Abstract
Background
The IL6R SNP rs4129267 has recently been identified as an asthma susceptibility locus in subjects of European ancestry but has not been characterized with respect to asthma severity. The SNP rs4129267 is in linkage disequilibrium (r2=1) with the IL6R coding SNP rs2228145 (Asp358Ala). This IL6R coding change increases IL6 receptor shedding and promotes IL6 transsignaling.
Objectives
To evaluate the IL6R SNP rs2228145 with respect to asthma severity phenotypes.
Methods
The IL6R SNP rs2228145 was evaluated in subjects of European ancestry with asthma from the Severe Asthma Research Program (SARP). Lung function associations were replicated in the Collaborative Study on the Genetics of Asthma (CSGA) cohort. Serum soluble IL6 receptor (sIL6R) levels were measured in subjects from SARP. Immunohistochemistry was used to qualitatively evaluate IL6R protein expression in BAL cells and endobronchial biopsies.
Results
The minor C allele of IL6R SNP rs2228145 was associated with lower ppFEV1 in the SARP cohort (p=0.005), the CSGA cohort (0.008), and in combined cohort analysis (p=0.003). Additional associations with ppFVC, FEV1/FVC, and PC20 were observed. The rs2228145 C allele (Ala358) was more frequent in severe asthma phenotypic clusters. Elevated serum sIL6R was associated with lower ppFEV1 (p=0.02) and lower ppFVC (p=0.008) (N=146). IL6R protein expression was observed in BAL macrophages, airway epithelium, vascular endothelium, and airway smooth muscle.
Conclusions
The IL6R coding SNP rs2228145 (Asp358Ala) is a potential modifier of lung function in asthma and may identify subjects at risk for more severe asthma. IL6 transsignaling may have a pathogenic role in the lung.
Keywords: soluble interleukin 6 receptor, sIL6R, interleukin 6, IL6, asthma, pulmonary lung function, severe asthma, IL6 transsignaling, genetic variation, SNP rs2228145
INTRODUCTION
In a recent genome wide association study (GWAS) of 57,800 subjects from multiple asthma cohorts, the minor T allele of the IL6R SNP rs4129267 (1q21.3) was identified as a novel asthma susceptibility loci (OR=1.09) in subjects of European ancestry 1. This SNP was not evaluated for associated asthma severity phenotypes including lung function in this GWAS study, but this SNP has been associated with differences in pulmonary function in normal subjects from the Framingham Heart Study 2.
The SNP rs4129267 is located in intron 8 of IL6R and is in linkage disequilibrium (LD; r2=1) with the IL6R coding SNP rs2228145 (Asp358Ala). The Ala358 variation modifies the IL6 receptor peptide structure adjacent to the exterior cell surface and significantly enhances proteolytic cleavage of IL6 receptor from cell surfaces into the extracellular space, termed IL6R “shedding” 3;4. IL6R shedding is increased in subjects who inherit the IL6R rs2228145 C allele (Ala358), and is easily measured in serum 5. The soluble IL6 receptor can be activated by IL6 and can form a complex with the ubiquitously expressed membrane bound glycoprotein 130 (gp130), resulting in activation of the IL6 signal transduction pathway in cells that do not express membrane bound IL6 receptor. This dysfunctional activation of IL6 signaling is termed IL6 transsignaling 6. During IL6 transsignaling, the sIL6R/gp130 complex induces tyrosine kinase JAK2 (Janus kinase 2) activity, which phosphorylates and activates the transcription factor Signal transducer and activator of transcription 3 (STAT3). Activated STAT3 translocates to the nucleus to affect a wide range of gene expression. IL6 transsignaling and has been implicated in a range of inflammatory diseases, including rheumatoid arthritis 7, Crohn’s disease 8, and inflammatory bowel disease 9, and is the target of anti-IL6R therapies 10.
In this study, the IL6R coding SNP rs2228145 (Asp358Ala) was evaluated in subjects of European white ancestry in two asthma cohorts to determine if the coding variation is associated with lung function, an important asthma severity phenotype. Because of the wide range of lung function in the Severe Asthma Research Program (SARP) cohort, the IL6R coding SNP was first evaluated in the SARP cohort 11. Lung function associations were then verified in the Collaborative Study on the Genetics of Asthma (CSGA) cohort and in a combined analysis of the SARP and CSGA cohorts. Because this coding SNP is associated with elevated serum levels of sIL6R, SARP subjects were evaluated to determine if correlations exist between serum sIL6R and lung function in subjects who inherit the IL6R Ala358 isoform. The IL6R coding SNP was also evaluated in SARP phenotypic asthma clusters 12 to evaluate potential associations with more severe asthma. Immunohistochemistry was used to evaluate lung biopsies and bronchial alveolar lavage (BAL) cells to determine if lung cells express IL6R protein and thus possible sources for IL6R shedding. Finally, BAL fluid was evaluated in a small subset of SARP subjects to determine if elevated BAL sIL6R was associated with inheriting the IL6R Ala358 isoform.
