Abstract
How centromeres are assembled and maintained remains one of the fundamental questions in cell biology. Over the past 20 years the idea of centromeres as precise genetic loci has been replaced by the realization that it is predominantly the protein complement that defines centromere localization and function. Thus, placement and maintenance of centromeres are excellent examples of epigenetic phenomena in the strict sense. In contrast, the highly derived “point centromeres” of the budding yeast Saccharomyces cerevisiae and its close relatives are counterexamples for this general principle of centromere maintenance. While we have learned much in the past decade, it remains unclear if mechanisms for epigenetic centromere placement and maintenance are shared amongst various groups of organisms. For that reason it seems prudent to examine species from many different phylogenetic groups with the aim to extract comparative information that will yield a more complete picture of cell division in all eukaryotes. This review addresses what has been learned by studying the centromeres of filamentous fungi, a large, heterogeneous group of organisms that includes important plant, animal and human pathogens, saprobes and symbionts that fulfill essential roles in the biosphere, as well as a growing number of taxa that have become indispensable for industrial use.
Keywords: CenH3/CENP-A, Centromere, Kinetochore, Epigenetics, Heterochromatin, Histone modification, Neurospora crassa, Fusarium graminearum
Introduction
Much of what is currently known about centromeres in fungi is based on studies with the ascomycetous yeasts S. cerevisiae, Schizosaccharomyces pombe, and Candida albicans, representatives of a relatively small group of fungi. Pioneering work with S. cerevisiae uncovered extremely short centromeres comprised of conserved DNA sequences that are recognized by specialized DNA-binding proteins, thus suggesting a requirement for specific DNA sequence to determine centromere position (reviewed in Cleveland et al., 2003, Meraldi et al., 2006). This “point centromere” model, however, did not hold true for S. pombe (Cam et al., 2005, Nakaseko et al., 1986, Scott et al., 2006), C. albicans (Baum et al., 2006, Sanyal et al., 2004), or other eukaryotes (Copenhaver et al., 1999, Dong et al., 1998, Nagaki et al., 2005, Richards et al., 1991, Rudd and Willard, 2004, Schueler et al., 2001, Sun et al., 2003, Yamamoto and Miklos, 1978, Zhang and Zhang, 2004), which form “regional centromeres” that are characterized by long stretches of repetitive DNA (Allshire, 1997, Black et al., 2007, Blower and Karpen, 2001, Blower et al., 2002, Karpen and Allshire, 1997, Nagaki et al., 2003, Shibata and Murata, 2004, Sullivan et al., 2001, Sullivan, 2001, Yan et al., 2005). In C. albicans, regional centromeres are composed of unique 3–4.5 kilobase (kb) sequences (Sanyal et al., 2004), and the overall size and position of these CEN sequences is conserved between different C. albicans strains (Mishra et al., 2007). Neocentromeres can form near repetitive DNA, often near the original centromere (Ketel et al., 2009). Nevertheless, DNA sequence alone does not establish centromere identity, as CEN DNA introduced on a plasmid was insufficient to establish centromeres de novo (Baum et al., 2006). The much larger centromeric regions of S. pombe chromosomes contain segments that constitute the 10–15 kb centromere cores (the imr, or “inner repeats”, and cc, or “central cores”) and 10–60 kb of repeats that generate the surrounding pericentric heterochromatin (the otr, or “outer repeats”) on each side of the three centromere cores (Fishel et al., 1988). Deletion of centromeric DNA caused neocentromere formation near telomeric heterochromatin or telomere-telomere fusions between the acentric and a normal chromosome (Ishii et al., 2008). Again, specific DNA segments alone were insufficient to induce de novo centromere assembly. Rather, functional RNAi and heterochromatin machineries are required to generate centromeres on plasmid-based minichromosomes, but neither pathway is required for maintenance of functional centromeres (Folco et al., 2008, Kagansky et al., 2009).
Identification of centromeric DNA in filamentous fungi
The nature of centromeric DNA in the filamentous fungi is paradoxically both a hindrance and an advantage for the study of centromeres in these organisms. In an era when whole genome sequencing has become seemingly trivial, discovery of centromeric DNA sequences in filamentous fungi still remains a substantial challenge. Like those of many higher eukaryotes, putative centromeric DNA sequences in the filamentous fungi are composed of a complex, heterogeneous set of repetitive, AT-rich sequences that can span between 30–450 kb. These sequences are inherently difficult to capture by traditional Sanger sequencing methods that require in vivo cloning, and a challenge for sequence assembly algorithms used with both traditional or newer high-throughput sequencing (HTS) technologies, which avoid cloning but generate typically short, 100–400 nucleotide reads.
The first centromeric DNA from a filamentous fungi to be cloned came from Neurospora crassa (Centola and Carbon, 1994) and was partially sequenced (Cambareri et al., 1998), aided by genetic data that placed centromere VII (Cen-VII) next to well-studied classical markers. Based on only 16 kb of sequence, it was concluded that Neurospora centromeres are composed of degenerate transposons, mostly retrotransposons, and simple sequence repeats. The degenerate nature of the transposons is due to the action of a premeiotic process called “Repeat-Induced Point mutation” (RIP), which through an unknown mechanism recognizes repeated DNA and mutates both copies, yielding numerous C:T and G:A transition mutations (Cambareri et al., 1989, Selker, 1990). RIP will continue in successive sexual cycles until sequence identity between two copies decreases below ~85% but it will begin again if such regions become re-duplicated (Cambareri et al., 1991). Presumably these cycles of duplication and mutagenesis can continue until no Cs remain. Combined with potential gene conversion or recombination events, RIP appears to provide an interesting mechanism for diversification of centromeric DNA in many filamentous fungi.
Nearly all centromeric DNA on the seven Neurospora chromosomes has been assembled (Borkovich et al., 2004, Galagan et al., 2003), partly because the genome has been sequenced to more than one thousand-fold coverage. The early conclusions still hold true but have been refined. The DNA component of each centromere is comprised of 175–300 kb of mutated degenerate transposons and other AT-rich sequence that is no longer recognizable as ancestral transposon because of the action of RIP (Smith et al., 2011). No specific recognizable pattern in the arrangement of these transposon relics has emerged to allow identification of segments that are functionally similar to the S. pombe otr and imr regions. Indeed, comparing segments of centromeric DNA to segments of DNA that are subject to heterochromatization reveals no obvious compositional or structural bias between core centromeric, pericentric and dispersed heterochromatic DNA in Neurospora (Smith et al., 2011).
Some uncertainties remain, however. Except for Cen-IV, centromeric DNA on the other chromosomes is still contained on up to eleven contigs that are separated by unknown sequence. This suggests that some centromeric DNA sequences are still missing, although based on an optical map this may be as little as 0.5 Mb for the entire genome. Secondly, even in the not completely identical, yet repeat-rich centromeric DNA of Neurospora there is potential for misassembly. One such example was identified on Cen-II, where a single expressed pseudogene that is also associated with active chromatin marks, i.e. dimethylation of histone H3 lysine 4 (H3K4me2), was erroneously assembled as part of the centromere (Smith et al., 2011). There is little doubt that there are additional minor inconsistencies, so that centromeric DNA assembly in N. crassa will need some further refinement, perhaps aided by the quickly advancing sequencing technologies that promise longer reads in the near future.
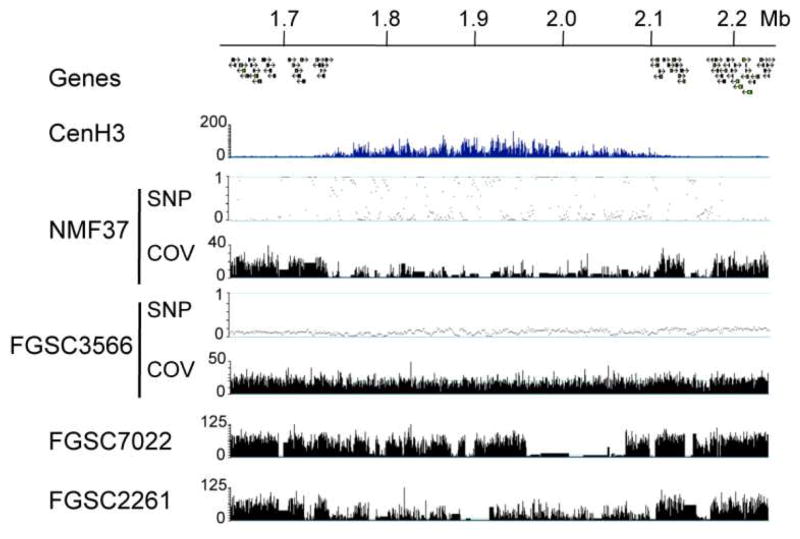
With the availability of over 20 N. crassa genomes (McCluskey et al., 2011, Pomraning et al., 2011) and those of at least three sister species (Ellison et al., 2011b, Nowrousian et al., 2010) we can ask how centromeric DNA has changed or is changing in real time. To address the second question, we compared several laboratory strains (N1961, N3011, and progeny from crosses) to the widely used laboratory wild type strain FGSC2489 (P. Phatale, K.M. Smith and M. Freitag, unpublished data). Both N1961 and N3011 were derived from FGSC2489 by transformation and genetic crosses with mixed background strains. As expected, coverage of Illumina sequence reads across the chromosome was fairly even in centromeric regions that had been previously identified by chromatin immunoprecipitation followed by HTS (“ChIP-seq”) with CenH3 (Smith et al., 2011), suggesting that no major rearrangements had occurred, similar to what we observed with FGSC3566 (Fig. 1). Single nucleotide polymorphisms (SNPs) were predominantly found outside the centromeric DNA. In stark contrast, centromeric DNA of the more distantly related Mauriceville laboratory strain (NMF37), which is frequently used for RFLP mapping (Metzenberg et al., 1984), shows low overall coverage in the centromeric regions but a relatively high number of SNPs in regions with similar DNA (Pomraning et al., 2011). In contrast to larger eukaryotic genomes, absence of coverage indicates that specific DNA segments are completely absent from the N. crassa genome. Intermediate coverage is based on the presence of SNPs. Analyzing a set of genome sequences that were generated to map classical mutations of N. crassa (McCluskey et al., 2011) revealed that centromeric DNA patterns in Neurospora lineages are not static (Fig. 1B). While FGSC3566 has almost identical Cen-VII sequences as the reference strain FGSC2489, FGSC2261 and FGSC7022 harbor significant differences. FGSC2261 and FGSC7022 were derived by crossing mixed-lineage strains into the lineage that eventually produced the reference strain.
