Abstract
A characteristic of sporadic and familial breast tumours is genomic instability, resulting from either inherited mutations in genes that control genome integrity or mutations that are acquired in somatic cells during development. It is well established that abnormal chromosome number and structural changes to chromosomes play an important role in the cause and progression of breast cancer. Familial BRCA1 breast tumours are characterised by basal-like phenotype and high-histological grade which are typically associated with increased genomic instability. Consistent with previous studies, the genomes with the greatest number of base pairs covered by copy number change were typically found in basal-like and/or high-histological grade breast tumours within our cohort. Moreover, we show that luminal A tumours that are high grade had significantly less copy number variant (CNV) coverage than the more clinically aggressive high-grade luminal B tumours, suggesting that chromosomal instability rather than cellular differentiation contributes to the aggressive nature of luminal B tumours. It has previously been proposed that germline CNVs may contribute to somatically acquired chromosome changes in the tumour, but this is the first study to address this idea in breast cancer. By comparing germline CNVs and tumour-specific CNVs in matched breast tumour and normal tissue using data from the Illumina Human CNV370 duo beadarray, we provide evidence that germline CNVs do not tend to act as a foundation on which larger chromosome copy number aberrations develop in tumour cells. Further studies are required with increased sequence resolution that will detect smaller CNVs and define CNV breakpoints to comprehensively assess the relationship between inherited genomic variation and genome evolution in breast cancer.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-012-2024-6) contains supplementary material, which is available to authorized users.
Keywords: Copy number variant, Familial breast cancer, Tumour genome, SNP array
Introduction
Familial and sporadic forms of breast cancer are now recognised to be a complex and heterogeneous disease at both the clinical and molecular levels [1–4]. The genome of a breast tumour typically represents a culmination of somatically acquired, poorly understood, genomic aberrations that functionally alter genes contributing to tumourigenesis [5]. The extent of genomic abberation in breast cancer has been shown to differ significantly in histological subtype, such as low- and high-grade tumours [6, 7]. Since patients with high-grade tumours generally have worse prognosis than those who have low-grade tumours [8], these findings indicate that there is an association between the mutational burden of tumours and tumour pathogenesis. Similarly, basal-like breast tumours are characterised by high levels of genomic aberrations in comparison to other tumour subtypes, such as the luminal A or luminal B subtypes [1], and are associated with poor prognosis [9]. A number of phenotypical and molecular features are shared by basal-like breast cancer and tumours arising in BRCA1 germline mutation carriers, including high grade and a high number of chromosome copy number changes [10]. This suggests a common tumourigenic pathway of the BRCA1 and basal-like subtypes; however, the biological mechanisms associated with increased frequency of chromosomal changes in these tumour types are currently poorly understood.
Studies of choroid plexus tumours in Li–Fraumeni Syndrome (LFS)-affected families [11] and of colon cancer-affected individuals [12] have suggested that constitutional copy number variants (CNVs) may act as a foundation on which chromosome copy number aberrations develop in tumour cells. These findings suggest a direct relationship between constitutional genomic variation and tumour genome evolution. However, the number of cancer cases, where matched normal tissue was also assessed, was relatively small for each of the choroid plexus tumour and colon cancer studies (n = 4 and 5, respectively) [11, 12]. Furthermore, it is currently unknown whether germline CNVs play a role in genomic instability associated with breast tumour grade and breast tumour subtype. To address this issue, we utilised single nucleotide polymorphism array data from a previously published study [1] to compare germline CNV and breast tumour-specific CNVs using 28 matched normal and tumour tissue pairs. Furthermore, we reassessed the association between pathological features, such as breast tumour subtype and histological grade, and the extent of CNV coverage in germline and tumour genomes.
Methods and materials
Patient material
Twenty-eight breast cancer affected women participating in this study were from multi-case families who had been recruited into the Kathleen Cuningham Foundation for Research into Breast Cancer (kConFab) [13]. Of these, 9 carried a mutation in BRCA1, 7 carried a mutation in BRCA2 and 12 were mutation negative (BRCAx) after full sequencing and multiplex ligation-dependant probe amplification (MLPA) analysis of BRCA1/2. The histological features of all tumours were reviewed by a pathologist who also scored the percentage of neoplasia in the specimen prior to DNA isolation, as previously described [1]. The molecular subtype (luminal A, luminal B, HER2, basal and normal breast-like) for each tumour had been earlier determined by Waddell et al. [1].
