Abstract
Zasmidium angulare, a novel species of Mycosphaerellaceae, and several novel taxa that reside in Dissoconiaceae, were identified from a collection of apples and Cucurbita maxima (cv. Blue Hubbard) from China and the USA that exhibited sooty blotch and flyspeck (SBFS) signs on their host substrata. Morphology on fruit surfaces and in culture, and phylogenetic analyses of the nuclear ribosomal DNAs 28S and internal transcribed spacer regions, as well as partial translation elongation factor 1-alpha gene sequences in some cases, were used to delineate seven previously unidentified species and three known species. Pseudoveronaea was established as a new genus of Dissoconiaceae, represented by two species, P. ellipsoidea and P. obclavata. Although Pseudoveronaea was morphologically similar to Veronaea, these fungi clustered with Dissoconiaceae (Capnodiales) rather than Chaetothyriales (Herpotrichiellaceae). Ramichloridium mali comb. nov., and three novel species, R. cucurbitae, R. luteum and R. punctatum were closely related with R. apiculatum, which together formed a distinct subclade in Dissoconiaceae. Species of Dissoconium s.lat. clustered in two well-supported clades supported by distinct morphological and cultural features. Subsequently Uwebraunia, a former synonym of Dissoconium, was resurrected for the one clade, with new combinations proposed for U. australiensis, U. commune, U. dekkeri and U. musae. Furthermore, we also reported that D. aciculare, Dissoconium sp., U. commune and U. dekkeri were associated with SBFS on apples.
Keywords: hyphomycetes, Malus, microfungi, SBFS, taxonomy
INTRODUCTION
Dissoconium, an anamorphic genus based on the type species Dissoconium aciculare (de Hoog et al. 1983), is known to have different ecological niches. Two species, D. aciculare and D. dekkeri, were reported as mycoparasitic fungi on Erysiphaceae (de Hoog et al. 1983, 1991). However, recent studies showed that D. dekkeri acts as a foliar pathogen of Eucalyptus (Crous 1998, Jackson et al. 2004). Other Dissoconium species were mostly reported from or associated with leaf spots. For example, D. australiensis, D. commune and D. eucalypti co-occurred with plant pathogenic species of Capnodiales on leaf spots of Eucalyptus spp. (Crous et al. 2004b, 2007a, b, 2009a, d). Two other species, D. protea and D. musae, colonised leaves of Protea sp. and Musa sp., respectively (Arzanlou et al. 2008, Crous et al. 2008). Although some species of ‘Dissoconium’ have a Mycosphaerella-like teleomorph, they were shown to cluster between Teratosphaeriaceae and Schizothyriaceae (Crous et al. 2004b). Species of Dissoconium, together with Ramichloridium apiculatum, formed a distinct clade, indicating they represented a group different from other members of Capnodiales (Crous et al. 2009b). A new family, Dissoconiaceae, was therefore established to accommodate this group (Crous et al. 2009b).
Several Dissoconium species contribute to a disease complex known as sooty blotch and flyspeck (SBFS) on the surface of several types of fruit (Batzer et al. 2005, Díaz Arias et al. 2010, Gleason et al. 2011). On apples, blemishes caused by SBFS result in substantial economic losses to growers in humid production areas worldwide (Colby 1920, Williamson & Sutton 2000, Batzer et al. 2005, Díaz Arias et al. 2010, Gleason et al. 2011). The SBFS fungal complex is highly diverse, comprising more than 80 species (Batzer et al. 2008, Frank et al. 2010, Yang et al. 2010, Gleason et al. 2011, Li et al. 2011). Several putative species of Dissoconium were first reported as SBFS fungi on apple in the USA (Batzer et al. 2005, Díaz Arias et al. 2010), and D. aciculare was also isolated from SBFS colonies on the fruit of pawpaw (Asimina triloba) (Hemnani et al. 2008). Additionally, D. mali was found to cause SBFS on apple and persimmon in China (Zhang et al. 2007, Sun et al. 2008).
Based on an extensive culture collection obtained from apples and winter squash in the United States and China, we aimed to resolve the taxonomy of this epiphytic fungal group, using both DNA sequence analyses and morphological comparisons.
MATERIAL AND METHODS
Isolates and scanning electron microscopy
Sixteen isolates were included in the present study. Two were obtained from apple fruit collected from Weifang City, Shandong Province, China in October 2006, following the method of Sun et al. (2003). Twelve isolates were obtained from surveys conducted in apple orchards in the eastern and midwestern USA in 2000 and 2005 (Batzer et al. 2005, Díaz Arias et al. 2010). The other two isolates were obtained from winter squash (Cucurbita maxima cv. Blue Hubbard) harvested from an Iowa State University’s Horticulture Research Farm near Gilbert, Iowa, USA, in 2009 (Mayfield et al. 2011). Colonies on fruit cuticles from which isolates were obtained were pressed between paper towels and photographed under a dissecting microscope. To further clarify the sporulation of a novel species of Ramichloridium (CPC 18961), the protocol of Zhang et al. (2009) was used to obtain scanning electron micrographs (SEM) of this isolate from China.
DNA isolation, amplification and phylogenetic analysis
Genomic DNA was isolated from fungal mycelium grown on potato-dextrose agar (PDA), using the PrepManTM Ultra Sample Preparation Reagent (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocols. The primers V9G (de Hoog & Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester 1990) were used to amplify a part of the nuclear rDNA operon spanning the 3′ end of the 18S nrRNA gene (SSU), the internal transcribed spacer 1, the 5.8S nrRNA gene, the internal transcribed spacer 2 (ITS) and the first 900 bases at the 5′ end of the 28S nrRNA gene (LSU). Besides V9G and LR5, primers ITS4 (White et al. 1990) and LR0R (Rehner & Samuels 1994) were also used as additional internal sequence primers to ensure good quality sequences over the entire length of the amplicon. The PCR reaction mixture and amplification conditions were the same as those described by Cheewangkoon et al. (2008). Additionally, part of the translation elongation factor 1-alpha gene (TEF) was amplified and sequenced as described by Bensch et al. (2010) to better resolve taxa showing identical or near identical ITS sequences.
Sequences generated from the present study were used as queries for a Blastn search in NCBIs GenBank nucleotide (nr) database, and those with high nucleotide identities were downloaded. The sequence alignment was performed in ClustalX v. 2.1 (Thompson et al. 1994, Larkin et al. 2007), followed by manual adjustment in BioEdit v. 7.0.5 (Hall 1999).
Phylogenetic analyses were conducted on the LSU and ITS alignments in PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003) by employing distance and maximum parsimony algorithms. Distance analyses employed the uncorrected (‘p’), the Kimura 2-parameter and the HKY85 substitution models, with the neighbour-joining search. Alignment gaps were treated as missing data, and ties were broken randomly when encountered. The neighbour-joining bootstrap support values (NJBP) equal or greater than 50 % are shown after the slash at nodes (generated from the HKY85 substitution model analysis). Parsimony analyses were performed using the heuristic search option with 100 replicates of random stepwise additions and tree bisection reconnection (TBR) as the branch-swapping algorithm. All characters were unordered and given equal weight and all equally most parsimonious trees were saved. The robustness of obtained trees was evaluated by 1 000 bootstrap replications. Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were also calculated. Phylogenetic trees were drawn in TreeView v. 1.6.6 (Page 1996) and Geneious v. 5.5.4 (Drummond et al. 2011) and layout was done in Adobe Illustrator CS 5.1. New sequences from this study were deposited in GenBank (Table 1) and the alignment and trees in TreeBASE (www.treebase.org). The results of the TEF sequences are discussed under the species notes where applicable.
Table 1.