Figure 1. Different Neurospora strains have vastly different DNA sequence at their centromeres.
The centromere region of LG VII is identified by the absence of genes and enrichment of CenH3 by ChIP-seq (“CenH3”). Genomic HTS of strains with diverse genetic backgrounds demonstrates DNA sequence differences in centromere regions. There are regions of missing sequence and high levels of SNPs in the centromere of NMF37, the Mauriceville wild-collected strain. Both FGSC3566 and FGSC7022 have mixed genetic backgrounds, and FGSC2261 was backcrossed into the St. Lawrence lineage two times. Tracks show SNPs per kb (“SNP”) and coverage (“COV”) as read coverage per base (if not specified, tracks show coverage).
In the near future, the genomes of a large number of wild N. crassa isolates from Louisiana will become available. This set of strains has already served as a resource to address fundamental questions connecting genotype to phenotype (Ellison et al., 2011a). It also offers the opportunity to study centromere evolution in a well-defined group of strains that appear more closely related than the Oak Ridge and Mauriceville wild types of N. crassa that are widely in use in laboratories. In the meantime, we have analyzed crosses between the Mauriceville (NMF37) and Oak Ridge (FGSC2489) strains for the presence of gene conversion or crossing over in centromeric regions. These studies are aided by a remarkable advantage of Neurospora sexual development, as meiosis and a subsequent mitosis result in ordered ascospores, where position of the eight resulting spores in the ascus directly reflects position of the eight strands of DNA that participate in meiosis. This gives us the opportunity to visualize the complete extent of crossing over and gene conversion along the whole genome from individual meioses by HTS.
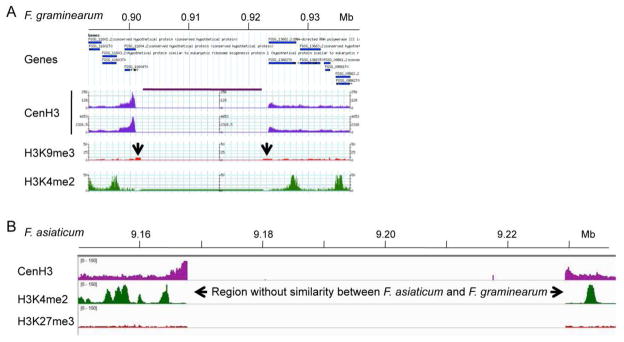
While we generated a first draft of the Mauriceville genome sequence (Pomraning et al., 2011), the gold standard has to be a finished sequence that contains precisely mapped and assembled centromeric DNA. This is an achievable goal for genome sequencing projects of filamentous fungi. The most recent studies with short-read HTS suggest that centromeric DNA is more easily captured by HTS than by traditional Sanger sequencing, likely because no in vivo cloning is required. Many fungal genomes that were sequenced pre-HTS (e.g. several Aspergillus and Fusarium species) are missing most or all centromeric DNA sequences from the currently available assemblies. For example, while Fusarium graminearum DNA sequences located on the chromosome arms are almost completely assembled, all four centromeric DNA segments remain unknown. ChIP-seq with CenH3 showed mapping to the edges of 20-kb spacers that were inserted as placeholders into the genome sequence (Fig. 2A). Curiously, all eight boundaries contain 1–1.5 kb of assembled sequence that cannot be amplified from the genome, suggesting that assembly programs “stalled” at these points (K.R. Pomraning, K.M. Smith and M. Freitag, unpublished data). A perhaps less likely alternative for the apparent lack of centromeric DNA from otherwise well-assembled fungal genomes is that centromeric DNA may not be recognizable because it is uncharacteristically not particularly AT- or repeat-rich.
Figure 2. Mapping of Fusarium centromeres by ChIP-seq with CenH3.
A. Region that contains the presumed centromeric DNA on Chromosome 1 of Fusarium graminearum. Genes flanking the centromere are shown in blue, CenH3 ChIP-seq-derived reads are shown in purple (two replicates), enrichment for the silencing H3K9me3 modification is shown in red, and the activating H3K4me2 modification is shown in green. The region under the purple line above the two CenH3 tracks contains only “N”s. The two arrows indicate position of misassembled DNA that cannot be amplified from the genome by PCR. B. Mapping of F. graminearum CenH3 ChIP-seq reads to the F. asiaticum Cen1 region. Only the edges of the centromeric regions are identified. The core region is so divergent that essentially no reads map to this AT- and repeat-rich region. One silencing (H3K27me3, red) and one activating (H3K4me2, green) histone modification are also shown. In both A. and B. CenH3 enrichment was observed at the edges of the presumed centromeric regions (and even overlapping genes, as in A.), likely because shearing by sonication can result in arrays of several nucleosomes.
Early de novo genome assemblies based on short-read HTS, like that of Sordaria macrospora (Nowrousian et al., 2010), also lack completely assembled centromeric DNA; even the second Sordaria assembly still has over 1500 contigs. Both sequencing and assembly from short-read HTS are constantly improving, however, and several more recently started genome projects that relied on either Illumina or 454 HTS (or a mixture of both) generated supercontigs that contain 30–100 kb stretches of AT- and repeat-rich DNA. In several cases (e.g. Fusarium asiaticum and F. fujikuroi) such long uninterrupted segments occur only once on large supercontigs. Their edges are often syntenic with F. graminearum sequence. When mapping F. graminearum reads from CenH3 ChIP-seq to the F. asiaticum genome, edges were enriched for CenH3 but the intervening AT-rich DNA showed no or little similarity to any DNA immunoprecipitated with F. graminearum CenH3, suggesting that the centromeric DNA of these species is diverged, similar to what we found by analyzing centromeric DNA from several Neurospora strains (Fig. 2B; L.R. Connolly, K.M. Smith and M. Freitag, unpublished data). For all fungal genomes, no matter if Sanger- or HTS-derived, the best assay for centromeric DNA will be to identify the specialized nucleosomes that contain the centromere-specific histone H3, CenH3, which is today’s litmus test for centromere placement. Among filamentous fungi this has been achieved so far only in Neurospora.
Centromere identity is determined by a centromere-specific histone H3, CenH3
Independent of DNA sequence, the universal “centromere identifier” in all organisms studied so far is a specialized histone H3 variant called CENP-A in mammals (Palmer et al., 1987), Cid in Drosophila (James and Elgin, 1986), CenH3 in N. crassa (Smith et al., 2011), Cnp1CenpA in S. pombe (Takahashi et al., 2000), Cse4 in S. cerevisiae (Stoler et al., 1995) and C. albicans (Sanyal and Carbon, 2002), and HRT12 in Arabidopsis thaliana (Talbert et al., 2002). The nomenclature for fungal homologs of centromere and kinetochore proteins is currently not settled. For Neurospora, we propose “CEN” followed by the letter code used for mammalian proteins; this is based on the requirement for three-letter code for fungal gene names. Thus, if we use CENP-B we mean the animal protein, if we use CEN-B we mean the Neurospora homolog specifically. CenH3 sequences from fungi show high variability in length and sequence of the N-terminal tail and loop I region within the histone fold domain (Baker and Rogers, 2006). The region surrounding loop I, the “CENP-A targeting domain” (CATD), proved sufficient to target CenH3 (or heterologous H3 with a CATD) into centromeric nucleosomes (Black et al., 2007), although this was not the case in Arabidopsis thaliana (Ravi et al., 2010). Ongoing studies with Neurospora suggest that the CATD is conserved in filamentous fungi, as CenH3 proteins from several filamentous fungi yield the expected chromocenter localization when the native Neurospora CenH3 gene is replaced with heterologous genes (P.A. Phatale, K.M. Smith, L.R. Connolly and M. Freitag, unpublished data).
Centromere proteins bind centromeric DNA with a non-uniform but stable pattern
The availability of N. crassa centromeric DNA sequences has allowed detailed mapping of proteins associated with these segments by ChIP-seq. For this, centromere proteins or modified histones were immunoprecipitated, the associated DNA was sequenced and mapped back to the N. crassa reference genome (Smith et al., 2011). Because centromere “repeats” of N. crassa are non-identical, reads can be mapped to centromeric segments with great precision, something not yet possible in most plants and mammals with homogeneous repeats that are nearly identical across most centromeres.
