Abstract
Separation of the single anterior foregut tube into the esophagus and trachea involves cell proliferation and differentiation, as well as dynamic changes in cell-cell adhesion and migration. These biological processes are regulated and coordinated at multiple levels through the interplay of the epithelium and mesenchyme. Genetic studies and in vitro modeling have shed light on relevant regulatory networks that include a number of transcription factors and signaling pathways. These signaling molecules exhibit unique expression patterns and play specific functions in their respective territories before the separation process occurs. Disruption of regulatory networks inevitably leads to defective separation and malformation of the trachea and esophagus and results in the formation of a relatively common birth defect, esophageal atresia with or without tracheoesophageal fistula (EA/TEF). Significantly, some of the signaling pathways and transcription factors involved in anterior foregut separation continue to play important roles in the morphogenesis of the individual organs. In this review, we will focus on new findings related to these different developmental processes and discuss them in the context of developmental disorders (or birth defects) commonly seen in clinics.
Keywords: EA/TEF, Sox2, BMP, Esophageal Muscle, Myenteric System
The newborn infant quickly needs to fill its lungs with air and later, its stomach with food. The two tubes that enable these substances to enter the body - the trachea and the esophagus - are both derived from a developmental intermediate called the anterior foregut. Separation of this single tube into two separate tubes requires coordinated cellular and molecular events that are orchestrated by multiple signaling pathways and their downstream effectors, including several transcription factors. Abnormalities in these regulatory networks lead to defective separation processes resulting in birth defects such as esophageal atresia with or without tracheoesophageal fistula (EA/TEF), a condition commonly seen in clinics (de Jong et al., 2010; Williamson et al., 2006). We have previously summarized the mutations in genes encoding components of multiple signaling pathways that lead to the formation of EA/TEF in both human patients and mouse models (Que et al., 2006). In recent years, an array of new findings has been added to the list and new pathways involved in the regulation of foregut morphogenesis have been identified (Table 1). Significantly, recent studies have shown that signaling pathways that regulate anterior foregut separation continue to play essential roles in the subsequent organogenesis of the trachea and the esophagus. In this review, we will focus first on recent advances that further our understanding of how the dorsal-ventral patterning of transcription factors and signaling molecules regulates the separation process. We will then review how these regulatory elements participate in the subsequent development of the two tubes, with a focus on the esophagus. In addition, we will also discuss new findings related to the innervation of the esophagus.
Table 1.
Genes Associated with Defects in Tracheoesophgeal Development in Mouse and Human.
| Mouse gene | Foregut malformations (mouse) | EA/TEF (human) | Reference |
|---|---|---|---|
| Shh−/− | EA/TEF, rudimentary lung buds | EA/TEF in some SHH+/− patients | (Litingtung et al., 1998; Spilde et al., 2003) |
| Gli2−/−;Gli3+/− | EA/TEF, abnormal lungs | EA/TEF in some patients with GLI3 mutation | (Johnston et al., 2005; Motoyama et al., 1998) |
| Gli2−/−;Gli3−/− | No esophagus, trachea and lungs | ||
| Foxf1+/− | Narrow esophagus or TEF, lung hypoplasia | EA/TEF with the deletion of locus containing FOXF1 gene | (Mahlapuu et al., 2001; Stankiewicz et al., 2009) |
| RARα−/−;RARβ2−/− | EA/TEF, lung hypoplasia or agenesis | Unknown | (Luo et al., 1996) (Kastner et al., 1997) |
| RARα1−/−;RARβ−/− | (Luo et al., 1996) | ||
| Nkx2.1−/− | TEF, rudimentary lung buds | Unknown | (Minoo et al., 1999) |
| Sox2GFP/COND hypomorph | EA/TEF, abnormal lung epithelial differentiation | EA/TEF in SOX2+/− patients | (Que et al., 2007; Williamson et al., 2006) |
| Noggin−/− | EA/TEF | EA/TEF with the deletion of locus containing NOG gene | (Li et al., 2007; Marsh et al., 2000; Que et al., 2006) |
| HoxC4−/− | Blocked esophageal lumen with abnormal musculature | Unknown | (Boulet and Capecchi, 1996) |
| PCSK5Vcc/Vcc* | TEF, lung hypoplasia | Unknown | (Szumska et al., 2008) |
Mutations of MYCN, CHD7, and MID1.2 have been implicated in the cause of syndromic EA/TEF in humans. However, mouse genetic deletion models reveal no EA/TEF.
Vcc is an Ethylnitrosourea (ENU)-induced mouse mutation which predicts a C470R amino acid change. (Adapted from (de Jong et al., 2010; Que et al., 2006))
Abbreviation: EA, esophageal atresia; TEF, tracheoesophageal fistula.
Overview of the Separation of the Anterior Foregut Tube into the Trachea and Esophagus
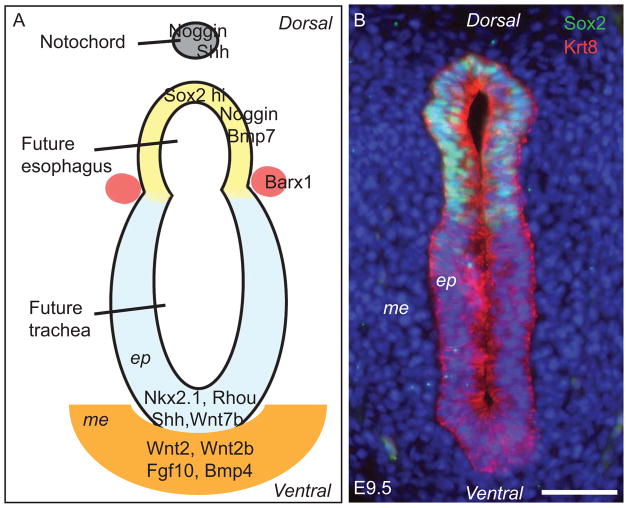
Much of our understanding of foregut separation comes from experimental and genetic manipulations that have revealed the importance of reciprocal signaling between the endoderm and mesoderm during early development (Grapin-Botton and Melton, 2000; Lewis and Tam, 2006; Morrisey and Hogan, 2010). The foregut tube is derived from the ventral folding of the endodermal epithelial sheet during gastrulation at about embryonic (E) day 8.0 in the mouse (Sherwood et al., 2009; Wells and Melton, 1999). This folding is accompanied at around E9.0 by a process in which the rod-like notochord delaminates from the endoderm and becomes more closely associated with the neural tube (Jurand, 1974). Signals emanating from the notochord are essential for the dorsal-ventral patterning of the neural tube and its subsequent tissue morphogenesis (Chamberlain et al., 2008). In recent years, studies have established that once the foregut tube has formed it also exhibits dorsal-ventral patterning of signaling molecules and transcription factors in both the epithelium and the surrounding mesenchyme (Figure 1A). This dorsal-ventral expression pattern is required for normal anterior foregut morphogenesis (E9.5 –E11.5), in which the dorsal region of the tube gives rise to the esophagus and the ventral region forms the trachea and lung buds.
Figure 1.
Dorsal-ventral patterning of the E9.5 anterior foregut. (A) Schematic section through the unseparated anterior foregut tube showing high levels of Sox2, Noggin, Bmp7 in the dorsal epithelium, which will give rise to the esophagus. Conversely, the transcription factor Nkx2.1 and signaling molecules Shh and Wnt7b, along with the Rho GTPase family member Rhou, are highly expressed in the ventral epithelium, which will contribute to the formation of the trachea. The homeobox gene Barx-1 is expressed predominantly in the mesenchyme demarcating the separation site of the dorsal and ventral foregut. Wnt2, Wnt2b, Fgf10 and Bmp4 are enriched in the ventral mesenchyme and are important for gene expression in the underlying epithelium. Mutation of Sox2, Nkx2.1 or Rhou or defects in the Shh, Wnt or Bmp signaling pathways leads to abnormal foregut development, including the formation of esophageal atresia with/without tracheoesophageal fistula (EA/TEF). (B) Immunostained section through the E9.5 foregut tube showing high levels of Sox2 protein in the dorsal epithelium. The cytoskeleton protein Keratin 8 (Krt8) is expressed in both the dorsal and ventral pseudostratified epithelium. Nuclei are counterstained with DAPI. Scale bar: 50μm. ep, epithelium; me, mesenchyme.
