlapping peaks on chromosome 13, where the mutation has been previously mapped. (B) Sequence pileup of the clo m39 allele confirms the previously established deletion and reveals the boundaries of the deletion interval. (C) Log likelihood scoring of the clo s5 alleles achieves a higher resolution of the mutation region with a narrower interval than the homozygosity scoring method.
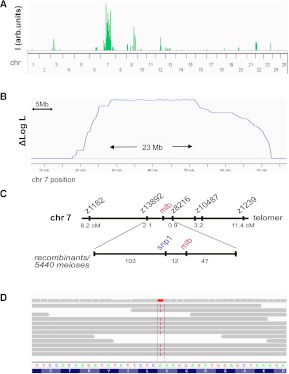
It is important to note that identification of all ENU-induced or novel variants within the mutation region can directly reveal causal mutations, given sufficient sequencing coverage. As an example, we performed NGS-based SNP analysis of a spontaneous zebrafish cilial mutant ca1 and localized the likely recombinant interval to chromosome 3 by homozygosity score (Fig. 3A). HMM log likelihood analysis further defined this region to an ∼7-Mb interval, while the homozygosity score missed the mutation region due to very low SNP frequency in some areas of the interval (Fig. 3B). Targeted PCR sequencing of candidate SNPs revealed a premature stop codon in exon 6 of the fleer (flr) gene, identifying this ca1 mutant line as a new allele of fleer (Fig. 3C; Pathak et al. 2007). In a similar fashion, NGS of the red blood cell mutant malbec (mlb bw306) resulted in eight- to ninefold genomic coverage (see Supplemental Table ST1) and identified a mutation region on chromosome 7 (Fig. 4A,B), consistent with data obtained from prior microsatellite-based mapping analysis of 5440 meioses (Fig. 4C; BH Paw, pers. comm.). Here, instead of comparing the mutant genome to unaffected siblings, wild-type parental siblings (aunts/uncles) were used. Further analysis of the SNPs within the mutation region revealed a nonsense mutation in the gene (Fig. 4D). Allele-specific oligonucleotide hybridization with probes corresponding to the respective wild-type and nonsense mutant alleles showed complete linkage with the mlb phenotype (Supplemental Fig. S5). These examples demonstrate that our method enables direct identification of mutation-causing SNPs.
Figure 3.
Mapping of ca1. (A) Homozygosity scoring reveals a single peak on chromosome 3. (B) Log likelihood analysis (red line) returns a ∼7-Mb interval for the presumptive mutation, while the homozygosity score (green graph) misses the mutation region due to very low SNP frequency in some areas of the interval. (C) Sequencing of exon 6 of the flr gene reveals a premature stop codon.
Figure 4.
Mapping of malbec. (A) Homozygosity scoring maps the mutation to chromosome 7. (B) Log likelihood calculation reveals a ∼23-Mb high-scoring interval. (C) Meiotic map of the mlb locus on chromosome 7. The tightly linked genetic marker snp1 was identified by a traditional chromosomal walk. (D) NGS sequence analysis of mlb and control embryos identified a nonsense mutation in a candidate gene disrupted in mlb. This nonsense mutation is nonrecombinant with the mutant phenotype and resolved all remaining 12 genetic recombinants to a resolution <1/5440 meioses.
One surprising outcome of our analysis was the observation of unexpected patterns in homozygous allele distribution, which appeared to be caused by reference genome misassembly (Fig. 5A). Areas with absolute homozygosity suddenly were interrupted by areas of high heterozygosity in the mutant pool, followed again by regions of complete homozygosity. The HMM score also showed nonmonotonous behavior with severe drops and following hikes back (Figs. 2C, 5B). Inspection of these aberrant patterns and their position with respect to scaffolds showed that they were perfectly delineated by the scaffold boundaries, suggesting the misplacement of several scaffolds in the assembly. Removal of erroneously placed scaffolds (Fig. 5B) resulted in the improvement of the HMM log likelihood curve. Similarly, the incorporation of unplaced scaffolds into the homozygous region could be accomplished by means of the homozygosity score (Supplemental Fig. S6).
Figure 5.
Improving the genome assembly using NGS-based SNP mapping. (A) Analysis of homozygous and heterozygous SNPs in the clo m39 mutation region demonstrates abrupt changes in the frequency of heterozygous SNPs, corresponding with three distinct scaffolds. (B) Removal of the scaffolds indicated above improves the HMM log likelihood score.