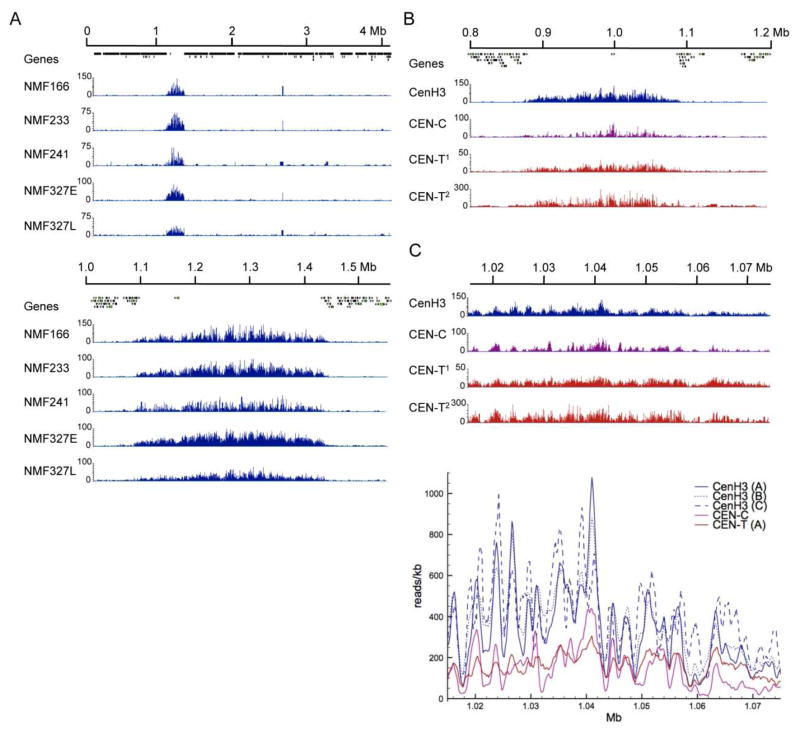
As mentioned above, DNA composition is indistinguishable between the centromere and other regions of AT-rich sequence dispersed along the chromosome arms and near the telomeres, yet CenH3 is only associated with centromeric DNA (Smith et al., 2011). If centromere boundaries were precise and CenH3 occupancy was completely uniform in each of the many nuclei analyzed, one would expect a rectangular shape as readout of CenH3 localization. Instead, we found a distribution that suggests that CenH3 occupancy is variable within a defined region on each chromosome (Fig. 3A). This distribution is stable, as essentially the same result was obtained from three independently derived strains (Smith et al., 2011). An additional strain, NMF327, was grown for extended periods of time through Ryan or “racetubes”, which is a convenient way to analyze Neurospora linear growth over a long time (Davis, 2000). These experiments confirmed that CenH3 binds with a regular pattern that is maintained throughout multiple mitotic divisions. As the four strains resulted from different crosses, distribution of CenH3 is also stable through meiosis or at least reproducibly reestablished after meiotic divisions (P.A. Phatale, K.M. Smith and M. Freitag, unpublished results).
Figure 3. Centromere proteins bind centromeric DNA with a non-uniform but stable pattern.
A. CenH3 localization at the centromere is stable through many rounds of mitotic and meiotic cell divisions. The entire LG II (top) or 0.5 Mb including Cen II (bottom) shows distribution of CenH3 in four different strains. Localization of CenH3 is similar in each strain and stable even following continued growth of NMF327 in race tubes. E, early time point, and L, late time point. B. The inner kinetochore proteins CEN-C and CEN-T colocalize with CenH3. LG IV and Cen IV coverage by CenH3, CEN-C and two replicates of CEN-T (CEN-T1 and CEN-T2) ChIP-seq. C. Binning CenH3 enrichment as reads per kb reveals periodicity in enrichment for three CenH3 replicates (CenH3 A–C), and to a lesser extent for CEN-C. CEN-T seems to be more evenly distributed.
CenH3 co-localizes with two inner kinetochore proteins, CEN-C and CEN-T (Smith et al., 2011) (Fig. 3B and Table 1). Plotting reads per kb revealed discrete peaks and valleys for CenH3 distribution, on average 1.0–1.2 kb (or roughly five to six nucelosomes) between inflection points (Fig. 3C). Like that for CenH3, enrichment of CEN-C shows patterns of peaks and valleys, but CEN-T binding appears more uniform across the centromere. We first discounted this periodicity, or “phasing”, as a product of preferred shearing sites in preparation for ChIP, but the observed reproducibility, especially of the CenH3 mapping for numerous replicates (Fig. 3C), suggests the presence of preferred regions of CenH3 nucleosome occupancy. How may this phasing be generated? There are no obvious differences in the underlying DNA sequence (Smith et al., 2011). We considered additional possibilities: (1) phasing is generated by centromere-specific histone modifications, or (2) additional centromere proteins, e.g. CEN-B, CenH3 chaperones such as CEN-C and SCM3/HJURP, or the CEN-T-W-S-X complex, are involved in generating ordered arrays of CenH3 and H3 nucleosomes. We will discuss these possibilities below.
Table 1. Centromere and kinetochore proteins in filamentous fungi.
Proteins were identified by Blast searches with the indicated human or S.pombe protein sequences as bait. Nomenclature for fungal homologs is currently uncertain; for N. crassa we propose “CEN-” and for Fusarium we propose “Cen”, in both cases followed by the letter code used for mammalian proteins. Here we provide current locus numbers based on annotations that can be found at the Fungal Genome Initiative website at the Broad Institute (http://www.broadinstitute.org/scientific-community/data). Many of these homologs had been previously identified (Meraldi et al., 2006) but annotations have been updated. “None”, no homolog was identified by sequence similarity.
| H. sapiens | S. pombe | N. crassa | F. graminearum | A. nidulans | M. oryzae |
|---|---|---|---|---|---|
| CENP-A | Cnp1 | NCU00145 | FGSG_02602.3 | ANID_06554.1 | MGG_06445.7 |
| HJURP | Scm3 | NCU03123 | FGSG_00678.3 | ANID_01514.1 | MGG_03695.7 |
| NPM1 | None | None | None | None | None |
|
| |||||
| CENP-B | None | None | None | None | None |
| None | None | NCU06592 | FGSG_05264.3 | None | MGG_01698.7 |
| None | None | NCU00392 | FGSG_07243.3 | ANID_04077.1 | MGG_13165.7 |
| Abp1 | ANID_09495.1 | ||||
| Cbh2 | ANID_08717.1 | ||||
| None | Cbh1 | None | None | ANID_06156.1 | None |
| INCENP | None | NCU05211 | FGSG_05106.3 | None | MGG_01835.7 |
| Aurora B | Ark1 | NCU00108 | FGSG_06959.3 | ANID_05815.1 | MGG_00479.7 |
|
| |||||
| CENP-C | Cnp3 | NCU09609 | FGSG_11834.3 | ANID_05115.1 | MGG_06960.7 |
| Mis18α/Mis18β | Mis18 | None | None | None | None |
| RbAp48/RbAp46 | Mis16 | NCU06679 | FGSG_06798.3: | ANID_08187.1 | MGG_07323.7 |
| M18BP1 | None | None | None | None | None |
| MgcRacGAP | None | None | None | None | None |
|
| |||||
| CENP-T | Cnp20 | NCU02161 | FGSG_08859.3 | ANID_04752.1 | MGG_02732.7 |
| CENP-X | Mhf2 | NCU09478 | FGSG_06022.3 | ANID_11893.1 | MGG_16148.7 |
| CENP-W | SPAC17G8.15 | NCU03400 | FGSG_10821.3 | ANID_10643.1 | MGG_11869.7 |
| CENP-S | SPBC2D10.16 | NCU03629 | FGSG_07161.3 | ANID_00598.1 | MGG_15064.7 |
|
| |||||
| CENP-H | Fta3 | NCU09996 | FGSG_00383.3 | ANID_02886.1 | MGG_04487.7 |
| CENP-I | Mis6 | NCU04131 | FGSG_07166.3 | ANID_06600.1 | MGG_09521.7 |
| CENP-K | Sim4 | NCU09238 | FGSG_01054.3 | ANID_02088.1 | MGG_17440.7 |
|
| |||||
| CENP-L | Fta1 | NCU07591 | FGSG_04225.3 | None | MGG_10701.7 |
| CENP-M | Mis17 | None | None | None | None |
| CENP-N | Mis15 | NCU03537 | FGSG_05172.3 | ANID_04360.1 | MGG_16236.7 |
|
| |||||
| CENP-O | Mal2 | NCU09100 | FGSG_08806.3 | ANID_04835.1 | MGG_12605.7 |
| CENP-P | Fta2 | NCU02135 | None | None | None |
| CENP-Q | Fta7 | NCU06791 | FGSG_10762.3 | None | MGG_06713.7 |
| CENP-R | None | None | None | None | None |
| CENP-U | Fta4 | NCU01005 | FGSG_02626.3 | ANID_01011.1 | MGG_15811.7 |
|
| |||||
| hMis12 | Mis12 | NCU06463 | FGSG_02525.3 | ANID_00960.1 | MGG_06304.7 |
| DSN1 | Mis13 | NCU01344 | FGSG_10141.3 | ANID_03187.1 | MGG_08211.7 |
| NNF1 | Nnf1 | NCU07571 | FGSG_05590.3 | ANID_08263.1 | MGG_04669.7 |
| NSL1 | Mis14 | NCU02262 | FGSG_10778.3 | ANID_04996.1 | MGG_00906.7 |
|
| |||||
| NDC80 | Ndc80 | NCU03899 | FGSG_09262.3 | ANID_04969.1 | MGG_01027.7 |
| NUF2 | Nuf2 | NCU06568 | FGSG_05288.3 | ANID_00166.1 | MGG_01848.7 |
| SPC24 | Spc24 | NCU05312 | FGSG_05313.1 | ANID_06224.1 | MGG_06076.7 |
| SPC25 | Spc25 | NCU11159 | FGSG_01538.3 | ANID_01392.1 | MGG_08566.7 |
|
| |||||
| KNL1 | Spc7 | NCU03103 | FGSG_00672.3 | ANID_05221.1 | MGG_03693.7 |
|
| |||||
| CENP-E | None | NCU02626 | FGSG_01004.3 | ANID_08286.1 | MGG_04366.7 |
| CENP-F | None | NCU03621 | FGSG_10111.3 | ANID_05829.1 | MGG_07274.7 |
Neurospora centromeres are heterochromatic
Centromeric chromatin has long been thought of as constitutively silenced and thus described as a heterochromatic domain. This view of centromeres has been challenged by cytological and biochemical analyses of rice, fly and mammalian chromatin (Sullivan and Karpen, 2004, Yan et al., 2005, Ma et al., 2007). In animals, production of RNA from centromeric DNA appears necessary for CenH3 targeting (Bergmann et al., 2010). The original notion still holds in Neurospora, however, where centromeric DNA is associated largely with nucleosomes that are modified by silencing histone modifications (Smith et al., 2011).