Dorsal-Ventral Patterning of the Transcription Factors Sox2 and Nkx2.1 in the Early Foregut
Sox2 is a member of the Sox family of conserved transcription factors, which are characterized by an Sry-related high mobility group (HMG) box. Sox2 is important for the development of multiple organs including the tongue, retina, hair follicles and inner ear and is also required for the self-renewal of embryonic stem (ES) cells (Driskell et al., 2009; Kiernan et al., 2005; Okubo et al., 2006; Taranova et al., 2006; Ura et al., 2011). Sox2 is preferentially expressed in the dorsal epithelial cells of the unseparated foregut tube at E9.5 (Figure 1B) in direct contrast to the transcription factor Nkx2.1 (also known as TTF1), which is expressed predominantly in the ventral epithelium (Harris-Johnson et al., 2009; Que et al., 2007). Proper dorsal-ventral patterning of these two transcription factors proves to be a central requirement for foregut morphogenesis. Significant downregulation of Sox2 protein to near 5% of the wildtype level leads to the formation of EA/TEF in Sox2GFP/COND hypomorphic mutants (Que et al., 2007). By contrast, deletion of Nkx2.1 also results in defects in foregut separation and the formation of EA/TEF with high Sox2 expression in the epithelium of the TEF (Minoo et al., 1999; Que et al., 2007). Conversely, the epithelial cells in the fistula of Sox2GFP/COND hypomorphic mutants express high levels of Nkx2.1 suggesting that in the absence of a sufficiently high level of Sox2, Nkx2.1 expression expands dorsally and reprograms the dorsal epithelium to a respiratory fate (Que et al., 2007). These findings suggest that the dorsal-ventral arrangement of Sox2 and Nkx2.1 is required for foregut separation and the subsequent differentiation of epithelial progenitor cells into esophageal and tracheal epithelium, respectively.
SOX2 has been shown to bind the promoter region of the NKX2.1 gene and inhibit its transcription in human embryonic stem cells (Boyer et al., 2005). Nevertheless, it remains to be determined if a similar regulatory mechanism is active during foregut morphogenesis. We have previously shown using in vitro organ culture that Fgf10 inhibits Sox2 expression in the mouse foregut, but it remains to be determined whether this inhibition is mediated by Nkx2.1 (Que et al., 2007). Fgf10 is enriched in the mesenchyme of the ventral foregut before separation occurs (Figure 1A). However, foregut separation proceeds normally in Fgf10 null mutants despite a lack of lung morphogenesis, suggesting that there are other signaling molecules involved in the regulation of Sox2/Nkx2.1 patterning in the early foregut (Min et al., 1998; Que et al., 2007).
Wnt/β-catenin Signaling Regulates Foregut Morphogenesis
Wnt signaling is highly conserved from nematodes to humans and is a critical mediator of cell-cell signaling events during embryogenesis. Depending on the ligand engagement, Wnt signaling can be transduced through either the canonical Wnt/β-catenin pathway or non-canonical β-catenin independent pathways. In the canonical pathway, Wnt ligands (e.g. Wnt3a, Wnt7b) bind to receptors Frizzled (Fzd) and LDL-receptor-related proteins (Lrp) -5 or -6 and inhibit the phosphorylation of β-catenin by glycogen synthase kinase 3 (GSK3) and casein kinase 1 (CK1). This stabilization of β-catenin promotes its accumulation and subsequent translocation into the nucleus where it interacts with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family to activate the transcription of target genes (Angers and Moon, 2009; Logan and Nusse, 2004). These genes are important regulators of diverse cellular processes including differentiation, proliferation and adhesion. By contrast, the non-canonical pathways are important in regulating cell polarity and asymmetric cell division. These pathways are mediated by the binding of Wnt ligands (e.g. Wnt5a, Wnt11) to Fzd or alternative receptors (e.g. Ror2) to activate the β-catenin–independent Wnt/PKC/Ca2+ and Wnt/JNK polarity pathways (Oishi et al., 2003; Yamamoto et al., 2007; Yamanaka et al., 2002). While recent studies have begun to shed light on the function of canonical Wnt/β-catenin signaling in foregut morphogenesis, non-canonical Wnt signaling remains largely unstudied in this context.
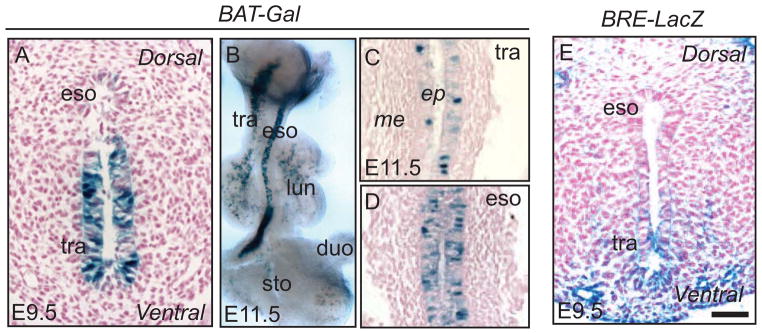
Canonical Wnt/β-catenin signaling activity exhibits a dynamic pattern in the anterior foregut region before and after separation processes occur (Figure 2A–D). At E9.5, Wnt signaling is active in the ventral side of the unseparated foregut tube, where Wnt ligands Wnt2 and Wnt2b proteins are enriched (Figure 1A) (Goss et al., 2009; Harris-Johnson et al., 2009). Interestingly, Wnt2 and 2b are expressed in the mesenchyme of the ventral foregut while X-gal positive staining in the BAT-Gal canonical Wnt signaling reporter mouse line is limited to the epithelium (Figure 2A). This suggests that the Wnt signal receiving cells are located in the epithelium. In line with this notion, deletion of β-catenin in the epithelium using Shh-Cre results in abnormal separation of the foregut tube and complete lung agenesis (Goss et al., 2009; Harris-Johnson et al., 2009). It has further been shown that at the cellular level, Wnt/β-catenin abrogation reduces cell proliferation by diminishing Cyclin D1 protein levels (Goss et al., 2009). Moreover, dorsal-ventral patterning of Sox2/Nkx2.1 is disrupted in Shh-Cre;β-cateninloxp/loxp mutants in which high levels of Sox2 protein are expressed in the ventral region at the expense of the Nkx2.1+ve domain. Accordingly, the resulting fistula expresses high levels of Sox2 (Harris-Johnson et al., 2009). Consistent with the importance of mesenchymal Wnt expression in foregut separation, the combined deletion of Wnt2/2b results in similar phenotypic changes (Goss et al., 2009). Notably, Wnt7b is expressed in the endoderm of the early foregut and its deletion results in irregular lung branching morphogenesis and vasculature development but does not disrupt foregut separation (Shu et al., 2002).
Figure 2.
Dynamic signaling activities in the early anterior foregut tube before and after separation. (A–D) Wnt signaling indicated by the BAT-Gal reporter line. (A) Wnt activity is high in the ventral foregut epithelium at E9.5 as shown by X-gal staining. (B–D) Wnt signaling activities are observed in the epithelium lining both tubes after the anterior foregut separates into the trachea and esophagus at E11.5. (C,D) Sagittal sections. (E) Bmp signaling activity (as reported by Bmp reporter line BRE-LacZ) is high in the epithelium and mesenchyme of the ventral side of the unseparated foregut, consistent with the presence of high levels of Bmp4 and absence of the antagonist Noggin, as shown in Figure 1A. Scale bar: 50μm. eso, esophagus; tra, trachea; lun, lung; duo, duodenum; sto, stomach; ep, epithelium; me, mesenchyme.
The homeobox gene Barx1 is highly expressed in the mesenchyme adjacent to the groove where the future trachea and esophagus split (Figure 1A). Genetic evidence suggests that Barx1 functions to suppress Wnt signaling activity in this region and limits it to the ventral territory prior to the separation of the trachea and esophagus. Deletion of Barx1 leads to a dorsal shift of the domain of Wnt activity accompanied by dorsal expansion of Nkx2.1, resulting in separation defects (Woo et al., 2011).
