Abstract
Three new species of Phyllosticta, P. hostae on Hosta plantaginea (China), P. schimae on Schima superba (China), and P. ilicis-aquifolii on Ilex aquifolium (UK), are described and illustrated in this study. They are compared with morphologically similar and phylogenetically closely related species. A polyphasic approach using phylogeny, host association, disease symptoms, colony and morphological characteristics, is employed to justify the introduction of the new taxa. Phylogenetic relationships of the new species with other Phyllosticta species are revealed by DNA sequence analyses based on the nrDNA-internal transcribed spacer (ITS) regions and a combined multilocus alignment of the ITS, partial translation elongation factor 1-alpha (TEF1), actin (ACT), and glyceraldehyde 3-phosphate dehydrogenase (GPDH) gene regions.
Keywords: molecular, morphology, phylogeny, systematics, taxonomy
INTRODUCTION
Many Phyllosticta (teleomorph Guignardia) species cause plant diseases such as leaf spots, leaf blotch, as well as black spots and lesions on fruits of various plants (van der Aa & Vanev 2002). These plant pathogenic fungi may cause serious damage to the host plant through reduced photosynthetic ability and premature leaf or fruit fall (Glienke-Blanco et al. 2002, Baldassari et al. 2008). Phyllosticta species have also been recorded as endophytes and saprobes on a wide range of host plants (Baayen et al. 2002, van der Aa & Vanev 2002, Okane et al. 2003, Wulandari et al. 2009, Glienke et al. 2011).
The generic circumscription of Phyllosticta as defined by van der Aa (1973) has been widely accepted (Bissett 1979, 1986, Yip 1989, Crous et al. 2006, Motohashi et al. 2008, 2009, Glienke et al. 2011, Wikee et al. 2011). The main characters are: pycnidial conidiomata, holoblastic conidiogenous cells with percurrent proliferation, conidia aseptate, surrounded by a mucilaginous sheath, and provided with an apical extracellular appendage, a Guignardia sexual state, and Leptodothiorella spermatial state (van der Aa 1973, van der Aa & Vanev 2002). According to these criteria, van der Aa & Vanev (2002) reconsidered 2 936 names in Phyllosticta, accepting 141 species based on original literature and a re-examination of herbarium specimens. About 50 % of the species were reclassified in Phoma, 20 % in Asteromella, 5 % in Phomopsis and c. 18 % in other coelomycetous genera or other taxonomic groups. Some Phyllosticta species have been linked to their teleomorph states, for example, P. ampelicida is the anamorph of G. bidwellii (van der Aa 1973), but most appear to be asexual. Recent changes to the rules that govern fungal nomenclature require that only one name for a single biological species should be used instead of different names for different morphs (Hawksworth et al. 2011, Wingfield et al. 2012). The earlier and well-known generic name Phyllosticta (Persoon 1818), thus has priority over Guignardia (Viala & Ravaz 1892), as followed by Glienke et al. (2011).
The systematics of Phyllosticta species has long been problematic because of the limited morphological characters and the unreliable use of host-association based nomenclature. Polyphasic approaches combining morphological characters and phylogenetic relationships can resolve species relationships, based on which a natural classification could be established (Wulandari et al. 2009, Glienke et al. 2011). Although the rDNA-internal transcribed spacer (ITS) locus has some resolution at species level, it is insufficient for separating cryptic species in Phyllosticta (Wulandari et al. 2009, Glienke et al. 2011). Therefore, multilocus phylogenetic analyses have been increasingly used for species discrimination in this genus (Wulandari et al. 2009, Glienke et al. 2011, Wang et al. 2012). For example, it was shown that G. mangiferae is a distinct taxon from P. capitalensis, which is a species complex awaiting more detailed phylogenetic study (Glienke et al. 2011).
In the present study, three new species of Phyllosticta are described based on morphological characters and phylogenies derived from ITS and combined multilocus gene sequences.
MATERIALS AND METHODS
Isolates
Phyllosticta species were isolated from diseased leaves of ornamental or forest plant species from China and the United Kingdom. Infected leaves were incubated in moist chambers at room temperature to induce sporulation. Pure cultures were obtained by single spore isolation as described by Choi et al. (1999). Alternatively, 5 × 5 mm pieces of surface-sterilised tissue were taken from the margin of leaf lesions and were consecutively immersed in 70 % ethanol solution for 1 min, sodium hypochlorite solution with 3 % available chlorine for 2 min, rinsed in sterile distilled water, blotted dry in sterile paper towels and incubated on 2 % potato-dextrose agar (PDA) (Cai et al. 2009).