Genome-wide SNP genotyping
SNP genotyping data were generated from 28 matched tumour and normal tissue using Illumina arrays containing 370,000 SNPs. Tissue samples of the primary tumour and matching normal tissue were obtained by macrodissectioning frozen sections of 10 μm with a needle, ensuring that the neoplastic content was >75%. DNA was extracted by the salting-out method followed by a phenol chloroform extraction. The Infinium II assay protocol (Illumina Inc., San Diego, CA) was used to perform whole genome amplification. DNA of 750 ng was fragmented and hybridised to Illumina Human CNV370 duo beadarrays [1].
CNV calling and data analysis
Germline and tumour-specific CNVs were determined using the SNP array data for all 28 matching tissue pairs. Data were imported and visualised in GenomeStudio Software v2010.3, and the B-allele frequencies and logR ratios were exported for each sample. The software package R was used to perform SOMATICS [14] to identify regions containing CNVs, as previously described [1]. SOMATICS was used as it can analyse SNP data from tissues which are heterogeneous due to the presence of stromal contamination or multiple tumour clones. For technical validation of predicted CNVs, comparative qPCR was performed as described previously [1]. Galaxy genome analysis tools [15] were utilised for mapping CNVs to the hg18 build of the human genome and to perform intersection and subtraction, to compute base coverage and to obtain flanking sequences.
In this study, we defined tumour-specific CNV regions as those which show copy number change in the tumour genome but do not overlap germline CNV regions identified in the genome of the matched normal tissue. To account for the possibility that contiguous CNVs called by SOMATICS may represent a single larger CNV, especially in the tumour genome, we have measured the base pair coverage of these variants as opposed to their frequency. Our hypothesis that germline CNVs are ‘hotspots’ for tumour-specific CNVs, and hence that the fraction of tumour-specific CNVs located in proximity to germline CNVs is higher than expected was tested in silico. First, we estimated the expected fraction of genomic DNA containing tumour-specific CNVs in proximity to germline CNVs. For each sample, mock CNVs of the same size as the observed set of tumour-specific CNVs were randomly distributed over the human genome. Using these data, we computed the fraction of randomly placed CNVs in proximity to a germline CNV (overlapping or less than 1,000 bp downstream or upstream). This simulation was repeated 2,000 times for each sample. The average fraction of randomly placed CNVs in proximity to a germline CNV was used as the expected fraction. Second, we tested the hypothesis ‘the fraction of tumour-specific CNVs located in proximity to a germline CNV is higher than expected’ with the paired-Wilcoxon rank test (P < 0.05). Genomic coordinates corresponding to germline CNVs and tumour-specific CNVs used in this analysis are listed in Table S1 (Supplementary material). A schematic representation of the experimental design is illustrated in Fig. S1 (Supplementary material).
Results
DNA copy number profiles of matched breast tumour and normal tissue pairs
The germline DNA copy number profiles of our study cohort show that on average, 2.6% (range 0.6–7.0%) of a haploid genome was affected by copy number change (Table S2, Supplementary material). By comparison, analysis of DNA from matched breast tumour tissue revealed copy number changes that covered 37.6% (range 10.5–70.1%) of the haploid genome (Table S2, Supplementary material) and tumour-specific copy number (germline CNV regions excluded) changes covering 36.4% (range 10.0–69.3%) of the haploid genome. These results therefore show that germline CNVs covering an average of 1.4% of the genome are not detected in the tumour genome. There are several potential explanations for this difference, including the possibility that (1) the genomic regions containing germline CNVs may change copy number status after further rearrangement of the tumour; (2) the complexity of the structural rearrangements in the tumour genome results in a failure to call some tumour CNVs by SOMATICS and (3) a proportion of CNVs in the germline DNA are miscalled.
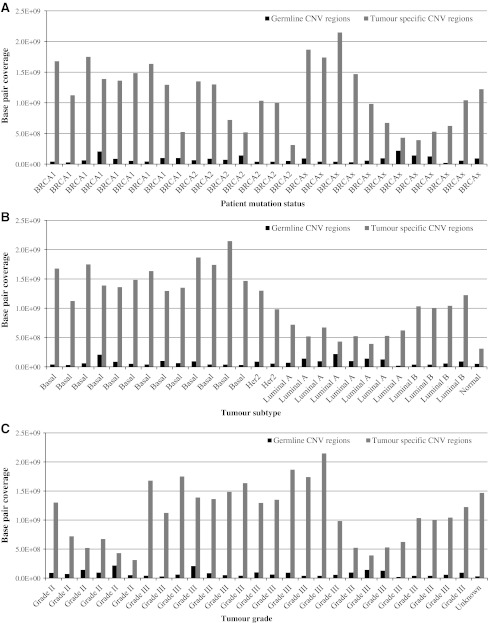
No significant difference was observed in the number of base pairs affected by germline CNVs when comparing cases by mutation status, histological grade of the tumour and basal/non-basal subtype (Table 1). However, a comparison of germline CNV base pair coverage between cases with luminal A and luminal B tumours did show a twofold increase in coverage for luminal A cases that was moderately significant (P = 0.04; Table 1).