Foregut separation requires coordinated cytoskeletal rearrangement and cell shape changes and culminates with the division of a single lumen tube into two. Non-canonical Wnt signaling is known to be an important regulator of these cellular processes (Gros et al., 2009; Roszko et al., 2009). However, single deletion of Wnt5a or Wnt11 has no reported foregut separation defects, possibly due to functional redundancy between these genes (Li et al., 2002; Majumdar et al., 2003). It will be interesting to determine if a combined deletion of Wnt5a and Wnt11 induces defects in the separation process. It is noteworthy that Rhou, a Cdc42-related atypical Rho GTPase, has recently been identified as an upstream regulator of the Wnt5a/JNK/PCP pathway (Loebel et al., 2011). In the early foregut, Rhou expression is limited to the ventral and lateral foregut endoderm. In vitro knockdown of Rhou disrupts the differentiation and morphogenesis of foregut derivatives in cultured embryos. Reduced Rhou activity also attenuates the apical accumulation of F-actin and affects cellular morphology and cytoskeletal organization, disrupting the normal conversion of simple columnar epithelium into pseudostratified epithelium during foregut morphogenesis (Loebel et al., 2011). Interestingly, this epithelial conversion also occurs during foregut separation when the ventral side of the tube develops into the lung and trachea [(Loebel et al., 2011) and Que J unpublished observation]. The effect of Rhou deletion on the separation process in vivo remains to be determined.
New Findings on the Roles of Bmp Signaling in Foregut Morphogenesis
We have previously shown that Bmp signaling is required for foregut separation. In the unseparated foregut tube, the Bmp ligand Bmp4 is preferentially expressed in the ventral mesenchyme while Bmp7 and the Bmp inhibitor Noggin are enriched in the dorsal endoderm (Que et al., 2006). Consistent with this dorsal-ventral patterning scheme, Bmp signaling activity is limited to the ventral side of the foregut in the BRE-LacZ (Bmp signaling reporter) embryos (Figure 2E). Disruption of dorsal-ventral patterning by Noggin deletion leads to the formation of EA/TEF in ~70% of the mutants. Similar to the developing skeleton and heart (Brunet et al., 1998; Choi et al., 2007), Noggin deletion leads to increased Bmp signaling in the foregut. Removal of one copy of Bmp4 or Bmp7 in the Noggin null background rescues separation defects (Li et al., 2007; Que et al., 2006). In addition, Noggin deletion also induces abnormal delamination of the notochord from the early definite endoderm epithelial sheet, resulting in epithelial cells of endodermal origin being present in the kinky notochord (Li et al., 2007). These findings suggest an abnormal notochord has a two-fold contribution to defective foregut separation: (1) Abnormal delamination diminishes the quantity of endodermal cells to the point where there are not enough cells to form an intact esophagus and (2) The deformed notochord is unable to provide sufficient signaling to support tissue morphogenesis.
Further support for the importance of dorsal-ventral Bmp signaling comes from findings from the tissue specific ablation of Bmp4. Deletion of Bmp4 using Foxg1-Cre results in tracheal agenesis accompanied by reduced cellular proliferation in both the epithelial and mesenchymal compartments. Significantly, although the trachea does not separate from the foregut, expression of the tracheal lineage marker Nkx2.1 is preserved in the ventral endodermal epithelium, suggesting that Bmp4-mediated signaling is required for separation but not for the initial specification of the tracheal epithelium (Li et al., 2008). In line with these findings, deletion of Bmp receptors 1a and 1b in Shhcre/+; Bmpr1afl/−; Bmpr1b−/− compound mutants also leads to tracheal agenesis, reduction of Nkx2.1 and ventral expansion of Sox2, which is associated with the abrogation of Bmp signaling. Interestingly, Wnt signaling in the foregut of these mutants is not altered, indicating that Wnt signaling does not operate downstream of Bmp during foregut separation. A genetic complementation study showed that removal of the Sox2 gene in a Shhcre/+; Bmpr1afl/−; Bmpr1b−/− background rescues the separation defect, further emphasizing that a dorsal-ventral distribution of signaling and transcription factors is required for foregut separation (Domyan et al., 2011).
Smad proteins, including Smad1/5/8 and Smad4, are key mediators of Bmp signaling. Upon Bmp ligand engagement, Smad1/5/8 are phosphorylated and associate with Smad4, followed by nuclear translocation and activation of downstream target gene transcription (Conidi et al., 2011). Significantly, no foregut separation defects result from the deletion of Smad4 using Nkx2.5-Cre, which is active at ~ E9.5 in both the mesenchyme and epithelium in the ventral foregut [(Que et al., 2009) and Que J unpublished observation]. This could be due to the fact that Smad4 mediates both Bmp and Tgfβ signaling, suggesting that simultaneous loss of these two signals rescues foregut separation defects. In this vein, it will be interesting to determine how Tgfβ signaling is involved in foregut morphogenesis.
Summary of Foregut Tube Separation
Sox2 and Nkx2.1 maintain reciprocal domains of expression in the early foregut endoderm prior to separation. This patterning is controlled by a signaling network consisting of the Fgf, Wnt and Bmp pathways (Figure 1A). The disruption of signaling networks or transcription factor distribution affects cellular proliferation, differentiation and cytoskeletal rearrangement and results in separation abnormalities. Thus far, we have gained considerable insight into the patterning of the ventral side of the foregut, which is characterized by active Nkx2.1 expression and suppressed Sox2 expression. By contrast, we know little about the signaling pathways mediating epithelial-mesenchymal interaction in the dorsal region. This discrepancy is due in part to a deeper understanding of the respiratory derivatives of the ventral foregut than of the development the dorsal foregut derivative, the esophagus. Therefore, more studies in esophageal development are necessary to broaden our understanding of the signaling pathways that act on the dorsal foregut during separation.
Overview of Esophageal Development
Once the esophagus separates from the foregut, it undergoes extensive morphogenesis to become a functional tube that is ensheathed by layers of muscle and lined with stratified squamous epithelium. At the time of foregut separation (~E11.0) the lumen is comprised of a ciliated simple columnar epithelium. It is then gradually replaced by a stratified squamous epithelium that consists of an undifferentiated basal progenitor layer and several differentiated suprabasal layers (Figure 3B)(Yu et al., 2005). Meanwhile, the mesenchymal cells surrounding the nascent esophagus proliferate and differentiate into multiple layers of muscle cells. Although our understanding of esophageal morphogenesis comes primarily from studies of mouse models, there are some important structural differences between the human and mouse esophagi (Table 2). It is noteworthy that in mice the epithelium of both the anterior stomach (also known as the forestomach/proximal stomach) and the esophagus is stratified and keratinized. Recent studies have shown that transcription factors and signaling pathways that are important for foregut separation continue to be critical in the subsequent esophageal epithelial morphogenesis (Table 3). Moreover, new genetic tools have revealed surprising findings about the mechanism by which the mesenchyme develops into muscle cells and their connection to neural networks, and these findings will be reviewed in the context of human esophageal diseases.
Figure 3.
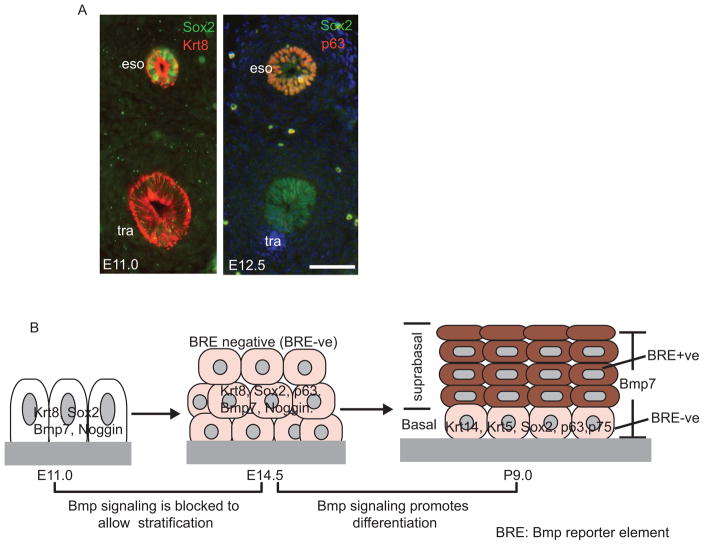
Conversion of simple columnar to stratified squamous epithelium in the esophagus involving dynamic Bmp signaling activities. (A) Immunostained cross sections of E11.0 and E12.5 trachea and esophagus. Krt8 is expressed in the epithelium of both the esophagus and trachea after their formation from the foregut. Sox2 remains highly expressed in the epithelium of the dorsal, foregut-derived esophagus at E11.0 and E12.5. Note the conversion from single to multi-layered epithelium in the esophagus from E11.0 to E12.5, shown by co-immunostaining with p63 and Sox2 antibodies. (B) Schematic of the stratification and differentiation of esophageal epithelium through a two-stage Bmp signaling pattern. Stratification of the epithelium from E11.0 to E14.5 correlates with Noggin-mediated suppression of Bmp signaling. Ectopic Bmp activity in Noggin null and Shh-Cre; Rosa26caBmpr1a mutants inhibits the stratification process. Differentiation of the top layers of epithelium at E14.5-P9.0 requires activation of the Bmp signaling pathway while basal progenitor cells remain negative for Bmp signaling. Deletion of Bmpr1a in Shh-Cre; Bmpr1aloxp/loxp mutants inhibits the differentiation of suprabasal cells. Scale bar: 50μm. eso, esophagus; tra, trachea.