Morphology
Cultures were grown on PDA for microscopic examination. Fungal structures were mounted on glass slides in clear lactic acid, and studied by means of a light microscope. Colony morphologies were assessed after 7 d growth on PDA, and colours rated according to the colour charts of Rayner (1970).
DNA extraction, PCR amplification and sequencing
Mycelial discs were taken from actively sporulating areas near the growing edge of 10 d old cultures and transferred to PDA. Genomic DNA was extracted with a Biospin Fungus Genomic DNA Extraction Kit (Bioer. Technology Co., Ltd., Hangzhou, P.R. China) according to the manufacturer’s protocol. Quality and quantity of DNA were estimated visually by staining with GelRed after 1 % agarose gel electrophoresis. The ITS1 and ITS4 primer pair (White et al. 1990) was used to amplify the ITS region following the procedure described by White et al. (1990). The primers EF1-728F and EF1-986R (Carbone & Kohn 1999) were used to amplify a partial fragment of the translation elongation factor 1-α gene (TEF1); the primers ACT-512F and ACT-783R (Carbone & Kohn 1999) were used to amplify a partial fragment of the actin gene (ACT); the primers GDF1 (Guerber et al. 2003) and Gpd2-LM (Myllys et al. 2002) or GDR1 (Guerber et al. 2003) were used to amplify a partial fragment of the glyceraldehyde 3-phosphate dehydrogenase gene (GPDH). Amplification conditions followed Arzanlou et al. (2008). DNA sequencing was performed at the SinoGenoMax Company Limited, Beijing.
Sequence alignment and phylogenetic analyses
Sequences from forward and reverse primers were aligned to obtain a consensus sequence. Sequences of our isolates, together with reference sequences obtained from GenBank (Table 1), were aligned using Clustal X (Thompson et al. 1997). The separate ITS and the combined multilocus alignments were manually optimised in BioEdit 7.0.9.0 for maximum alignment and minimum gaps (Hall 1999). Both these alignments were subjected to phylogenetic analyses.
Table 1.
Sources of isolates and GenBank accession numbers used in this study. The newly generated sequences in this study are shown in bold.
1 ATCC: American Type Culture Collection, Virginia, USA; CBS: CBS Fungal Biodiversity Centre, Utrecht, The Netherlands; CGMCC: China General Microbial Culture Collection; CPC: Culture collection of P.W. Crous, housed at CBS; IFO: Institute for Fermentation, Osaka, Japan; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, UK; LGMF: Culture collection of Laboratory of Genetics of Microorganisms, Federal University of Parana, Curitiba, Brazil; PD: Plant Protection Service, Wageningen, The Netherlands; VIC: Culture collection of Federal University of Viçosa, Viçosa, Brazil.
* indicates the ex-type cultures.
2 ITS: Internal transcribed spacers 1 and 2 together with 5.8S nrDNA; TEF1: partial translation elongation factor 1-alpha gene; ACT: partial actin gene; GPDH: partial glyceraldehyde-3-phosphate dehydrogenase gene.
Phylogenetic analyses were performed using PAUP v. 4.0b10 (Swofford 2003). Ambiguously aligned regions were excluded from all analyses. An unweighted parsimony (UP) analysis was performed. Trees were inferred using the heuristic search option with TBR branch swapping and 1 000 random sequence additions, branches of zero length were collapsed and all equally most parsimonious trees were saved. Descriptive tree statistics such as tree length [TL], consistency index [CI], retention index [RI], rescaled consistency index [RC], and homoplasy index [HI], were calculated for trees generated. Clade stability was assessed in a bootstrap analysis with 1 000 replicates, each with 10 replicates of random stepwise addition of taxa. A Shimodaira-Hasegawa test (SH test) (Shimodaira & Hasegawa 1999) was performed in order to determine whether trees were significantly different. Trees were visualised in TreeView v. 1.6.6 (Page 1996).
For the Bayesian analyses, the models of evolution were estimated by using MrModeltest v. 2.3 (Nylander 2004). Posterior probabilities (PP) (Rannala & Yang 1996, Zhaxybayeva & Gogarten 2002) were determined by Markov Chain Monte Carlo sampling (BMCMC) in MrBayes v. 3.0b4 (Huelsenbeck & Ronquist 2001), under the estimated model of evolution. Six simultaneous Markov chains were run for 1 000 000 generations and trees were sampled every 100th generation (resulting in 10 000 total trees). The first 2 000 trees, representing the burn-in phase of the analyses, were discarded and the remaining 8 000 trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree. Novel sequence data were deposited in GenBank (Table 1), alignments in TreeBASE (www.treebase.org, submission no.: 12430), and taxonomic novelties in MycoBank (Crous et al. 2004).