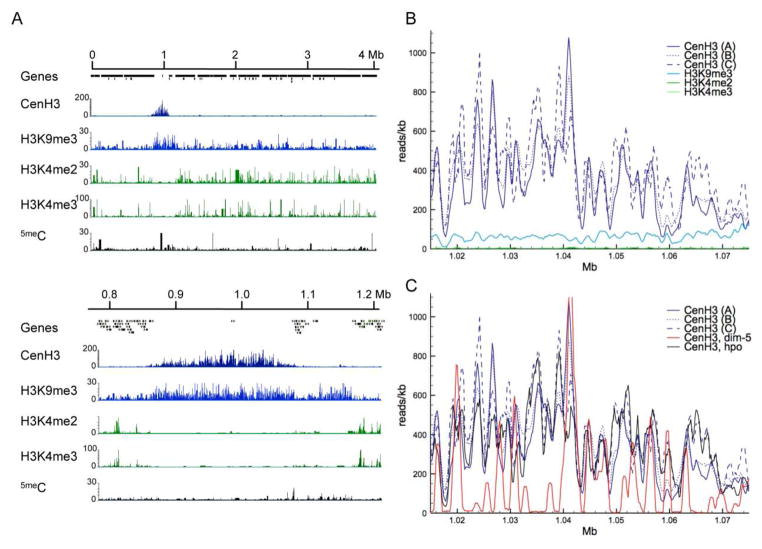
In humans, Drosophila, and S. pombe centromeres, CenH3 nucleosomes are interspersed between nucleosomes containing canonical H3 that is di- or trimethylated at lysine 4 (H3K4me2 or -me3) (Sullivan and Karpen, 2004, Cam et al., 2005). In contrast, ChIP of H3K4me2 or H3K4me3 in N. crassa showed that histones with these modifications localize to coding regions and are absent from centromeres under standard laboratory growth conditions (Smith et al., 2011) (Fig. 4A). In their place H3K9me3 was found to co-localize with the CenH3 signal. Indeed, H3K9me3 signals extend further than CenH3 into the chromosome arms, thus–together with the presence of methylated cytosines–delineating the relatively small pericentric regions of Neurospora chromosomes (Fig. 4A). These findings are similar to those from studies with mice, where CenH3 has been co-localized with H3K9me2 (Guenatri et al., 2004), and with chicken, where a high-resolution map of kinetochores suggests co-existence of H3K4me2, H3K9me3, and CenH3 in centromere cores (Ribeiro et al., 2010).
Figure 4. Neurospora centromeres are heterochromatic.
A. The first 4 Mb of LG IV (top) and Cen-IV (bottom) show colocalization of CenH3 and H3K9me3 at the centromere. Together with cytosine DNA methylation (5meC), H3K9me3 is also found in relatively short pericentric regions directly adjacent to the regions enriched for CenH3, and in dispersed regions of heterochromatin (e.g. around 1.1 Mb). H3K4me2 and –me3 are localized in gene-rich regions and excluded from the centromere. B. Binning CenH3 enrichment (reads/kb) reveals periodicity that is essentially absent from H3K9me3 enrichment. C. Phasing observed in WT strains (CenH3, A–C) is altered in dim-5 strains that lack H3K9me3 (CenH3, dim-5) but similar to WT in hpo strains (CenH3, hpo).
We next asked if heterochromatin is essential for centromere localization and function in Neurospora. In N. crassa, heterochromatin is formed by trimethylation of H3K9 by the histone methyltransferase, DIM-5, and trimethylated H3K9 is bound by the chromo domain of Heterochromatin Protein 1 (HP1) (Tamaru and Selker, 2001, Freitag et al., 2004a). Previous studies in S. pombe showed that RNAi-directed heterochromatin assembly via the H3K9 trimethylase Clr4/DIM-5/Suv39h and Swi6/HP1 at the otr is required to target CenH3 to the imr and cc segments (Folco et al., 2008). Tethering Clr4 to minichromosomes induced synthetic heterochromatin, which was sufficient to recruit CenH3 and form a functional neocentromere (Kagansky et al., 2009). Thus targeting of Clr4 abolished the need for the RNAi pathway. Heterochromatin is, however, not required for the inheritance of functional, native S. pombe centromeres (Folco et al., 2008, Kagansky et al., 2009). In contrast, both DIM-5 and HP-1 are required for proper localization of CenH3 in Neurospora. In absence of either protein, the regions occupied by CenH3 shrink on most chromosomes, to as little as 55% in a dim-5 mutant and 27% in an hpo mutant (Smith et al., 2011). Nevertheless, CenH3 distribution remains within the boundaries of the “normal” centromere. Centromeric regions that are free of H3K9me3 do not acquire H3K4me2 (Smith et al., 2011), but other potential histone modifications have not been tested yet. Phenotypically, dim-5 CenH3-GFP strains are more impaired in growth than dim-5 mutants, suggesting a slight synergistic effect. So far, the requirement for heterochromatin to maintain CenH3 localization is unique to Neurospora but we expect that this pathway will be conserved in other filamentous fungi.
Unlike for CenH3, however, the ChIP signal from H3K9me3 is uniform throughout the Neurospora centromere (Fig. 4B). H3K9me3 is also found at dispersed and subtelomeric regions of heterochromatic AT-rich DNA (Lewis et al., 2009, Smith et al., 2011). We hypothesized that CenH3 phasing may be caused by specific histone modifications and may thus also be altered in dim-5 or hpo mutants. If blocks of H3 and CenH3 nucleosomes would form the basis for the phasing in CenH3 distribution observed (Figs. 3C and 4B) one would expect that H3 or H3K9me3 distribution would be out of phase compared to that of CenH3, but this is not observed (Fig. 4B). While the overall enrichment of CenH3 within the normal centromeric regions as well as CenH3 phasing was altered in dim-5, only CenH3 distribution was changed in hpo, suggesting that heterochromatin may contribute partially to the phasing effect (Fig. 4C). It is still unknown if other chromo domain proteins can recognize H3K9me3 if HP1 is absent.
Proteins that are involved in targeting DIM-5 and HP1 to regions that are destined to become heterochromatic should show subtle centromere defects. DIM-5 is part of at least one complex, the DCDC (DIM-5, -7, -9, CUL-4, and DDB-1 Complex) that recruits HP1 and the DNA methyltransferase DIM-2 (Lewis et al., 2010a, Lewis et al., 2010b, Zhao et al., 2010). DIM-7 targets DIM-5 to the appropriate regions but the activity of DIM-5 is dependent on the entire DCDC. DIM-7 is required for the interaction between DIM-5 and DIM-9 (or DDB-1/CUL-4 Associated Factor, DCAF) (Lewis et al., 2010a, Xu et al., 2010). DDB-1 and CUL-4 are part of a conserved E3 ubiquitin ligase complex (Petroski and Deshaies, 2005), yet the substrate responsible for the control of heterochromatin formation remains unknown. Thus cullins may control the activity or stability of DIM-5 and CenH3 directly.
Two additional complexes are involved in heterochromatin regulation in or near centromeric regions in Neurospora. A newly identified histone deacetylase complex with two chromo domain proteins, “HCHC” (HP1, CDP-2, HDA-1, CHAP), can generate heterochromatin. Defects in HCHC result in histone hyperacetylation and DNA hypermethylation in centromeric regions, the latter based on increased accessibility of the chromatin to DIM-2 (Honda et al., 2012). A second complex, “DMM” (DNA Methylation Modulator), contains a jumonji domain protein and limits spread of heterochomatin and subsequent DNA methylation from AT-rich regions into genes (Honda et al., 2010).
Ongoing studies address de novo assembly of centromeres on minichromosomes in N. crassa and F. graminearum. So far artificial chromosomes have not been generated in any filamentous fungus, and plasmids with autonomously replicating sequences have been recalcitrant to stabilization by integration of centromeric DNA. These experiments need to be carried out to establish if heterochromatin is required for neocentromere formation in filamentous fungi. Similarly, mutants in core RNA silencing components retain normal heterochromatin and DNA methylation (Freitag et al., 2004b, Lewis et al., 2009). Thus, while small RNA is produced from the centromeric DNA in Neurospora (Chicas et al., 2004, Lee et al., 2010), it remains unclear if these or other non-coding RNAs are required for proper centromere function, as has been seen in maize for long single-stranded RNA species (Du et al., 2010).
Fungal centromere proteins: CEN-B, CEN-C, and the CEN-T-W-S-X complex
Most centromere and kinetochore proteins that have been identified in S. pombe and vertebrates have homologs in filamentous fungi, albeit with fairly divergent amino acid sequences (Table 1). Many of these homologs had been described previously (Meraldi et al., 2006) and we provide here updated locus numbers for four species, two organisms used widely as reference, or “model”, species (N. crassa and Aspergillus nidulans) and two important plant pathogens, on wheat and maize (F. graminearum) or rice (Magnaporthe oryzae). The genomes of almost 200 filamentous fungi have now been sequenced and, with few exceptions, similar lists can be assembled for most of the taxa using the provided locus numbers. Homologs that are still missing from Table 1 are presumed to be either acquired more recently in animals (e.g. NPM1, M18BP1, MgcRacGAP), or may simply not be recognizable by similarity alone (e.g., Mis17, CENP-R). Within the fungi, there is wide variation, so that certain homologs appear to be missing from certain taxa (e.g., INCENP from S. pombe and A. nidulans, CEN-L and CEN-Q from A. nidulans), while they are recognizable in other taxa.
Experimentally, centromeric proteins were first identified using immune serum from individuals with scleroderma as antibodies from these patients specifically recognized centromeric regions of chromosomes (Moroi et al., 1980). Immunoprecipitation performed with these antibodies resulted in the identification of three proteins, CENP-A (CenH3), CENP-B and CENP-C (Earnshaw and Migeon, 1985). CENP-B was cloned in the late 1980s (Earnshaw et al., 1987) but a true function for this protein is still unknown. Human CENP-B consists of an N-terminal DNA-binding domain, which binds to 17-bp stretches (“CENP-B boxes”) in alpha-satellite repeats (Masumoto et al., 1989), and a C-terminal dimerization domain. Electron microscopy revealed that CENP-B can contort centromeric DNA into loops, presumably by bringing two CENP-B boxes together upon CENP-B dimerization, and that CENP-B can alter the positioning of nucleosomes in vitro (Yoda et al., 1998). This suggested that CENP-B plays a key role in determining the structure of the centromere and kinetochore, but multiple observations argue against a central role for CENP-B in centromere formation. CENP-B knockout mice have only mild growth abnormalities and have no detectable mitotic or meiotic defects (Hudson et al., 1998, Perez-Castro et al., 1998, Fowler et al., 2000, Fowler et al., 2004). Moreover, CENP-B and CENP-B boxes are apparently absent from the Y chromosome centromere and neocentromeres (Choo, 2001).