Table 2.
Comparison of the Human and Mouse Esophagus.
| Mouse | Human | |
|---|---|---|
| Length | Adult 1.0–1.5 cm | Adult 18–26cm |
| Keratin layer | Yes | No |
| Thickness of epithelium | 3–5 cells | 20–30 cells |
| Muscularis externa | Cervical and majority of thoracic segments are striated. The lower thoracic and distal segments are smooth muscle. | Cervical region (upper third): skeletal muscle Thoracic region (middle third): skeletal and smooth muscle Abdominal region (lower third): smooth muscle |
| Submucosal glands | No | Yes |
| Epithelium in the forestomach | Non-glandular, stratified squamous epithelium | Simple columnar glandular epithelium |
Table 3.
Transcription Factors Relevant to the Development of Epithelium and Mesenchyme in the Esophagus.
| Gene | Expression Pattern | Esophageal Phenotype of Deletion Mutant | Reference |
|---|---|---|---|
| Sox2 | Epithelium | Mucous metaplasia in the esophagus and forestomach, and the epithelial stratification is disrupted. | (Que et al., 2007) |
| P63 | Epithelium | Mucous metaplasia in the esophagus and forestomach, and the epithelial stratification is disrupted. | (Daniely et al., 2004; Wang et al., 2011) |
| Keap1 | Epithelium | Thickened cornification (keratin layer) in the esophagus and forestomach. | (Wakabayashi et al., 2003) |
| Foxp1/Foxp2 | Foxp1 (epithelium and mesenchyme); Foxp2 (mesenchyme) | Foxp1+/−; Foxp2−/− mutants have a complete absence of esophageal skeletal muscle. | (Shu et al., 2007) |
| HoxC4 | Mesenchyme | Disorganized musculature. Complete esophageal blockage due to epithelial proliferation. | (Boulet and Capecchi, 1996) |
Transcription Factors Sox2, p63 and Nrf2 Regulate Epithelial Morphogenesis in the Developing Esophagus
Sox2 remains highly expressed in the epithelial cells of the esophagus and the forestomach after foregut separation is completed (Figure 3A). Genetic evidence has shown that Sox2 is required for the stratification and lineage differentiation of progenitor cells of both during their development. Reduced Sox2 protein levels in Sox2GFP/COND hypomorphic mutants blocks the formation of a stratified squamous epithelium, resulting in simple columnar epithelial cells that secrete a large amount of mucin (Que et al., 2007). These phenotypic changes resemble mucous metaplasia in the lower esophagus, a pathological condition commonly seen in clinics. This metaplasia partially recaptures phenotypic changes in Barrett’s esophagus (BE, also called intestinal metaplasia), in which simple columnar intestinal-like cells replace the stratified squamous epithelium of the esophagus (Souza et al., 2011). Of note is that Sox2 protein levels are dramatically decreased or completely lost in human BE biopsies (Chen et al., 2008). Moreover, recent studies have shown that SOX2 gene amplification and increased SOX2 protein levels are associated with esophageal squamous cancer (Bass et al., 2009; Gen et al., 2010). Sox2 is exclusively expressed in the basal progenitor cells of the adult esophagus (Arnold et al., 2011), but whether the gain- or loss- of -Sox2 function initiates the pathological conditions remains to be determined.
The transcription factor Trp-63 (p63) is a member of the p53 family, which also includes p73. p63 has two isoforms, TAp63 and ΔNp63 which are transcribed from different promoters and have distinct properties and expression patterns (Candi et al., 2007). While TAp63 is highly expressed in oocytes and is considered a “guardian of the female germline” (Laurikkala et al., 2006), ΔNp63 is the main isoform in the stratified epithelium and is critical to stratification processes (Shalom-Feuerstein et al., 2011). Deletion of the p63 gene affects all stratified epithelia, including the skin and esophagus (Daniely et al., 2004; Mills et al., 1999; Yang et al., 1999). In mutants, the esophageal epithelium fails to stratify and remains simple-columnar with multiple cilia on the apical surface (Daniely et al., 2004). Interestingly, epithelial cells in the forestomach switch on genes normally expressed by mucous-producing cells, suggesting that p63 deletion not only abrogates stratification but also affects epithelial differentiation (Wang et al., 2011). Pertinent to these findings, p63 expression has been shown to be low or completely absent in Barrett’s esophagus (Daniely et al., 2004; Wang et al., 2011).
The transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) is pivotal for mounting cellular defense against oxidative stress via the induction of cytoprotective proteins including NAD(P)H quinone oxidoreductase 1(Nqo1) and glutathione S-transferase (GST) family members (Nguyen et al., 2009). Nrf2 is targeted for degradation by Cullin-3 (Cul-3) based ubiquitin E3 ligase through the substrate adaptor Keap1 (Kobayashi et al., 2004). The cytoprotective function of Nrf2 protein in the developing esophagus is revealed by the study of mutants lacking Keap1, which die at weaning from occlusion of the upper digestive tract by keratin overproduction (Chen et al., 2012; Wakabayashi et al., 2003). The mutants have increased expression of the differentiation markers Involucrin and Loricrin despite no changes in epithelial proliferation. In addition, high levels of Nrf2 protein accumulate in the nuclei of Keap1 mutants and initiate the transcription of Nqo1 and GSTs. Deletion of Nrf2 in a Keap1 null background rescues the hyperkeratosis phenotype (Wakabayashi et al., 2003). These studies provide novel links between the Nrf2/Keap1 pathway and esophageal development but leave open questions of how this pathway or oxidative stress regulates progenitor cell differentiation in the esophagus at the cellular and molecular levels.
Dual Roles of Bmp Signaling for Epithelial Morphogenesis in the Mouse Esophagus and Forestomach
Bmp signaling activity displays a dynamic pattern in the developing esophagus and forestomach. From E11.0~E14.5 the highly proliferative stratifying epithelium remains negative for Bmp activity. However, at E15.0 Bmp signaling is detected in the esophageal suprabasal cells of Bmp reporter (BRE) embryos (Rodriguez et al., 2010). This two-stage presentation of Bmp signaling correlates with Bmp function in the regulation of epithelial development (Figure 3B). In Noggin null mutants, Bmp signaling is ectopically activated during the first stage and results in a simple columnar epithelium that contains a decreased number of p63 positive cells and forms convoluted glandular mucin-secreting pits. In the second stage, activation of Bmp signaling in the suprabasal cells is accompanied by epithelial differentiation. Deletion of Bmp receptor IA with Shh-Cre perturbs lineage differentiation, resulting in Sox2 and p63 expression in the top layers of the epithelium (Rodriguez et al., 2010). During foregut separation Bmp negatively regulates Sox2 transcription (Domyan et al., 2011). It will be interesting to determine whether a similar regulatory mechanism exists in the developing esophagus.
Mesenchymal Differentiation into Muscle Cells Involves the Regulatory Roles of Myogenic Regulatory Factors and Homeobox Genes Foxp1 and Foxp2
Although it has been known for some time that muscle is the major mesenchymal derivative in the developing esophagus, the mechanisms regulating its differentiation are just beginning to be unraveled. The adult mouse esophagus has three muscle layers: the muscularis mucosae (smooth muscle) and two muscularis externae layers (longitudinal and circumferential skeletal muscle in the thoracic segment) (Samarasinghe, 1972; Sang and Young, 1997) (Table 2). In the developing esophagus the outer two muscle layers are composed entirely of smooth muscle cells that are subsequently converted to striated muscle in a craniocaudal direction from E15.5-P21 (Kablar et al., 2000). Earlier studies regarding the formation of the muscle layers have been contradictory, but the introduction of genetic tracing tools has provided new insight into this process (Rishniw et al., 2003).