RESULTS
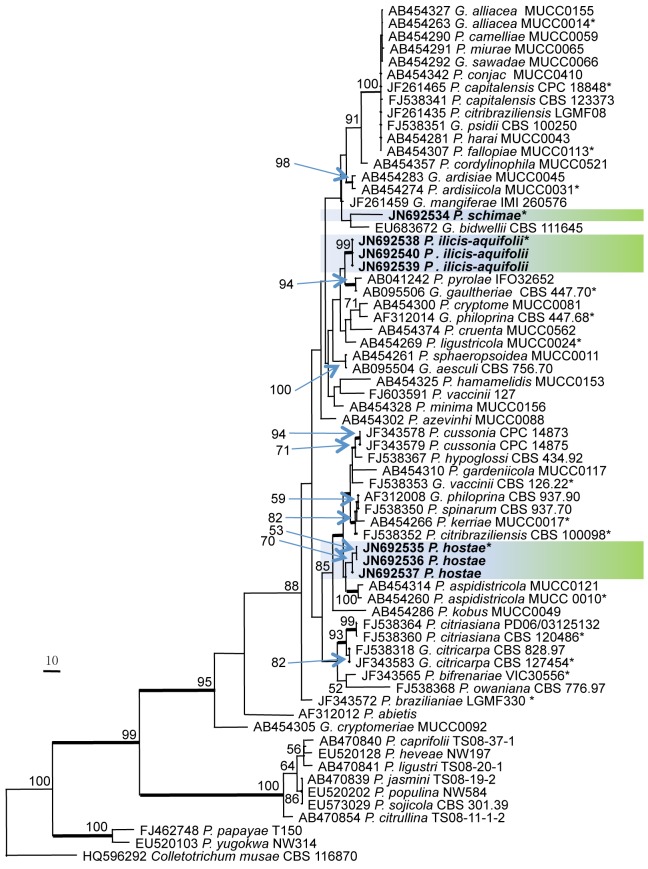
Phylogenetic relationships were inferred using the ITS alignment, and the combined ITS, TEF1, GPDH, and ACT sequence alignment. The 67 ITS sequence dataset from 52 taxa comprised 517 characters after alignment. Of these, 252 characters were parsimony informative, 47 were variable and parsimony-uninformative, and 218 were constant. Parsimony analysis generated two trees, and one of the equally most parsimonious trees with shorter tree length (TL = 935, CI = 0.539, RI = 0.827, RC = 0.446, HI = 0.461) was selected and shown in Fig. 1. For the Bayesian analyses, model (GTR+I+G) was selected in MrModeltest 2.3. The branches with significant Bayesian posterior probability (≥ 95 %) were thickened in the phylogenetic tree. All three species described as new in this manuscript appear in distinct lineages (Fig. 1).
Fig. 1.
Phylogenetic tree generated from a maximum parsimony analysis based on the ITS nrDNA sequence alignment. Values above the branches represent parsimony bootstrap support values (> 50 %). Thickened branches represent significant Bayesian posterior probability values (≥ 95 %). Novel sequences are printed in bold and the scale bar indicates 10 changes. The tree is rooted to Colletotrichum musae. An asterisk (*) indicates the ex-type strains.
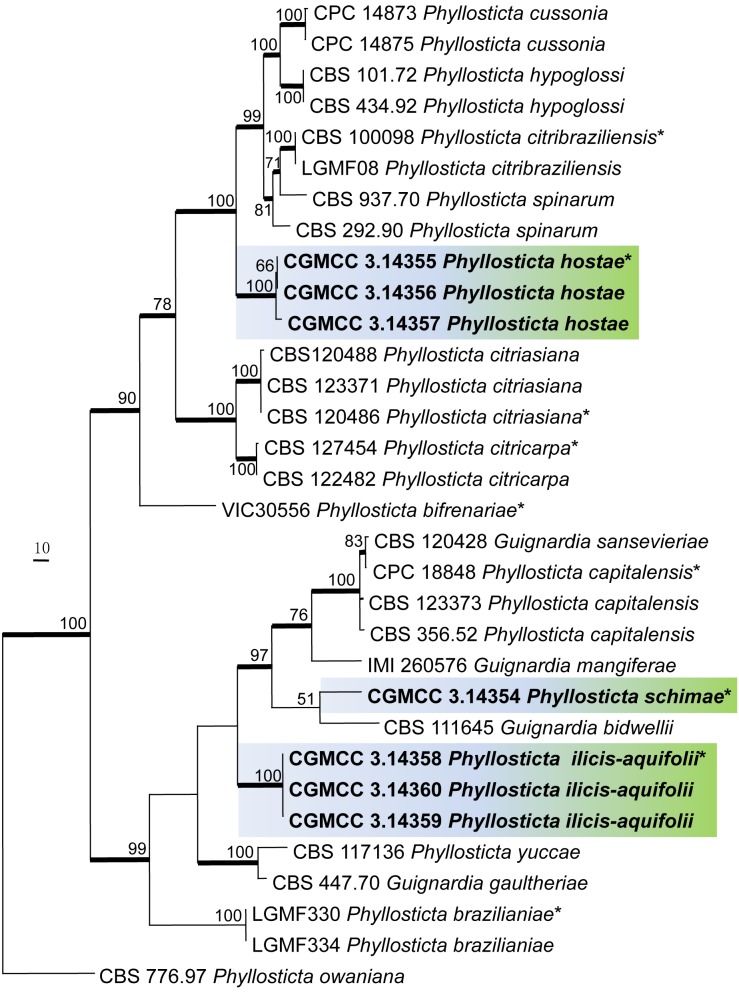
The combined datasets of ITS, TEF1, GPDH, and ACT contained 32 combined sequences from 18 taxa and comprised 1 791 characters after alignment. Of these, 407 characters were parsimony informative; 129 were variable and parsimony-uninformative, and 1 255 were constant. The parsimony analysis generated three equally most parsimonious trees and the tree with shortest tree length (TL = 1051, CI = 0.669, RI = 0.841, RC = 0.562, HI = 0.331) was selected and shown in Fig. 2. For the Bayesian analyses, the best-fit model (GTR+I+G) was selected in MrModeltest 2.3. The branches with significant Bayesian posterior probability (≥ 95 %) were thickened in the phylogenetic tree. Similarly all three species appear in distinct lineages (Fig. 2).
