Abstract
Neocentromeres are ectopic sites where new functional kinetochores assemble and permit chromosome segregation. Neocentromeres usually form following genomic alterations that remove or disrupt centromere function. The ability to form neocentromeres is conserved in eukaryotes ranging from fungi to mammals. Neocentromeres that rescue chromosome fragments in cells with gross chromosomal rearrangements are found in several types of human cancers, and in patients with developmental disabilities. In this review, we discuss the importance of neocentromeres to human health and evaluate recently developed model systems to study neocentromere formation, maintenance, and function in chromosome segregation. Additionally, studies of neocentromeres provide insight into native centromeres; analysis of neocentromeres found in human clinical samples and induced in model organisms distinguishes features of centromeres that are dependent on centromere DNA from features that are epigenetically inherited together with the formation of a functional kinetochore.
Keywords: neocentromere, centromere, kinetochore, chromosome segregation
Centromeres, the regions on each chromosome where kinetochores assemble and promote the attachment of spindle microtubules, are essential for chromosome segregation and genome integrity. In humans and almost all other eukaryotes, kinetochore assembly is not dependent on primary sequence, and centromeres are inherited epigenetically (Allshire and Karpen, 2008). The kinetochore protein CenH3/CENP-A, a histone H3 variant that replaces canonical H3 at centromeric chromatin, is a defining feature of centromeres (Buscaino et al., 2010). At least 100 kinetochore proteins have been associated with centromeres in humans (Liu et al., 2006) and many of these are conserved among eukaryotes. CENP-A is at or near the top of the kinetochore assembly dependency pathway (Liu et al., 2006, Mendiburo et al., 2011) and is critically important for proper centromere formation and function.
Neocentromeres are new sites of assembly of functional kinetochores at ectopic loci that rescue chromosome fragments in cells with gross chromosomal rearrangements (Marshall et al., 2008). Centromere positions are generally stable, but genomic alterations can remove or disrupt centromere function; neocentromeres that form elsewhere on the remaining chromosome restore the ability of that chromosome to segregate efficiently (Blom et al., 2010). In rare cases, neocentromeres form on otherwise normal chromosomes, without physical deletion of the native centromere, presumably following inactivation of the native centromere through unknown mechanisms (Amor et al., 2004, Liehr et al., 2010). “Evolutionary new centromeres” are important steps in speciation that involve centromere repositioning events that become fixed in the population (Ventura et al., 2007, Capozzi et al., 2009). In this review, we will discuss the importance of neocentromeres to human health and recently developed model systems that are used to study neocentromere formation and function.
Analysis of neocentromeres from clinical samples has revealed some characteristics of human neocentromeres, but this is necessarily limited to retrospective analysis. Insights into the mechanisms of neocentromere formation and function are more readily achieved from direct experiments using model organisms. Recently, induced neocentromere formation has been reported in Drosophila melanogaster (fruit flies), the fission yeast Schizosaccharomyces pombe, and Candida albicans, a multimorphic and pathogenic yeast. We will discuss the strengths of each model organism for understanding neocentromere formation and function as well as how they help us identify which centromere features are dependent on the native context and which features are epigenetically inherited together with kinetochore assembly. Finally, we will highlight some of the important open questions in the field.
Neocentromeres in human health
The majority of neocentromeres reported in clinical samples rescue acentric chromosome fragments associated with duplications or chromosomal rearrangements found in patients with developmental disabilities (Marshall et al., 2008). Such neocentromeres are more likely to be observed clinically because of phenotypes associated with the accompanying amplifications, deletions, or gene disruptions (Marshall et al., 2008). Neocentromeres stabilize genomic rearrangements that form small supernumerary marker chromosomes. Marker chromosomes are abnormal chromosomes formed by rearrangements or amplifications of specific genomic regions. Phenotypes associated with marker chromosomes stabilized by neocentromeres include: facial dysmorphisms, renal defects, short stature, and developmental delays (Mascarenhas et al., 2008, Burnside et al., 2011). Prenatal identification of neocentric marker chromosomes can be achieved by cytogenetic analysis of chromosomes from amniotic fluid and fetal blood cells (Mascarenhas et al., 2008).
In several types of human cancers, neocentromeres have rescued chromosome fragments that arose via gross chromosomal rearrangements (Sirvent et al., 2000, Blom et al., 2010). Solid tumors in cases of well-differentiated liposarcomas are cytogenetically characterized by the presence of a supernumerary ring or giant chromosome with amplified material from the 12q14–21 region combined with DNA from other chromosomes and a neocentromere (Italiano et al., 2009). Other types of human cancers associated with neocentromeres include: retinoblastoma (Morrissette et al., 2001), non-Hodgkin’s lymphoma (Blom et al., 2010), acute myeloid leukemia (de Figueiredo et al., 2009), and lung cancer (Italiano et al., 2006). Interestingly, CENP-A is overexpressed in several types of cancer cells (Tomonaga et al., 2003, Amato et al., 2009) and high CENP-A expression correlated with shorter survival times in lung cancer patients (Wu et al., 2012). While overexpression of CENP-A does not appear to be sufficient for ectopic kinetochore assembly in the presence of native centromeres (Van Hooser et al., 2001), excess CENP-A may enhance extracentromeric CENP-A incorporation and thereby facilitate neocentromere formation on acentric supernumerary chromosomes. We suggest that neocentromeres may be underappreciated as a mechanism of stabilizing genomic alterations in cancer cells as only a small proportion of tumor samples are analyzed by cytogenetic techniques capable of detecting neocentromeres (Fletcher, 2005).