METHODS
Subject Recruitment
Subjects of European ancestry from nine clinical research centers that comprise the NHLBI sponsored Severe Asthma Research Program (SARP) study were characterized with spirometry and were phenotyped using comprehensive, standardized approaches previously described 11;12. Current smokers and subjects with > 5 pack years smoking were ineligible for this study. In brief, subjects were defined to have severe asthma using the American Thoracic Society (ATS) workshop criteria for refractory asthma 13. For comparison, additional subjects with non-severe asthma were studied who did not meet the ATS workshop criteria. After appropriate withholding of bronchodilators, spirometry was performed according to ATS guidelines 14. Hankinson values were used to calculate FEV1 % predicted 15. In order to qualify for testing for bronchial responsiveness to methacholine (PC20), subjects were required to have a FEV1 % predicted of >55% 11. Bronchodilator reversibility was testing using a maximal protocol of up to 8 puffs of albuterol. This study was approved by the Institutional Review Board at all study sites and all patients provided informed consent to participate in this study. The replication population consisted of subjects of European ancestry from the Collaborative Study of the Genetics of Asthma at Wake Forest School of Medicine 16;17. These subjects were studied using a similar protocol to SARP for phenotype characterization 11. SARP controls used for serum analysis had no history of asthma and no first-degree relatives with asthma.
SNP Genotyping and Assessment
All IL6R genotypes were acquired from the Illumina Human1M-Duo DNA BeadChip from our previous GWAS study 18. In addition to primary analysis of rs2228145, the eight additional IL6R SNPs rs6427641, rs1386821, rs6684439, rs4845618, rs8192282, rs4845371, rs4129267, rs4240872, and rs2229238 were selected for analysis based on linkage disequilibrium (r2) values from the HapMap database (http://hapmap.ncbi.nlm.nih.gov/). The genotyping efficiency on the Illumina Human1M-Duo DNA BeadChip for all IL6R SNPs >99%, and all SNPs were in Hardy-Weinberg equilibrium.
Serological and Immunohistochemistry
Investigative bronchoscopy was performed on a subset of SARP subjects with all levels of asthma severity 11. All bronchoscopy subjects were self selected for this portion of the SARP study and biopsies analyzed were randomly selected without a priori selection based on asthma severity. Endocronchial biopsies (N=14) from formalin-fixed, paraffin-embedded sections (5u) were deparaffinized (Histoclear II) and rehydrated through decreasing ethanol solutions. The airway tissue was treated for antigen retrieval (Dako) and blocked prior to incubation steps with primary IL6Ra monoclonal antibody (specific to Arg387–Arg468, R&D Systems), secondary biotin-labeled anti-mouse Ig (GE Healthcare), streptavidin-alkaline phosphatase (Roche) and development with Vector Red substrate (Vector). Sections were counterstained with Mayer’s hematoxylin.
Bronchoalveolar lavage cells (N=22) were air dried on cytospin slides, fixed with 10% formalin-for 15 min, and washed once with phosphate buffered saline (PBS) before storage at −20°C. BAL cells were randomly selected from subjects that had undergone bronchoscopy, and the subset of cells were analyzed without a priori selection based on asthma severity. Cells were blocked with 100% fetal bovine serum (FBS) in PBS with 0.1% saponin, avidin and biotin separately (Vector Blocking Kit) before incubation with primary IL6Ra monoclonal antibody (R&D Systems), secondary biotin-labeled anti-mouse Ig (GE Healthcare), streptavidin-alkaline phosphatase (Roche) and development with BCIP/NBT substrate (DAKO) containing 1/100 dilution levamisole (Sigma).
Serum and BAL sIL6R were measured using the sIL-6R DuoSet (R&D Systems, Minneapolis, MN) ELISA kit and reported ng/ml. All serum samples were diluted 1:200.