Collection details and GenBank accession numbers of isolates for which novel sequences were generated in this study.
| Species | Strain number |
Substrate | Country | Collector | GenBank Accession no. |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CBSa | CPCb | CMGc | ITSd | LSUe | TEFf | ||||||
| Dissoconium aciculare | 132079 | 18965 | MA1 10B1a | Malus domestica fruit surface | Massachusetts, USA | D. Cooley | JQ622082 | JQ622090 | JQ622107 | ||
| 132080 | 18966 | PEB1a | Malus domestica fruit surface | Iowa, USA | M. Gleason | JQ622083 | JQ622091 | JQ622108 | |||
| 132081 | 18967 | CUB2c | Malus domestica fruit surface | Illinois, USA | M. Gleason | AY598874 | JQ622097 | JQ622114 | |||
| 132082 | 18968 | MSTB4b | Malus domestica fruit surface | Wisconsin, USA | P. McManus | JQ622081 | JQ622089 | JQ622106 | |||
| 132083 | 18973 | UMB4b | Malus domestica fruit surface | Missouri, USA | M. Gleason | AY598875 | JQ622098 | JQ622115 | |||
| Dissoconium sp. | 132084 | 18969 | KY4 19.1B2 | Malus domestica fruit surface | Kentucky, USA | J. Hartman | JQ622084 | JQ622092 | JQ622109 | ||
| Pseudoveronaea ellipsoidea | 132085* | 18970* | MI3 34F1a* | Malus domestica fruit surface | Michigan, USA | G. Sundin | FJ425205 | JQ622103 | JQ622120 | ||
| Pseudoveronaea obclavata | 132086* | 18972* | UIF3a* | Malus domestica fruit surface | Illinois, USA | M. Gleason | AY598877 | JQ622102 | JQ622119 | ||
| Ramichloridium cucurbitae | 132087* | 19423* | BHF50a3* | Cucurbita maxima fruit surface | Gilbert, Iowa, USA | D. Mayfield | JQ622087 | JQ622095 | JQ622112 | ||
| Ramichloridium luteum | 132088* | 18961* | ZXRSD2* | Malus domestica fruit surface | Weifang, Shandong Province, China | G.Y. Sun | EU329730 | JQ622099 | JQ622116 | ||
| 132089 | 18962 | ZXRSD5 | Malus domestica fruit surface | Weifang, Shandong Province, China | G.Y. Sun | EU329731 | JQ622100 | JQ622117 | |||
| Ramichloridium punctatum | 132090* | 18974* | BHE35b1* | Cucurbita maxima fruit surface | Gilbert, Iowa, USA | D. Mayfield | JQ622086 | JQ622094 | JQ622111 | ||
| Uwebraunia commune | 132091 | 18963 | NC1 32C1d | Malus domestica fruit surface | North Carolina, USA | T. Sutton | JQ622085 | JQ622093 | JQ622110 | ||
| 132092 | 18971 | MSTF3b | Malus domestica fruit surface | Wisconsin, USA | P. McManus | AY598876 | JQ622101 | JQ622118 | |||
| Uwebraunia dekkeri | 132093 | 18964 | OH3 37E1d | Malus domestica fruit surface | Ohio, USA | M. Ellis | FJ425204 | JQ622104 | JQ622121 | ||
| Zasmidium angulare | 132094* | 19042* | GA2 27B1a* | Malus domestica fruit surface | Georgia, USA | M. Wheeler | JQ622088 | JQ622096 | JQ622113 | ||
* Ex-type strains are indicated with an asterisk.
a CBS: CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands.
b CPC: Culture collection of P.W. Crous, housed at CBS.
c CMG: Culture collection of M. Gleason, housed at Iowa State University, Ames, Iowa, the USA.
d ITS: Internal transcribed spacers 1 and 2 together with 5.8S nrDNA.
e LSU: 28S nrDNA.
f TEF: partial translation elongation factor 1-alpha gene sequence.
Morphology
Isolates were established on PDA, synthetic nutrient-poor agar (SNA) and oatmeal agar (OA; Crous et al. 2009e), and subsequently incubated at 25 °C under near-ultraviolet light (300–400 nm) to promote sporulation. Preparations from cultured fungal colonies on SNA were mounted on glass slides with Shear’s solution using transparent adhesive tape (Titan Ultra Clear Tape, Conglom Inc., Toronto, Canada) as explained by Schubert et al. (2007) for microscopic examination after 1 mo of incubation. Thirty measurements per relevant microscopic structure were made where possible. Colony colours on PDA and OA (surface and reverse) were determined using the colour charts of Rayner (1970) after 1 mo at 25 °C in the dark. Reference strains are maintained in the culture collection of the Centraalbureau voor Schimmelcultures (CBS-KNAW), Utrecht, The Netherlands, the working collection of P.W. Crous (CPC), and at Iowa State University (Table 1). Nomenclatural novelties and descriptions were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004a).
RESULTS
Phylogenetic analysis
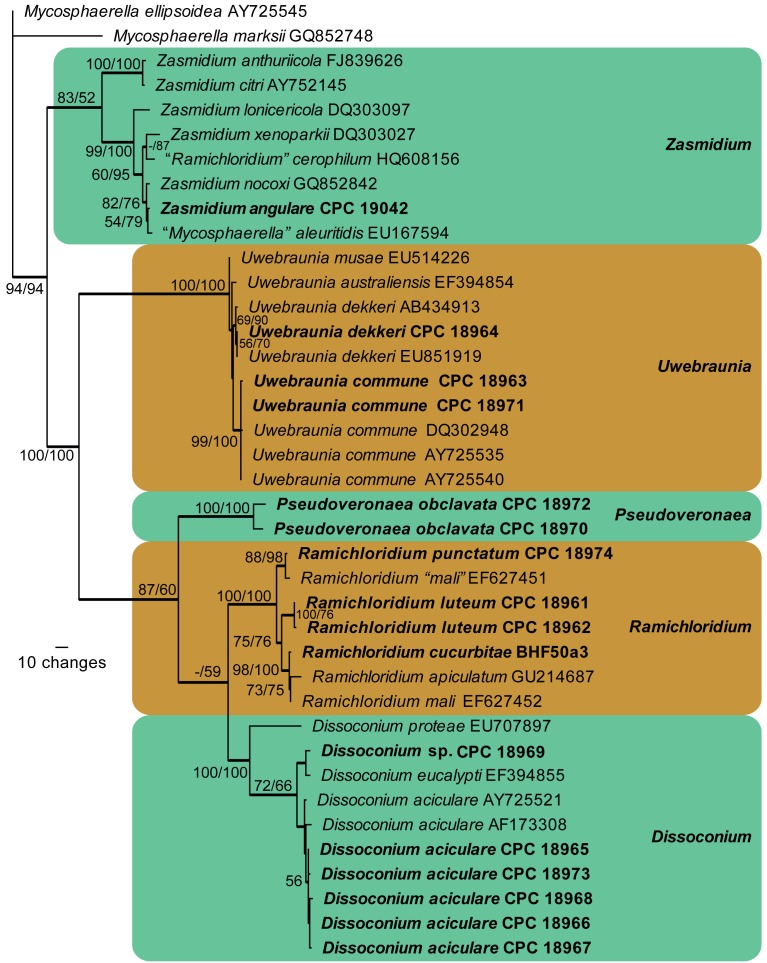
The LSU alignment consisted of 74 taxa (including two outgroups) and 753 characters including alignment gaps. Of these characters, 478 were constant, 28 were variable and parsimony-uninformative, and 247 were parsimony-informative. The first of 48 equally most parsimonious trees generated from maximum parsimony analysis is shown in Fig. 1 (TL = 862 steps; CI = 0.491; RI = 0.855; RC = 0.420). The neighbour-joining analyses using three substitution models yielded trees with topologies and bootstrap support values similar to those of MP analysis (data not shown). From the LSU phylogenetic inference, the strain CPC 19042 clustered together with Zasmidium nocoxi and Mycosphaerella aleuritidis (93 % MPBP, 92 % NJBP), indicating it is a member of Mycosphaerellaceae. The other 15 strains all clustered in Dissoconiaceae (95 % MPBP, 100 % NJBP) and cluster in four subclades. The first subclade consisted of two strains, CPC 18970 and CPC 18972, with 100 % bootstrap support from both MP and NJ analyses, suggesting that they represent a novel genus of Dissoconiaceae. Four strains including BHF50a3, CPC 18974, CPC 18961 and CPC 18962 grouped with Ramichloridium apiculatum (98 % MPBP, 100 % NJBP), indicating they are closely related with R. apiculatum. All Dissoconium species grouped in two subclades. One was composed of the former Dissoconium species D. australiensis, D. dekkeri and D. commune (97 % MPBP, 98 % NJBP; here combined into Uwebraunia). The other subclade (92 % MPBP, 98 % NJBP) included D. aciculare, D. eucalypti, Dissoconium sp. and D. proteae (Fig. 1).
Fig. 1.