Some neocentromeres form in the absence of obvious chromosome rearrangements, and these have very little direct impact on human health. Importantly, these centromere repositioning events are stable through meiosis, as they have been inherited from one generation to the next. In two documented cases, the new centromere position was maintained through at least three generations (Tyler-Smith et al., 1999, Capozzi et al., 2009). While such centromere repositioning is rarely detected (Amor et al., 2004, Ventura et al., 2004, Capozzi et al., 2009, Hasson et al., 2011), it is likely that the rate of centromere repositioning events are underreported, since they are only identified by cytogenetic screening and cause no significant phenotypes.
“Evolutionary new centromeres” – centromere repositioning on an evolutionary scale
Evolutionary new centromeres are essentially neocentromeres that are inherited through many generations and become fixed in the population. Centromere repositioning via ectopic kinetochore assembly is a key step in the formation of evolutionary new centromeres. Evolutionary new centromeres often arise in gene deserts (Lomiento et al., 2008) and accumulate repetitive sequences (Ventura et al., 2007). These evolutionary events, detected via altered synteny of centromere-associated satellite DNA as well as through karyotype analysis, require that kinetochore proteins assemble at a new locus and no longer assemble at the old locus. Six human chromosomes have evolutionary new centromeres (Rocchi et al., 2012). For example, human centromere 6 repositioned from an ancestral location to its current location in a common ancestor of hominids (Capozzi et al., 2009). Comparison of the human and macaque genome reveals that 9 of 20 macaque chromosomes have evolutionary new centromeres that accumulated over the comparatively short time period of approximately 14 million years (Ventura et al., 2007). The large fraction of chromosomes with evolutionary new centromeres in humans and macaques indicates that, on an evolutionary time scale, centromere repositioning is a relatively frequent event. Evolutionary new centromeres are also found in other mammals (Rocchi et al., 2012) as well as in plants (e.g., rice (Nagaki et al., 2004) and cucurbits (Han et al., 2009)).
Retrospective analysis of human clinical neocentromere samples
Most of our current knowledge regarding neocentromere formation and function in chromosome segregation is derived from analysis of neocentromeres isolated from human clinical samples. More than 100 neocentromeres have been characterized in humans (Marshall et al., 2008, Alonso et al., 2010, Klein et al., 2012) and these neocentromeres show remarkable diversity in chromosome position and associated DNA sequences. Neocentromeres have been identified on 21 of 22 autosomic chromosomes as well as on the X and Y sex chromosomes (Marshall et al., 2008, Liehr et al., 2010). Neocentromeres are particularly common on specific chromosome regions including 3q, 13q and 15q (Marshall et al., 2008, Liehr et al., 2010). However, even among neocentromeres found in the same chromosomal band, the exact DNA sequences bound by CENP-A are unique in each case examined to date (Alonso et al., 2003, Marshall et al., 2008, Hasson et al., 2011). Sequences from the characteristic amplification of 12q14–21 in four supernumerary ring chromosomes from well-differentiated liposarcoma cases are not the sites of neocentromere formation, rather neocentromeres formed on amplified sequences from other chromosomes that are also part of the ring chromosome. The neocentromere sequences differed in each patient analyzed (Italiano et al., 2009).
Human centromeres are always found in highly repetitive regions containing alpha-satellite DNA (Vafa and Sullivan, 1997), yet neocentromeres form on very diverse DNA sequences and are not associated with alpha-satellite DNA. Some neocentromeres formed in repetitive DNA regions (Hasson et al., 2011), but many neocentromeres are not associated with a significant amount of repetitive DNA (Alonso et al., 2010). Human neocentromeres form either in gene deserts or in regions that include actively transcribed genes (Marshall et al., 2008, Alonso et al., 2010). LINE retrotransposon sequences appear to be important for maintaining neocentric chromatin at some neocentromeres, as knockdown of transposon transcript levels impaired neocentromere function during mitosis (Chueh et al., 2009), but this has only been tested for one neocentromere thus far. In at least one instance, a neocentromere formed very close to the breakpoint of a chromosomal rearrangement (Hasson et al., 2011). This result is intriguing as CENP-A has been shown to bind transiently to sites of double-stranded DNA breaks (Zeitlin et al., 2009), suggesting that sites of DNA damage bound by CENP-A may initiate neocentromere formation.
Chromatin immunoprecipitation (ChIP) analyses have failed to define common features of human neocentromeres (Rocchi et al., 2012). Human centromeres have distinct chromatin domains. Centromeric chromatin is hypoacetylated and contains CENP-A nucleosomes interspersed with nucleosomes containing histone H3 dimethylated at lysine 4. The CENP-A core is surrounded by pericentric heterochromatin (Sullivan and Karpen, 2004). Proximal heterochromatic regions, identified by ChIP of histone H3 methylated at lysine 9 (H3K9me) and heterochromatin protein 1 (HP1), are not required either for neocentromere formation or for function in chromosome segregation (Alonso et al., 2010). However, the authors suggest that heterochromatin may contribute to chromosome cohesion, because neocentromeres with little or no detectable heterochromatin exhibited slight cohesion defects (Alonso et al., 2010).
Some neocentromeres also are more prone to chromosome missegregation than native centromeres. Neocentromere mosaicism (presence of the neocentromere in a subset of somatic cells) suggests that the chromosome carrying the neocentromere was lost in a subpopulation of the cells. This implies that some neocentromeres are less stable than native centromeres in vivo (Marshall et al., 2008). Human neocentromeres also have defects in the localization of Aurora B kinase, an essential regulator of kinetochore-microtubule attachments, and in error correction (Bassett et al., 2010). Determinants of the chromosome segregation efficiency of different neocentromere positions remain to be identified. One possible explanation for the reduced function in chromosome segregation is that some neocentromeres in humans have reduced incorporation of CENP-A relative to normal centromeres (Irvine et al., 2004). Additionally, human neocentromeres are frequently smaller than native centromeres with CENP-A binding regions of ~100kb at neocentromeres and CENP-A binding regions of ~200 kb to 1.5 Mb at native centromeres (Sullivan et al., 2011). In human artificial chromosomes, the length of chromosome regions surrounding the alpha-satellite DNA affects chromosome segregation accuracy (Rudd et al., 2003), potentially by contributing to the polar ejection forces necessary for chromosome alignment (Ke et al., 2009). Because neocentromeres are frequently found on chromosomes with short chromosome arms, some of the missegregation of chromosomes with neocentromeres may be attributed to the reduced polar ejection forces acting on these chromosomes.