Initial immunostaining studies showed that markers for smooth muscle (MLCK) and skeletal muscle (MHC) were transiently co-localized in single cells of the esophageal mesenchyme at E15.5 (Patapoutian et al., 1995). Therefore, it was thought that skeletal muscle is derived from the transdifferentiation of the two outer layers of smooth muscle cells (Kablar et al., 2000; Patapoutian et al., 1995; Sang and Young, 1997). Recently, however, genetic lineage-tracing studies using smooth muscle myosin heavy chain-Cre (SmMHC-Cre) mice revealed that transdifferentiation of smooth muscle cells into skeletal muscle cells does not occur in the developing esophagus. Rather, the adult skeletal muscularis externae (longitudinal and circumferential layers) and the initial outer layers of embryonic smooth muscle cells have distinct precursor origins (Rishniw et al., 2003). These findings were further confirmed by a selective gene deletion strategy in which SmMHC-Cre was used to delete loxP-flanked myogenin, a gene essential for striated myogenesis. Despite robust SmMHC-Cre expression in all smooth muscles of the embryonic esophagus, striated myogenesis progresses normally in the esophagus of SmMHC-Cre; myogeninloxp/loxp mutants (Rishniw et al., 2011).
Myogenin belongs to the myogenic regulatory factor (MRF) family, which also includes Myf5, MyoD and MRF4. MRFs are a group of basic helix-loop-helix (bHLH) transcription factors that play essential regulatory functions in the development of skeletal muscle in multiple tissues, including the esophagus (Kablar et al., 2000; Kassar-Duchossoy et al., 2004; Rudnicki et al., 1993; Valdez et al., 2000). Loss of Myf5 but not MyoD in the E17.5 esophagus results in the loss of skeletal muscle and outer muscle layers that remain positive for smooth muscle actin (Kablar et al., 2000). Shh and its downstream transcription factor Gli have been shown to directly regulate Myf5 transcription (Borello et al., 2006; Gustafsson et al., 2002). Consistently, severe defects of myotomal components within the somite have been reported in Shh−/− mutants along with decreased expression of Myf5 and MyoD (Chiang et al., 1996). It remains to be investigated whether Shh regulates skeletal development in the esophagus through similar interactions. Due to the formation of EA/TEF in Shh−/− null mutants (Litingtung et al., 1998), a tissue specific Shh ablation will be needed.
A recent genetic study showed that the homeobox genes Foxp1 and Foxp2 are also required for the differentiation of the mesenchyme into the skeletal muscle of the esophagus (Shu et al., 2007). Foxp1 and Foxp2 are both expressed in the muscular component of the E14.5 esophagus while Foxp1 is also expressed in the epithelium. Foxp1+/−; Foxp2−/− mutants die at birth and have a complete absence of esophageal skeletal muscle. Interestingly, these mutants have only one outer layer of muscle, which remains as smooth muscle at E18.5 (Shu et al., 2007), suggesting that Foxp1 and Foxp2 cooperatively regulate the specification of skeletal muscle.
The Neuronal Innervation of Muscles Requires Close Interaction of Neural Progenitor Cells with Microenvironmental Factors
The striated muscle in the adult esophagus is innervated by both intrinsic and extrinsic neurons (Neuhuber et al., 2006; Sang and Young, 1998), whereas the smooth muscle in the muscularis mucosae is directly innervated mostly by intrinsic neurons (Kamikawa and Shimo, 1979; Storr et al., 2001; Worl et al., 2002). The extrinsic neurons include the vagal nerve which contains sensory and motor fibers (Chang et al., 2003; Powley and Phillips, 2002) and mediates communication between the central and the intrinsic nervous systems (enteric nervous system, ENS), including the myenteric and the submucosal plexi (Aziz and Thompson, 1998). Previous dye-labeling lineage tracing experiments identified vagal sensory fibers innervating the developing mouse esophagus at around E12.0 (Ratcliffe et al., 2006). By contrast, ENS development in the esophagus initiates at E9.0 and proceeds postnatally until around two weeks after birth as a result of extensive proliferation and differentiation of precursor cells migrating from the neural crest (Breuer et al., 2004; Durbec et al., 1996; Sang and Young, 1997; Taraviras and Pachnis, 1999). Along the route of travel the crest-derived precursor cells interact closely with microenvironmental signaling factors including growth factors and extracellular matrix components to sequentially switch on genes necessary for ENS development.
The transcription factor Sox10 is expressed in the pre-migratory neural crest cells destined to colonize the esophagus and other parts of the gut. The role of Sox10 in ENS development was identified by positional cloning of the Dominant megacolon (Dom) locus of mice. Animals homozygous for this mutation (Dom/Dom) die during embryogenesis (60% die at E12–E13) and lack enteric neurons and their precursors from the entire length of the esophagus and gastrointestinal tract (Kapur, 1999). Prior to entry into the foregut, crest-derived progenitor cells start to express c-Ret, a receptor for glial cell line-derived neurotrophic factor (Gdnf) (Taraviras et al., 1999; Taraviras and Pachnis, 1999). Gdnf is enriched in the gut muscle and serves as a potent chemo-attractant for the migratory crest progenitor cells. c-Ret, a member of receptor tyrosine kinase, regulates the survival, proliferation and differentiation of progenitor cells during early stages of ENS development. Disruption of Gdnf/Ret signaling in Ret null mutants reduces the number of enteric neurons in the esophagus and forestomach and leads to aganglionosis in other parts of gastrointestinal tract (Durbec et al., 1996; Yan et al., 2004). Consistently, combined deletion of Sulf1 and Sulf2, sulfotransferases that modify the binding of Gdnf to extracellular matrix and c-Ret, also leads to reduced esophageal innervation (Ai et al., 2007). Shortly after the expression of c-Ret the crest progenitor cells start to express Mash1, a basic helix-loop-helix (b-HLH) transcription factor. Mash1 is critical for the generation of sublineages of enteric neurons. Deletion of Mash1 leads to the loss of serotonin- and nitric oxide synthase (NOS)-containing neurons (Blaugrund et al., 1996; Guillemot et al., 1993; Sang et al., 1999). Interestingly, while Sox10 is not required for the expression of Mash1 in neural crest progenitor cells in vitro, it regulates Mash1 induction in vivo and Mash1 expression is lost in Dom/Dom mutants (Kim et al., 2003). Mash1 induction by Sox10 imparts multipotent neural crest progenitors with neural differentiation capability. However, once neural differentiation is initiated, Mash1 attenuates Sox10 expression, supporting a negative-feedback loop in ENS development (Kim et al., 2003).
A functional extrinsic nervous system is critical for the generation of peristaltic movement in the striated muscle portion of the esophagus. By contrast, the intrinsic nervous system fine-tunes peristalsis by regulating the contractility of smooth muscle cells in response to inputs from the extrinsic system. In humans, the ENS also modulates the activity of the submucosal glands in the esophagus. Neural innervation defects can lead to neonatal death (Mash1−/−) and esophageal disorders, such as megaesophagus (Sulf1−/−; Sulf2−/−) (Guillemot et al., 1993; Neuhuber et al., 2006; van der Weyden et al., 2009). Defects in the generation of distal esophageal inhibitory neurons (NOS+ve) can also lead to achalasia, a motility disorder in which the lower esophageal sphincter (LES) fails to relax (Francis and Katzka, 2010). Mutant mice deficient in neuronal NOS (nNOS), Lsc/p115 (Rho guanine nucleotide exchange factor 1) or Rassf1 (Ras association family member 1) have impaired relaxation of the LES (Goyal and Chaudhury, 2010; Sivarao et al., 2001; van der Weyden et al., 2009; Zizer et al., 2010). Interestingly, although it has been proposed that the inhibitory neurons mediate LES relaxation through intramuscular interstitial cells of Cajal (ICC-IM), achalasia is not observed in W/Wv mutants in which ICC-IM cells are ablated, which suggests that this unique cell population is not directly involved in controlling muscle tone in the esophagus (Sivarao et al., 2001).
Summary of the Genetic and Cellular Mechanisms Underlying Esophageal Development
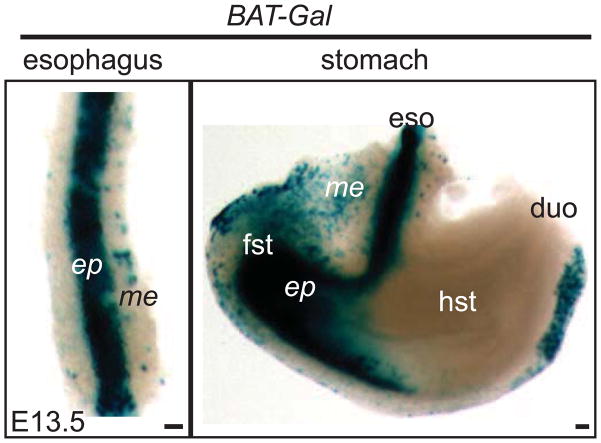
Conversion of the simple columnar epithelium of the embryonic esophagus into a stratified squamous epithelium requires the continued participation of Sox2 and Bmp signaling. p63 also plays an important role in epithelial morphogenesis and is essential for epithelial stratification. In future studies, it will be interesting to examine the regulatory relationship between these transcription factors. While Wnt signaling is known to be active in the esophagus after its separation from the foregut (Figure 4)(Chen et al., 2012), its role in esophageal development remains to be discerned. In addition, the transformation of the thin mesenchyme into multiple layers of muscle infiltrated by nerves and blood vessels requires microenvironmental factors including growth factors and elements of the extracellular matrix.