Statistical Analysis
Analysis was performed using a linear additive model adjusting for age and sex for each population and in a combined dataset for the asthma quantitative traits % predicted FEV1 [ppFEV1], % predicted FVC [ppFVC], FEV1/FVC, and PC20 using ANOVA (two-tailed p-values) as implemented in SAS/Genetics. Serum sIL6R measurements were analyzed by a general linear model and were log transformed to normalize sample distribution.
RESULTS
Study Population Characteristics
The demographic characteristics of SARP subjects with asthma in this study are described in Table I and recently have recently been extensively reported 11. DNA from subjects of European white ancestry with asthma (N=510; 44% with severe asthma) and without asthma (N=45) were genotyped. Severe asthma subjects met ATS criteria for severe asthma and were all treated with high dose inhaled or oral corticosteroids at recruitment, while 60% of subjects with non-severe asthma were treated with inhaled corticosteroids. Spirometry was performed after withholding short acting bronchodilators for 6 hours and long acting bronchodilators for 12 hours. Characteristics of subjects from the CSGA population (Table 1) have been previously described in detail and consisted of subjects of European ancestry with a spectrum of asthma severity (N=239) 16;17.
Table I.
Characteristics of Asthma Populations
| SARP
|
CSGA
|
||
|---|---|---|---|
| No asthma (N=45) | Asthma (N=510) | Asthma (N=239) | |
| Male Sex % | 28 | 39 | 39 |
| Age at Enrollment | 33 ± 14 | 37 ± 14 | 25 ± 14 |
| FEV1 % predicted* | 98 ± 10 | 74 ± 23 | 86 ± 18 |
| FEV1/FVC* | .83 ± .06 | .70 ± .13 | .78 ± .12 |
| Log PC20±SD | 0.12 ± 0.68 | 0.11 ± 0.71 | |
| BMI | 25.6 ± 6.1 | 28.5 ± 7.6 | 26.0 ± 7.8 |
| Patients qualifying for BHR** | 329 | 219 | |
Bronchodilator withhold measurement.
Subjects with % predicted FEV1 >55%
The IL6R SNP rs2228145 Is Associated with Lung Function and Hyperresponsiveness
Genotypes for the IL6R SNP rs4129267 and rs2228145 were available from the same GWAS study 18 and these two SNPs are in complete LD (r2=1). Since lower lung function is an important phenotype for asthma 13, we first evaluated the SNP rs2228145 for association with lung function first in the SARP cohort, replicated independently in CSGA, and then investigated these phenotypes in a combined cohort analysis (SARP+CSGA)(Table II). After adjusting for age of enrollment and sex, the C allele of rs2228145 was associated with lower ppFEV1, lower ppFVC, and lower FEV1/FVC in the SARP cohort. In the CSGA cohort, the C allele of rs228145 was associated with lower ppFEV1. In a combined cohort analysis (SARP+CSGA), the rs2228145 C allele was associated with lower ppFEV1, lower ppFVC, and lower FEV1/FVC. In the combined analysis, subjects who were homozygous for the rs2228145 C allele (Ala358) had the lowest mean ppFEV1 (71.90 %), lowest ppFVC (84.23%), and lowest FEV1/FVC (0.69). Bronchial hyperresponsiveness (BHR; PC20) was not significantly associated with rs2228145 in the SARP cohort; however the rs2228145 C allele (Ala358) was associated with methacholine responsiveness in the CSGA and combined cohort analyses.
Table II.