Overall, chromosome regions that maintain neocentromere function show little similarity, and unlike native human neocentromeres, none of them are associated with alpha-satellite DNA. Currently, identification of common neocentromere features is limited by the low resolution of microscopy approaches and the limited number of neocentromere positions that have been analyzed with molecular biology techniques. High resolution analysis with tiling ChIP-chip or ChIP-seq of CENP-A binding regions in many neocentromere strains together with the use of model systems to develop testable hypothesis that can be tested in human cells should provide additional insight in the future.
Model organisms to understand mechanisms of neocentromere formation
The inability to manipulate neocentromeres in human cells limits the ability to understand the process of neocentromeres formation. The conservation of neocentromere formation from humans to plants to fungi justifies the study of neocentromeres in diverse organisms and suggests that such studies can provide significant insight into neocentromere formation and function.
In plants the term neocentromere is used in two different ways: “rescue” neocentromeres that, like human neocentromeres, appear following centromeric deletions or rearrangements, and neocentromere “knobs” that do not involve canonical kinetochore proteins (Nasuda et al., 2005, Topp et al., 2009). Neocentromere “knobs” drive selection of specific chromosomes during meiosis (Mroczek et al., 2006). Thus, their mechanisms of formation and transient maintenance are not likely to be representative of other types of neocentromeres (Dawe and Cande, 1996). For the purposes of this review, we will focus on plant “rescue” neocentromeres that are more similar to those found in humans. Neocentromeres that rescue chromosomes following inactivation of a native centromere or a chromosomal rearrangement occur in divergent plant species, including barley, maize and rice (Nasuda et al., 2005, Gong et al., 2009, Topp et al., 2009). Plant neocentromeres form either proximal to the native centromere (Nasuda et al., 2005) or at distal loci on the chromosome arms (Topp et al., 2009).
Kinetochores assembled at maize neocentromeres exhibit substantial variation in the binding of CENP-A (frequently called CenH3 in plants) among different isolates. Very low levels of CENP-A correlated with decreased chromosome stability and the amount of CENP-A at the neocentromere increased in subsequent generations. Chromosome stability also increased in later generations, suggesting that the neocentromere became more functional as it was inherited (Topp et al., 2009). The observation in plants that neocentromeres stabilize over time suggests that initial CENP-A binding and neocentromere initiation may happen at many locations, and that stable neocentromere formation requires locations where more CENP-A can incorporate so that a neocentromere of substantive size can be maintained.
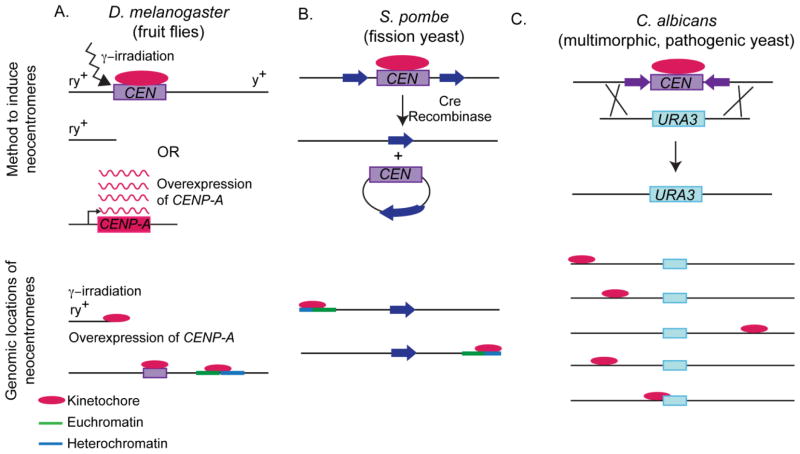
Fruit flies (D. melanogaster) are a classic model for neocentromere formation. Ectopic centromeres can be induced by γ-irradiation-induced chromosome breakage or by overproduction of CENP-A (called CID in D. melanogaster) (Williams et al., 1998, Maggert and Karpen, 2001, Olszak et al., 2011) (Figure 1A). γ-irradiation-induced chromosome breakage resulted in formation of neocentromeres in the pericentric region (Maggert and Karpen, 2001). Overproduction of CENP-A resulted in ectopic kinetochore assembly at the boundaries of heterochromatin and euchromatin (Olszak et al., 2011). Additionally, tethering of CENP-A with a LacI/LacO system is sufficient for ectopic kinetochore assembly in fruit flies, and this ectopic kinetochore position is maintained even when the plasmid containing the LacI-CENP-A fusion is lost (Mendiburo et al., 2011). This is a powerful system to direct kinetochore assembly to specific loci in D. melanogaster.
Figure 1. Models to induce neocentromere formation.
(A) D. melanogaster ectopic kinetochores can be induced by γ-irradiation or overproduction of CENP-A. Following, γ-irradiation, chromosome fragments were stabilized by neocentromeres if the fragment was adjacent to the native centromere. Following CENP-A overexpression, ectopic kinetochores assembled at the border of euchromatin and heterochromatin, resulting in dicentric chromosomes. (B) S. pombe neocentromeres can be induced by recombination-mediated deletion of the native centromere. All neocentromeres formed near subtelomeres. (C) C. albicans neocentromeres can be induced by replacing the native centromere with a selectable marker through by DNA transformation. Neocentromeres formed proximal to the native centromere or distal to the native centromeres on chromosome arms. Drawings not to scale.