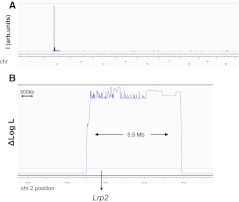
To demonstrate the utility of the method across vertebrate organisms, we applied it to mapping an ENU-induced mutation in a mouse with a holoprosencephaly phenotype (Line 27SH). Craniofacial defects in affected mice include cleft palate, abnormal snout shape, round forehead shape, and abnormal or absent eyes. Gross morphological defects of the brain, smaller or single olfactory bulb, and abnormal interhemispheric fissure, were often observed. Histological analysis revealed that the mutants failed to form a nasal septum or to develop normal forebrain with bilateral hemispheres, and often had fused lateral ventricles (Supplemental Fig. S7). A pool of genomic DNA obtained from nine affected Line 27SH mice was analyzed by NGS, resulting in ∼131 million 75-bp paired-end reads (∼7.6× genomic coverage), which was subsequently analyzed by homozygosity score and HMM. Fully characterized genetic variation in parental strains available from public databases was used instead of sequencing unaffected siblings. This analysis identified a single peak (∼5.5 Mb), confirming and narrowing the independent mapping using SNP genotyping (Fig. 6A,B). We observed one nonsynonymous change that distinguishes mutants from both parental strains in gene Lrp2. Lrp2 is an excellent biological candidate, as a targeted knock-out mutation results in a holopropsencephaly phenotype similar to that observed in our ENU-induced mutant line (Willnow et al. 1996). These results demonstrate full applicability of our method to large mammalian genomes.
Figure 6.
Application to ENU-induced mouse mutant. (A) Homozygosity scoring reveals a single peak on chromosome 2. (B) Log likelihood analysis highlights a 5.5-Mb interval harboring a single nonsynonymous change in the candidate gene Lrp2.
We implemented the entire variant-calling pipeline together with the gene-mapping method in a web-based software tool, SNPtrack. The SNPtrack resource is available online at http://genetics.bwh.harvard.edu/snptrack/. Paired files containing sequencing reads (in fastq format) are uploaded as SNPtrack input. Sequencing reads are automatically aligned by BWA (Li and Durbin 2009) and sequence variants are called using GATK (DePristo et al. 2011). The log likelihood score is then returned to remotely located users. The interactive software enables visualization of the mutation region and direct examination of individual SNPs in the interval. It directly links to the UCSC and Ensembl genome browsers to readily highlight annotated genes in the mutation region, which enables rapid biological assessment and potential validation of putative causal loci by PCR sequencing, cRNA complementation, and morpholino-mediated knockdown.
Discussion
SNPtrack enables rapid and accurate mapping of mutants in model organisms and facilitates immediate identification of causative mutations. In the analysis of fish data, where strains maintain natural genetic variation, we used direct comparison of mutant and unaffected siblings, increasing the density of informative SNPs and reducing mapping errors due to fixed regions of homozygosity in the parental strains. In contrast to prior approaches, we developed a robust and precise methodology by calculating the log likelihood based on a hidden Markov model of recombination breakpoints to define the mutation region. The frequency of intrastrain SNP variation in zebrafish combined with the sensitivity of the analysis obviates the need for outcrossing into polymorphic strains, thereby saving considerable time and resources in the mutant mapping process. We demonstrated that SNPtrack is equally applicable to other species including mice, where the availability of extensively sequenced reference strains results in the complete knowledge of informative SNPs, allowing sequencing of the mutant strains alone. In addition to facilitating gene mapping online, SNPtrack offers the opportunity to compile and compare a large number of sequenced genomes centrally and will serve as an additional resource to further improve and annotate the existing zebrafish genome assembly: This method can improve the assembly of—until now—not fully completed genomes, by enabling the identification of misassembled contigs through the homozygosity score calculated as part of the mapping process. Use of the SNPtrack software by multiple users will allow the accumulation of these informative data to enhance the accuracy of existing assemblies.