Table 1.
Genomic coverage of germline and tumour-specific CNV regions
| n | Germline CNV regions | Tumour-specific CNV regions | |||
|---|---|---|---|---|---|
| Average base pair coverage | P a | Average base pair coverage | P a | ||
| Mutation status | |||||
| BRCA1 | 9 | 79,801,545 | 0.70(BRCA1 vs BRCA2) | 1,359,740,736 | 0.03(BRCA1 vs BRCA2) |
| BRCA2 | 7 | 70,879,436 | 0.52(BRCA2 vs BRCAx) | 890,264,516 | 0.39(BRCA2 vs BRCAx) |
| BRCAx | 12 | 84,758,924 | 0.84(BRCA1 vs BRCAx) | 1,092,785,789 | 0.23(BRCA1 vs BRCAx) |
| Molecular subtype | |||||
| Basal | 13 | 68,410,342 | 0.27(Basal vs all other subtypes) | 1,560,362,706 | 10−7 (Basal vs all other subtypes) |
| Her2 | 2 | 73,290,002 | 1,141,773,431 | ||
| Luminal A | 8 | 114,241,923 | 550,866,350 | ||
| Luminal B | 4 | 57,603,536 | 0.04(LumA vs LumB) | 1,074,264,196 | 10−4 (LumA vs LumB) |
| Normal | 1 | 51,213,066 | 310,698,090 | ||
| Histological grade | |||||
| Grade II | 6 | 111,118,574 | 658,829,402 | ||
| Grade III | 21 | 73,035,723 | 0.20(Grade II vs Grade III) | 1,245,855,304 | 0.007(Grade II vs Grade III) |
aTwo-tailed Student’s t test
In contrast to germline CNVs, and as previously demonstrated [1], the average number of base pairs affected by copy number change within the tumours differed significantly between molecular subtypes and to a lesser degree, by mutation status (Fig. 1a, b; Table 1). This difference was most striking (twofold, P = 10−7) when comparing genomic profiles from basal and non-basal breast tumours (Fig. 1b; Table 1). The average number of base pairs affected by copy number change also differed significantly (twofold, P = 0.007) when classified by histological grade (Fig. 1c; Table 1). This finding is concordant with previous studies that have reported the frequency of genomic aberrations in grade III breast tumours to be greater than those found in grade I or II tumours [7, 16–18]. Sample numbers limited the number of statistical comparisons that could be carried out between the various molecular subtypes. However, we were able to compare the genomic profiles of luminal A and luminal B tumours, and found that the CNV coverage in luminal B tumours was twofold greater than that in luminal A tumours (P < 10−4; Table 1). This difference is not explained by the observation that four of the eight luminal A tumours were also grade II, as a comparison of luminal A and luminal B grade III tumours also showed a significant difference in base pair coverage (P = 0.0002).
Fig. 1.