Three S. pombe homologs of CENP-B (Abp1, Cbh1, Cbh2) have been identified (Murakami et al., 1996, Lee et al., 1997, Irelan et al., 2001) but none has been found in S. cerevisiae. While all single deletion strains survive, triple deletion of all CENP-B homologs is lethal (Baum and Clarke, 2000, Irelan et al., 2001). The S. pombe CENP-B homologs bind to centromeric DNA (Lee et al., 1997), but have also been found associated with the yeast DNA replication machinery (Murakami et al., 1996), the silencing of retrotransposons (Cam et al., 2008), and genome maintenance during replication of repetitive DNA (Zaratiegui et al., 2011). Taken together, these results suggest that the roles for CENP-B proteins in fission yeast are different from that of human CENP-B.
The idea for distinct functions of fission yeast and human CENP-B is supported by results that suggest that CENP-B genes are of independent origins. Phylogenetic analysis supports the notion that S. pombe and human CENP-B genes evolved independently from distinct clades of pogo-like transposons (Casola et al., 2008), providing an excellent example of convergent evolution as there seems to be a strong selective advantage to employing CENP-B homologs in genome maintenance. While pogo-like transposons are widespread in fungi, it has been suggested that the domestication event that produced the S. pombe CENP-B homologs has occurred fairly recently, and true orthologs of Abp1, Cbh1, and Cbh2 should thus be found only in closely related species (Casola et al., 2008). Likewise, pogo-like transposons can be found in many animals, but direct orthologs of human CENP-B are presumably found only in mammals.
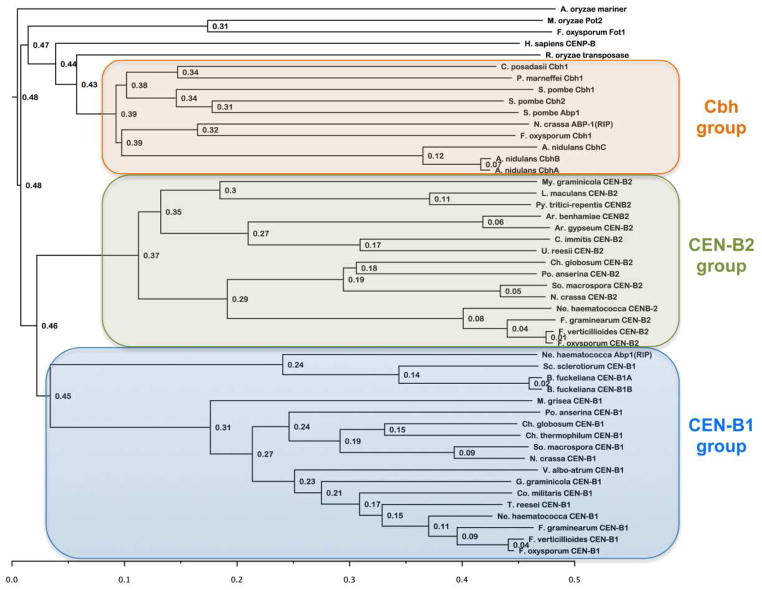
The evolutionary history of the human and S. pombe CENP-B homologs discussed above makes it difficult to compare results in these two systems. Study of a third, independently evolved homolog of CENP-B should help to clarify the role of these proteins in genome maintenance. We discovered two N. crassa homologs of CENP-B, named CEN-B1 (NCU06592) and CEN-B2 (NCU00392). Both genes appear to be derived from the domestication of pogo-like transposons, and these events were apparently independent of those leading to either the human or the S. pombe CENP-B homologs (Fig. 5). Curiously, a small family of Abp1-like proteins that occur in Neurospora has been inactivated by RIP (Fig. 5; ABP-1RIP), suggesting that the precursor of Abp1/Cbh1/Cbh2 or even a similar CENP-B-like protein existed at one time in Neurospora but has undergone mutation by RIP because of gene duplications. We searched the genomes of other filamentous fungi for putative CEN-B homologs. Different lineages - not necessarily coincident with accepted phylogeny - have different CEN-B homologs. This suggests that at least three groups of fungal CEN-B proteins exist (Fig. 5). The first group comprises homologues of S. pombe Abp1/Cbh1/Cbh2, and this contains CENP-B homologs from all Penicillium and Aspergillus species. Most Aspergillus species have three CEN-Bs, which we call CbhA, CbhB and CbhC, respectively. In contrast, most Penicillium species appear to have a single CEN-B homolog, which we call CbhA. The second and third groups comprise homologs of Neurospora CEN-B1 and CEN-B2, respectively, and are largely found in the filamentous fungi. For all groups, pogo-like transposons may have been founding members as Aspergillus mariner, Fusarium Fot1, Magnaporthe Pot2 and a Rhizopus transposase map with human CENP-B as an outgroup to all three CENP-B groups of fungi. Several basidiomycetes (e.g. Serpula and Laccaria) and ascomycetes (e.g. Tuber) have no recognizable CENP-B homologs but carry several groups of pogo transposons (data not shown). In Botrytinia fuckeliana duplication of CEN-B1 occurred relatively recently, yielding CEN-B1A and CEN-B1B (Fig. 5). Dothideomycetes, an important group of grass pathogens (e.g. Leptosphaeria maculans, Pyrenophora tritici-repentis, Stagonospora nodorum and Mycosphaerella graminicola) appear to lack CEN-B1, as do animal pathogens of the genus Coccidioides (Fig. 5).
Figure 5. Phylogeny of fungal CENPB homologs.
Putative CEN-B homologs from S. pombe and filamentous fungi were aligned with ClustalW and phylogenies constructed on Biology Workbench (http://seqtool.sdsc.edu). Phylip alignments were rendered with FigTree (version 1.3.1.; http://tree.bio.ed.ac.uk/software/figtree/). A consensus tree with node ages is shown. This tree suggests the existence of three CEN-B clades in the fungi: (1) Homologs of S. pombe Abp1/Cbh1/Cbh2, the heterogeneous “Cbh group”; (2) homologs of Neurospora CEN-B1 (“CEN-B1 group”); and (3) homologs of Neurospora CEN-B2 (“CEN-B2 group”). On this tree, fungal pogo-like transposases, the Cbh group and human CENP-B reside on a different branch than the CEN-B homologs of filamentous fungi.
By analyses with GlobPlot (Linding et al., 2003), Neurospora CEN-B1 and CEN-B2 are predicted to have disordered regions at their immediate N-terminus, followed by globular domains (aa 101–238 or 124–282, respectively), which are followed by proline-rich regions that are also predicted to be disordered (J.M. Galazka and M. Freitag, unpublished data). CEN-B2 is predicted to have a globular C-terminus (aa 493–573). Both proteins contain a Tc5 transposase-like DNA-binding domain, consisting of a helix-turn-helix structural motif, of which the second helix is predicted to make specific contacts with the major groove of DNA (Brennan and Matthews, 1989). CEN-B2 is also predicted to have an N-terminal “CENP-B N–terminal DNA-binding domain”. The CENP-B N-terminal DNA-binding domain consists of two linked helix-turn-helix domains that bind to the major groove of DNA (Tanaka et al., 2001).
Preliminary experimental evidence revealed peptides corresponding to CEN-B1 after mass spectrometry of immunoprecipitated Neurospora CenH3, which suggests that at least CEN-B1 localizes to the centromeres of N. crassa (S. Honda, P.A. Phatale, M. Freitag and E.U. Selker, unpublished results). ChIP-seq with CenH3 in CEN-B1 and CEN-B2 single or double mutants is ongoing and will test our central hypothesis regarding CEN-B, namely that the Neurospora CEN-B proteins are responsible for maintaining the phasing of CenH3 (Fig. 3C). We hypothesize that CenH3 is located at the “peaks” of the period because CEN-B1 and/or CEN-B2 are located at the “troughs” of the period. Our hypothesis is validated if deletion of CENP-B1 and/or CENP-B2 eliminates or modifies the CenH3 periodicity.
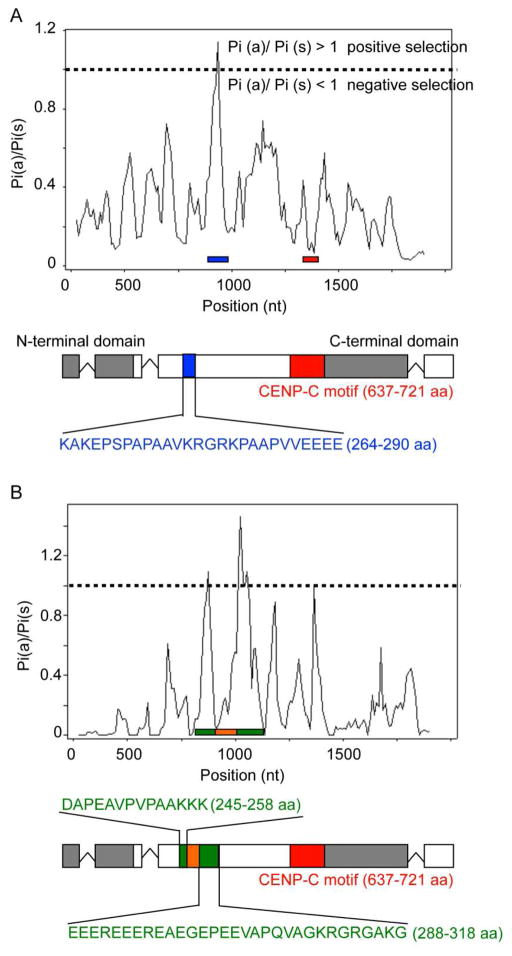
CENP-C plays an important role in linking centromeres to the kinetochore (Perpelescu and Fukagawa, 2011). Phylogenetic analyses of a small sample of animal, fungal and plant CENP-C proteins showed that regions under positive or negative selection are dispersed throughout the entire protein sequences. The C-terminal dimerization domain and “cupin-fold” (Dunwell et al., 2001, Trazzi et al., 2009) and CENP-B interaction domain of animal CENP-C is conserved, as is the C-terminus of plant CENP-C. The N-terminal and central domains show more extensive stretches of variable regions (Talbert et al., 2004, Schueler et al., 2010). Both N- and C-terminal regions interact with the outer kinetochore Mis12 complex, bridging the inner kinetochore (Schueler et al., 2010, Screpanti et al., 2011). The CENP-C motif, adjacent to a more centrally localized DNA-binding domain, is a small stretch of 10–15 amino acid residues, and is the most conserved motif in all CENP-C sequences in animals, fungi and plants ( Song et al., 2002, Talbert et al., 2004, Schueler et al., 2010, Screpanti et al., 2011). In S. cerevisiae, this motif interacts with the CBF1 complex, which binds directly to one of the conserved centromeric DNA regions, CDEI (Meluh and Koshland, 1995), and this motif is considered the centromere targeting domain of human CENP-C (Song et al., 2002).