Fission yeast (S. pombe) is unicellular model for studying neocentromere specification and kinetochore assembly. A useful feature is that it has only 3 chromosomes in its haploid genome. Inducible deletion of centromere DNA via Cre-Lox excision resulted in neocentromere formation adjacent to the telomeres but not at internal loci (Ishii et al., 2008) (Figure 1B). Similar to ectopic centromeres induced by overproduction of CENP-A in Drosophila, S. pombe neocentromeres are found at the borders of euchromatin and heterochromatin (Ishii et al., 2008). Furthermore, neocentromere formation in S. pombe requires functional heterochromatin proteins. In the absence of RNA interference-dependent heterochromatin, the frequency of chromosome rescue via neocentromere formation was much lower than an alternative mechanism of chromosome rescue (Ishii et al., 2008). Thus, chromatin context strongly influences neocentromere formation in S. pombe and D. melanogaster. Some human neocentromeres also form at the boundaries of euchromatin and heterochromatin (Wong et al., 2006), however analysis of clinical human neocentromeres indicates that many chromosome contexts may be permissive for human neocentromeres.
Recently, Candida albicans, a human fungal pathogen, has emerged as a promising model organism for studying neocentromere formation. C. albicans has a diploid genome and small (~3–4 kb/central core) regional centromeres (Baum et al., 2006, Ketel et al., 2009). Neocentromere formation can be induced in C. albicans by deleting the native centromere and replacing it with a selectable marker (Ketel et al., 2009) (Figure 1C). Neocentromeres form proximal to the deleted native centromere locus or at distal positions along the chromosome arms (Ketel et al., 2009) as has been observed for human neocentromeres (Marshall et al., 2008, Ketel et al., 2009). Five neocentromere locations on chromosome 5 in C. albicans have been described (Ketel et al., 2009). An additional 17 neocentromere positions on chromosome 5 have been identified (unpublished observations). Similar to some human neocentromeres (and unlike native human centromeres), C. albicans neocentromeres are not associated with heterochromatin (Alonso et al., 2010). Additionally, neocentromeres in C. albicans are associated with larger than average intergenic regions, but can form in both transcriptionally active and in intergenic non-expressed regions (Ketel et al., 2009). Interestingly, both human neocentromeres (Saffery et al., 2003, Wong et al., 2006, Alonso et al., 2010) and plant centromeres can form on transcriptionally active regions (Nagaki et al., 2004). Recent work also suggests that active transcription is important for mammalian centromere function (Chan et al., 2012).
Epigenetically inherited features of centromeres
In both human cells and model systems, neocentromeres are a powerful tool for distinguishing epigenetically inherited features of centromeres from proteins recruited by centromere DNA sequences. A pseudo-dicentric human chromosome (containing a neocentromere with a functional kinetochore, as well as an inactivated native centromere alpha-satellite sequences) has been used to identify features associated with the functional kinetochore, with alpha-satellite DNA sequences or with both (Amor et al., 2004, Bassett et al., 2010). HP1α, a marker of heterochromatin, localized to both the inactivated alpha-satellite sequences and the functional kinetochore (Amor et al., 2004). Further analysis of this pseudo-dicentric chromosome found that all characterized kinetochore proteins, except for CENP-B, associated with the functional kinetochore, rather than with the vestigial alpha-satellite sequences (Bassett et al., 2010). CENP-B is recruited to the centromere in a sequence-dependent manner (Ohzeki et al., 2002) by alpha-satellite sequences not found at neocentromeres, suggesting CENP-B is not necessary for kinetochore assembly. Aurora B kinase is recruited to the functional kinetochore, although its localization pattern is altered. The altered localization pattern results in the reduced ability of Aurora B kinase to detect and correct chromosome-spindle misattachments (Bassett et al., 2010).
In fungi, cohesin binding is enriched near centromeres in S. cerevisiae (Weber et al., 2004). Pericentric cohesin binding is dependent on the presence of a functional kinetochore and has been proposed to enhance chromosome segregation by stabilizing a looped DNA structure around the centromere (Yeh et al., 2008). In S. pombe cohesin binds to pericentric regions and this binding is dependent on heterochromatin proteins (Nonaka et al., 2002). Pericentric cohesin binding in C. albicans appears to be linked with the functional epigenetic neocentromere rather than the native centromere (unpublished observations), but the roles of cohesin in neocentromere formation and function are not yet fully characterized.