Association of IL6R Mutation Asp358Ala with Lung Function in Subjects with Asthma in SARP, CSGA, and Combined Analyses
| % Predicted FEV1 | % Predicted FVC | FEV1/FVC | LogPC20 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| N | Mean (SD) | p-Value | N | Mean (SD) | p-Value | N | Mean (SD) | p-Value | N | Mean (SD) | p- Value | ||
|
SARP rs2228145 |
AA | 160 | 74.9 ± 23.4 | 0.005 | 154 | 85.5 ± 18.6 | 0.047 | 156 | 0.71 ± 0.13 | 0.01 | 102 | 0.14 ± 0.8 | 0.58 |
| AC | 241 | 75.9 ± 22.6 | 225 | 87.5 ± 18.5 | 235 | 0.71 ± 0.13 | 167 | 0.14 ± 0.7 | |||||
| CC | 98 | 67.7 ± 23.8 | 90 | 81.7 ± 19.1 | 95 | 0.67 ± 0.13 | 59 | 0.04 ± 0.6 | |||||
|
CSGA rs2228145 |
AA | 86 | 89.8 ± 17.14 | 0.008 | 85 | 94.5 ± 14.8 | 0.15 | 85 | 0.80 ± 0.01 | 0.07 | 87 | 0.26 ± 0.7 | 0.017 |
| AC | 100 | 83.3 ± 15.43 | 99 | 90.8 ± 13.1 | 99 | 0.77 ± 0.12 | 96 | 0.06 ± 0.7 | |||||
| CC | 39 | 82.4 ± 25.01 | 38 | 90.3 ± 19.9 | 38 | 0.76 ± 0.15 | 35 | −0.13 ± 0.6 | |||||
|
COMBINED rs2228145 |
AA | 246 | 80.1 ± 22.5 | 0.003 | 239 | 88.7 ± 17.8 | 0.08 | 241 | 0.74 ± 0.13 | 0.006 | 189 | 0.20 ± 0.7 | 0.033 |
| AC | 341 | 78.1 ± 21.0 | 324 | 88.5 ± 17.1 | 334 | 0.73 ± 0.13 | 263 | 0.11 ± 0.7 | |||||
| CC | 137 | 71.9 ± 24.96 | 128 | 84.2 ± 19.7 | 133 | 0.69 ± 0.14 | 94 | −0.02 ± 0.6 | |||||
After evaluation of rs2228145, additional IL6R SNPs available on the Illumina 1M Duo chip were evaluated. The results for the combine cohort analysis are shown in Table S1 on the Online Supplement. In addition to rs2228145, several additional IL6R SNPs were associated with ppFEV1, ppFVC, FEV1/FVC, and PC20.
IL6R SNP rs2228145 is Associated with Severe Asthma Clusters
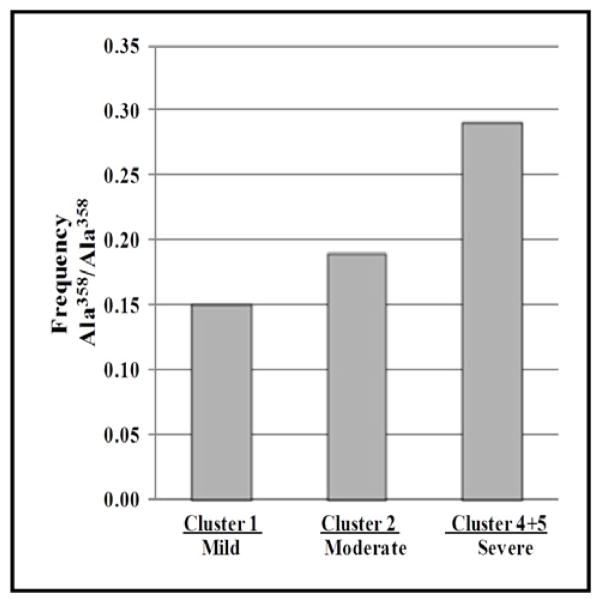
The SNP rs2228145 was further evaluated in phenotypic severity Clusters recently described for the SARP cohort 12. SARP Clusters 1 (mild asthma), 2 (moderate asthma), and 4+5 (severe asthma) were evaluated. A fifth Cluster (Cluster 3) representing a moderate to severe phenotype with late onset asthma was not used in this analysis to better define the severity classes. The rs2228145 C allele was most frequent (0.48) in the combined 4+5 SARP Cluster (Table III) (p=0.003). A secondary analysis comparing rs2228145 CC genotype frequency vs AA+AC genotype frequency showed that subjects with the rs2228145 CC genotype (Ala358/Ala358) were more frequent in the SARP 4+5 cluster (frequency = 0.29) compared to the mean frequency among all five clusters (Freq= 0.20)(Figure 1).
Table III.
Evaluation of IL6R rs2228145 (Asp358Ala) and Serum sIL6R in SARP Asthma Phenotypic Clusters
| Cluster 1 | Cluster 2 | Clusters 4+5 | p-value | ||
|---|---|---|---|---|---|
| Mild | Moderate | Severe | |||
|
| |||||
| N | N | N | |||
| rs2228145 (Asp358Ala) | AA | 23 | 48 | 41 | 0.003 |
| AC | 40 | 80 | 49 | ||
| CC | 7 | 30 | 37 | ||
| Log Serum sIL6R | 1.58 ± 0.11 (n=21) | 1.64 ± 0.14 (N=62) | 1.69 ± 0.13 (N=40) | 0.016* | |
| Mean ppFEV1 | 99.1% | 83.4% | 54.3% | <0.0001* | |
General linear model
Figure 1. Frequency of Subjects Homozygous IL6R Ala358 Allele In SARP Phenotypic Asthma Clusters.