Fig. 2.
Phylogenetic tree generated from a maximum parsimony analysis based on the combined ITS, EF, GPDH, and ACT sequence alignment, showing the phylogenetic relationships of the three new species. Values above the branches represent parsimony bootstrap support values (> 50 %). Thickened branches represent significant Bayesian posterior probability (≥ 95 %). Novel sequences are printed in bold and the scale bar indicates 10 changes. The tree is rooted to Phyllosticta owaniana. An asterisk (*) indicates the ex-type strains.
TAXONOMY
Phyllosticta hostae Y.Y. Su & L. Cai, sp. nov. — MycoBank MB564904; Fig. 3
Fig. 3.
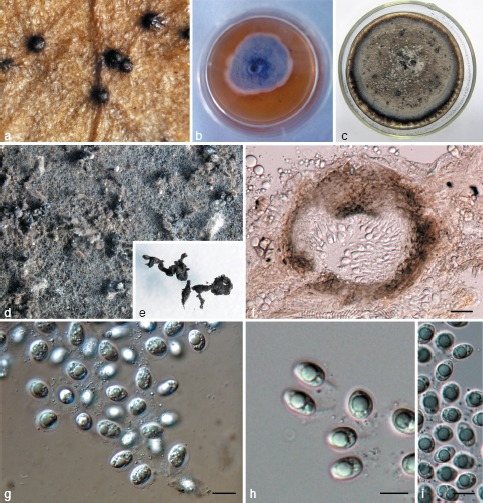
Phyllosticta hostae. a. Appearance of conidiomata on host leaf surface; b. colony on PDA 7 d after inoculation; c. colony on PDA 1 mo after inoculation; d, e. pycnidia forming on PDA; f. vertical section of pycnidium in leaf tissue; g–i. conidia. — Scale bars: f = 20 μm; g–i = 10 μm.
Etymology. Named after its host, Hosta plantaginea.
Leaf spots ellipsoid or circular to somewhat irregular, yellow to pale brown, surrounded by dark brown border. Pycnidia black, subepidermal, globose, 40–150 μm diam. Pycnidial wall composed of depressed or irregular cells in 2–3 layers, brown to dark brown, darker around ostiole, hyaline or pale and flattened towards the inside. Conidiogenous cells 7–22 × 2–5 μm, holoblastic, phialidic, cylindrical, subcylindrical to ampulliform, hyaline, thin-walled, smooth. Conidia 8–15 × 5–9 μm (𝑥̅ = 10.9 ± 1.4 × 7.6 ± 0.8, n = 30), unicellular, thin- and smooth-walled, ellipsoid, subglobose to obovoid, with a large central guttule, truncate at the base when young, later rounded at both ends, enclosed in a 1–3 μm thick mucilaginous sheath, and bearing a hyaline, mucoid apical appendage, 4–8 × 1–3 μm, straight to flexible, unbranched, tapering towards an acute tip.
Culture characteristics — Colonies on PDA flat, surface greenish grey in centre, white-grey at margin when young, becoming leaden-grey in centre, lavender-grey at margin after 2 wk.