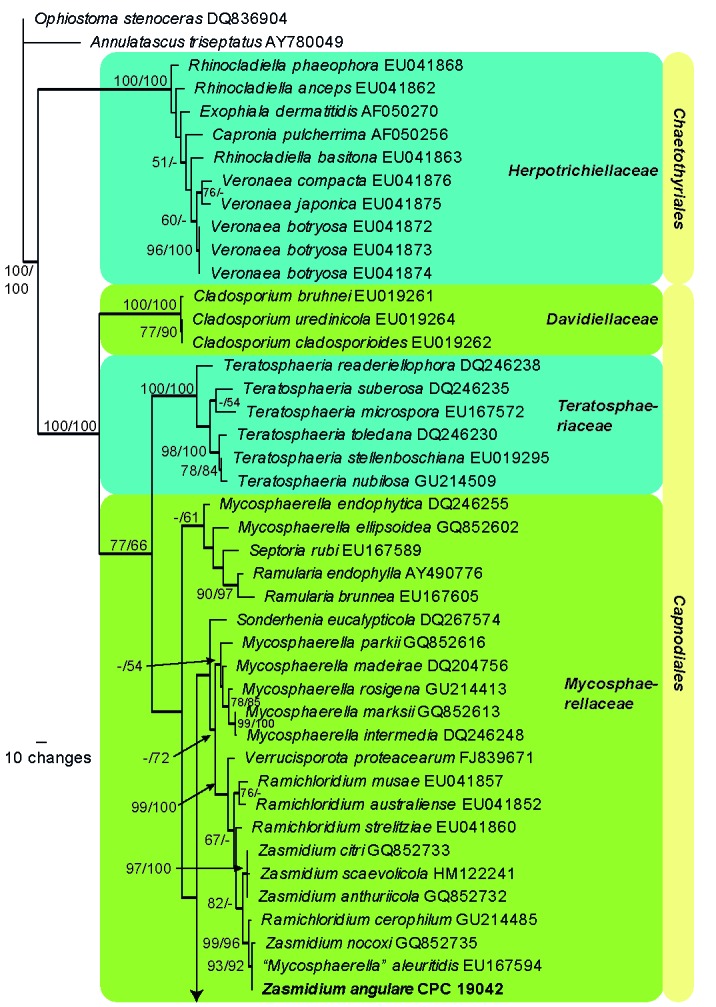
The first of 48 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the LSU sequence alignment. Thickened lines indicate branches present in the strict consensus tree. The scale bar indicates 10 changes and bootstrap support values from 1 000 replicates are shown at the nodes, followed by bootstrap support values from 1 000 replicates obtained by a neighbour-joining analysis using the HKY85 substitution model on the same alignment. Our SBFS isolates treated in this study were indicated in bold. The tree was rooted to Annulatascus triseptatus (GenBank AY780049) (Annulatascaceae, Sordariomycetes) and Ophiostoma stenoceras (GenBank DQ836904) (Ophiostomataceae, Ophiostomatales, Sordariomycetes).
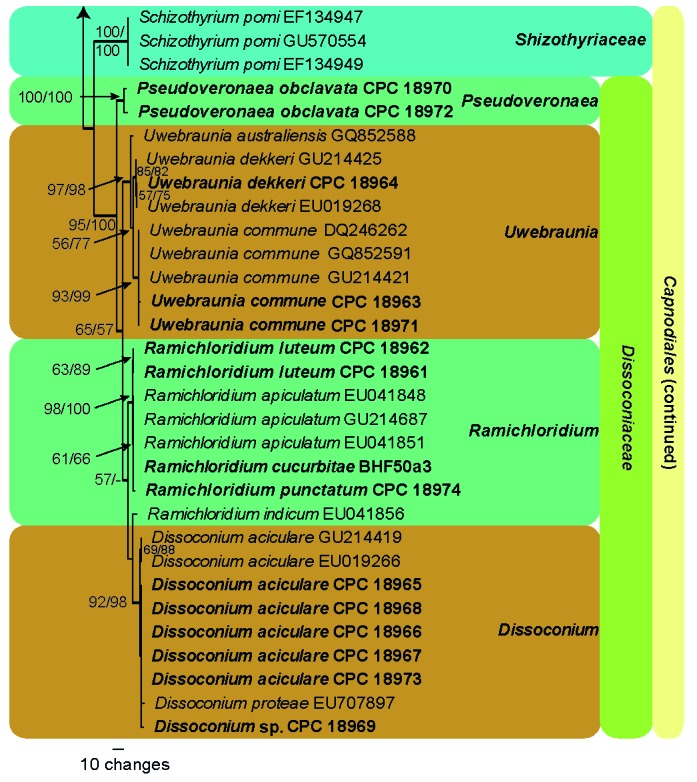
The ITS alignment contained 39 taxa (including two outgroups) and 519 characters including alignment gaps. Of these characters, 271 were constant, 27 were variable and parsimony-uninformative, and 221 were parsimony-informative. Five equally most parsimonious trees were saved from the maximum parsimony analysis, the first of which shown in Fig. 2 (TL = 576 steps; CI = 0.727; RI = 0.927; RC = 0.674). The neighbour-joining analysis of the ITS sequence alignment also yielded similar tree topologies and bootstrap support with the MP algorithm. Most species were strongly supported in the ITS tree (Fig. 2), including Zasmidium angulare, Pseudoveronaea spp., Ramichloridium spp. and D. commune (as Uwebraunia commune below). Several other species had moderate or no bootstrap support in the ITS analysis, including D. aciculare, D. eucalypti, Dissoconium sp. and D. dekkeri (as Uwebraunia dekkeri below) (Fig. 2). The clustering of isolate CPC 18969 with D. eucalypti was not supported in any of the analyses. Isolates of D. aciculare, together with D. eucalypti, had moderate bootstrap support (72 % MPBP, 66 % NJBP). Dissoconium dekkeri (as Uwebraunia dekkeri below) grouped with D. australiensis (as Uwebraunia australiensis below), U. commune and D. musae (as Uwebraunia musae below) (100 % MPBP, 100 % NJBP). Uwebraunia dekkeri was better supported in the distance analysis (69 % MPBP, 90 % NJBP), whereas U. commune was well-supported in both analyses (99 % MPBP, 100 % NJBP).
Fig. 2.
The first of five equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of ITS sequence alignment. Thickened lines indicated branches present in the strict consensus tree. The scale bar indicates 10 changes and bootstrap support values from 1 000 replicates are shown at the nodes, followed by bootstrap support values from 1 000 replicates obtained by a neighbour-joining analysis using the HKY85 substitution model on the same alignment. Our SBFS isolates treated in this study were indicated in bold. The tree was rooted to Mycosphaerella ellipsoidea (GenBank AY725545) and Mycosphaerella marksii (GenBank GQ852748).
Taxonomy
Several species of Dissoconium, Ramichloridium, Uwebraunia, one species of Zasmidium, and members of an undescribed genus were found to be associated with SBFS blemishes on fruit surfaces of apple and winter squash (Fig. 3). These taxa are treated below.
Fig. 3.
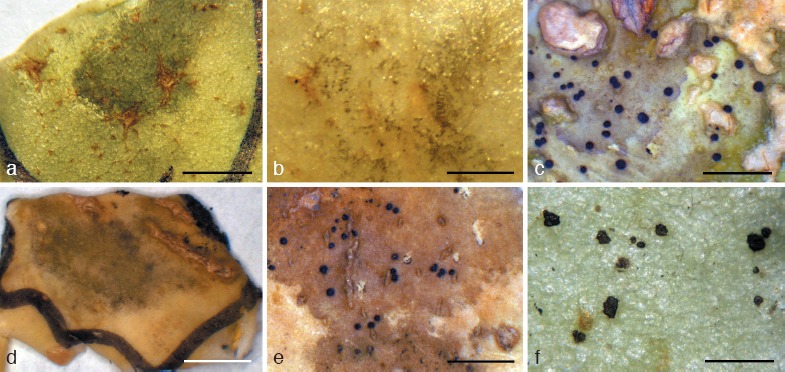
SBFS signs on fruit surface. a. Pseudoveronaea obclavata on apple; b. Pseudoveronaea ellipsoidea on apple; c. Ramichloridium punctatum on winter squash; d. Ramichloridium cucurbitae on winter squash; e. Ramichloridium luteum on apple; f. Zasmidium angulare on apple. — Scale bars: a, d = 5 mm, b, c, e, f = 2 mm.
Dissoconium and Uwebraunia
The genus Dissoconium was established based on D. aciculare, a suspected hyperparasite on Erysiphe (de Hoog et al. 1983). In contrast, Uwebraunia was described as an anamorph genus for three species with Mycosphaerella-like teleomorphs associated with leaf spot diseases of Eucalyptus spp. (Crous & Wingfield 1996, Crous 1998). Uwebraunia was based on U. juvenis, a species incorrectly linked to ‘Mycosphaerella’ juvenis (Crous et al. 2004b). Based on DNA sequence data and type studies, strains of the latter taxon were later shown to be heterogeneous, ‘Mycosphaerella’ juvenis was considered as a synonym of Teratosphaeria nubilosa, a major foliar pathogen of Eucalyptus (Crous 2009, Hunter et al. 2009, 2011), and segregated from T. ohnowa (Crous et al. 2004b, 2007a). Furthermore, molecular data revealed a second species, U. lateralis, to be synonymous with the earlier described D. dekkeri (Crous et al. 1999).