This method also allows mapping of mutations when it is not feasible to provide hundreds or thousands of mutants typically required for classical mapping panels, as we only used as low as five or 20 embryos per pool in our studies; this will be of particular interest for hard-to-score phenotypes such as behavioral phenotypes or adult phenotypes, such as cancer susceptibility. Importantly, the presumptive recombination break points are inferred with high resolution, which maximizes the informative content of even limited numbers of characterized meioses. In addition, our method can support innovative strategies to elucidate oligogenic traits such as modifier or synthetic lethal screens. Also, it is possible to envision pursuing mutagenesis on fully inbred backgrounds (i.e., without a cross to a mapping strain) using the mutagen-induced sequence variants as informative markers, which will be directly applicable to mice. Further studies are needed to systematically determine the optimal mapping protocol and optimize parameters of the experiment, including the number of individuals per pool, sequencing coverage depth, and the length of sequence reads. Sequencing technology is rapidly evolving and will lead to faster and cheaper sequencing of longer reads, further enhancing the impact and accuracy of our approach.
Methods
DNA from pooled zebrafish embryos and from mutant mice was isolated using DNeasy Blood+Tissue kits (QIAGEN) and sheared (Covaris) to ∼150 bases (for single-end reads) and ∼350 base length (for paired-end reads). Libraries were constructed using Illumina PE-adapter, NEBNext DNA Library Prep kits, and standard Illumina protocols. Invitrogen 2% SizeSelect E-gel was used to purify and size select the library after the adapter ligation step and as the final post-PCR purification step. A total of 1 μg of DNA per pool was sufficient, indicating the applicability of this method for sequencing of zebrafish embryos as early as 2–3 days post fertilization; the amount can further be lowered by special library preparation protocols. NGS was performed on an Illumina HiSeq2000 apparatus using one lane per pool with 100-bp single-end or 75-bp paired-end reads. Returned read number varied between 90 and 132 million, resulting in about six- to 10-fold coverage of the genome (see Supplemental Table ST1). Sequence read files in fastq format were aligned with the zebrafish and mouse reference genome using the Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009), and SNPs were identified by the Genome Analysis Tool Kit (GATK) (DePristo et al. 2011). Analysis of read alignments and subsequent visualization of scores was accomplished using the Integrative Genomics Viewer (IGV) (Robinson et al. 2011). The crude SNP sets were further quality controlled on mapping accuracy and genomic coverage as well as controlled for possible PCR errors prior to running HMM analysis. Initial homozygosity scoring was performed on the whole-genome scale as described above and the HMM algorithm that used the parredhmmlib (Nielsen and Sand 2011) library was run on the highest scoring chromosome giving an accurate measure of the mutation position.
The HMM architecture is shown in Figure 1. Transition probabilities between states k and k+1 within the IBD region are determined by recombination rate and are given by
where r is per nucleotide recombination rate, assumed to be constant across the genome, and l is the distance between SNPs. For computational feasibility instead of computing transition probability for each and every SNP-to-SNP distance, it is valid to take an average fixed distance (currently 100 bp), and if there is more than one informative SNP inside interval l—dispose the rest, when there is none—an empty SNP (with coverage 0) should be used to maintain the correct distance from mutation. It is worth noting that during this process a small amount of informative SNPs can be lost, though a lot of the SNPs clustered in <100 bp are misalignment errors that are beneficial to avoid. Transition probabilities to the “random” state are chosen so the length of the IBD region would be ∼20 Mb. Certainly the size of IBD region significantly depends on the breeding history of a specific strain, but this value will not affect the HMM behavior greatly if set somewhat incorrectly (the main information comes from the emission of events). Transition probability to the error state was set to 10−5 and the probability to stay in the error state was set to 0.01 to account for clustered errors. Emission probabilities for the states within the IBD region are given by Equation 1. Emission probabilities for the “random” state outside the IBD region are given by a randomized binomial distribution.
Further analysis of variants in the region was performed by the use of Variant Effect Predictor (Ensembl) (McLaren et al. 2010) and snpEff tools (Cingolani 2012). All computational steps are fully automated in the web-based SNPtrack software.
Prior to the mouse mapping experiment, A/J male mice were mutagenized using ENU, and outcrossed with C57BL6/N females to generate the first generation (G1) progeny. The line 127 G1 male was outcrossed with C57BL/6N females to obtain G2 progeny; G2 females were backcrossed with the line 127 G1 male, and mutant G3 progeny with a holoprosencephaly phenotype were identified. Mutants used for WGS were obtained from three different intercrosses using G2 and G3 parents. Genomic DNA was prepared from embryonic liver or skin of a total of nine mutants. NGS and computational analysis was done as described above.