Figure 4.
Wnt signaling activity as shown by the BAT-Gal reporter in the E13.5 esophagus and stomach. The stratified epithelium in the esophagus and forestomach is strongly positive for X-gal staining. Sporadic X-gal positive cells are also present in the mesenchyme of both the esophagus and forestomach. Note that part of the mesenchyme in the hindstomach is also positive for X-gal staining. Scale bar: 50μm. ep, epithelium; me, mesenchyme; fst, forestomach; eso, esophagus; hst, hindstomach; duo, duodenum.
Conclusion and Future Directions
The separation of the anterior foregut into the trachea and esophagus and the subsequent development of the esophagus involve reciprocal interactions between the epithelium and the mesenchyme that are mediated by signaling molecules and transcription factors. Still, how morphogenetic processes such as cell proliferation, differentiation and migration are controlled and regulated at the cellular level remains largely unexplored. Gaining a greater understanding of these processes will require more genetic and molecular studies using new mouse lines for gain and loss-of-function experiments. Biochemical tools in conjunction with in vivo genetic manipulation will be helpful in providing further insights into pertinent molecular mechanisms. Techniques including chromatin immunoprecipiation (ChIP) and ChIP-sequencing that have been instrumental in studying the function of transcription factors in the development of other tissues will be especially important. One issue that has not been well-studied is the morphogenesis of the submucosal glands, whose secretory products are critical for esophageal luminal clearance and tissue resistance (Long and Orlando, 1999). In addition, the expansion of gland ductal cells has been associated with re-epithelialization in Barrett’s esophagus patients after laser and photodynamic therapy (Biddlestone et al., 1998). However, we know little about the morphogenesis of these unique glands due to the lack of proper animal models. We don’t even know when exactly they are generated or whether they are generated from ingrowths of surface squamous epithelium or as direct extensions of the oropharyngeal minor salivary glands (Johns, 1952; Krause et al., 1976). The recently introduced Zinc-finger nuclease (ZFN) technology will enable genetic engineering in species other than mouse and will likely provide answers to these outstanding questions through studies in species that do have submucosal glands, such as swine and opossum (Carroll, 2011; Watanabe et al., 2010). This new tool will be particularly useful in targeting candidate genes that may potentially regulate gland initiation and the specification of progenitor cells into different lineages within glands (Abdulnour-Nakhoul et al., 2007; Long and Orlando, 1999; Watanabe et al., 2010).
Advances in the understanding of morphogenetic processes during embryonic development will promote greater insights into the pathophysiology of esophageal diseases. We and others have shown that abnormal levels of Sox2 and Bmps are associated with esophageal diseases including Barrett’s esophagus and cancers (Bass et al., 2009; Chen et al., 2008; Milano et al., 2007; Que et al., 2007). A better characterization of relevant mechanisms will help in devising therapeutic strategies for targeting these diseases. Finally, we expect that better knowledge of esophageal development will lend support to future mechanistic studies of foregut separation as a whole.
Highlights.
An updated overview of foregut morphogenesis and esophageal development.
Dorsal-ventral patterning of signaling and transcription factors in early foregut.
Disruption of D-V patterning affects the separation of the trachea and esophagus.
Same molecules are essential for subsequent esophageal development.
Overview of muscle development and innervation in the esophagus.
Acknowledgments
We thank members of our laboratory and Dr. Brigid Hogan, Department of Cell Biology at Duke University and Dr. Roy Orlando, Department of Medicine at the University of North Carolina (Chapel Hill) for critical reading of the manuscript and helpful discussion. Research in the Que lab is partly supported by NIH K99/R00 Independent Pathway Award DK082650 and March of Dimes Basil O’Connor Starter Scholar Research Award. Figure 2E is reproduced with permission from Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulnour-Nakhoul S, Nakhoul NL, Wheeler SA, Haque S, Wang P, Brown K, Orlando G, Orlando RC. Characterization of esophageal submucosal glands in pig tissue and cultures. Dig Dis Sci. 2007;52:3054–3065. doi: 10.1007/s10620-006-9739-3. [DOI] [PubMed] [Google Scholar]
- Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz Q, Thompson DG. Brain-gut axis in health and disease. Gastroenterology. 1998;114:559–578. doi: 10.1016/s0016-5085(98)70540-2. [DOI] [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O’Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, Jr, UR, Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlestone LR, Barham CP, Wilkinson SP, Barr H, Shepherd NA. The histopathology of treated Barrett’s esophagus: squamous reepithelialization after acid suppression and laser and photodynamic therapy. Am J Surg Pathol. 1998;22:239–245. doi: 10.1097/00000478-199802000-00013. [DOI] [PubMed] [Google Scholar]
- Blaugrund E, Pham TD, Tennyson VM, Lo L, Sommer L, Anderson DJ, Gershon MD. Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympathoadrenal lineage markers and Mash-1-dependence. Development. 1996;122:309–320. doi: 10.1242/dev.122.1.309. [DOI] [PubMed] [Google Scholar]
- Borello U, Berarducci B, Murphy P, Bajard L, Buffa V, Piccolo S, Buckingham M, Cossu G. The Wnt/beta-catenin pathway regulates Gli-mediated Myf5 expression during somitogenesis. Development. 2006;133:3723–3732. doi: 10.1242/dev.02517. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR. Targeted disruption of hoxc-4 causes esophageal defects and vertebral transformations. Dev Biol. 1996;177:232–249. doi: 10.1006/dbio.1996.0159. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C, Neuhuber WL, Worl J. Development of neuromuscular junctions in the mouse esophagus: morphology suggests a role for enteric coinnervation during maturation of vagal myoneural contacts. The Journal of comparative neurology. 2004;475:47–69. doi: 10.1002/cne.20156. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Candi E, Dinsdale D, Rufini A, Salomoni P, Knight RA, Mueller M, Krammer PH, Melino G. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 2007;6:274–285. doi: 10.4161/cc.6.3.3797. [DOI] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain CE, Jeong J, Guo C, Allen BL, McMahon AP. Notochord-derived Shh concentrates in close association with the apically positioned basal body in neural target cells and forms a dynamic gradient during neural patterning. Development. 2008;135:1097–1106. doi: 10.1242/dev.013086. [DOI] [PubMed] [Google Scholar]
- Chang HY, Mashimo H, Goyal RK. Musings on the wanderer: what’s new in our understanding of vago-vagal reflex? IV. Current concepts of vagal efferent projections to the gut. Am J Physiol Gastrointest Liver Physiol. 2003;284:G357–366. doi: 10.1152/ajpgi.00478.2002. [DOI] [PubMed] [Google Scholar]
- Chen H, Li J, Li H, Hu Y, Tevebaugh W, Yamamoto M, Que J, Chen X. Transcript Profiling Identifies Dynamic Gene Expression Patterns and an Important Role for Nrf2/Keap1 Pathway in the Developing Mouse Esophagus. PLoS One. 2012;7:e36504. doi: 10.1371/journal.pone.0036504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Qin R, Liu B, Ma Y, Su Y, Yang CS, Glickman JN, Odze RD, Shaheen NJ. Multilayered epithelium in a rat model and human Barrett’s esophagus: similar expression patterns of transcription factors and differentiation markers. BMC Gastroenterol. 2008;8:1. doi: 10.1186/1471-230X-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Choi M, Stottmann RW, Yang YP, Meyers EN, Klingensmith J. The bone morphogenetic protein antagonist noggin regulates mammalian cardiac morphogenesis. Circ Res. 2007;100:220–228. doi: 10.1161/01.RES.0000257780.60484.6a. [DOI] [PubMed] [Google Scholar]