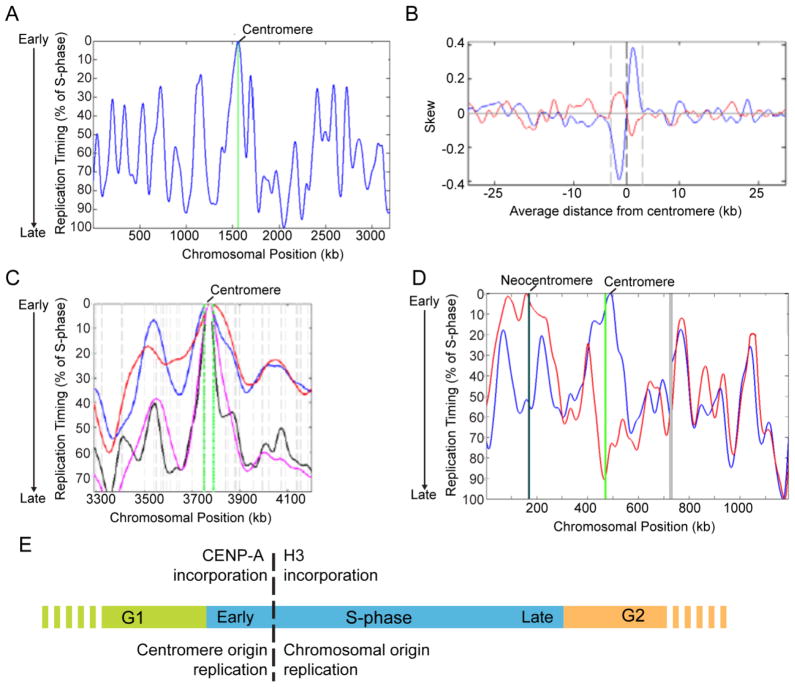
Neocentromeres have provided particularly valuable insight into the link between DNA replication and centromeres. A specific replication timing pattern for centromeres is an appealing mechanism for facilitating the maintenance of centromere specification. Initial studies in flies and humans did not detect a specific timing pattern for centromeres (Shelby et al., 2000, Ahmad and Henikoff, 2001, Lo et al., 2001, Sullivan and Karpen, 2001), however these experiments were limited by the low resolution of microscopy experiments. Using higher resolution techniques, such as microarrays, fungal centromeres have been shown to replicate early in S. cerevisiae (Raghuraman et al., 2001), C. albicans and S. pombe (Koren et al., 2010) (Figure 2). Centromeres in C. albicans were the earliest and most efficient origins in the genome (Koren et al., 2010), and they contain evolutionarily conserved sequence biases (e.g., GC-skew (Sernova and Gelfand, 2008)) indicative of constitutive origin firing (Figure 2B and (Koren et al., 2010)). To determine if efficient replication was determined by the chromosomal context or by the presence of a functional neocentromere, replication timing was measured in a C. albicans strain with a homozygous neocentromere. Neocentromere formation dramatically shifted the replication timing pattern on the chromosome such that the neocentromere region now contained the earliest and most efficient origin (Koren et al., 2010) (Figure 2D). Origin recognition complex (ORC) did not bind the locus prior to neocentromere formation, but was recruited following neocentromere formation (Koren et al., 2010). Increasing CENP-A (called Cse4 in C. albicans) levels results in increased recruitment of kinetochore proteins (Burrack et al., 2011) and ORC (Koren et al., 2010) to centromere DNA, further suggesting that a kinetochore component recruits ORC and enhances replication efficiency of centromeres and neocentromeres.
Figure 2. Efficient DNA replication at fungal centromeres.
(A) Centromeres are the earliest and most efficient origins on each C. albicans chromosome. The replication timing profile for chromosome in wild-type cells is indicated in blue. The position of CEN1 is indicated with a green line. (B) Centromere DNA in C. albicans and related species contains GC-skew, a replication-dependent strand-bias sequence pattern characteristic of constitutively active replication origins. Mean GC-skew (blue) and AT-skew (red) skew at the C. albicans centromere regions from all chromosomes are shown. (C) In S. pombe, CEN1 replicates earliest of all loci on chromosome 1. The centromere is indicated in green, while black, magenta, blue and red lines indicate data from different S. pombe replication timing microarray experiments (Feng et al., 2006, Heichinger et al., 2006, Mickle et al., 2007). (D) Neocentromere loci become the earliest, most efficient origins following kinetochore assembly, indicating that the replication pattern is determined by the presence of a function kinetochore. The native centromere (green line) is the earliest replicating region in wild-type cells (blue line), while the neocentromere locus approximately 170kb from the left telomere (black line) is the earliest replicating region in cells with this homozygous neocentromere (red line). Gray lines indicate a gap in data from the multiple repeat sequences in C. albicans on the right arm of chromosome 5. (E) Model for coordination of replication timing and maintenance of centromere specification. Adapted from (Koren et al., 2010).
Early replication of centromeres in fungi provides a compelling model for the coordination of CENP-A incorporation and DNA replication (Figure 2E). CENP-A is incorporated in S-phase in S. cerevisiae (Pearson et al., 2004) and in S- and G2-phase in S. pombe (Takayama et al., 2008). Coordinated replication of centromeres and incorporation of CENP-A early in S-phase may help limit extracentromeric incorporation of CENP-A and enhance the inheritance of centromeres in these fungi. CENP-A is incorporated during mitosis in D. melanogaster cells (Mellone et al., 2011) and in G1 in human cells (Dunleavy et al., 2009, Foltz et al., 2009). The lack of apparent coordination of replication timing among centromeres in humans and flies may be due to insufficient resolution or may be attributed to differences in the timing of CENP-A incorporation in metazoans. Further high-resolution studies of metazoan centromeres, especially using neocentromeres where repetitive sequences are not as prevalent, will help clarify whether specific replication timing is also a feature of metazoan centromeres.
Open questions and future directions
While recent studies have begun to explore the mechanisms of neocentromere formation, inheritance and function, many open questions remain. A better understanding of neocentromeres will inform our general understanding of centromere and kinetochore assembly and maintenance as well as potentially open up new diagnostic and/or treatment options for patients with congenital neocentromeres or cancers involving neocentromeres.
The mechanism that initiates neocentromere formation is a particularly interesting topic for future study. CENP-A is primarily found at centromere regions, but CENP-A also binds ectopic locations in response to DNA damage (Zeitlin et al., 2009) or upon overexpression of CENP-A (Van Hooser et al., 2001, Heun et al., 2006). One current hypothesis for neocentromere formation is that it is may be initiated by small extracentromeric binding of CENP-A. Overexpression of CENP-A in S. pombe (Chen et al., 2003, Joglekar et al., 2008) and C. albicans (Burrack et al., 2011) results in increased binding of CENP-A to centromere DNA sequences. While overexpression of CENP-A in the absence of disruption to the native centromere does not result in significant changes in extracentromeric incorporation of CENP-A in S. pombe (Song et al., 2008) or in C. albicans ((Burrack et al., 2011)), small amounts of CENP-A binding may be sufficient in the absence of a native centromere. The three-dimensional organization of the genome within the nucleus influences diverse biological processes including genome rearrangements (Zhang et al., 2012) and gene expression (Gheldof et al., 2010). Chromosome conformation, looping, and position within the nucleus potentially influence neocentromere site selection, as neocentromeres may preferentially form in close physical proximity to CENP-A loading regions. C. albicans and S. pombe may be excellent model systems to test the hypothesis that excess CENP-A incorporation facilitates neocentromere formation when the native centromere is inactivated and to explore the relationship between genome organization and neocentromere formation.