Although Uwebraunia was seen as representing a genus of plant pathogens (Crous & Wingfield 1996, Crous et al. 1999, Jackson et al. 2004), compared to the hyperparasitic Dissoconium (de Hoog et al. 1983), there was no robust support for retaining them as separate genera in Dissoconiaceae (Crous et al. 2009b, Seifert et al. 2011). Preference was thus given to Dissoconium, which was the older name.
Since these initial studies several species have been added to the Dissoconium complex (Crous et al. 2004b, 2006, 2007b, 2008, Arzanlou et al. 2008), which is shown to cluster in two well-supported clades, separated by Ramichloridium s.str. (Fig. 1, 2). Although little is known about their ecology, several morphological differences support generic segregation of Uwebraunia from Dissoconium. The genus Dissoconium (D. aciculare, D. eucalypti and D. protea) has large, obclavate to ellipsoid microconidia, and produces sclerotia as well as a yellow pigment in culture. In contrast, the genus Uwebraunia (U. australiensis, U. commune, U. dekkeri and U. musae) has small, pyriform microconidia, does not form sclerotia or any yellow pigment in culture, and is associated with a Mycosphaerella-like teleomorph. Based on these differences and the robust phylogenetic support for separating these genera (Fig. 1, 2), we choose to resurrect the generic name Uwebraunia for the second clade resolved in this study.
Dissoconium de Hoog, Oorschot & Hijwegen, Proc. Kon. Ned. Akad. Wetensch. C 86, 2: 198. 1983.
Type species. Dissoconium aciculare de Hoog, Oorschot & Hijwegen.
Mycelium internal and external, consisting of branched, septate, smooth, hyaline to pale brown hyphae, that anastomose, forming nets in culture. Conidiophores separate, arising from hyphae, subcylindrical, subulate or lageniform to cylindrical, tapering to a bluntly rounded or truncate apex, straight to once geniculate, smooth, medium brown, 0–2-septate; loci terminal and lateral, visible as slightly thickened, darkened scars on a rachis; proliferation sympodial but also appearing to be percurrent. Primary conidia solitary, pale olivaceous-brown, smooth, ellipsoid to obclavate, 1-septate; hila somewhat darkened. Secondary conidia developing adjacent to primary conidia, pale olivaceous to subhyaline, aseptate, smooth, obclavate to ellipsoid; conidium discharge active, usually with both conidial types being discharged simultaneously. One or more secondary conidia anastomosing with primary conidium once discharged. Sexual state unknown, producing yellow pigment and black sclerotia in culture.
Notes — Three species are presently accepted in Dissoconium based on DNA data and morphology, which are listed here along with their ex-type strains.
Dissoconium aciculare de Hoog, Oorschot & Hijwegen, Proc. Kon. Ned. Akad. Wetensch. C 86, 2: 198. 1983.
Specimens examined. Germany, on Medicago lupulina, CBS 342.82 ex-type. – USA, Illinois, on fruit surface of Malus domestica, Oct. 2000, M. Gleason, CPC 18967 = CUB2c = CBS 132081; Iowa, on fruit surface of Malus domestica, Oct. 2000, M. Gleason, CPC 18966 = PEB1a = CBS 132080; Massachusetts, on fruit surface of Malus domestica, Oct. 2005, D. Cooley, CPC 18965 = MA110B1a = CBS 132079; Missouri, on fruit surface of Malus domestica, Oct. 2000, M. Gleason, CPC 18973 = UMB4b = CBS 132083; Wisconsin, on fruit surface of Malus domestica, Oct. 2000, P. McManus, CPC 18968 = MSTB4b = CBS 132082.
Notes — TEF sequence data support the taxon resolution suggested on the basis of ITS sequences (Fig. 2).
Dissoconium eucalypti Crous & Carnegie, Fung. Diversity 26: 157. 2007.
Specimen examined. Australia, New South Wales, Morpeth Park, Plantation, Bonalbo, E152°36′47″, S28°46′3″, on leaves of Eucalyptus tereticornis, 8 Feb. 2006, A. Carnegie, holotype CBS-H 19770, cultures ex-type CPC 13004 = CBS 120039, CPC 13005–13006.
Dissoconium protea Crous, Persoonia 20: 68. 2008.
Specimen examined. Canary Islands, Tenerife, on leaves of Protea sp., 1 Mar. 2007, P.W. Crous, holotype CBS H-20091, culture ex-type CPC 13853 = CBS 122900.
Dissoconium sp.
Specimen examined. USA, Kentucky, on fruit surface of Malus domestica, Sept. 2005, J. Hartman, CPC 18969 = KY4 19.1B2 = CBS 132084.
Notes — Both ITS (Identities = 484/489 (99 %), Gaps = 3/489 (1 %)) and TEF (Identities = 168/225 (75 %), Gaps = 26/225 (12 %)) sequences suggested a degree of similarity between CBS 132084 and D. eucalypti (CBS 120039). However, these two isolates were not identical and it is possible that the isolates from Eucalyptus and Malus represent distinct species (Fig. 2). More isolates from these hosts should be collected to test this hypothesis.
Pseudoveronaea Crous & Batzer, gen. nov. — MycoBank MB564667
Type species. Pseudoveronaea obclavata Batzer & Crous.
Etymology. Named after its morphological similarity to Veronaea.
Colonies flat, spreading, olivaceous grey, either slow or moderately fast growing. Submerged hyphae hyaline to pale olivaceous, smooth; aerial hyphae hyaline to brown, smooth to warty. Conidiophores erect, straight or flexuose, unbranched or occasionally branched at apex, smooth-walled, medium-brown to brown. Conidiogenous cells terminal or lateral, integrated, pale brown to brown, smooth, subcylindrical, but with apical taper, terminal and intercalary, proliferating sympodially, forming a rachis with crowded, flat to slightly prominent, somewhat darkened, unthickened scars, 0.5–1 μm diam. Conidia solitary, finely verruculose to verruculose, obclavate to ellipsoid, 0–2-septate, apex with or without mucoid appendage; base truncate, darkened, somewhat thickened, not refractive, 1–1.5 μm diam; conidial secession schizolytic.
Notes — Using the key of Arzanlou et al. (2007) Pseudoveronaea has conidiophores similar to those of both Veronaea (up to 200 μm in length) or Veronaeopsis (up to 60 μm in length); its conidiogenous cells lack denticles, and are more Veronaea-like than those of Veronaeopsis (Venturiaceae). Morphologically it is similar to Veronaea, except that the latter is a genus of Chaetothyriales (Herpotrichiellaceae), whereas Pseudoveronaea represents a new genus in Dissoconiaceae (Capnodiales), characterised by having septate conidia.
Pseudoveronaea ellipsoidea Batzer & Crous, sp. nov. — MycoBank MB564668; Fig. 4
Fig. 4.

Pseudoveronaea ellipsoidea (CPC 18970). a. Colony on synthetic nutrient poor agar (SNA); b. aerial mycelium on SNA; c–i. macroconidiophores showing rachis; j. curling lateral hyphae on SNA; k. conidia. — Scale bars = 10 μm.
Etymology. Named after the shape of its conidia, which are ellipsoid.
Mycelium consisting of septate, branched, hyaline, smooth, or brown and warty, 2–4 μm diam hyphae. Conidiophores erect, solitary, arising as lateral branches on superficial hyphae, 2–15-septate, straight to flexuous, subcylindrical, brown, smooth, thick-walled, unbranched or with fertile lateral branches close to apex, 35–130 × 3–5 μm. Conidiogenous cells terminal or lateral, integrated, pale brown to brown, smooth, subcylindrical, but with apical taper to acutely rounded pale brown apex, terminal and intercalary, 6–20 × 3–5 μm; proliferating sympodially, forming a rachis with crowded, flat to slightly prominent, somewhat darkened, unthickened scars, 0.5–1 μm diam. Conidia (6−)8–10(−15) × (3−)3.5–4(−5) μm, 0–1-septate, solitary, finely verruculose, pale brown, granular, ellipsoid to obclavate (in larger conidia), apex subobtuse, base truncate, slightly darkened and thickened, 1–1.5 μm diam.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, with sparse aerial mycelium and lobate, feathery margin; reaching 15–25 mm diam. On SNA olivaceous grey; on PDA iron-grey on surface and reverse; on OA olivaceous grey.
Appearance on natural substratum — On apple surfaces forming a fuliginous appearance (Gleason et al. 2011) characterised by olive-grey, smooth mycelial mats with distinct, but uneven edges.