Base pair coverage of germline and tumour-specific CNV regions as stratified by a patient mutation status, b breast tumour subtype and c breast tumour grade
Correlating the location of germline CNVs with tumour-specific CNVs in paired tumour tissue
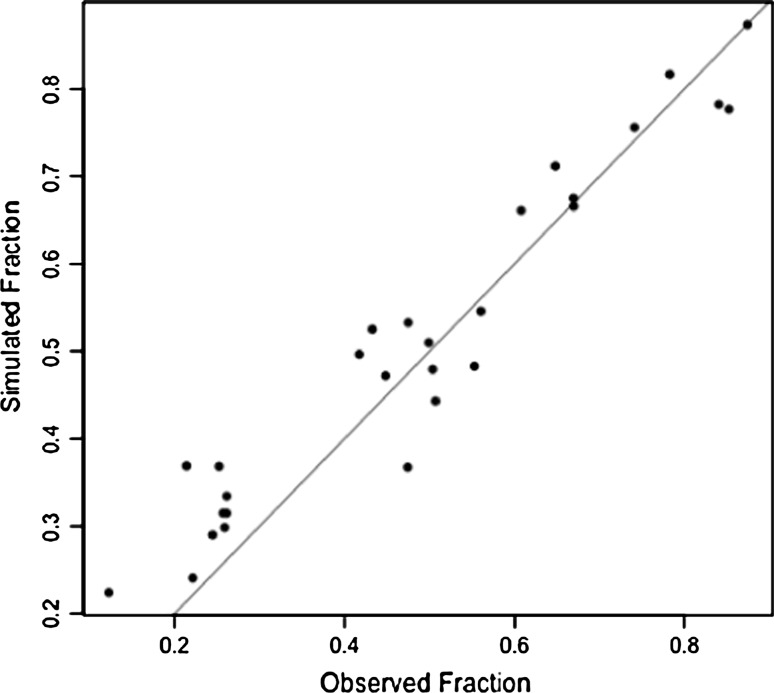
To assess whether germline CNVs act as ‘hotspots’ for tumour-specific CNVs, we tested the hypothesis that tumour-specific CNVs are preferentially located in proximity to germline CNVs. On average, 18.8% (range 1.8–38.7%) of haploid genomes from the breast tumours evaluated in this study consisted of tumour-specific CNVs that either overlapped or were located within 1 kb of a germline CNV. This equates to approximately half of the total number of base pairs that are affected by tumour-specific CNVs (36.4%, range 10.0–69.3%). To determine whether these germline CNVs are hotspots for tumour-specific CNVs, we simulated events using observed CNV sizes and calculated the fraction of randomly placed tumour-specific CNVs in proximity to a germline CNV. We found that the overall difference between actual and simulated events was not significant (P = 0.97; Fig. 2; Table 2). Only 10 of 28 cases showed greater proximity between the actual germline CNVs and tumour-specific CNVs and represented all tumour subtypes (Table 2). By comparison, the proximity between randomised intervals of germline CNVs and tumour-specific CNVs in 18 of the 28 cases was slightly greater than that between actual intervals of germline CNVs and tumour-specific CNVs (Table 2). This was also the case for 9 of the 13 basal-like tumours suggesting that germline CNVs in these tumours are not located in genomic regions that give rise to somatically acquired changes. Together these findings suggest that the location of tumour-specific CNVs is not biased by germline CNVs.
Fig. 2.
Proximity of tumour-specific CNVs to germline CNVs from actual and simulated data. Each dot represents for one patient the fraction of observed and expected (randomly placed) tumour CNVs in proximity to a germline CNV
Table 2.
Proximity of tumour-specific CNVs to germline CNVs from actual and simulated data (2000 replications per sample)
| Sample ID | Mutation status | Original bp fraction in proximity (x) | Simulated bp fraction in proximity | Difference (x − y) | ||
|---|---|---|---|---|---|---|
| Average fraction (y) | Minimum fraction | Maximum fraction | ||||
| B7 | BRCA1 | 0.78 | 0.82 | 0.69 | 0.90 | −0.03 |
| B8 | BRCA1 | 0.65 | 0.71 | 0.44 | 0.84 | −0.07 |
| B9 | BRCA1 | 0.12 | 0.22 | 0.10 | 0.38 | −0.10 |
| B11 | BRCA1 | 0.47 | 0.37 | 0.20 | 0.53 | 0.11 |
| B15 | BRCA1 | 0.43 | 0.52 | 0.35 | 0.68 | −0.09 |
| B16 | BRCA1 | 0.67 | 0.67 | 0.43 | 0.82 | 0.00 |
| B19 | BRCA1 | 0.51 | 0.44 | 0.23 | 0.64 | 0.06 |
| B21 | BRCA1 | 0.25 | 0.36 | 0.16 | 0.54 | −0.11 |
| B22 | BRCA1 | 0.50 | 0.48 | 0.30 | 0.64 | 0.03 |
| B2 | BRCA2 | 0.47 | 0.54 | 0.37 | 0.74 | −0.06 |
| B3 | BRCA2 | 0.26 | 0.34 | 0.08 | 0.54 | −0.08 |
| B5 | BRCA2 | 0.50 | 0.50 | 0.27 | 0.67 | −0.01 |
| B12 | BRCA2 | 0.22 | 0.25 | 0.04 | 0.44 | −0.02 |
| B13 | BRCA2 | 0.42 | 0.50 | 0.27 | 0.69 | −0.08 |
| B17 | BRCA2 | 0.21 | 0.38 | 0.00 | 0.71 | −0.16 |
| B27 | BRCA2 | 0.84 | 0.78 | 0.62 | 0.86 | 0.06 |
| B1 | BRCAx | 0.87 | 0.87 | 0.70 | 0.92 | 0.00 |
| B4 | BRCAx | 0.24 | 0.29 | 0.13 | 0.45 | −0.04 |
| B6 | BRCAx | 0.74 | 0.75 | 0.54 | 0.84 | −0.01 |
| B10 | BRCAx | 0.61 | 0.66 | 0.52 | 0.77 | −0.05 |
| B14 | BRCAx | 0.26 | 0.32 | 0.10 | 0.54 | −0.06 |
| B18 | BRCAx | 0.56 | 0.55 | 0.30 | 0.74 | 0.01 |
| B20 | BRCAx | 0.67 | 0.67 | 0.50 | 0.79 | 0.00 |
| B23 | BRCAx | 0.55 | 0.47 | 0.03 | 0.70 | 0.08 |
| B24 | BRCAx | 0.85 | 0.78 | 0.62 | 0.87 | 0.08 |
| B25 | BRCAx | 0.45 | 0.47 | 0.28 | 0.64 | −0.02 |
| B26 | BRCAx | 0.26 | 0.29 | 0.08 | 0.49 | −0.03 |