Another important question is whether the control of neocentromere formation occurs at the level of neocentromere maintenance. This is based on the idea that potential neocentromere initiation occurs constantly by CENP-A incorporation at ectopic sites, but that establishment and maintenance of neocentromere formation is normally inhibited through a dominant negative effect exerted by the presence of a functional native centromere elsewhere on the chromosome (Dalal et al., 2007). Dicentric chromosomes, especially those with centromeres far apart from each other, are more prone to breakage during segregation than monocentric chromosomes (Koshland et al., 1987). Human dicentric chromosomes can be stably maintained during mitosis, but inactivation of one centromere is observed both in vivo and in vitro, suggesting that functionally dicentric chromosomes can be detected and inactivated (Stimpson et al., 2010). Alternatively, does neocentromere formation initiate only after a native centromere has been inactivated? Several model systems for understanding the inactivation of dicentric chromosomes may provide insight into the mechanisms that result in the formation of one, and only one neocentromere per chromosome. In addition to providing insight into regions favorable for neocentromere formation, CENP-A overproduction in Drosophila may be a particularly advantageous model for understanding how dicentric centromeres are selected (Olszak et al., 2011). Recent studies in S. pombe (Sato et al., 2012) and humans (Stimpson et al., 2010) have also provided insight into mechanisms that inactivate dicentric chromosomes. An interesting area of future research will be to induce neocentromere formation while manipulating factors important for centromere inactivation on dicentric chromosomes. This will determine whether the factors that inactivate native centromeres are the same factors preventing the simultaneous formation of multiple neocentromeres.
In fungi, early DNA replication conferred by kinetochore assembly may enhance neocentromere inheritance, but overall, the mechanisms allowing neocentromeres to be maintained once they form at a specific location have not been well characterized. In the model system C. albicans, both short-range and long-range neocentromere movements were observed (Ketel et al., 2009), indicating that the inheritance of neocentromere position on the DNA is impaired relative to that of native centromeres that have been maintained at the same loci over millions of years of evolution (Padmanabhan et al., 2008).
Changes in chromatin structure and histone modification patterns can also induce neocentromere movement in humans (Craig et al., 2003). Comparing histone modifications associated with native centromeres to those associated with neocentromeres may provide further insight into the mechanisms promoting the stable inheritance of centromere position. The role of CENP-A assembly by HJURP (also known as Scm3 in fungi), which is required for CENP-A incorporation at native centromeres (Dunleavy et al., 2009, Foltz et al., 2009), in neocentromere assembly and maintenance has not yet been addressed. Another open question is whether HJURP/Scm3 association with neocentromeres is similar to that at native centromeres. Additionally, are all neocentromeres equally likely to move to new chromosomal positions or are some more positionally stable than others?
Once neocentromeres form, how do they alter the chromosomal locus and what DNA features are associated with the loci at which neocentromeres form? The LacI/LacO tethering system in D. melanogaster permits targeting of neocentromere formation to specific chromosomal regions (Mendiburo et al., 2011). This model may be useful for determining how neocentromere formation alters the surrounding chromosomal region. Additionally, C. albicans could be an especially useful model to identify factors that determine the location of neocentromere formation on the chromosome because of its small centromere size (~3–4 kb). Native centromeres in C. albicans each have a unique sequence and, unlike most regional centromeres, have been sequenced completely (Sanyal et al., 2004), permitting detailed comparison of DNA features at native centromere and neocentromeres and facilitating a comparison of the levels of kinetochore proteins and other biological markers (such as histone modifications) associated with neocentromeres and native centromeres by ChIP.
Transcriptional activity has been detected at some human neocentromeres (Saffery et al., 2003, Wong et al., 2006, Alonso et al., 2010), as well as at some plant centromeres (Nagaki et al., 2004). In C. albicans, the ability to compare high resolution transcript profiles at neocentromere loci in isogenic strains with native centromeres and neocentromeres will determine the positive and/or negative effects of transcription on neocentromere formation and function in chromosome segregation.
As described above, different human neocentromeres have different chromosome segregation efficiencies (Marshall et al., 2008) and error correction abilities (Bassett et al., 2010). While Aurora B localization and cohesion defects contribute to neocentromere chromosome segregation fidelity (Alonso et al., 2010, Bassett et al., 2010), they do not account for the broad range of mosaicism that is observed for human chromosomes with neocentromere. Thus, a current challenge is to identify the other factors that contribute to the segregation accuracy of chromosomes with neocentromeres. Neocentromeres in C. albicans are marked with a counter-selectable marker, allowing selection both for and against the presence of the neocentromere. This makes C. albicans a particularly useful model organism for the analysis of the segregation fidelity of chromosomes with neocentromeres.
The study of neocentromeres is advancing very rapidly. Methods to analyze human neocentromeres in greater numbers and at higher resolution will improve our ability to understand the events that gave rise to human neocentromeres in clinical isolates. Additionally, model systems provide the ability to induce and manipulate neocentromeres, permitting direct hypothesis testing and providing mechanistic models that can subsequently be tested in human isolates. This iterative process of testing hypotheses (often inspired by observations in clinical isolates) in model organisms, developing mechanistic models that explain the results, and then developing methods to test these mechanistic models in clinical isolates should greatly increase our understanding of neocentromere formation and maintenance.