Specimen examined. USA, Michigan, on fruit surface of Malus domestica, Oct. 2005, G. Sundin, holotype CBS H-20926, ex-type cultures CBS 132085 = CPC 18970 = MI3 34F1a.
Notes — Pseudoveronaea ellipsoidea is distinct from P. obclavata (described below) by having smaller, 0–1-septate conidia that lack apical appendages.
Pseudoveronaea obclavata Batzer & Crous, sp. nov. — MycoBank MB564669; Fig. 5
Fig. 5.
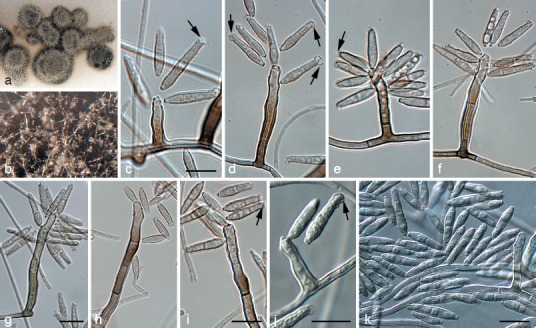
Pseudoveronaea obclavata (CPC 18972). a. Colony on oatmeal agar; b. aerial mycelium of colony on malt extract agar; c–i. macroconidiophores showing rachis; j. conidiophore reduced to conidiogenous cell; k. conidia. — Scale bars = 10 μm.
Etymology. Named after the shape of its conidia, which are obclavate.
Mycelium consisting of subhyaline, smooth, septate, branched hyphae, 1.5–2.5 μm diam. Conidiophores erect, solitary, arising as lateral branches on superficial hyphae, 1–6-septate, unbranched, mostly straight to flexuous to geniculate-sinuous, subcylindrical, brown, smooth, thick-walled, 10–70 × 3–4 μm. Conidiogenous cells terminal, integrated, brown, subcylindrical, even in width or with slight apical taper, 10–25 × 2–4 μm; proliferating sympodially, forming a rachis with crowded, flat to slightly prominent, somewhat darkened, unthickened scars, 0.5 μm diam. Conidia (10−)13–18(−27) × (2.5−)3(−3.5) μm, solitary, finely verruculose to verruculose, obclavate, 0–2-septate, apex subobtuse with globose mucoid apical appendage (2–4 μm diam); base truncate, darkened, somewhat thickened, not refractive, 1 μm diam.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, with sparse aerial mycelium and lobate, smooth margin; reaching 3–6 mm diam. On SNA olivaceous grey; on PDA olivaceous grey, iron-grey in reverse; on OA olivaceous grey.
Appearance on natural substratum — On apple surfaces forming a fuliginous appearance (Gleason et al. 2011) characterised by olive-grey smooth mycelial mats with distinct, but uneven edges.
Specimen examined. USA, Illinois, on fruit surface of Malus domestica, Oct. 2000, M. Gleason, holotype CBS H-20927, ex-type cultures CBS 132086 = CPC 18972 = UIF3a.
Notes — Pseudoveronaea obclavata is characterised by having conidia that are obclavate, 0–2-septate, and have a globose, apical mucoid appendage.
Ramichloridium Stahel ex de Hoog, Stud. Mycol. 15: 59. 1977.
Type species. Ramichloridium apiculatum (J.H. Mill., Giddens & A.A. Foster) de Hoog.
Ramichloridium cucurbitae Mayfield, Batzer & Crous, sp. nov. — MycoBank MB564670; Fig. 6
Fig. 6.
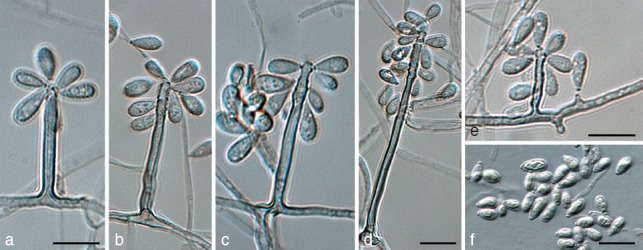
Ramichloridium cucurbitae (BHF50a3). a–d. Macroconidiophores showing rachis; e. conidiophores reduced to conidiogenous cells; f. conidia. — Scale bars = 10 μm.
Etymology. Named after the host genus from which it was collected, Cucurbita.
Mycelium consisting of smooth, subhyaline to pale brown, septate, branched, 1.5–2 μm diam hyphae. Conidiophores erect, arising as lateral branches on superficial hyphae, 0–3-septate or reduced to intercalary conidiogenous cells, unbranched, straight to gently curved, subcylindrical, with apical taper in upper fertile part, brown, smooth, 3–90 × 2–3 μm. Conidiogenous cells terminal, integrated, tapering towards an acutely rounded apex, pale to medium brown, subcylindrical, 3–50 × 1.5–2.5 μm; proliferating sympodially, forming a rachis with slightly thickened and darkened, circular, somewhat protruding scars, ± 0.5 μm diam. Conidia (4−)5–6(−7) × (2−)2.5–3(−3.5) μm, solitary, aseptate, pale brown, smooth, clavate, apex obtuse, base truncate, slightly darkened and thickened, not refractive, 0.5 μm diam.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, with sparse to moderate aerial mycelium and smooth, even margin; reaching 25–30 mm diam. On SNA smoke-grey; on PDA smoke-grey to pale olivaceous grey, olivaceous grey in reverse; on OA pale olivaceous grey.
Appearance on natural substratum — On squash surfaces (Fig 3d) and re-inoculated apple; fuliginous signs (Gleason et al. 2011) characterised by uniform mats of mycelia with abrupt to feathered edges. As described in Batzer et al. (2005), a modified version of Koch’s postulates was performed to verify the signs observed on squash are also observed on apple.
Specimen examined. USA, Iowa, Gilbert, on fruit surface of Cucurbita maxima (cv. Blue Hubbard), Oct. 2009, D. Mayfield, holotype CBS H-20928, ex-type cultures CBS 132087 = CPC 19423 = BHF50a3.
Notes — Ramichloridium cucurbitae is phylogenetically closely related to R. apiculatum, but distinct in having conidiophores that can be reduced to conidiogenous cells. Its conidiogenous cells are slightly narrower than those of R. apiculatum (2–3.5 μm; Arzanlou et al. 2007). Generally colonies of R. apiculatum are also fast-growing, reaching 35 mm after 2 wk, which is not the case for the slower growing R. cucurbitae, which only reaches this diameter after 1 mo of incubation.
Ramichloridium luteum G.Y. Sun, H.Y. Li & Crous, sp. nov. — MycoBank MB564671; Fig. 7, 8
Fig. 7.
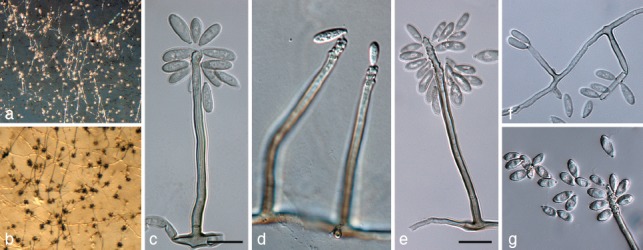
Ramichloridium luteum (CPC 18961). a, b. Sporulating colonies on potato-dextrose agar; c–e. macroconidiophores showing rachis; f. conidiophores reduced to conidiogenous cells; g. conidia. — Scale bars = 10 μm.
Fig. 8.

Ramichloridium luteum (CPC 18961). a–e. Scanning electron micrographs showing sympodial proliferation with scars on conidiogenous cells. — Scale bars: a, c = 5 μm; b, d = 2 μm; d = 1 μm.
Etymology. Named after the luteous pigment produced when cultivated on artificial medium.
Mycelium consisting of subhyaline to pale brown, smooth, septate, branched, 1.5–3 μm diam hyphae. Conidiophores erect, arising as lateral branches on superficial hyphae, 1–3-septate, unbranched, mostly straight to gently curved, subcylindrical, brown, smooth, 25–80 × 2–3 μm. Conidiogenous cells terminal, integrated, tapering towards an acutely rounded apex, pale to medium brown, subcylindrical, 15–30 × 2–3 μm; fertile part as wide as lower part of conidiogenous cell, with taper towards apex; proliferating sympodially, forming a rachis with slightly thickened and darkened, circular, somewhat protruding scars, ± 0.5 μm diam. Conidia (6−)7–10(−13) × (2−)3–4(−4.5) μm, solitary, aseptate, pale brown, finely verruculose, oblong to ellipsoid, apex subobtuse, base truncate, slightly darkened and thickened, not refractive, 0.5 μm diam.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, with sparse aerial mycelium and lobate, feathery margin; reaching 20–30 mm diam. On SNA grey olivaceous; on PDA grey olivaceous, iron-grey in reverse; on OA grey olivaceous, reverse iron-grey, with pale luteous pigment diffusing into agar.