Genotypic analysis performed for 355 subjects of European ancestry with asthma.
Serum sIL6R is Elevated in Severe Asthma Clusters and Associated with Lung Function
Serum sIL6R levels were measured in 146 SARP subjects of European ancestry with asthma (40 with severe asthma) and 46 controls. Standard curves measuring sIL6R or sIL6R saturated with IL6 were not significantly different (r2>0.95), indicating that IL6 did not alter the specificity of the anti-sIL6R antibody in the ELISA. The IL6R mutation Ala358 was strongly associated with elevated sIL6R in subjects with asthma (p<10−31) (Figure 2A.) and without asthma (p<0.0001; data not shown). There was no significant difference in mean sIL6R serum between subjects with asthma (Log sIL6R=1.64 ± 0.14) and controls (Log sIL6R=1.61 ± 0.18). In subjects with asthma, elevated serum sIL6R was associated with lower ppFEV1 (p=0.02) and ppFVC (p=0.008) (Figure 2B. and 2C., respectively). Serum sIL6R levels were significantly higher in the SARP 4+5 cluster (p=0.016) (Table III). The combined SARP 4+5 Cluster also had the lowest mean ppFEV1 (54.3%). There was no association between serum sIL6R levels and lung function in normal subjects.
Figure 2. Evaluation of Serum sIL6R in SARP Subjects with Asthma.
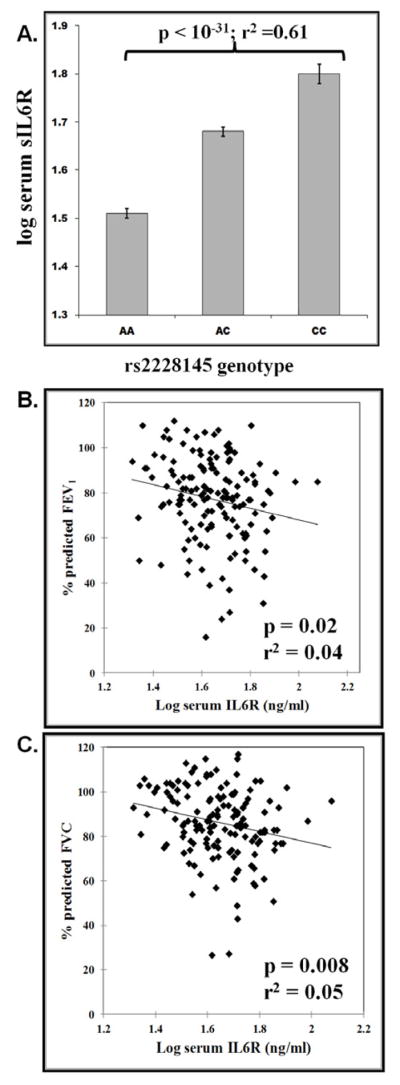
A.) Association of the IL6R coding SNP rs2228145 (Asp358Ala) with serum sIL6R; B.) Serum sIL6R vs % Predicted FEV1; B.) Serum sIL6R vs % Predicted FVC. sIL6R analyses were performed from serum collected from 146 subjects of European ancestry.
IL6 Receptor Protein is Expressed in Bronchoalveolar Lavage and Lung Epithelial Cells
To determine if IL6 receptor protein was expressed in the lung, endobronchial biopsies (N=14) and BAL cells (N=22) from subjects with asthma were immunostained with a monoclonal antibody specific for membrane-bound IL-6Ra (Arg387–Arg468). IL6 receptor staining was observed in airway epithelium in all samples. Additional IL6 receptor was observed vascular endothelium, airway smooth muscle, and some individual cells in the submucosa. A representative stained endobronchial biopsy is shown in Figure 3A. In BAL cells (Figure 3B), IL6Ra staining was observed predominantly in macrophages, although some poly-morphonucleated granulocytes showed lower amounts of IL6Ra expression (upper right of Figure 3B.). Lack of BAL cell permeabilization during antibody incubation steps reduced specific IL6Ra immunostain intensity, indicating that the intracellular portion of the IL6 receptor was targeted by this antibody.
Figure 3. Representative Immunostaining of Endobronchial Biopsy and Bronchoalveolar Lavage (BAL) Cells with Anti-IL6 Receptor-A In Subjects with Asthma.