Acknowledgments
We apologize to authors whose work we did not have room to cite due to space limitations. We thank Berman lab members for helpful discussions, especially Shelly Applen and Matthew Z. Anderson for helpful comments on the manuscript. This work is supported by a Ruth L. Kirschstein NRSA Fellowship F32 AI800742 and the 2011 Williston Postdoctoral Fellowship, Grant #PF-12-108-01-CCG from the American Cancer Society to L.S.B. and by NIH/NIAID AI075096 to J.B.
Abbreviations
- CEN
centromere
- CENP-A
centromere protein A, a centromere-specific histone H3 variant (also known as CenH3)
- CENP-B
centromere protein B
- ChIP
chromatin immunoprecipitation
- ChIP-chip
chromatin immunoprecipitation with microarray analysis
- ChIP-seq
chromatin immunoprecipitation with high-throughput sequencing analysis
- H3K9me
methylated lysine 9 on histone H3
- HJURP
Holliday junction recognition protein (also known as Scm3 in fungi)
- HP1
heterochromatin protein 1
- LacI
lac (lactose) repressor
- LacO
lac (lactose) operator
- LINE
long interspersed nuclear elements
- ORC
origin recognition complexl
- Scm3
suppressor of chromosome missegregation 3 (also known as HJURP in metazoans)
References
- Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J Cell Biol. 2001;153:101–10. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–37. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Hasson D, Cheung F, Warburton PE. A paucity of heterochromatin at functional human neocentromeres. Epigenetics Chromatin. 2010;3:6. doi: 10.1186/1756-8935-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32 neocentromeres. Hum Mol Genet. 2003;12:2711–21. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A. 2004;101:6542–7. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE. Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010;190:177–85. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:14877–82. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom E, Heyning FH, Kroes WG. A case of angioimmunoblastic T-cell non-Hodgkin lymphoma with a neocentric inv dup(1) Cancer Genet Cytogenet. 2010;202:38–42. doi: 10.1016/j.cancergencyto.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Burnside RD, Ibrahim J, Flora C, Schwartz S, Tepperberg JH, Papenhausen PR, Warburton PE. Interstitial deletion of proximal 8q including part of the centromere from unbalanced segregation of a paternal deletion/marker karyotype with neocentromere formation at 8p22. Cytogenet Genome Res. 2011;132:227–32. doi: 10.1159/000322815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Applen SE, Berman J. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol. 2011;21:889–96. doi: 10.1016/j.cub.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, Pidoux A. Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20:118–26. doi: 10.1016/j.gde.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Capozzi O, Purgato S, D’addabbo P, Archidiacono N, Battaglia P, Baroncini A, Capucci A, Stanyon R, Della Valle G, Rocchi M. Evolutionary descent of a human chromosome 6 neocentromere: a jump back to 17 million years ago. Genome Res. 2009;19:778–84. doi: 10.1101/gr.085688.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A. 2012;109:1979–84. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Saitoh S, Yanagida M, Takahashi K. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell. 2003;11:175–87. doi: 10.1016/s1097-2765(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, Wong LH. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5:e1000354. doi: 10.1371/journal.pgen.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JM, Wong LH, Lo AWI, Earle E, Choo KHA. Centromeric chromatin pliability and memory at a human neocentromere. The EMBO Journal. 2003;22:2495–504. doi: 10.1093/emboj/cdg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:15974–81. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe RK, Cande WZ. Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc Natl Acad Sci U S A. 1996;93:8512–7. doi: 10.1073/pnas.93.16.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Figueiredo AF, Mkrtchyan H, Liehr T, Soares Ventura EM, De Jesus Marques-Salles T, Santos N, Ribeiro RC, Abdelhay E, Macedo Silva ML. A case of childhood acute myeloid leukemia AML (M5) with a neocentric chromosome neo(1)(qter-->q23 approximately 24::q23 approximately 24-->q43-->neo-->q43-->qter) and tetrasomy of chromosomes 8 and 21. Cancer Genet Cytogenet. 2009;193:123–6. doi: 10.1016/j.cancergencyto.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–97. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol. 2006;8:148–55. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JA. Cytogenetics of Solid Tumors. In: Keagle MB, Gersen SL, editors. The Principles of Clinical Cytogenetics. Totowa, New Jersey: Humana Press; 2005. [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–84. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–36. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Yu H, Huang J, Yi C, Gu M. Unstable transmission of rice chromosomes without functional centromeric repeats in asexual propagation. Chromosome Res. 2009;17:863–72. doi: 10.1007/s10577-009-9073-7. [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W. Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci U S A. 2009;106:14937–41. doi: 10.1073/pnas.0904833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Alonso A, Cheung F, Tepperberg JH, Papenhausen PR, Engelen JJ, Warburton PE. Formation of novel CENP-A domains on tandem repetitive DNA and across chromosome breakpoints on human chromosome 8q21 neocentromeres. Chromosoma. 2011 doi: 10.1007/s00412-011-0337-6. [DOI] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006;25:5171–9. doi: 10.1038/sj.emboj.7601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–15. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KH, Saffery R. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 2004;12:805–15. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Italiano A, Attias R, Aurias A, Perot G, Burel-Vandenbos F, Otto J, Venissac N, Pedeutour F. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23–p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167:122–30. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Italiano A, Maire G, Sirvent N, Nuin PA, Keslair F, Foa C, Louis C, Aurias A, Pedeutour F. Variability of origin for the neocentromeric sequences in analphoid supernumerary marker chromosomes of well-differentiated liposarcomas. Cancer Lett. 2009;273:323–30. doi: 10.1016/j.canlet.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–94. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke K, Cheng J, Hunt AJ. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr Biol. 2009;19:807–15. doi: 10.1016/j.cub.