Specimens examined. China, Shandong Province, Weifang City, on fruit surface of Malus domestica, Oct. 2006, G.Y. Sun, holotype CBS H-20929, ex-type cultures CBS 132088 = CPC 18961 = ZXRSD2; Weifang City, on fruit surface of M. domestica, Oct. 2006, G.Y. Sun, CPC 18962 = ZXRSD5 = CBS 132089.
Notes — Morphologically R. luteum resembles R. apiculatum in conidiophore and conidium morphology, though conidia are larger than those of R. apiculatum (3–7.5 × 2–4 μm; Arzanlou et al. 2007).
Ramichloridium mali (G.Y. Sun, Z. Zhang & Rong Zhang) G.Y. Sun, H.Y. Li & Crous, comb. nov. — MycoBank MB564672
Basionym. Dissoconium mali G.Y. Sun, Z. Zhang & Rong Zhang, Mycotaxon 101: 167. 2007.
Specimen examined. China, Shaanxi, Liquan, on fruit surface of Malus pumila, holotype HMUABO 822500, culture ex-type LQ73.
Notes — Based on its well-developed rachis, mode of conidiogenesis (Zhang et al. 2007) and phylogeny, this species is better accommodated in Ramichloridium. A second isolate identified as D. mali (strain LQ45, GenBank EF627451) appears to be allied with R. punctatum rather than R. mali (Fig. 2; see R. punctatum species notes below).
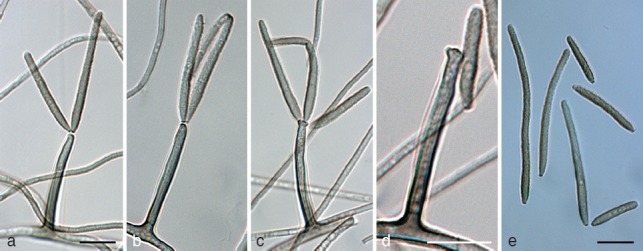
Ramichloridium punctatum Mayfield, Batzer & Crous, sp. nov. — MycoBank MB564673; Fig. 9
Fig. 9.

Ramichloridium punctatum (CPC 18974). a, b. Sporulating colonies on potato-dextrose agar; c–h. macroconidiophores showing rachis; i, j. rachis showing pimple-like denticles; k. conidia. — Scale bars = 10 μm.
Etymology. Named after the speckled appearance of colonies on squash cuticles.
Mycelium consisting of smooth, brown, septate, branched, pale to medium brown, 1.5–2 μm diam hyphae. Conidiophores erect, arising as lateral branches on superficial hyphae, straight, unbranched, subcylindrical, 1–3-septate, thin-walled, brown, smooth, 20–90 × 2–2.5 μm. Conidiogenous cells terminal, subcylindrical or somewhat clavate, uniformly subcylindrical with subobtuse apex, or with a series of irregular, nodulose swellings along the length of the conidiogenous cell, 20–50 × 2–5 μm, brown, becoming paler brown towards apex; fertile part wider than basal part, proliferating sympodially, forming a short rachis with slightly thickened and darkened, circular, somewhat protruding scars, 1 μm diam. Conidia solitary, aseptate, guttulate, finely verruculose, pale brown, clavate to ellipsoid, apex obtuse, base truncate, 1 μm diam, slightly darkened and thickened, not refractive, (5−)8–10(−12) × (3−)3.5–4.5(−5) μm.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, with sparse aerial mycelium and lobate, feathery margin; reaching 20–30 mm diam. On SNA pale mouse-grey; on PDA olivaceous grey in centre, iron-grey in outer margin and underneath; on OA iron-grey.
Appearance on natural substratum — Appearance on squash (Fig. 3c) and re-inoculated apple; ramose signs (Gleason et al. 2011) characterized by olive-grey, smooth mycelial mat with uneven edges consisting of shiny, black, flattened, round sclerotium-like bodies (30–500 μm diam), densely (52–68/cm2) to sparsely (6–38/cm2) distributed on apple and squash, respectively. As described in Batzer et al. (2005), a modified version of Koch’s postulates was performed to verify the signs observed on squash are also observed on apple.
Specimen examined. USA, Iowa, Gilbert, on fruit surface of Cucurbita maxima (cv. Blue Hubbard), Oct. 2009, D. Mayfield, holotype CBS H-20930, ex-type cultures CBS 132090 = CPC 18974 = BHE35b1.
Notes — Phylogenetically, R. punctatum is closely related to an isolate previously identified as R. mali (strain LQ45, GenBank EF627451) (Fig. 2). However, conidia of R. mali were reported as being 3.4–6.2 × 1.6–3 μm (aseptate) and 6–12.6 × 2.1–2.8 μm (1-septate) (Zhang et al. 2007), thus being morphologically quite distinct from those of R. punctatum. The ex-type culture of R. mali (LQ73) does not appear to be conspecific to the second strain identified as R. mali (LQ45), which clusters closer to R. apiculatum. Unfortunately, neither LQ45 nor LQ73 are available for morphological examination, and thus we describe R. punctatum as new, noting that it may be the same species represented by LQ45.
Uwebraunia Crous & M.J. Wingf., Mycologia 88: 446. 1996.
Type species. Uwebraunia juvenis Crous & M.J. Wingf.
Ascomata pseudothecial, immersed, globose, unilocular, papillate, ostiolate, canal periphysate; wall consisting of 3–4 layers of brown textura angularis; inner layer of flattened, hyaline cells. Pseudoparaphyses absent. Asci fasciculate, 8-spored, bitunicate. Ascospores ellipsoid-fusoid, 1-septate, hyaline, with or without mucoid sheath. Mycelium internal and external, consisting of branched, septate, smooth, hyaline to pale brown hyphae, that anastomose, forming nets in culture. Conidiophores separate, arising from hyphae, subcylindrical, subulate or lageniform to cylindrical, tapering to a bluntly rounded or truncate apex, straight to once geniculate, smooth, medium brown, 0–2-septate; loci terminal and lateral, visible as slightly thickened, darkened scars on a rachis; proliferation sympodial but also appearing to be percurrent. Primary conidia solitary, pale olivaceous-brown, smooth, ellipsoid to obclavate, 1-septate; hila somewhat darkened. Secondary conidia developing adjacent to primary conidia, pale olivaceous to subhyaline, aseptate, smooth, pyriform; conidium discharge active, usually with both conidial types being discharged simultaneously. One or more secondary conidia anastomosing with primary conidium once discharged. Colonies not forming yellow pigment, nor sclerotia in culture.
Specimen examined. South Africa, KwaZulu-Natal, Pietermaritzburg, leaves of Eucalyptus nitens, Jan. 1995, M.J. Wingfield, PREM 51910.
Notes — The proposed link between U. juvenis and Tetatosphaeria nubilosa (as Mycosphaerella juvenis) was incorrect (Crous et al. 2004b, 2007a), and all ex-type strains of M. juvenis retained (CPC 932–934) were representatives of T. nubilosa, and none formed U. juvenis in culture. Presently there are no living strains of U. juvenis.
Uwebraunia australiensis (Crous & Summerell) Crous, comb. nov. — MycoBank MB564674
Basionym. Dissoconium australiensis Crous & Summerell, Fung. Diversity 26: 156. 2007.
Specimen examined. Australia, Queensland, Cairns, nr Kuranda, S16°56′23.3″, E145°32′34.6″, on leaves of Eucalyptus platyphylla, 26 Aug. 2006, P.W. Crous, holotype CBS H-19837, culture ex-type CPC 13282 = CBS 120729.
Uwebraunia commune (Crous & Mansilla) Crous, comb. nov. — MycoBank MB564675
Basionym. Dissoconium commune Crous & Mansilla, Stud. Mycol 50: 203. 2004.
= Mycosphaerella communis Crous & Mansilla, Stud. Mycol 50: 203. 2004.
Specimens examined. Spain, Pontevedra, Lourizán, Areeiro, on leaves of E. globulus, Dec. 2002, J.P. Mansilla, CBS H-9900, holotype of M. communis and D. commune, culture ex-type CBS 114238 = CPC 10440. – USA, North Carolina, on fruit surface of Malus domestica, Aug. 2005, T. Sutton, CPC 18963 = NC132C1d = CBS 132091; Wisconsin, on fruit surface of M. domestica, Oct. 2000, P. McManus, CPC 18971 = MSTF3b = CBS 132092.