Specimens examined. China, Beijing, Botanical Garden, on leaf of Hosta plantaginea, 10 Sept. 2010, L. Cai, HMAS242924 (holotype); ex-type culture CGMCC3.14355; ibid. SYY572, culture CGMCC3.14356; ibid. SYY573, culture CGMCC3.14357.
Notes — Phyllosticta hostae was isolated from Hosta plantaginea (Liliaceae), which is grown as a common ornamental plant in China and many Asian countries. There are several reports of fungal pathogens isolated from H. plantaginea, e.g., Alternaria asphodeli (Zhang 1999), Botrytis cinerea (Zhang 2006), and Colletotrichum omnivorum (Cho & Shin 2004). To date, P. hostae is the only species of Phyllosticta described from the plant genus Hosta. However nine Phyllosticta species are currently known on Liliaceae, i.e. P. aspidistricola, P. cruenta, P. crypta, P. cumminsii, P. hemerocallidis, P. hypoglossi, P. subeffusa, P. uvulariae, and P. yuccae (van der Aa & Vanev 2002, Motohashi et al. 2008). A comparison of their morphological characters with P. hostae is given in Table 2.
Table 2.
Synopsis of characters of Phyllosticta species from Liliaceae.
| Phyllosticta species | Pycnidial size (μm) | Pycnidial wall | Conidiogenous cells (μm) | Conidia (μm) | Sheath diam (μm) | Appendage size (μm) | References |
|---|---|---|---|---|---|---|---|
| P. aspidistricola | 61–118 × 86–110 | – | 7–12.5 × 1.2–2.5 | 9.5–12.5 × 8.5–10 | – | 17–24.5 | Motohashi et al. (2008) |
| P. cruenta | 80–200, mostly 120–160 | 1–4 cells, 7–20 μm thick | 7–12 × 2–4 | 12–21 × 5–10, mostly 16–19 × 8–10 | – | 4–17 × 3 | van der Aa (1973) |
| P. crypta | 70–130 × 45–95 | 4–14 μm thick | 5–12 × 2–3.5 | 5.4–8.9 × 3.8–6.2 | 0.3–1 | 3–8 × 0.5–1 | Bissett (1979) |
| P. cumminsii | 75–140 | 4–19 μm thick | 3.5–14 × 3–6 | 9–19 × 6.7–10.5 | 1–2 | 5–20 × 1.6–4 | Bissett (1979) |
| P. hemerocallidis | 84–139 | – | – | 8–13 × 3–5 (av. 10 × 3.5) | – | 3–10 | van der Aa & Vanev (2002) |
| P. hostae | 40–150 | 2–3 layers | 7–22 × 2–5 | 8–15 × 5–9 (av. 10.9 × 7.6) | 1–3 | 4–8 × 1–3 | Present study |
| P. hypoglossi | 120–250 | 2–4 cells, 12–30 μm thick | 4–10 × 2–3.5 | 8–15(−18) × 6–10 | – | 10, up to 35 | van der Aa (1973) |
| P. subeffusa | 90–150 ×120–140 | 5–16 μm thick | 5–8 × 3–4.5 | 7–13 × 7–10 | – | 5–7, up to 15 | van der Aa (1973) |
| P. uvulariae | 65–120 (usually 90–75) | 5–12 μm thick | 4–13 × 2.5–6 | 5–8.6 × 3.9–6.6 (av. 6.5–5.5) | less than 0.8 | 4–14 × 0.3–0.8 | Bissett (1979) |
| P. yuccae | 90–150 | 3–7 layers, 14–38 μm thick | 5.4–9.8 × 2.7–6 | 7.5–15.4 × 6–9.5 (av. 10–7.3) | 1 | 4–15 | Bissett (1986) |
The phylogenetic tree generated from a multilocus sequence alignment showed that the three strains of P. hostae constituted a distinct lineage with 100 % bootstrap support (Fig. 2). DNA sequence analysis showed that P. hostae was most closely related to P. citribraziliensis, P. cussonia, P. hypoglossi, P. spi-narum, and P. vaccinii (teleomorph Guignardia vaccinii). Of these species, P. vaccinii is morphologically most similar, but the ex-type strain (CBS 126.22) shares only 94 % identity to P. hostae in ITS sequence. Phyllosticta vaccinii was isolated from the leaves of Vaccinium arboretum. The pycnidia of P. vaccinii are larger (80–175 μm vs 40–150 μm) than that of P. hostae, and the conidia are slightly smaller (8–12 × 5–8 μm vs 8–15 × 5–9 μm). In addition, the appendages of P. vaccinii can be up to 17 μm long, while that of P. hostae is less than 8 μm (van der Aa 1973).