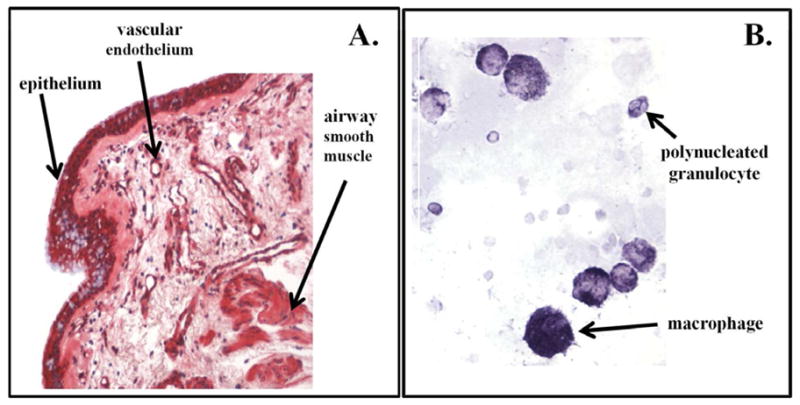
A). Airway epithelium, smooth muscle and vascular endothelium stained positive for IL6Ra in the biopsy; B.) Predominantly macrophages stained positive for IL6Ra in lavage samples, although some poly-morphonucleated granulocytes showed lower amounts of IL6Ra stain as seen in the upper right.
BAL fluid was evaluated in 16 subjects with asthma. The highest levels of BAL sIL6R were observed in the rs2228145 CC genotype (Ala358/Ala358) (Log sIL6R =2.1 ± 0.12; N=4) compared to the AA genotype (Asp358/Asp358) (Log sIL6R =1. ± 0.12; N=8) and the AC genotype (Asp358/Ala358) (Log sIL6R =1.8 ± 0.09; N=4); however the trend was not statistically significant.
DISCUSSION
In this study, we have identified the IL6R coding SNP rs2228145 (Asp358Ala) as a potential genetic modifier of lung function in asthma and as a potential novel genetic marker of asthma severity. In separate and combined analyses of the SARP and CSGA European ancestry asthma cohorts, the minor C allele of rs2228145 (Ala358) was consistently associated with lower ppFEV1 and lower FEV1/FVC, and the highest level of methacholine responsiveness. In addition, the frequency of the rs2228145 C allele frequency was found to be higher in SARP phenotypic asthma clusters consisting of subjects with more severe asthma. We have also shown for the first time that copious IL6 receptor protein is expressed in multiple cell types in the lung.
These observations are important in light of the recent GWAS study of ~58,000 subjects that identified IL6R as an asthma risk gene 1. In this GWAS, a single IL6R SNP (rs4129267) was identified as conferring asthma risk. Because of the small sizes of the SARP and CSGA cohorts used in this study, we were not able replicate the asthma risk assessment performed in the large GWAS. In addition, direct comparison of the data from this study with the large GWAS was not possible since the GWAS study did not evaluate lung function and presented no information on the proportion of subjects with severe asthma. Because our genotyping data was derived from the Illumina Human1M-Duo DNA BeadChip, addition IL6R SNP genotypes, including the risk SNP rs4129267, could be analyzed. We were thus able to show that rs4129267 was a tagging SNP (r2=1) for the coding SNP rs2228145 (Asp358Ala). We were also able to show that additional IL6R SNPs were associated with lung function, including the SNP rs4129267 from the original GWAS. However, the SNP rs228145 was the only non-synonymous coding change genotyped in our study and is the only known IL6R SNP that affects IL6R protein function. Based on our observations and the strong linkage disequilibrium between rs2228145 and rs4129267, we propose that the coding SNP rs2228145 is most probably the genetic variant conferring asthma risk in the large asthma GWAS.
In a random subset of SARP subjects, the rs2228145 C allele (Ala358) was strongly associated with elevated serum sIL6R, which is consistent with previous observations 5. We did not, however, observe a statistically significant difference in serum sIL6R levels between subjects with asthma and unaffected controls, has had been previously reported in a smaller sampling of Japanese subjects (N=38) 19. The discrepancy between our results and the Japanese study could be explained by ethnic differences; however this discrepancy is most likely the result of smaller sample size used in the Japanese study.