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C, Wang HS, Mcclellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein E, Rocchi M, Ovens-Raeder A, Kosyakova N, Weise A, Ziegler M, Meins M, Morlot S, Fischer W, Volleth M, Polityko A, Ogilvie CM, Kraus C, Liehr T. Five Novel Locations of Neocentromeres in Human: 18q22.1, Xq27.1 approximately 27.2, Acro p13, Acro p12, and Heterochromatin of Unknown Origin. Cytogenet Genome Res. 2012 doi: 10.1159/000336648. [DOI] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–12. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Liehr T, Kosyakova N, Weise A, Ziegler M, Raabe-Meyer G. First case of a neocentromere formation in an otherwise normal chromosome 7. Cytogenet Genome Res. 2010;128:189–91. doi: 10.1159/000271471. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Craig JM, Saffery R, Kalitsis P, Irvine DV, Earle E, Magliano DJ, Choo KH. A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 2001;20:2087–96. doi: 10.1093/emboj/20.8.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomiento M, Jiang Z, D’addabbo P, Eichler EE, Rocchi M. Evolutionary-new centromeres preferentially emerge within gene deserts. Genome Biol. 2008;9:R173. doi: 10.1186/gb-2008-9-12-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–28. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–82. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas A, Matoso E, Saraiva J, Tonnies H, Gerlach A, Juliao MJ, Melo JB, Carreira IM. First prenatally detected small supernumerary neocentromeric derivative chromosome 13 resulting in a non-mosaic partial tetrasomy 13q. Cytogenet Genome Res. 2008;121:293–7. doi: 10.1159/000138901. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–90. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Mickle KL, Ramanathan S, Rosebrock A, Oliva A, Chaudari A, Yompakdee C, Scott D, Leatherwood J, Huberman JA. Checkpoint independence of most DNA replication origins in fission yeast. BMC Mol Biol. 2007;8:112. doi: 10.1186/1471-2199-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette JD, Celle L, Owens NL, Shields CL, Zackai EH, Spinner NB. Boy with bilateral retinoblastoma due to an unusual ring chromosome 13 with activation of a latent centromere. Am J Med Genet. 2001;99:21–8. doi: 10.1002/1096-8628(20010215)99:1<21::aid-ajmg1122>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Mroczek RJ, Melo JR, Luce AC, Hiatt EN, Dawe RK. The maize Ab10 meiotic drive system maps to supernumerary sequences in a large complex haplotype. Genetics. 2006;174:145–54. doi: 10.1534/genetics.105.048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J. Sequencing of a rice centromere uncovers active genes. Nat Genet. 2004;36:138–45. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci U S A. 2005;102:9842–7. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–75. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak AM, Van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci U S A. 2008;105:19797–802. doi: 10.1073/pnas.0809770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–7. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–21. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R. Centromere repositioning in mammals. Heredity. 2012;108:59–67. doi: 10.1038/hdy.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Mays RW, Schwartz S, Willard HF. Human artificial chromosomes with alpha satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol Cell Biol. 2003;23:7689–97. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH. Transcription within a functional human centromere. Mol Cell. 2003;12:509–16. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A. 2004;101:11374–9. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Masuda F, Takayama Y, Takahashi K, Saitoh S. Epigenetic Inactivation and Subsequent Heterochromatinization of a Centromere Stabilize Dicentric Chromosomes. Curr Biol. 2012 doi: 10.1016/j.cub.2012.02.062. [DOI] [PubMed] [Google Scholar]
- Sernova NV, Gelfand MS. Identification of replication origins in prokaryotic genomes. Brief Bioinform. 2008;9:376–91. doi: 10.1093/bib/bbn031. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–8. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirvent N, Forus A, Lescaut W, Burel F, Benzaken S, Chazal M, Bourgeon A, Vermeesch JR, Myklebost O, Turc-Carel C, Ayraud N, Coindre JM, Pedeutour F. Characterization of centromere alterations in liposarcomas. Genes Chromosomes Cancer. 2000;29:117–29. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1014>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Song JS, Liu X, Liu XS, He X. A high-resolution map of nucleosome positioning on a fission yeast centromere. Genome Res. 2008;18:1064–72. doi: 10.1101/gr.075374.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA. Telomere disruption results in non-random formation of de novo dicentric chromosomes involving acrocentric human chromosomes. PLoS Genet. 2010;6(8):e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;154:683–90. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–83. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–70. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–90. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–6. [PubMed] [Google Scholar]
- Topp CN, Okagaki RJ, Melo JR, Kynast RG, Phillips RL, Dawe RK. Identification of a maize neocentromere in an oat-maize addition line. Cytogenet Genome Res. 2009;124:228–38. doi: 10.1159/000218128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C, Gimelli G, Giglio S, Floridia G, Pandya A, Terzoli G, Warburton PE, Earnshaw WC, Zuffardi O. Transmission of a fully functional human neocentromere through three generations. Am J Hum Genet. 1999;64:1440–4. doi: 10.1086/302380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski Ii, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–42. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D’addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. Evolutionary formation of new centromeres in macaque. Science. 2007;316:243–6. doi: 10.1126/science.1140615. [DOI] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D’addabbo P, Wandall A, Bjorck E, De Jong PJ, She X, Eichler EE, Archidiacono N, Rocchi M. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SA, Gerton JL, Polancic JE, Derisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Murphy TD, Goldberg ML, Karpen GH. Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nat Genet. 1998;18:30–7. doi: 10.1038/ng0198-30. [DOI] [PubMed] [Google Scholar]
- Wong NC, Wong LH, Quach JM, Canham P, Craig JM, Song JZ, Clark SJ, Choo KHA. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet. 2006;2:e17. doi: 10.1371/journal.pgen.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012 doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–7. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mccord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]