| B28 | BRCAx | 0.26 | 0.32 | 0.00 | 0.54 | −0.06 |
bp Base pair
Discussion
Similar to sporadic breast tumours, familial breast tumours can be classified into at least five molecular subtypes that are clinically distinct [1]. Of these subtypes, the basal-like tumour subtype, which is common in BRCA1 mutation carriers, is characterised by a high number of chromosomal copy number changes. Genomic instability is also a molecular feature of tumours with high-histological grade that, analogous to basal-like tumours, are typically aggressive lesions. The ability to predict which breast cancer patients will develop tumours with extensive genomic instability will undoubtedly be a valuable tool in clinical diagnostics. To our knowledge, this is the first study to test the hypothesis that variation in genotype as a result of inherited copy number changes contributes to genomic instability within breast tumour cells. In contrast to the previous small studies [11, 12] that evaluated choroid plexus and colon carcinomas, our results using 28 matched breast tumour and normal tissue suggest that, at a whole genome level, there is no evidence of an association between the genomic location of germline CNVs and breast tumour-specific CNVs using matched normal and tumour pairs.
Consistent with a previous study of familial breast tumours, we found that CNV base pair coverage in luminal B tumours was significantly greater than that in luminal A tumours [19]. However, we show that this difference was independent of breast tumour grade. Compared to luminal A tumours, luminal B tumours are known to have higher cellular proliferation and confer poorer prognosis [20]. Our results suggest that the chromosomal instability phenotype, but not the differentiated state of the tumour cells, contributes to the aggressive nature of luminal B tumours.
A previous study found a significant association between known CNV loci and de novo chromosome breaks in colon cancer [12], suggesting that germline CNVs and tumour-specific CNVs are likely to be located at chromosome regions that are most prone to breakage. However, it remains to be determined whether sequences at these loci predispose to further genomic instability after copy number change has occurred in the germline DNA. Our study went some way to investigate this issue; however, there are notable limitations with the current dataset. First, the exact boundaries of predicted CNVs cannot be precisely determined using microarray-based platforms, and it is therefore impossible to characterise the breakpoint sequences that are involved in the copy number change without further sequence analysis. Second, it is unclear as to the precise location of duplicated or amplified CNVs in that some CNV units may not be located in tandem but may map to different chromosomes entirely; confounding studies assessing their contribution to somatically acquired genomic events. Whole genome high-resolution technologies, such as next-generation sequencing, will be required to accurately map the location of each CNV amplicon to better ascertain which sequences contribute to tumour genome instability. Third, SOMATICS was not able to discriminate heterozygous loss and gain for some predicted CNVs. Thus, we were only able to identify copy number variable regions that had undergone copy number change but were unable to classify many of these regions by exact copy number status. Forth, germline DNA used by Waddell et al. [1] was obtained from breast tissue that appears histologically normal but may potentially harbour somatically acquired copy number changes. Although DNA from peripheral blood cells or buccal cells would have prevented such possibility, recent evidence suggests that the detection of clonal changes in normal tissue found adjacent breast tumours would be unlikely [21, 22].