An important advantage of this current study was the use of the SARP cohort. SARP consists of an asthma cohort enriched for severe asthma, and unlike other asthma cohorts, has subjects with a wide range of lung function and extensive phenotypic characterization that are complemented with comprehensive genetic studies 11;12. The extensive phenotypes collected in the SARP cohort 11 have allowed development of phenotypic asthma clusters 12 that better describe asthma heterogeneity and severity. Since there IL6R Ala358 variant occurred at a higher frequency in the severe asthma clusters, subjects in these clusters may be important in evaluating the long-term effects of the IL6R coding SNP rs2228145 (Asp358Ala), perhaps providing one reason why subjects in asthma clusters 4 and 5 develop lower lung function and more severe asthma. SARP is a cross-sectional study that is currently evolving into a longitudinal cohort, thus we will be able to determine if our IL6R observations can be used to predict changes in lung function over time. Unfortunately, we are not aware of other well-characterized asthma cohorts with longitudinal data and a wide range of lung function in which these data can be replicated.
Based on our observations and reports linking IL6 transsignaling to other inflammatory diseases such as inflammatory bowel disease 9 and rheumatoid arthritis 7, we propose that IL6 transsignaling also has a pathogenic effect in the lung. The immunochemistry data from this study shows that membrane bound IL6R is expressed in both BAL cells and lung epithelial cells, providing ample sources of IL6R shedding in the lung that may be associated with increase bronchial inflammation. During inflammation, concurrent increases in IL6 and IL6R shedding would create an optimal environment for increased IL6 transsignaling 7. In subjects with mild, episodic asthma, the pathological effects of IL6 transsignaling would be short lived as IL6 levels return to normal when lung inflammation resolves. In subjects with more severe asthma and persistent bronchial inflammation, chronically elevated IL6 levels in the lung could sustain IL6 transsignaling in the presence of high levels of sIL6R. The pathogenic effects of short-term and long-term IL6 transsignaling in the lung have not been investigated.
The identification of the IL6R coding SNP rs2228145 (Asp358Ala) as a risk variant for lower lung function indicates that dysfunctional IL6 transsignaling may be a potentially important therapeutic target in severe asthma. Currently, therapies targeting IL6 transsignaling are under development for treating several inflammatory diseases 8;10;20–24, and the results of this study may serve as the focus for the evaluation of this therapeutic class of drug in severe asthma. In the absence of a clinical genetic test for the IL6R mutation (Asp358Ala), it is possible that elevated serum sIL6R may be a surrogate biomarker to identify subjects at risk for more severe asthma and for who may benefit from anti-IL6R therapy.
Clinical Implications.
IL6 transsignaling may have a pathogenic role in airways relating to asthma severity. There are currently anti-IL6R therapies that block IL6 transsignaling and may have potential therapeutic value in asthma
Acknowledgments
Grant support: HL69116, HL69130, HL69155, HL69167, HL69170, HL69174, HL69349, HL091762, HL087665, HL101487, KL2RR025009, M01 RR02635, M01 RR03186, M01 RR007122-14, 1UL1RR024153, 1UL1RR024989, 1UL1RR024992, 1UL1RR025008, 1UL1RR025011, Children’s Healthcare of Atlanta Center for Developmental Lung Biology.
We want to thank Dr. Lilly Zheng, Brian Rector, Shelly Smith, Carla Marsh, Catherine Brewer, and Abdoulaye Diallo for their technical help, and Dr. Elizabeth Ampleford for help in data analysis. We also want to thank all of the participants in the Severe Asthma Research Program and Collaborative Study on the Genetics of Asthma.