A recent study on the evolutionary history of CENP-C within primates suggested conservation of numerous domains but also the presence of 76 specific residues that are under positive selection (Schueler et al., 2010). An earlier study included only two yeasts, S cerevisiae and S. paradoxus (Talbert et al., 2004). Thus, we asked if filamentous fungi show evidence for positive selection in CENP-C. With MEGA (Tamura et al., 2007), we generated multiple sequence alignments of CENP-C homologues, whose sequences were retrieved from GenBank and the Broad Institute Fungal Genome Initiative (http://www.broadinstitute.org/scientific-community/data) using the Neurospora CEN-C sequence as bait. Fungal CEN-C trees were built by maximum likelihood analysis (Felsenstein, 1993). We estimated the ratio of non-synonymous to synonymous substitutions, Pi(a)/Pi(s), of amino acids (Rozas and Rozas, 1995). Overall similar branching patterns were observed for the CEN-C tree and the tree based on a six-gene phylogeny for the fungal kingdom (James et al., 2006), suggesting that clades in the CEN-C tree matched evolutionary relationships. CEN-C sequences of selected sister taxa of N. crassa (i.e., Fusarium graminearum, Fusarium verticillioides, Fusarium oxysporum, Magnaporthe grisea, Chaetomium globosum and Podospora anserina) were used for determining Pi(a)/Pi(s) ratios. We found one region (at 264–290 aa in the N. crassa sequence) that appears under slight positive selection [Pi(a)/Pi(s) ratio > 1], whereas the N- and C-terminus and the CENP-C motif were under purifying selection. This pattern more closely resembles what has been found in plants and animals (Talbert et al., 2004, Schueler et al., 2010), rather than with the two Saccharomyces species (Talbert et al., 2004). To study CEN-C evolution in more detail, we amplified cen-c genes from seven N. crassa, three N. tetrasperma, two N. sitophila, and one N.africana, N.terricola, and Gelasinospora tetrasperma wildtype strains (P.A. Phatale, K.M. Smith and M. Freitag, unpublished data). Signatures of positive selection were found in the central region (from 245–258 and 288–318 aa), interrupted by a short region under negative selection. Based on the residues found, we hypothesize that this high variability region interacts with centromeric DNA and may support increased rate of centromeric DNA evolution, as has been proposed previously (reviewed in Dawe and Henikoff, 2006). Alternatively, this region may interact with other centromere proteins, as does fission yeast Moa1, which is recruited by Cnp3/CENP-C (Tanaka et al., 2009). So far, however, no homologue of Moa1 has been uncovered in filamentous fungi by sequence similarity alone. In summary, CEN-C proteins of filamentous fungi show evidence of negative selection along most of their separate domains, but small regions that may be under positive selection have been identified, as is the case for CenH3 (P.A. Phatale, K.M. Smith and M. Freitag, unpublished data; Schueler et al., 2010). Whether, however, these potentially adaptively evolving regions are involved in protein-protein or protein-DNA interactions remains to be determined. Thus, we are currently studying important regions of CEN-C by genetic means (e.g. domain swaps of the type that we and others have carried out with CenH3), and we anticipate that functional analyses, together with refined phylogenetic analyses will improve the resolution provided by analyses provide here.
To allow assembly of CenH3 chromatin, CEN-C recruits specific proteins, for example the Mis12/MIND complex and the Mis18 complex protein M18BP1 (Milks et al., 2009, Moree et al., 2011). M18BP1/KNL2 has been shown to interact with a GTPase-activating protein, MgcRacGAP, which stabilizes newly formed CenH3 chromatin (Lagana et al., 2010). While there are predicted Mis12 homologs in filamentous fungi (Table 1), there are no data to suggest that M18BP1/KNL2 or MgcRacGAP are conserved in filamentous fungi.
Conversely, homologs for the CenH3 chaperone or “receptor”, Scm3/HJURP, found in the yeasts (Stoler et al., 2007, Camahort et al., 2007, Mizuguchi et al., 2007, Pidoux et al., 2009, Williams et al., 2009, Shivaraju et al., 2011, Zhou et al., 2011) and in mammals (Dunleavy et al., 2009, Foltz et al., 2009, Sanchez-Pulido et al., 2009, Bergmann et al., 2010, Hu et al., 2011) are present in filamentous fungi. While a functionally related protein, CAL1, has been identified from flies (Erhardt et al., 2008, Mellone et al., 2011), no Scm3/HJURP homolog has been found in plants yet. The predicted SCM3 proteins in the filamentous fungi are much longer than the yeast homologs (from 800 to 1,300 aa, instead of 223 and 336 aa for S. cerevisiae and S. pombe, respectively) and show no sequence similarity outside of the short N-terminal Scm3/HJURP motif (P.A. Phatale, K.M. Smith and M. Freitag, unpublished data). Functional tests are underway but it remains unclear how similar these proteins are to other Scm3/HJURP proteins.
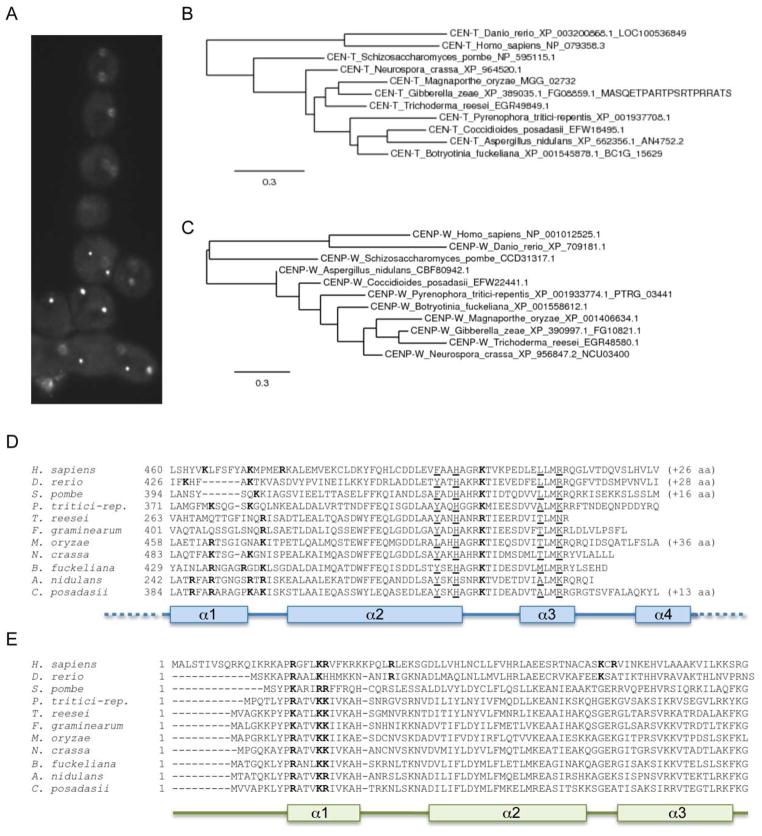
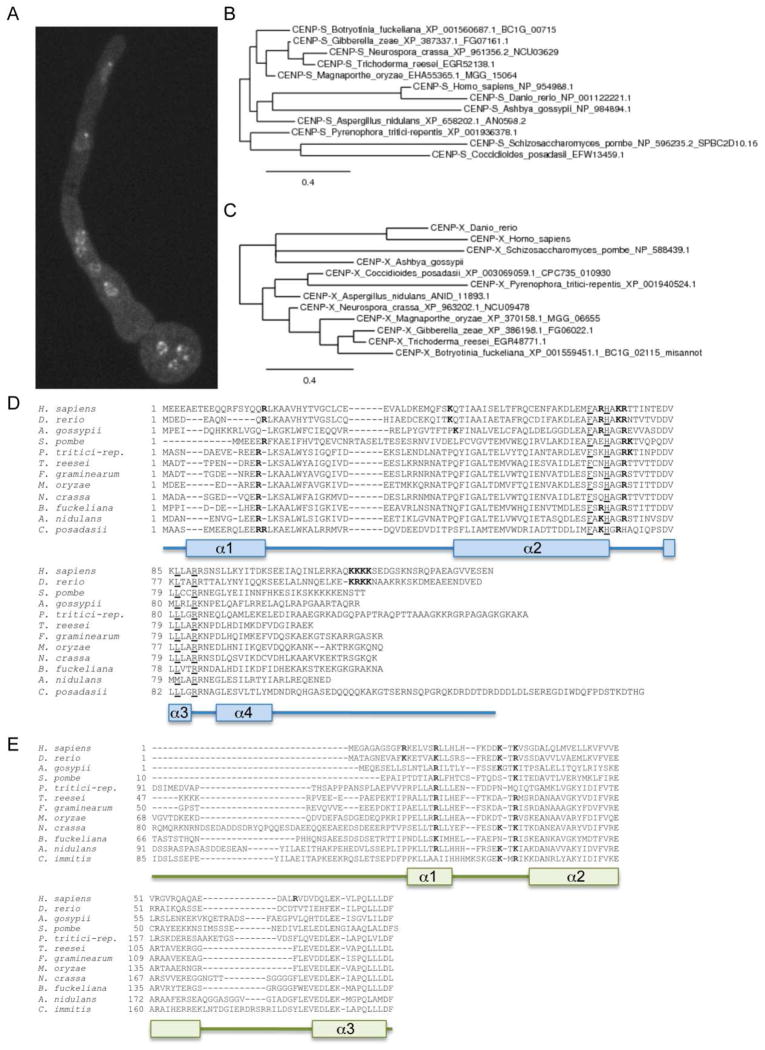
Recent studies with metazoan CENP-T, CENP-W, CENP-S, and CENP-X revealed that these proteins appear to be present in a tetrameric complex that may mimic canonical nucleosomes (Nishino et al., 2012). Tethering experiments revealed that the CENP-T-W-S-X complex is sufficient to nucleate at least partially functional kinetochores, even in the absence of CenH3 (Gascoigne et al., 2011). How similar are animal CENP-T-W-S-X to putative functional homologs from fungi? Homologs for CENP-S and CENP-X can be found in yeasts (e.g., S. cerevisiae CENP-S, Ashbya gossypii CENP-S and CENP-X); in S. pombe all four proteins are recognizable by sequence similarity (Figs. 7 and 8, Table 1). Little is known about these proteins in filamentous fungi. Neurospora CEN-T (NCU02161), a 599 amino acid protein, maps to the same chromosomal regions as CenH3 (Smith et al., 2011) (Fig. 7A), and is predicted to serve as a bridge from the centromere to outer kinetochore proteins. Under standard laboratory growth conditions little transcription from this gene has been observed, and the gene structure remains unclear (K.M. Smith and M. Freitag, unpublished data). The histone H4-like CEN-S (NCU03629), 112 amino acids long, is also localized to chromocenters (Fig. 8A), and the predicted gene structure in Neurospora is supported by RNA-seq data (K.M. Smith and M. Freitag, unpublished data). CEN-W (NCU03400) and CEN-X (NCU09478) are short proteins of 77 and 203 amino acids, respectively, that are difficult to identify by sequence similarity alone. For both CEN-W and CEN-X predicted gene structures are supported by transcripts that were identified by RNA-seq (K.M. Smith, E.L. Bredeweg and M. Freitag, unpublished data). Homologs of all four proteins are found in other filamentous fungi as well, and several conserved, histone-fold-like motifs can be discerned in all of them. These regions largely overlap with motifs that had been recognized to be functionally important for targeting of the metazoan homologs to centromeric DNA (Gascoigne et al., 2011, Nishino et al., 2012). Phylogenies of the four proteins derived with representative fungi mirror established relationships between the species (Fig. 7 and 8, panels B, C), and regions considered to be important for the proposed CEN-T-W-S-X tetramer formation or DNA-binding (Nishino et al., 2012) appear conserved between human, zebrafish and fungal CEN-T and CEN-S (Fig. 7 and 8, panels D, E). Genetic studies to uncover the function of this putative complex in Neurospora and Fusarium are underway, and in combination with ongoing studies on the true function of centromeric heterochromatin, the CEN-B, CEN-C and SCM-3 homologs of N. crassa, will hopefully provide further mechanistic insights into the CenH3 phasing effect observed by ChIP-seq.
Figure 7. Phylogeny of fungal CENP-T and CENP-W.
A. Neurospora CEN-T-GFP localizes to chromocenters, here shown in a cluster of asexual spores. Spores typically contain 2 – 3 nuclei and each nucleus has a single bright focus. Phylogenies for CEN-T (B) and CEN-W (C) were constructed from predicted protein sequences of representative filamentous fungi as well as S. pombe, zebrafish and human. Phylogenetic relationships between the taxa are maintained in these single-protein trees. Alignments of the conserved histone-like domain of CEN-T homologs (D) or the whole CEN-W proteins (E) from selected filamentous fungi as well as S. pombe, zebrafish and human. Residues to be predicted as necessary for tetramer formation (Nishino et al., 2012) are underlined and appear conserved between animal and fungal CEN-T. Residues that may be involved in DNA-binding are shown in bold.
Figure 8. Phylogeny of fungal CENP-S and CENP-X.
A. Neurospora CEN-S-GFP localizes to chromocenters, here shown in a germinating asexual spore (lower right). Chromocenters are oriented towards the growing tip (top center), and each nucleus has one single bright CEN-S-GFP focus. Phylogenies for CEN-S (B) and CEN-X (C) were constructed from predicted protein sequences of representative filamentous fungi as well as A. gossypii, S. pombe, zebrafish and human. Phylogenetic relationships between the taxa are largely maintained in these single-protein trees. Alignments of CEN-S homologs (D) or the histone-like domain from CEN-X proteins (E) from selected filamentous fungi as well as A. gossypii,S. pombe, zebrafish and human. Residues to be predicted as necessary for tetramer formation (Nishino et al., 2012) are underlined and appear conserved between animal and fungal CEN-T. Residues that may be involved in DNA-binding are shown in bold.
Concluding remarks
Filamentous fungi appear to span the gap from the smaller or relatively regularly constructed regional centromeres of C. albicans and S. pombe to both animals and plants with relatively chaotic centromeric DNA assemblies. This group of organisms offers experimental advantages for the analysis of centromeric DNA and the examination of co-evolving DNA and protein in real time. Questions relating to recombination suppression near centromeres (Talbert and Henikoff, 2010) and the existence of gene conversion events within centromeric DNA segments (Shi et al., 2010), for example, can be addressed in N. crassa by HTS of progeny deriving from a single meiosis because of its ordered ascospores. One of the main themes of our future centromere research on filamentous fungi will be to consider if what is true for the best-studied filamentous fungus, N. crassa, also holds for other taxa in this large, heterogeneous group of organisms. For this reason, we have begun to assemble the centromeric DNA sequences of several filamentous fungi in the genera Aspergillus, Fusarium and Mycosphaerella, and–as in Neurospora–we are currently mapping the distribution of centromere proteins in these taxa by ChIP-seq. Our hope is that such comparative studies will help to distinguish between general and specific findings, and that they will lend context for findings from other, more divergent taxa of eukaryotes. This is not just an intellectually interesting exercise, as filamentous fungi are exceedingly successful pathogens, especially on plants, but also increasingly on animals and humans. Few antifungals are truly “fungus-specific”–most act on many eukaryotes. If centromeric DNA and proteins do indeed co-evolve more quickly than other systems, there may be hope for designing small-molecule drugs to interfere with centromere and kinetochore function in only certain species.
Figure 6. Evolution of fungal CENPC homologs.
A. Analysis of CEN-C proteins identified from representative fungal genomes suggests that CEN-C has one short region (blue line) that may be under positive selection, while the N- and C-terminal regions are under negative selection (gray), as is the CEN-C motif (red line). Numerous charged amino acid residues (shown in blue) in the putatively adaptive region may be involved in DNA-binding by CEN-C. B. Neurospora CEN-C proteins have two short regions (green lines) that may be under positive selection and that surround a less variable region (orange line). This region mostly coincides with the putative motif under positive selection when comparing representative fungal taxa (see A.). Alignments were done by MEGA (Tamura et al., 2007) or MEME (Bailey et al., 2009) and the graphs show Pi(a)/Pi(s), the ratio of non-synonymous to synonymous mutations calculated by analyzing sequences in DNAsp (Rozas and Rozas, 1995).
Acknowledgments
We thank Mark Dasenko, Steve Drake, Matthew Peterson and Chris Sullivan at the OSU CGRB core facility for assistance with Illumina sequencing, and Shinji Honda and Eric Selker for sharing preliminary data. Centromere research in our lab is supported by grants from the American Cancer Society (RSG-08-030-01-CCG) and the NIH (R01GM097637).