Data access
Next-generation sequencing data used in this work have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRA051382.
Acknowledgments
This work was supported by a Junior Faculty Grant from the Harvard Stem Cell Institute and institutional start-up funds to W.G., American Heart Association grants to J.D.C. and M.J.K., Sigrid Juselius Foundation grant to H.A., March of Dimes grant to B.H.P., and NIH grants 1R01DK090311 to W.G., 5R01MH084676 to S.S., R01HD036404 and R01MH081187 to D.R.B, R01DK070838 and P01HL032262 to B.H.P.
Author contributions: I.L. and S.S. designed and I.L. performed the bioinformatic analysis and created the webtool. K.A., P.K., and M.K.G. isolated mutants and performed DNA isolation and library construction. I.A. assisted with the bioinformatic analysis. C.A.A. and I.A.D. isolated and performed manual sequencing analysis of the fleer mutant allele. J.D.C., M.J.K., H.A., and B.H.P. isolated and performed confirmation analysis of the malbec (mlb bw306) mutant. R.W.S. and S.H. identified, characterized, and provided DNA for the mouse mutant. T.E.N. provided m39 and s5 cloche mutants and DNA and contributed to study design. D.R.B, S.S., and W.G. conceived and supervised the project and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.135541.111.
References
- Arnold CN, Xia Y, Lin P, Ross C, Schwander M, Smart NG, Muller U, Beutler B 2011. Rapid identification of a disease allele in mouse through whole genome sequencing and bulk segregation analysis. Genetics 187: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376 doi: 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314–331 [PMC free article] [PubMed] [Google Scholar]
- Bradley KM, Elmore JB, Breyer JP, Yaspan BL, Jessen JR, Knapik EW, Smith JR 2007. A major zebrafish polymorphism resource for genetic mapping. Genome Biol 8: R55 doi. 10.1186/pb-2007-8-4-r55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani, P. 2012 snpEff: Variant effect prediction. http://snpeff.sourceforge.net. [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O 2010. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 5: e15435 doi: 10.1371/journal.pone.0015435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryev V, Koudijs MJ, Berezikov E, Johnson SL, Plasterk RH, van Eeden FJ, Cuppen E 2006. Genetic variation in the zebrafish. Genome Res 16: 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS, Sprunt AD, Haldane NM 1915. Reduplication in mice. J Genet 5: 133–135 [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Ho CY, Yan YL, Postlethwait J, Stainier DY 2000. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development 127: 4303–4313 [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M, Nijman IJ, van Dijken A, Benjamins R, Heidstra R, Scheres B, Cuppen E 2011. Identification of factors required for meristem function in Arabidopsis using a novel next generation sequencing fast forward genetics approach. BMC Genomics 12: 256 doi: 10.1186/1471-2164-12-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, Manning DK, Bjork BC, Li C, Montgomery K, Siepka SM, et al. 2006. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res 16: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan TH 1911. Random segregation versus coupling in Mendelian inheritance. Science 34: 384 doi: 10.1126/science.34.873.384 [DOI] [PubMed] [Google Scholar]
- Nielsen J, Sand A 2011. Algorithms for a parallel implementation of hidden Markov models with a small state space. IEEE International Parallel & Distributed Processing Symposium, pp. 447–454. IEEE Computer Society, Washington, DC [Google Scholar]
- Pathak N, Obara T, Mangos S, Liu Y, Drummond IA 2007. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell 18: 4353–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Prabhu S, O'Meara MM, Pe'er I, Hobert O 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods 5: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU 2009. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Stainier DY, Weinstein BM, Detrich HW III, Zon LI, Fishman MC 1995. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121: 3141–3150 [DOI] [PubMed] [Google Scholar]
- Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, et al. 1996. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 123: 285–292 [DOI] [PubMed] [Google Scholar]
- Stickney HL, Schmutz J, Woods IG, Holtzer CC, Dickson MC, Kelly PD, Myers RM, Talbot WS 2002. Rapid mapping of zebrafish mutations with SNPs and oligonucleotide microarrays. Genome Res 12: 1929–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, Burns DK, Herz J 1996. Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci 93: 8460–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JW, Yu Q, Zhang J, Mably JD 2008. An acyltransferase controls the generation of hematopoietic and endothelial lineages in zebrafish. Circ Res 102: 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]