- Conidi A, Cazzola S, Beets K, Coddens K, Collart C, Cornelis F, Cox L, Joke D, Dobreva MP, Dries R, Esguerra C, Francis A, Ibrahimi A, Kroes R, Lesage F, Maas E, Moya I, Pereira PN, Stappers E, Stryjewska A, van den Berghe V, Vermeire L, Verstappen G, Seuntjens E, Umans L, Zwijsen A, Huylebroeck D. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine & growth factor reviews. 2011;22:287–300. doi: 10.1016/j.cytogfr.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- de Jong EM, Felix JF, de Klein A, Tibboel D. Etiology of esophageal atresia and tracheoesophageal fistula: “mind the gap”. Curr Gastroenterol Rep. 2010;12:215–222. doi: 10.1007/s11894-010-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development. 2011;138:971–981. doi: 10.1242/dev.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Giangreco A, Jensen KB, Mulder KW, Watt FM. Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development. 2009;136:2815–2823. doi: 10.1242/dev.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbec PL, Larsson-Blomberg LB, Schuchardt A, Costantini F, Pachnis V. Common origin and developmental dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development. 1996;122:349–358. doi: 10.1242/dev.122.1.349. [DOI] [PubMed] [Google Scholar]
- Francis DL, Katzka DA. Achalasia: update on the disease and its treatment. Gastroenterology. 2010;139:369–374. doi: 10.1053/j.gastro.2010.06.024. [DOI] [PubMed] [Google Scholar]
- Gen Y, Yasui K, Zen Y, Zen K, Dohi O, Endo M, Tsuji K, Wakabayashi N, Itoh Y, Naito Y, Taniwaki M, Nakanuma Y, Okanoue T, Yoshikawa T. SOX2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma. Cancer genetics and cytogenetics. 2010;202:82–93. doi: 10.1016/j.cancergencyto.2010.01.023. [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal RK, Chaudhury A. Pathogenesis of achalasia: lessons from mutant mice. Gastroenterology. 2010;139:1086–1090. doi: 10.1053/j.gastro.2010.08.013. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Melton DA. Endoderm development: from patterning to organogenesis. Trends Genet. 2000;16:124–130. doi: 10.1016/s0168-9525(99)01957-5. [DOI] [PubMed] [Google Scholar]
- Gros J, Serralbo O, Marcelle C. WNT11 acts as a directional cue to organize the elongation of early muscle fibres. Nature. 2009;457:589–593. doi: 10.1038/nature07564. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Gustafsson MK, Pan H, Pinney DF, Liu Y, Lewandowski A, Epstein DJ, Emerson CP., Jr Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification. Genes Dev. 2002;16:114–126. doi: 10.1101/gad.940702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns BA. Developmental changes in the oesophageal epithelium in man. Journal of anatomy. 1952;86:431–442. [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, Booth C, Curry CJ, David A, Dinulos MB, Flannery DB, Fox MA, Graham JM, Grange DK, Guttmacher AE, Hannibal MC, Henn W, Hennekam RC, Holmes LB, Hoyme HE, Leppig KA, Lin AE, Macleod P, Manchester DK, Marcelis C, Mazzanti L, McCann E, McDonald MT, Mendelsohn NJ, Moeschler JB, Moghaddam B, Neri G, Newbury-Ecob R, Pagon RA, Phillips JA, Sadler LS, Stoler JM, Tilstra D, Walsh Vockley CM, Zackai EH, Zadeh TM, Brueton L, Black GC, Biesecker LG. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. American journal of human genetics. 2005;76:609–622. doi: 10.1086/429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurand A. Some aspects of the development of the notochord in mouse embryos. Journal of embryology and experimental morphology. 1974;32:1–33. [PubMed] [Google Scholar]
- Kablar B, Tajbakhsh S, Rudnicki MA. Transdifferentiation of esophageal smooth to skeletal muscle is myogenic bHLH factor-dependent. Development. 2000;127:1627–1639. doi: 10.1242/dev.127.8.1627. [DOI] [PubMed] [Google Scholar]
- Kamikawa Y, Shimo Y. Cholinergic and adrenergic innervations of the muscularis mucosae in guinea-pig esophagus. Archives internationales de pharmacodynamie et de therapie. 1979;238:220–232. [PubMed] [Google Scholar]
- Kapur RP. Early death of neural crest cells is responsible for total enteric aganglionosis in Sox10(Dom)/Sox10(Dom) mouse embryos. Pediatric and developmental pathology: the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society. 1999;2:559–569. doi: 10.1007/s100249900162. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kastner P, Mark M, Ghyselinck N, Krezel W, Dupe V, Grondona JM, Chambon P. Genetic evidence that the retinoid signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development. 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Molecular and cellular biology. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause WJ, Cutts JH, Leeson CR. The postnatal development of the alimentary canal in the opossum. I. Oesophagus. Journal of anatomy. 1976;122:293–314. [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Lewis SL, Tam PP. Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn. 2006;235:2315–2329. doi: 10.1002/dvdy.20846. [DOI] [PubMed] [Google Scholar]
- Li C, Xiao J, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Developmental biology. 2002;248:68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- Li Y, Gordon J, Manley NR, Litingtung Y, Chiang C. Bmp4 is required for tracheal formation: a novel mouse model for tracheal agenesis. Dev Biol. 2008;322:145–155. doi: 10.1016/j.ydbio.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Litingtung Y, Ten Dijke P, Chiang C. Aberrant Bmp signaling and notochord delamination in the pathogenesis of esophageal atresia. Dev Dyn. 2007;236:746–754. doi: 10.1002/dvdy.21075. [DOI] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- Loebel DA, Studdert JB, Power M, Radziewic T, Jones V, Coultas L, Jackson Y, Rao RS, Steiner K, Fossat N, Robb L, Tam PP. Rhou maintains the epithelial architecture and facilitates differentiation of the foregut endoderm. Development. 2011;138:4511–4522. doi: 10.1242/dev.063867. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annual review of cell and developmental biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Long JD, Orlando RC. Esophageal submucosal glands: structure and function. Am J Gastroenterol. 1999;94:2818–2824. doi: 10.1111/j.1572-0241.1999.1422_b.x. [DOI] [PubMed] [Google Scholar]
- Luo J, Sucov HM, Bader JA, Evans RM, Giguere V. Compound mutants for retinoic acid receptor (RAR) beta and RAR alpha 1 reveal developmental functions for multiple RAR beta isoforms. Mechanisms of development. 1996;55:33–44. doi: 10.1016/0925-4773(95)00488-2. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Vainio S, Kispert A, McMahon J, McMahon AP. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 2003;130:3175–3185. doi: 10.1242/dev.00520. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Enerback S, Carlsson P. Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development. 2001;128:2397–2406. doi: 10.1242/dev.128.12.2397. [DOI] [PubMed] [Google Scholar]
- Marsh AJ, Wellesley D, Burge D, Ashton M, Browne C, Dennis NR, Temple K. Interstitial deletion of chromosome 17 (del(17)(q22q23.3)) confirms a link with oesophageal atresia. Journal of medical genetics. 2000;37:701–704. doi: 10.1136/jmg.37.9.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano F, van Baal JW, Buttar NS, Rygiel AM, de Kort F, DeMars CJ, Rosmolen WD, Bergman JJ, JVAM, Wang KK, Peppelenbosch MP, Krishnadath KK. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132:2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- Neuhuber WL, Raab M, Berthoud HR, Worl J. Innervation of the mammalian esophagus. Advances in anatomy, embryology, and cell biology. 2006;185:1–73. back cover. [PubMed] [Google Scholar]
- Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Suzuki H, Onishi N, Takada R, Kani S, Ohkawara B, Koshida I, Suzuki K, Yamada G, Schwabe GC, Mundlos S, Shibuya H, Takada S, Minami Y. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes to cells: devoted to molecular & cellular mechanisms. 2003;8:645–654. doi: 10.1046/j.1365-2443.2003.00662.x. [DOI] [PubMed] [Google Scholar]
- Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patapoutian A, Wold BJ, Wagner RA. Evidence for developmentally programmed transdifferentiation in mouse esophageal muscle. Science. 1995;270:1818–1821. doi: 10.1126/science.270.5243.1818. [DOI] [PubMed] [Google Scholar]
- Powley TL, Phillips RJ. Musings on the wanderer: what’s new in our understanding of vago-vagal reflexes? I. Morphology and topography of vagal afferents innervating the GI tract. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1217–1225. doi: 10.1152/ajpgi.00249.2002. [DOI] [PubMed] [Google Scholar]
- Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006;74:422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe EM, Setru SU, Chen JJ, Li ZS, D’Autreaux F, Gershon MD. Netrin/DCC-mediated attraction of vagal sensory axons to the fetal mouse gut. The Journal of comparative neurology. 2006;498:567–580. doi: 10.1002/cne.21027. [DOI] [PubMed] [Google Scholar]
- Rishniw M, Rodriguez P, Que J, Burke ZD, Tosh D, Chen H, Chen X. Molecular aspects of esophageal development. Annals of the New York Academy of Sciences. 2011;1232:309–315. doi: 10.1111/j.1749-6632.2011.06071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishniw M, Xin HB, Deng KY, Kotlikoff MI. Skeletal myogenesis in the mouse esophagus does not occur through transdifferentiation. Genesis. 2003;36:81–82. doi: 10.1002/gene.10198. [DOI] [PubMed] [Google Scholar]
- Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137:4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszko I, Sawada A, Solnica-Krezel L. Regulation of convergence and extension movements during vertebrate gastrulation by the Wnt/PCP pathway. Seminars in cell & developmental biology. 2009;20:986–997. doi: 10.1016/j.semcdb.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Samarasinghe DD. Some observations on the innervation of the striated muscle in the mouse oesophagus--an electron microscopy study. Journal of anatomy. 1972;112:173–184. [PMC free article] [PubMed] [Google Scholar]
- Sang Q, Ciampoli D, Greferath U, Sommer L, Young HM. Innervation of the esophagus in mice that lack MASH1. The Journal of comparative neurology. 1999;408:1–10. doi: 10.1002/(sici)1096-9861(19990524)408:1<1::aid-cne1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Sang Q, Young HM. Development of nicotinic receptor clusters and innervation accompanying the change in muscle phenotype in the mouse esophagus. The Journal of comparative neurology. 1997;386:119–136. doi: 10.1002/(sici)1096-9861(19970915)386:1<119::aid-cne11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Sang Q, Young HM. The origin and development of the vagal and spinal innervation of the external muscle of the mouse esophagus. Brain research. 1998;809:253–268. doi: 10.1016/s0006-8993(98)00893-2. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E. [Delta]Np63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Jiang YQ, Lu MM, Morrisey EE. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Sivarao DV, Mashimo HL, Thatte HS, Goyal RK. Lower esophageal sphincter is achalasic in nNOS(−/−) and hypotensive in W/W(v) mutant mice. Gastroenterology. 2001;121:34–42. doi: 10.1053/gast.2001.25541. [DOI] [PubMed] [Google Scholar]
- Souza RF, Freschi G, Taddei A, Ringressi MN, Bechi P, Castiglione F, Rossi Degl’Innocenti D, Triadafilopoulos G, Wang JS, Chang AC, Barr H, Bajpai M, Das KM, Schneider PM, Krishnadath KK, Malhotra U, Lynch JP. Barrett’s esophagus: genetic and cell changes. Annals of the New York Academy of Sciences. 2011;1232:18–35. doi: 10.1111/j.1749-6632.2011.06043.x. [DOI] [PubMed] [Google Scholar]
- Spilde T, Bhatia A, Ostlie D, Marosky J, Holcomb G, 3rd, Snyder C, Gittes G. A role for sonic hedgehog signaling in the pathogenesis of human tracheoesophageal fistula. Journal of pediatric surgery. 2003;38:465–468. doi: 10.1053/jpsu.2003.50080. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, Zackai EH, McDonald-McGinn D, Nowaczyk MM, Murray M, Hustead V, Mascotti K, Schultz R, Hallam L, McRae D, Nicholson AG, Newbury R, Durham-O’Donnell J, Knight G, Kini U, Shaikh TH, Martin V, Tyreman M, Simonic I, Willatt L, Paterson J, Mehta S, Rajan D, Fitzgerald T, Gribble S, Prigmore E, Patel A, Shaffer LG, Carter NP, Cheung SW, Langston C, Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. American journal of human genetics. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storr M, Geisler F, Neuhuber WL, Schusdziarra V, Allescher HD. Characterization of vagal input to the rat esophageal muscle. Autonomic neuroscience: basic & clinical. 2001;91:1–9. doi: 10.1016/S1566-0702(01)00290-9. [DOI] [PubMed] [Google Scholar]
- Szumska D, Pieles G, Essalmani R, Bilski M, Mesnard D, Kaur K, Franklyn A, El Omari K, Jefferis J, Bentham J, Taylor JM, Schneider JE, Arnold SJ, Johnson P, Tymowska-Lalanne Z, Stammers D, Clarke K, Neubauer S, Morris A, Brown SD, Shaw-Smith C, Cama A, Capra V, Ragoussis J, Constam D, Seidah NG, Prat A, Bhattacharya S. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008;22:1465–1477. doi: 10.1101/gad.479408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraviras S, Marcos-Gutierrez CV, Durbec P, Jani H, Grigoriou M, Sukumaran M, Wang LC, Hynes M, Raisman G, Pachnis V. Signalling by the RET receptor tyrosine kinase and its role in the development of the mammalian enteric nervous system. Development. 1999;126:2785–2797. doi: 10.1242/dev.126.12.2785. [DOI] [PubMed] [Google Scholar]
- Taraviras S, Pachnis V. Development of the mammalian enteric nervous system. Curr Opin Genet Dev. 1999;9:321–327. doi: 10.1016/s0959-437x(99)80048-3. [DOI] [PubMed] [Google Scholar]
- Ura H, Murakami K, Akagi T, Kinoshita K, Yamaguchi S, Masui S, Niwa H, Koide H, Yokota T. Eed/Sox2 regulatory loop controls ES cell self-renewal through histone methylation and acetylation. EMBO J. 2011;30:2190–2204. doi: 10.1038/emboj.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, Happerfield L, Arends MJ, Adams DJ. Megaoesophagus in Rassf1a-null mice. International journal of experimental pathology. 2009;90:101–108. doi: 10.1111/j.1365-2613.2008.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wang X, Ouyang H, Yamamoto Y, Kumar PA, Wei TS, Dagher R, Vincent M, Lu X, Bellizzi AM, Ho KY, Crum CP, Xian W, McKeon F. Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell. 2011;145:1023–1035. doi: 10.1016/j.cell.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun. 2010;402:14–18. doi: 10.1016/j.bbrc.2010.09.092. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Vertebrate endoderm development. Annu Rev Cell Dev Biol. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- Williamson KA, Hever AM, Rainger J, Rogers RC, Magee A, Fiedler Z, Keng WT, Sharkey FH, McGill N, Hill CJ, Schneider A, Messina M, Turnpenny PD, Fantes JA, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum Mol Genet. 2006;15:1413–1422. doi: 10.1093/hmg/ddl064. [DOI] [PubMed] [Google Scholar]
- Woo J, Miletich I, Kim BM, Sharpe PT, Shivdasani RA. Barx1-mediated inhibition of Wnt signaling in the mouse thoracic foregut controls tracheo-esophageal septation and epithelial differentiation. PLoS One. 2011;6:e22493. doi: 10.1371/journal.pone.0022493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worl J, Dutsch F, Neuhuber WL. Development of neuromuscular junctions in the mouse esophagus: focus on establishment and reduction of enteric co-innervation. Anatomy and embryology. 2002;205:141–152. doi: 10.1007/s00429-002-0239-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Yoo SK, Nishita M, Kikuchi A, Minami Y. Wnt5a modulates glycogen synthase kinase 3 to induce phosphorylation of receptor tyrosine kinase Ror2. Genes to cells: devoted to molecular & cellular mechanisms. 2007;12:1215–1223. doi: 10.1111/j.1365-2443.2007.01128.x. [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Moriguchi T, Masuyama N, Kusakabe M, Hanafusa H, Takada R, Takada S, Nishida E. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3:69–75. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bergner AJ, Enomoto H, Milbrandt J, Newgreen DF, Young HM. Neural cells in the esophagus respond to glial cell line-derived neurotrophic factor and neurturin, and are RET-dependent. Dev Biol. 2004;272:118–133. doi: 10.1016/j.ydbio.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yu WY, Slack JM, Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol. 2005;284:157–170. doi: 10.1016/j.ydbio.2005.04.042. [DOI] [PubMed] [Google Scholar]
- Zizer E, Beilke S, Bauerle T, Schilling K, Mohnle U, Adler G, Fischer KD, Wagner M. Loss of Lsc/p115 protein leads to neuronal hypoplasia in the esophagus and an achalasia-like phenotype in mice. Gastroenterology. 2010;139:1344–1354. doi: 10.1053/j.gastro.2010.06.041. [DOI] [PubMed] [Google Scholar]