Abbreviations
- Abp1
S. pombe centromere protein B
- ABP-1RIP
mutated centromere protein B homologue in N. crassa
- CAL1
chromosome alignment defect 1
- CATD
centromere protein A targeting domain
- CBF1
centromere binding factor 1
- Cbh1/Cbh2
S. pombe centromere protein B homologues
- CbhA to -C
Aspergillus nidulans centromere protein B homologues
- cc
central core of centromeric DNA in S. pombe
- CDEI
centromeric DNA element 1
- CDP-2
chromodomain protein 2 in filamentous fungi
- CEN1
centromere on chromosome 1 of C. albicans
- Cen1
centromere on chromosome 1 of F. graminearum
- Cen-I
centromere on chromosome 1 of N. crassa
- CEN-B
centromere protein B homologues in filamentous fungi
- CEN-B1
centromere protein B1 in N. crassa
- CEN-B2
centromere protein B2 in N. crassa
- CEN-C/cen-c
centromere protein C in filamentous fungi (genes are in italics)
- CenH3
centromere-specific histone H3 in filamentous fungi (CENP-A)
- CEN-L
centromere protein L in filamentous fungi
- CEN-Q
centromere protein Q in filamentous fungi
- CEN-T
centromere protein T in filamentous fungi
- CEN-T-W-S-X
complex of centromere proteins T, W, S, and X that mimics nucleosomes
- CENP-A
centromere-specific histone H3, centromere protein A
- CENP-C
centromere protein C
- CENP-R
centromere protein R
- CHAP
CDP-2- and HDA-1 associating protein in N. crassa
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput sequencing
- Cid
centromere-specific histone H3 in Drosophila, centromere identifier
- Cnp1CenpA
centromere-specific histone H3 in S. pombe
- Cnp3
centromere protein C in S. pombe
- Cse4
centromere-specific histone H3 in S. cerevisiae and C. albicans
- Cul4
cullin 4, part of ubiquitin ligase (E3) complex
- DCDC
complex of DIM-5, DIM-7, DIM-9, CUL-4 and DDB-1 in N. crassa
- DDB1
DNA damage-binding protein 1
- DIM-5
defective in DNA methylation 5
- H3K9me3
histone methyltransferase
- DIM-7
defective in DNA methylation 7, component of DCDC in N. crassa
- DIM-9
defective in DNA methylation 9, component of DCDC in N. crassa
- DMM
DNA methylation modulator complex in N. crassa
- Fot1
Fusarium oxysporum transposon family 1
- H3K4me2
dimethylated histone H3 lysine 4
- H3K9me2
trimethylated histone H3 lysine 9
- HCHC
complex of HP1, CDP-2, HDA-1 and CHAP in N. crassa
- HDA-1
histone deacetylase 1 in N. crassa
- HJURP
Holliday junction recognition protein
- HP1/hpo
heterochromatin protein 1 (N. crassa gene in italics)
- HRT12
centromere-specific histone H3 in Arabidopsis thaliana
- HTS
high-throughput sequencing
- imr
inner repeats of centromeric DNA in S. pombe
- INCENP
inner centromere protein
- kb
kilobasepair
- KNL2
kinetochore-null 2
- LG
linkage group
- M18BP1
Mis18 binding protein 1
- MEGA
molecular evolutionary genetics analysis, software package
- MEME
multiple em for motif elicitation, software package
- MgcRacGAP
human rac GTPase activating protein
- MIND
kinetochore subcomplex
- Mtw1
including Nnf1/Nsl1 and Dsn1
- Mis12
centromere protein Mis12 in S. pombe
- Mis17
centromere protein CENP-M in S. pombe
- Moa1
monopolar attachment 1, cohesin-associated protein
- NPM1
nucleophosmin 1
- otr
outer repeats of centromeric DNA in S. pombe
- Pi(a)/Pi(s)
ratio of non-synonymous to synonymous substitutions
- Pot2
DNA transposon from Magnaporthe grisea
- RFLP
restriction fragment length polymorphism
- RNAi
RNA interference
- RNA-seq
high-throughput sequencing of complementary DNA made from RNA
- RIP
Repeat-Induced Point mutation
- SCM3
suppressor of chromosome missegregation 3
- SNPs
single nucleotide polymorphisms
- Swi6
heterochromatin protein 1 in S. pombe
- Tc5
family of DNA transposons in C. elegans
Footnotes
The authors have no conflicting interests.
References
- Allshire RC. Centromeres, checkpoints and chromatid cohesion. Curr Opin Genet Dev. 1997;7:264–273. doi: 10.1016/s0959-437x(97)80137-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RE, Rogers K. Phylogenetic analysis of fungal centromere H3 proteins. Genetics. 2006;174:1481–1492. doi: 10.1534/genetics.106.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20:2852–2864. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2010;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–9. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, et al. Lessons from the Genome Sequence of Neurospora crassa: Tracing the Path from Genomic Blueprint to Multicellular Organism. Microbiol Mol Biol Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Matthews BW. The helix-turn-helix DNA binding motif. The J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–819. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Aisner R, Carbon J. Structure of the chromosome VII centromere region in Neurospora crassa: degenerate transposons and simple repeats. Mol Cell Biol. 1998;18:5465–5477. doi: 10.1128/mcb.18.9.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri EB, Jensen BC, Schabtach E, Selker EU. Repeat-induced G-C to A-T mutations in Neurospora. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- Cambareri EB, Singer MJ, Selker EU. Recurrence of repeat-induced point mutation (RIP) in Neurospora crassa. Genetics. 1991;127:699–710. doi: 10.1093/genetics/127.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centola M, Carbon J. Cloning and characterization of centromeric DNA from Neurospora crassa. Mol Cell Biol. 1994;14:1510–1519. doi: 10.1128/mcb.14.2.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Cogoni C, Macino G. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 2004;32:4237–4243. doi: 10.1093/nar/gkh764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH. Domain organization at the centromere and neocentromere. Developmental cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–74. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- Davis RH. Neurospora: Contributions of a Model Organism. Oxford University Press; 2000. [Google Scholar]
- Dawe RK, Henikoff S. Centromeres put epigenetics in the driver's seat. Trends Biochem Sci. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Dong F, Miller JT, Jackson SA, et al. Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci U S A. 1998;95:8135–8140. doi: 10.1073/pnas.95.14.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Evolution of functional diversity in the cupin superfamily. Trends Biochem Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, et al. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, Hall C, Kowbel D, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A. 2011a;108:2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CE, Stajich JE, Jacobson DJ, et al. Massive Changes in Genome Architecture Accompany the Transition to Self-fertility in the Filamentous Fungus Neurospora tetrasperma. Genetics. 2011b;189:55–69. doi: 10.1534/genetics.111.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Mellone BG, Betts CM, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. 1993. [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–7. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler KJ, Hudson DF, Salamonsen LA, et al. Uterine dysfunction and genetic modifiers in centromere protein B-deficient mice. Genome Res. 2000;10:30–41. [PMC free article] [PubMed] [Google Scholar]
- Fowler KJ, Wong LH, Griffiths BK, et al. Centromere protein b-null mice display decreasing reproductive performance through successive generations of breeding due to diminishing endometrial glands. Reproduction. 2004;127:367–77. doi: 10.1530/rep.1.00102. [DOI] [PubMed] [Google Scholar]
- Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in Neurospora. Mol Cell. 2004a;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Freitag M, Lee DW, Kothe GO, et al. DNA methylation is independent of RNA interference in Neurospora. Science. 2004b;304:1939. doi: 10.1126/science.1099709. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Lewis ZA, Huarte M, et al. The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa. Genes Dev. 2010;24:443–454. doi: 10.1101/gad.1893210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Lewis ZA, Shimada K, et al. Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat Struct Mol Biol. 2012;19:471–477. doi: 10.1038/nsmb.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Liu Y, Wang M, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. The Journal of cell biology. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irelan JT, Gutkin GI, Clarke L. Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics. 2001;157:1191–1203. doi: 10.1093/genetics/157.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, et al. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science. 2009;324:1716–1719. doi: 10.1126/science.1172026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Ketel C, Wang HS, McClellan M, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Dorn JF, De Rop V, et al. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. [DOI] [PubMed] [Google Scholar]
- Lee HC, Li L, Gu W, et al. Diverse Pathways Generate MicroRNA-like RNAs and Dicer-Independent Small Interfering RNAs in Fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1997;94:8427–8432. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Adhvaryu KK, Honda S, et al. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010a;6:e1001196. doi: 10.1371/journal.pgen.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Selker EU. Identification of DIM-7, a protein required to target the DIM-5 H3 methyltransferase to chromatin. Proc Natl Acad Sci U S A. 2010b;107:8310–8315. doi: 10.1073/pnas.1000328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis ZA, Honda S, Khlafallah TK, et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–437. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Wing RA, Bennetzen JL, Jackson SA. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 2007;23:134–139. doi: 10.1016/j.tig.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K, Wiest AE, Grigoriev IV, et al. Rediscovery by Whole Genome Sequencing: Classical Mutations and Genome Polymorphisms in Neurospora crassa. G3. 2011;1:303–316. doi: 10.1534/g3.111.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL, Stevens JN, Selker EU, Morzycka-Wroblewska E. A method for finding the genetic map position of cloned DNA fragments. Neurospora Newsletter. 1984;31:35–39. [Google Scholar]
- Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y, Peebles C, Fritzler MJ, Steigerwald J, Tan EM. Autoantibody to Centromere (Kinetochore) in Scleroderma Sera. Proc Natl Acad Sci U S A. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Huberman JA, Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1996;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Neumann P, Zhang D, et al. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol. 2005;22:845–855. doi: 10.1093/molbev/msi069. [DOI] [PubMed] [Google Scholar]
- Nagaki K, Talbert PB, Zhong CX, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163:1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M, Stajich JE, Chu M, et al. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 2010;6:e1000891. doi: 10.1371/journal.pgen.1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, et al. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol. 1998;201:135–43. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–46. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Euk Cell. 2011;10:724–733. doi: 10.1128/EC.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Kwong PN, Menorca RM, et al. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics. 2010;186:461–471. doi: 10.1534/genetics.110.120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Goodman HM, Ausubel FM. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci. 1995;11:621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci U S A. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 2010;27:1585–1597. doi: 10.1093/molbev/msq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16:119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, et al. Direct Binding of Cenp-C to the Mis12 Complex Joins the Inner and Outer Kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, et al. Widespread gene conversion in centromere cores. PLoS Biol. 2010;8:e1000327. doi: 10.1371/journal.pbio.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata F, Murata M. Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J Cell Sci. 2004;117:2963–2970. doi: 10.1242/jcs.01144. [DOI] [PubMed] [Google Scholar]
- Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 2011;31:2528–2542. doi: 10.1128/MCB.01285-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Gronemeyer B, Lu W, Eugster E, Tomkiel JE. Mutational analysis of the central centromere targeting domain of human centromere protein C (CENP-C) Exp Cell Res. 2002;275:81–91. doi: 10.1006/excr.2002.5495. [DOI] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Blower MD, Karpen GH. Determining centromere identity: cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF. A solid foundation: functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. doi: 10.1016/s0959-437x(00)00177-5. [DOI] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Centromeres convert but don't cross. PLoS Biol. 2010;8:e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone h3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nureki O, Kurumizaka H, et al. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S, Perini G, Bernardoni R, et al. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang J, Hu Q, et al. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet. 2010;6:e1001132. doi: 10.1371/journal.pgen.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Miklos GL. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma. 1978;66:71–98. doi: 10.1007/BF00285817. [DOI] [PubMed] [Google Scholar]
- Yan H, Jin W, Nagaki K, et al. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–3238. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/alpha-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Vaughn MW, Irvine DV, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang CT. Isochore structures in the genome of the plant Arabidopsis thaliana. J Mol Evol. 2004;59:227–238. doi: 10.1007/s00239-004-2617-8. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Shen Y, Yang S, et al. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem. 2010;285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]