Uwebraunia dekkeri (de Hoog & Hijwegen) Crous, comb. nov. — MycoBank MB564676
Basionym. Dissoconium dekkeri de Hoog & Hijwegen, Mycol. Res. 95: 679. 1991.
= Uwebraunia lateralis Crous & M.J. Wingf., Mycologia 88: 454. 1996.
= Mycosphaerella lateralis Crous & M.J. Wingf., Mycologia 88: 454. 1996.
= Mycosphaerella shimabarensis H.C. Evans & P.F. Cannon, Mycoscience 50: 187. 2009.
Specimens examined. Netherlands, Maarssen, on Juniperus chinensis, Nov. 1989, T. Hijwegen, ex-type of D. dekkeri, CBS 567.89. – South Africa, Northern Province, Tzaneen, Magoebaskloof, on leaves of E. grandis × saligna, Oct. 1994, G. Kemp, ex-type of U. lateralis, CPC 825 = CBS 110748. – USA, Ohio, on fruit surface of Malus domestica, Oct. 2005, M. Ellis, CPC 18964 = OH3 37E1d = CBS 132093.
Uwebraunia musae (Arzanlou & Crous) Crous, comb. nov. — MycoBank MB564677
Basionym. Dissoconium musae Arzanlou & Crous, Persoonia 20: 24. 2008.
Specimen examined. India, Tamil Nadu, Tiruchirapally, Musa cv. Nendran (Plantain) AAB, 2005, I. Buddenhagen, holotype CBS H-20036, culture ex-type X1021 = CBS 122453.
Zasmidium Fr., Summa Veg. Scand., Sect. Post (Stockholm): 407. 1849.
Type species. Zasmidium cellare (Pers.) Fr., Summa Veg. Scand., Sect. Post (Stockholm): 407. 1849.
Zasmidium angulare Batzer & Crous, sp. nov. — MycoBank MB564678; Fig. 10
Fig. 10.
Zasmidium angulare (CPC 18942). a–d. Macroconidiophores showing apical conidiogenous loci; e. conidia. — Scale bars = 10 μm.
Etymology. Named after the angular shaped sclerotium-like bodies observed on apple host.
Mycelium consisting of septate, branched, brown, verruculose, 1.5–2.5 μm diam hyphae. Conidiophores erect, solitary, arising as lateral branches on superficial hyphae, 0–2-septate, mostly straight to flexuous, subcylindrical, brown, smooth to verruculose, unbranched, 20–40 × 2–3 μm. Conidiogenous cells terminal, integrated, brown, smooth to finely verruculose, subcylindrical, straight, 10–15 × 1.5–2.5 μm; proliferating sympodially, with scars aggregated at clavate apex; truncate, somewhat darkened, thickened, 0.5–1 μm diam. Conidia (7−) 17–50(−130) × 2–3 μm, (0−)3–6(−20)-septate, solitary, verruculose, brown, occurring in branched chains, subcylindrical to narrowly obclavate, apex subobtuse, base long obconically truncate; hilum darkened and thickened, somewhat refractive, 0.5–1 μm diam.
Culture characteristics — Colonies after 1 mo at 25 °C in the dark flat, spreading, somewhat erumpent, with moderate aerial mycelium and lobate, even margin; reaching 20–25 mm diam. On SNA olivaceous; on PDA olivaceous grey on surface, iron-grey in reverse; on OA olivaceous grey, but iron-grey in areas where aerial mycelium has collapsed.
Appearance on natural substratum — On apple surfaces forming a discrete speck appearance (Gleason et al. 2011) consisting of shiny, black, irregular, sclerotium-like bodies, rounded to angular (30–630 μm diam), sparsely or densely distributed (6–41/cm2).
Specimen examined. USA, Georgia, on fruit surface of Malus domestica, Aug. 2005, M. Wheeler, holotype CBS H-20931, ex-type cultures CBS 132094 = CPC 19042 = GA227B1a.
Notes — Of the species of Zasmidium presently known in Mycosphaerellaceae (Arzanlou et al. 2007, Crous et al. 2007a, Braun et al. 2010, Kamal 2010), none have thus far been described from SBFS signs on apple. Phylogenetically, Z. angulare appears to be distinct from other species of Zasmidium presently deposited in GenBank, being closest to Z. nocoxi and ‘Mycosphaerella’ aleuritidis (Crous et al. 2009b). Morphologically, Z. angulare is different from Z. nocoxi by having shorter conidiophores and a slower growth rate in culture. Zasmidium nocoxi reaches 40 mm diam after 1 mo on MEA, with conidiophores up to 100 μm (Crous et al. 2009c). Zasmidium angulare only reaches 20–25 mm diam after 1 mo and conidiophores up to 40 μm tall. Furthermore, Z. nocoxi produces a synanamorph state in its aerial mycelium (Crous et al. 2009c), which is absent in Z. angulare.
DISCUSSION
We resolved the taxonomy of five new taxa that reside in Dissoconiaceae, as well as a novel species of Zasmidium, Z. angulare, based on a collection of apples and cv. Blue Hubbard squash with SBFS signs from China and the USA. The novel genus and species described here expanded Dissoconiaceae to include species of four genera. Pseudoveronaea, which is morphologically similar but phylogenetically distinct from Veronaea, was introduced as a new genus of Dissoconiaceae, and two species, P. ellipsoidea and P. obclavata, were described.
We proposed three novel species of Ramichloridium in this study, R. cucurbitae, R. luteum and R. punctatum, as well as a new combination, R. mali. The originally described Ramichloridum comprised a heterogeneous group of taxa with diverse life styles. Arzanlou et al. (2007) demonstrated that only species clustering in Capnodiales were true Ramichloridium, whereas other ‘Ramichloridium’ species segregated into different genera, including Rhinocladiella (Herpotrichiellaceae, Chaetothyriales), Pleurothecium (Chaetothyriales), Myrmecridium (Sordariomycetes) and Radulidium (incertae sedis). However, the phylogenetic inference of LSU sequence data from both Arzanlou et al. (2007) and the present study showed that R. indicum and R. apiculatum (Ramichloridium s.str.) clustered with Dissoconium species in Dissoconiaceae. By means of contrast, Pseudoramichloridium clustered in Teratosphaeriaceae (Cheewangkoon et al. 2009), and other Ramichloridium-like species grouped in Mycosphaerellaceae. The fact that Ramichloridium-like species clustered in both Dissoconiaceae and Mycosphaerellaceae suggests that it is still a heterogeneous genus and more molecular and morphological data are necessary to clarify the taxonomy of several of these species. The three novel Ramichloridium species described in our study are closely related to R. apiculatum in Dissoconiaceae, providing more taxa to further help delineate Ramichloridium s.str.
Our study further revealed that Dissoconium species clustered in two clades based on phylogenetic analyses of LSU and ITS sequences, indicating that Dissoconium is paraphyletic. Based on morphological differences and their distinct phylogeny, the name Uwebraunia was resurrected to accommodate taxa closely related to U. dekkeri. Furthermore, four species, namely D. aciculare, Dissoconium sp., U. dekkeri and U. commune are shown to be members of SBFS fungal complex, although they were originally described from different ecological niches, being either mycoparasitic or plant pathogenic (de Hoog et al. 1983, 1991, Crous et al. 2004b, 2007b). Batzer et al. (2005) and Díaz Arias et al. (2010) showed that these species caused characteristic signs on inoculated apples using a modified version of Koch’s postulates. SBFS fungal species within Dissoconiaceae may cause distinct mycelial types on the fruit surface. For example, D. aciculare causes discrete speck and U. commune causes a fuliginous mycelial type on apple. However, a single species will consistently express a single mycelial type on a fruit host. In addition, D. aciculare is tolerant to lower temperatures and grows considerably faster on the media than other SBFS fungal species (Batzer et al. 2010). Further collections are required to determine if the distinction between Dissoconium and Uwebraunia can also be correlated with differences in their ecology.
Acknowledgments
We are grateful to A. van Iperen, M. Vermaas, M. Starink (CBS, Utrecht) for providing technical assistance. This work was funded by National Natural Science Foundation of China (31170015, 31171797), the 111 Project from Education Ministry of China (B07049), and Top Talent Project of Northwest A&F University.