The notion that inherited CNVs may influence incidence of the various genomic copy number changes that occur during breast cancer progression has not only prognostic significance, but also important consequences for early decisions relating to clinical management. Although our findings suggest no association globally across the genome, it is still possible that some germline CNVs may indeed mark regions that are prone to further rearrangement in the breast tumour. Further work is therefore required using the latest genomic sequencing technologies to precisely map CNV breakpoints sequences across the genome to determine the relationship between inherited genomic variation and genome evolution in breast cancer. Moreover, studies with larger cohort size are warranted to assess our finding in familial breast tumours, that the genome of high-grade luminal A tumours had significantly less CNV coverage than the more clinically aggressive high-grade luminal B tumours.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We wish to thank Georgia Chenevix-Trench for sharing of the array data and helpful comments. We also wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded 2001-2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. LCW was supported by funding from the Genesis Oncology Trust, ABS is an NHMRC Senior Research Fellow and NW was supported by funding from the NHMRC.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
Ethical approvals were obtained from the Human Research Ethics Committees of the Queensland Institute of Medical Research and the Peter MacCallum Cancer Centre. Written informed consent was obtained from each participant.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Waddell N, Arnold J, Cocciardi S, da Silva L, Marsh A, Riley J, Johnstone CN, Orloff M, Assie G, Eng C, et al. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res Treat. 2010;123:661–677. doi: 10.1007/s10549-009-0653-1. [DOI] [PubMed] [Google Scholar]
- 2.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 3.Russnes HG, Navin N, Hicks J, Borresen-Dale AL. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121:3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin SF, Wang Y, Thorne NP, Teschendorff AE, Pinder SE, Vias M, Naderi A, Roberts I, Barbosa-Morais NL, Garcia MJ, et al. Using array-comparative genomic hybridization to define molecular portraits of primary breast cancers. Oncogene. 2007;26:1959–1970. doi: 10.1038/sj.onc.1209985. [DOI] [PubMed] [Google Scholar]
- 7.Walker LC, Harris GC, Wells JE, Robinson BA, Morris CM. Association of chromosome band 8q22 copy number gain with high grade invasive breast carcinomas by assessment of core needle biopsies. Genes Chromosom Cancer. 2008;47:405–417. doi: 10.1002/gcc.20545. [DOI] [PubMed] [Google Scholar]
- 8.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 9.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25:5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 11.Shlien A, Tabori U, Marshall CR, Pienkowska M, Feuk L, Novokmet A, Nanda S, Druker H, Scherer SW, Malkin D. Excessive genomic DNA copy number variation in the Li–Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2008;105:11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, Rodriguez V, Chandrasekharappa S, Chen Y, Difilippantonio MJ, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–1295. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann GJ, Thorne H, Balleine RL, Butow PN, Clarke CL, Edkins E, Evans GM, Fereday S, Haan E, Gattas M, et al. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006;8:R12. doi: 10.1186/bcr1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assie G, LaFramboise T, Platzer P, Bertherat J, Stratakis CA, Eng C. SNP arrays in heterogeneous tissue: highly accurate collection of both germline and somatic genetic information from unpaired single tumor samples. Am J Hum Genet. 2008;82:903–915. doi: 10.1016/j.ajhg.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roylance R, Gorman P, Harris W, Liebmann R, Barnes D, Hanby A, Sheer D. Comparative genomic hybridization of breast tumors stratified by histological grade reveals new insights into the biological progression of breast cancer. Cancer Res. 1999;59:1433–1436. [PubMed] [Google Scholar]
- 17.Bergamaschi A, Kim YH, Wang P, Sorlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Borresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosom Cancer. 2006;45:1033–1040. doi: 10.1002/gcc.20366. [DOI] [PubMed] [Google Scholar]
- 18.Richard F, Pacyna-Gengelbach M, Schluns K, Fleige B, Winzer KJ, Szymas J, Dietel M, Petersen I, Schwendel A. Patterns of chromosomal imbalances in invasive breast cancer. Int J Cancer. 2000;89:305–310. doi: 10.1002/1097-0215(20000520)89:3<305::AID-IJC15>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Melchor L, Honrado E, Garcia MJ, Alvarez S, Palacios J, Osorio A, Nathanson KL, Benitez J. Distinct genomic aberration patterns are found in familial breast cancer associated with different immunohistochemical subtypes. Oncogene. 2008;27:3165–3175. doi: 10.1038/sj.onc.1210975. [DOI] [PubMed] [Google Scholar]
- 20.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, Trivett M, Thompson ER, Ramakrishna M, Gorringe KL, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–655. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell I, Qiu W, Haviv I. Genetic changes in tumour microenvironments. J Pathol. 2011;223:450–458. doi: 10.1002/path.2842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.