Phyllosticta schimae Y.Y. Su & L. Cai, sp. nov. — MycoBank MB564905; Fig. 4
Fig. 4.
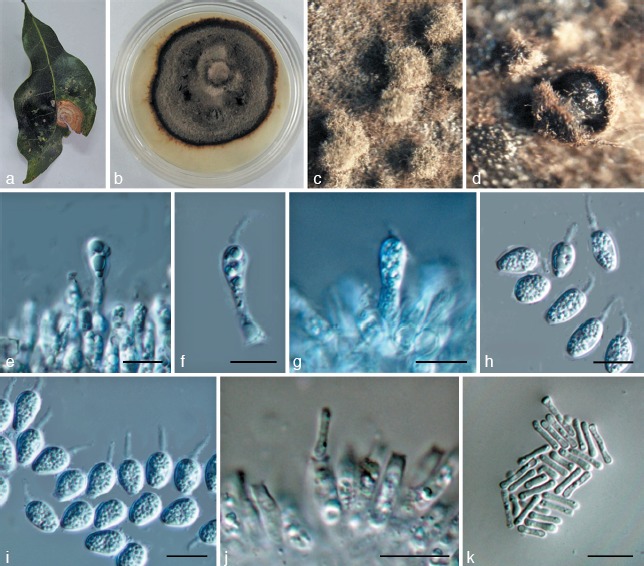
Phyllosticta schimae. a. Symptom on leaf of Schima superba; b. colony on PDA 1 mo after inoculation; c, d. pycnidia forming on PDA; e–g. conidiogenous cells giving rise to conidia; h, i. conidia; j. spermatogenous cells producing spermatia; k. spermatia. — Scale bars: e–k = 10 μm.
Etymology. Named after its host, Schima superba.
Leaf spots circular, somewhat irregular, yellow to pale brown, surrounded by dark brown borders, fruiting bodies not observed. Pycnidia on PDA grey to black, aggregated, superficial to erumpent, globose to ampulliform, 150–200 μm diam. Conidiogenous cells 8–30 × 2–4 μm, holoblastic, phialidic, short cylindrical, subcylindrical to ampulliform, hyaline, thin-walled, smooth. Conidia 7–13 × 4–7 μm (𝑥̅ = 9.5 ± 1.1 × 6.2 ± 0.4, n = 30), unicellular, thin- and smooth-walled, globose, ellipsoid to obovoid, truncate at the base when young, later rounded at both ends, enclosed in a mucilaginous sheath, and bearing a hyaline, mucoid apical appendage, 4–10 × 1–3 μm, straight to flexible, unbranched, tapering towards an acute tip. Spermatogenous cells subcylindrical to ampulliform, 11–25 × 2–4 μm. Spermatia aseptate, dumbbell-shaped, 7–11 × 1–2.5 μm (𝑥̅ = 8.3 ± 1.4 × 1.4 ± 0.3, n = 30).
Culture characteristics — Colonies on PDA flat, brown-black, with moderate aerial mycelium.
Specimens examined. China, Zhejiang, Gutianshan Nature Reserve, on leaf of Schima superba, 18 Aug. 2010, Y.-Y. Su, HMAS242923 (holotype); ex-type culture CGMCC3.14354.
Notes — Schima superba is one of the dominant tree species in evergreen broad leaf subtropical forests in China. Currently there are only two unnamed Phyllosticta species reported from Schima (Theaceae) (Kobayashi 2007). Only two species, P. plurivora and P. theacearum, were recorded in the plant family Theaceae (van der Aa & Vanev 2002), and the former has been considered a synonym of P. theacearum (van der Aa & Vanev 2002). Phyllosticta theacearum produces shorter conidiogenous cells (4–6 μm vs 8–30 μm) than that of P. schimae (van der Aa 1973, van der Aa & Vanev 2002). Phyllosticta schimae appears closely related to P. ampelicida (teleomorph G. bidwellii) (92 % identity in ITS sequence) (Fig. 1, 2), which was isolated from the leaves of Ampelopsis auinquefolia (van der Aa 1973). Morphologically, P. schimae produces larger pycnidia (150–200 μm vs 70–180 μm), and longer conidiogenous cells (8–30 × 2–3 μm vs 6 × 3 μm) than P. ampelicida (van der Aa 1973).
Phyllosticta ilicis-aquifolii Y.Y. Su & L. Cai, sp. nov. — MycoBank MB564906; Fig. 5
Fig. 5.