REFERENCES
- Arzanlou M, Groenewald JZ, Fullerton RA, Abeln EC, Carlier J , et al. 2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia 20: 19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzanlou M, Groenewald JZ, Gams W, Braun U, Shin H-D, Crous PW. 2007. Phylogenetic and morphotaxonomic revision of Ramichloridium and allied genera. Studies in Mycology 58: 57–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer JC, Díaz Arias MM, Harrington TC, Gleason ML, Groenewald JZ, Crous PW. 2008. Four species of Zygophiala (Schizothyriaceae, Capnodiales) are associated with the sooty blotch and flyspeck complex on apple. Mycologia 100: 246–258 [DOI] [PubMed] [Google Scholar]
- Batzer JC, Gleason ML, Harrington TC, Tiffany LH. 2005. Expansion of the sooty blotch and flyspeck complex on apples based on analysis of ribosomal DNA gene sequences and morphology. Mycologia 97: 1268–1286 [DOI] [PubMed] [Google Scholar]
- Batzer JC, Hernández Rincon S, Mueller DS, Petersen BJ, Le Corronc F, McManus PS, Dixon PM, Gleason ML. 2010. Effect of temperature and nutrient concentration on the growth of six species of sooty blotch and flyspeck fungi. Phytopathologia mediterranea 49: 3–10 [Google Scholar]
- Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, Shin H-D, Dugan FM, Schroers H-J, Braun U, Crous PW. 2010. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Studies in Mycology 67: 1–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Crous PW, Schubert K, Shin HD. 2010. Some reallocations of Stenella species to Zasmidium. Schlechtendalia 20: 99–104 [Google Scholar]
- Cheewangkoon R, Crous PW, Hyde KD, Groenewald JZ, To-anan C. 2008. Species of Mycosphaerella and related anamorphs on Eucalyptus leaves from Thailand. Persoonia 21: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheewangkoon R, Groenewald JZ, Summerell BA, Hyde KD, To-anun C, Crous PW. 2009. Myrtaceae, a cache of fungal biodiversity. Persoonia 23: 55–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby AS. 1920. Sooty blotch of pomaceous fruits. Transactions of the Illinois State Academy of Science 13: 139–179 [Google Scholar]
- Crous PW. 1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycologia Memoir 21: 1–170 APS Press, MN, USA [Google Scholar]
- Crous PW. 2009. Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Diversity 38: 1–24 [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007a. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004a. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Crous PW, Groenewald JZ, Mansilla JP, Hunter GC, Wingfield MJ. 2004b. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. Studies in Mycology 50: 195–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Summerell BA, Wingfield BD, Wingfield MJ. 2009a. Co-occurring species of Teratosphaeria on Eucalyptus. Persoonia 22: 38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hong L, Wingfield MJ, Wingfield BD, Kang J. 1999. Uwebraunia and Dissoconium, two morphologically similar anamorph genera with distinct teleomorph affinity. Sydowia 52: 155–166 [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C , et al. 2009b. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Groenewald JZ. 2009c. Novel species of Mycosphaerellaceae and Teratosphaeriaceae. Persoonia 23: 119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter GC , et al. 2009d. Unravelling Mycosphaerella: do you believe in genera? Persoonia 23: 99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Mohammed CAC, Himaman W, Groenewald JZ. 2007b. Foliicolous Mycosphaerella spp. and their anamorphs on Corymbia and Eucalyptus. Fungal Diversity 26: 143–185 [Google Scholar]
- Crous PW, Summerell BA, Mostert L, Groenewald JZ. 2008. Host specificity and speciation of Mycosphaerella and Teratosphaeria species associated with leaf spots of Proteaceae. Persoonia 20: 59–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, Samson RA. (eds). 2009e. Fungal Biodiversity. CBS Laboratory Manual Series 1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands [Google Scholar]
- Crous PW, Wingfield MJ. 1996. Species of Mycosphaerella and their anamorphs associated with leaf blotch disease of Eucalyptus in South Africa. Mycologia 88: 441–458 [Google Scholar]
- Crous PW, Wingfield MJ, Mansilla JP, Alfenas AC, Groenewald JZ. 2006. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. II. Studies in Mycology 55: 99–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Arias MM, Batzer JC, Harrington TC, Wong AW, Bost SC , et al. 2010. Diversity and biogeography of sooty blotch and flyspeck fungi on apple in the eastern and midwestern United States. Phytopathology 100: 345–355 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A , et al. 2011. Geneious v5.4, Available from www.geneious.com/ [Google Scholar]
- Frank J, Crous PW, Groenewald JZ, Oertel B, Hyde KD, Phengsintham P, Schroers HJ. 2010. Microcyclospora and Microcyclosporella: novel genera accommodating epiphytic fungi causing sooty blotch on apple. Persoonia 24: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason ML, Batzer JC, Sun GY, Zhang R, Arias MMD , et al. 2011. A new view of sooty blotch and flyspeck. Plant Disease 95: 368–383 [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. pp. 95- 98 [Google Scholar]
- Hemnani K, Malley PJO, Tanović B, Batzer JC, Gleason ML. 2008. First report of seven species of sooty blotch and flyspeck on Asimina triloba in Iowa. Plant Disease 92: 1366. [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41: 183–189 [DOI] [PubMed] [Google Scholar]
- Hoog GS de, Hijwegen T, Batenburg-van der Vegte WH. 1991. A new species of Dissoconium. Mycological Research 95: 679–682 [Google Scholar]
- Hoog GS de, Oorschot CAN van, Hijwegen T. 1983. Taxonomy of the Dactylaria complex. II. Dissoconium gen. nov. and Cordana preuss. Proceedings of the Koninklijke Nederlandse Akademie van Wetenschappen, Series C 86: 197–206 [Google Scholar]
- Hunter GC, Crous PW, Carnegie AJ, Burgess TI, Wingfield MJ. 2011. Mycosphaerella and Teratosphaeria diseases of Eucalyptus; easily confused and with serious consequences. Fungal Diversity 50: 145–166 [Google Scholar]
- Hunter GC, Crous PW, Carnegie AJ, Wingfield MJ. 2009. Teratosphaeria nubilosa, a serious leaf disease pathogen of Eucalyptus spp. in native and introduced areas. Molecular Plant Pathology 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Maxwell A, Neumeister-Kemp H, Dell B, Hardy G. 2004. Infection, hyperparasitism and conidiogenesis of Mycosphaerella lateralis on Eucalyptus globulus in Western Australia. Australasian Plant Pathology 33: 49–53 [Google Scholar]
- Kamal. 2010. Cercosporoid fungi of India. Bishen Singh Mahendra Pal Singh Press, Dehra Dun, India [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P , et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li HY, Sun GY, Batzer JC, Crous PW, Groenewald JZ , et al. 2011. Scleroramularia gen. nov. associated with sooty blotch and flyspeck of apple and pawpaw from the Northern Hemisphere. Fungal Diversity 46: 53–66 [Google Scholar]
- Mayfield DA, Batzer JC, Gleason ML. 2011. First report of sooty blotch and flyspeck fungi on cucurbit crop hosts (Abstract). Phytopathology 101, 10: S2.6 [Google Scholar]
- Page RDM. 1996. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society, Kew, Surrey, England [Google Scholar]
- Rehner S, Samuels G. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634 [Google Scholar]
- Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M , et al. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology 58: 105–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. 2011. The genera of Hyphomycetes. CBS Biodiversity Series 9: 1–997 CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands [Google Scholar]
- Sun GY, Li HY, Zhang R, Gleason ML. 2008. First report of Dissoconium mali associated with flyspeck signs on persimmon (Abstract). Phytopathology 98: S153 [Google Scholar]
- Sun GY, Zhang R, Zhang Z, Zhang M. 2003. Isolation of sooty blotch and flyspeck fungi from apple surface by picking up the thalli. Acta Phytopathologica Sinica 33: 479–480 [in Chinese]. [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4.0. Sinauer Associates, Sunderland, Massachusetts, USA [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172, 8: 4239–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee J, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA [Google Scholar]
- Williamson SM, Sutton TB. 2000. Sooty blotch and flyspeck of apple: Etiology, biology and control. Plant Disease 84: 714–724 [DOI] [PubMed] [Google Scholar]
- Yang HL, Sun GY, Batzer JC, Crous PW, Groenewald JZ, Gleason ML. 2010. Novel fungal genera and species associated with the sooty blotch and flyspeck complex on apple in China and the USA. Persoonia 24: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Yang HL, Sun GY, Li HY, Zhuang JL, Zhai XR, Gleason ML. 2009. Strelitziana mali, a new species causing sooty blotch on apple fruit. Mycotaxon 110: 477–485 [Google Scholar]
- Zhang R, Zhang Z, Zhai XR, Zhang M, Sun GY, Gleason ML. 2007. A new species of Dissoconium from China colonizing apples. Mycotaxon 101: 165–172 [Google Scholar]