Abbreviations
- ATS
American Thoracic Society
- BAL
bronchoalveolar lavage
- BHR
bronchial hyperresponsiveness
- CSGA
Collaborative Study on the Genetics of Asthma
- FEV1
forced expiratory volume in 1 second
- FEV1/FVC
forced expiratory volume in 1 second/forced volume vital capacity
- FVC
forced volume vital capacity
- gp130
glycoprotein 130
- IL6
interleukin 6
- JAK2
Janus kinase 2
- LD
linkage disequilibrium
- NHLBI
National Heart Lung Blood Institute
- PC20
provocative concentration of methacholine for 20% reduction in FEV1
- SARP
Severe Asthma Research Program
- sIL6R
soluble interleukin 6 receptor
- SNP
single nucleotide polymorphism
- STAT3
Signal transducer and activator of transcription 3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le SP, et al. Identification of IL6R and chromosome 11q13. 5 as risk loci for asthma. Lancet. 2011;378(9795):1006–14. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilk JB, Walter RE, Laramie JM, Gottlieb DJ, O’Connor GT. Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet. 2007;8 (Suppl 1):S8. doi: 10.1186/1471-2350-8-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;(1):43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- 4.Matthews V, Schuster B, Schutze S, Bussmeyer I, Ludwig A, Hundhausen C, et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J Biol Chem. 2003;278(40):38829–39. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 5.Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004;(6):513–6. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- 6.Jones SA, Novick D, Horiuchi S, Yamamoto N, Szalai AJ, Fuller GM. C-reactive protein: a physiological activator of interleukin 6 receptor shedding. J Exp Med. 1999;189(3):599–604. doi: 10.1084/jem.189.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Febbraio MA, Rose-John S, Pedersen BK. Is interleukin-6 receptor blockade the Holy Grail for inflammatory diseases? Clin Pharmacol Ther. 2010;87(4):396–8. doi: 10.1038/clpt.2010.1. [DOI] [PubMed] [Google Scholar]
- 8.Brulhart L, Nissen MJ, Chevallier P, Gabay C. Tocilizumab in a patient with ankylosing spondylitis and Crohn’s disease refractory to TNF antagonists. Joint Bone Spine. 2010;77(6):625–6. doi: 10.1016/j.jbspin.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Rose-John S, Mitsuyama K, Matsumoto S, Thaiss WM, Scheller J. Interleukin-6 trans-signaling and colonic cancer associated with inflammatory bowel disease. Curr Pharm Des. 2009;15(18):2095–103. doi: 10.2174/138161209788489140. [DOI] [PubMed] [Google Scholar]
- 10.Mima T, Nishimoto N. Clinical value of blocking IL-6 receptor. Curr Opin Rheumatol. 2009;21(3):224–30. doi: 10.1097/BOR.0b013e3283295fec. [DOI] [PubMed] [Google Scholar]
- 11.Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of Asthma Phenotypes using Cluster Analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2009;181:315–23. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. American Thoracic Society. Am J Respir Crit Care Med. 2000;162(6):2341–51. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 14.Standardization of Spirometry, 1994 Update. American Thoracic Society. Am J Respir Crit Care Med. 1995;152(3):1107–36. doi: 10.1164/ajrccm.152.3.7663792. [DOI] [PubMed] [Google Scholar]
- 15.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159(1):179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 16.Meyers DA, Wjst M, Ober C. Description of three data sets: Collaborative Study on the Genetics of Asthma (CSGA), the German Affected-Sib-Pair Study, and the Hutterites of South Dakota. Genet Epidemiol. 2001;21 (Suppl 1):S4–S8. doi: 10.1002/gepi.2001.21.s1.s4. [DOI] [PubMed] [Google Scholar]
- 17.Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, et al. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108(3):357–62. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Howard TD, Moore WC, Ampleford EJ, Li H, Busse WW, et al. Importance of hedgehog interacting protein and other lung function genes in asthma. J Allergy Clin Immunol. 2011;127(6):1457–65. doi: 10.1016/j.jaci.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoyama A, Kohno N, Sakai K, Kondo K, Hirasawa Y, Hiwada K. Circulating levels of soluble interleukin-6 receptor in patients with bronchial asthma. Am J Respir Crit Care Med. 1997;156(5):1688–91. doi: 10.1164/ajrccm.156.5.9610070. [DOI] [PubMed] [Google Scholar]
- 20.Kawabata H, Tomosugi N, Kanda J, Tanaka Y, Yoshizaki K, Uchiyama T. Anti-interleukin 6 receptor antibody tocilizumab reduces the level of serum hepcidin in patients with multicentric Castleman’s disease. Haematologica. 2007;92(6):857–8. doi: 10.3324/haematol.10794. [DOI] [PubMed] [Google Scholar]
- 21.Tocilizumab (Actemra) for rheumatoid arthritis. Med Lett Drugs Ther. 2010;52(1340):47–8. [PubMed] [Google Scholar]
- 22.Hagihara K, Kawase I, Tanaka T, Kishimoto T. Tocilizumab ameliorates clinical symptoms in polymyalgia rheumatica. J Rheumatol. 2010;37(5):1075–6. doi: 10.3899/jrheum.091185. [DOI] [PubMed] [Google Scholar]
- 23.Kluger N, Bessis D, Guillot B. Tocilizumab as a potential treatment in Schnitzler syndrome. Med Hypotheses. 2009;72(4):479–80. doi: 10.1016/j.mehy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 24.de BM, Saint-Marcoux B. Tocilizumab for multirefractory adult-onset Still’s disease. Ann Rheum Dis. 2009;68(1):153–4. doi: 10.1136/ard.2008.088179. [DOI] [PubMed] [Google Scholar]