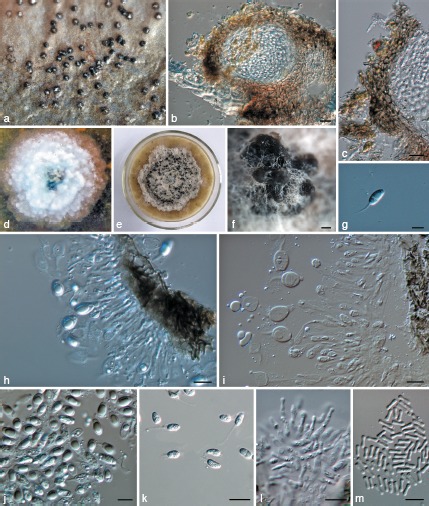
Phyllosticta ilicis-aquifolii. a. Appearance of conidiomata on host leaf surface; b. vertical section of pycnidium in leaf tissue; c. vertical section through the peridium; d. colony on PDA 7 d after inoculation; e. colony on PDA 1 mo after inoculation; f. pycnidia forming on PDA; g. conidium; h, i. conidiogenous cells giving rise to conidia; j, k. conidia; l. spermatogenous cells producing spermatia; m. spermatia. — Scale bars: b, c, k = 20 μm, f = 50 μm, g, h–j, l, m = 10 μm.
Etymology. Named after its host, Ilex aquifolium.
Leaf spots ellipsoid or circular to somewhat irregular, grey to pale brown, about 7 mm diam, surrounded by dark brown border. Pycnidia amphigenous, subepidermal, single, 70–230 μm diam. Pycnidial wall composed of depressed or irregular cells of 2–4 layers, brown to dark brown, darker around ostiole, hyaline or pale and flattened towards the inside. Conidiogenous cells (8−)12–17(−19) × (2−)3–4 μm, holoblastic, phialidic, cylindrical, subcylindrical to ampulliform, hyaline, thin-walled, smooth. Conidia 10–18 × 6–9 μm (𝑥̅ = 13.4 ± 1.8 × 7 ± 0.7, n = 30), unicellular, thin- and smooth-walled, globose, ellipsoid to obovoid, with a large central guttule, truncate at the base when young, later rounded at both ends, enclosed in a thick mucilaginous sheath, 1–3 μm thick, and bearing a hyaline, mucoid apical appendage, (9−)12–17(−30) × 2–3 μm, straight to flexible, unbranched, tapering towards an acute tip. Spermatiogenous cells subcylindrical to ampulliform, 5–17 × 1–4 μm. Spermatia aseptate, dumbbell-shaped, 5–8 × 1.5–2.5 μm (𝑥̅ = 6.7 ± 0.7 × 1.9 ± 0.2, n = 30).
Culture characteristics — Colonies on PDA flat, with irregular margin, surface white-grey when young; leaden-grey in centre, and white-grey at margin after 2 wk.
Specimens examined. UK, England, London, on leaf of Ilex aquifolium, 15 Aug. 2010, L. Cai, HMAS242922 (holotype), ex-type culture CGMCC3.14358; ibid. SYY590, culture CGMCC3.14359; ibid. SYY591, culture CGMCC3.14360. Three duplicate strains were deposited in IMI.
Notes — Phyllosticta ilicis-aquifolii was isolated from the common ornamental and hedge plant Ilex aquifolium. It is characterised by its large conidia that have a long mucoid appendage, which is distinct from most Phyllosticta species. Other Phyllosticta species reported from Aquifoliaceae include P. llimonae and P. concentrica (van der Aa & Vanev 2002). Phyllosticta ilicis-aquifolii differs from P. llimonae in producing shorter conidiogenous cells (12–17 vs 28–32 μm) (Bertault 1982), and from P. concentrica (teleomorph Guignardia philoprina) in producing larger spermatia (6–15 × 1.5–3 μm vs 5–8 × 1.5–2.5 μm) (van der Aa 1973). In addition, the ex-type strains Phyllosticta ilicis-aquifolii and G. philoprina shared 94 % identity in ITS sequence and clustered in different clades in the ITS phylogenetic tree (Fig. 1). Although there are 26 synonyms listed under P. concentrica (http://www.mycobank.org), the conidial sizes in these species descriptions are all smaller (shorter than 12 μm) than that of P. ilicis-aquifolii, except for Sphaeria taxi (20–22 × 10 μm vs 10–18 × 6–9 μm in P. ilicis-aquifolii) and Phoma ilicis (12–15 × 3 μm vs 10–18 × 6–9 μm).
Phyllosticta ilicis-aquifolii appears most closely related to P. gaultheriae (teleomorph G. gaultheriae) (94 % identity in ITS sequence) and P. pyrolae (95 % identity in ITS sequence) (Fig. 1). Morphologically, P. ilicis-aquifolii can be distinguished from these species by its larger conidia (10–18 × 6–9 μm vs 4–9 × 4–7 μm in P. gaultheriae and 4.5–7.5 × 4–9 μm in P. pyrolae) (van der Aa 1973).
DISCUSSION
In this paper we have described and named three new Phyllosticta species based on morphological and molecular characters. Each has morphological characters typical for Phyllosticta, i.e., stromatic conidiomata, holoblastic conidiogenesis, one-celled conidia provided with a surrounding mucoid layer and an apical appendage (van der Aa 1973, van der Aa & Vanev 2002).
Plant pathogenic Phyllosticta species are usually specific to host species or genera (van der Aa 1973, van der Aa & Vanev 2002, Motohashi et al. 2008, 2009, Wikee et al. 2011). Morphological comparisons of Phyllosticta spp. are often made with species reported from congeneric hosts (van der Aa & Vanev 2002, Motohashi et al. 2008, Wulandari et al. 2009, Glienke et al. 2011, Wang et al. 2012). In our study, the new species were compared with other species reported from the same host family and species that are morphologically and phylogenetically closely related. These results showed that the three species were distinct, representing novel taxa.
Jin (2011) reported that the conidial appendages of some Phyllosticta species might disappear with time or elongate when mounted in water. Therefore, fresh cultures were used for morphological observations, and the conidial appendages were not given undue significance in species delimitation. In this study, the morphological comparisons were made mainly based on other characters, e.g., the shape and size of conidia, pycnidia, and conidiogenous cells.
Although the generic concept of Phyllosticta as defined by van der Aa (1973) is extensively accepted, the identification of species is still difficult due to limited morphological characters that can be used for comparison. Recent molecular studies have revealed the ambiguity of taxonomy based on morphological characters and host associations (Wulandari et al. 2009, Glienke et al. 2011). Multilocus phylogenetic analysis has been shown to be more useful in predicting natural species relationships in the genus (Motohashi et al. 2009, Wulandari et al. 2009, Glienke et al. 2011). Traditionally applied phenotypic characters (host, symptom, colony characteristics, and morphology) should therefore be re-evaluated for their taxonomic usefulness in light of phylogenetic relationships (Hyde et al. 2010).
Acknowledgments
This work was financially funded by CAS KSCX2-YWZ-1026 and NSFC 31110103906. Drs Kevin D. Hyde and Roger G. Shivas are thanked for providing valuable comments on this manuscript.
REFERENCES
- Aa HA van der. 1973. Studies in Phyllosticta I. Studies in Mycology 5: 1–110 [Google Scholar]
- Aa HA van der, Vanev S. 2002. A revision of the species described in Phyllosticta. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands [Google Scholar]
- Arzanlou M, Groenewald JZ, Fullerton RA, Abeln ECA, Carlier J, Zapater MF , et al. 2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia 20: 19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baayen RP, Bonants PJM, Verkley G, Carroll GC, Aa HA van der , et al. 2002. Nonpathogenic isolates of the Citrus Black Spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of woody plants, G. mangiferae (Phyllosticta capitalensis). Phytopathology 92: 464–477 [DOI] [PubMed] [Google Scholar]
- Baldassari RB, Wickart E, Goes A de.2008. Pathogenicity, colony morphology and diversity of isolates of Guignardia citricarpa and G. mangiferae isolated from Citrus spp. European Journal of Plant Pathology 120: 103–110 [Google Scholar]
- Bertault R. 1982. Contribution a la flore mycologique de la Catalogne. Acta Botanica Barcinonensia 34: 5–35 [Google Scholar]
- Bissett J. 1979. Coelomycetes on Liliales: the genus Phyllosticta. Canadian Journal of Botany 57: 2082–2095 [Google Scholar]
- Bissett J. 1986. Discochora yuccae sp. nov. with Phyllosticta and Leptodothiorella synanamorphs. Canadian Journal of Botany 64: 1720–1726 [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, Weir B, Waller J, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204 [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556 [Google Scholar]
- Cho WD, Shin HD. (eds). 2004. List of plant diseases in Korea. Fourth edition. Korean Society of Plant Pathology [Google Scholar]
- Choi YW, Hyde KD, Ho WH. 1999. Single spore isolation of fungi. Fungal Diversity 3: 29–38 [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Reeder J, Marasas WFO , et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glienke C, Pereira OL, Stringari D, Fabris J, Kava-Cordeiro V , et al. 2011. Endophytic and pathogenic Phyllosticta species, with reference to those associated with Citrus Black Spot. Persoonia 26: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glienke-Blanco C, Aguilar-Vildoso CI, Vieira MLC, Barroso PAV, Azevedo JL. 2002. Genetic variability in the endophytic fungus Guignardia citricarpa isolated from citrus plants. Genetics and Molecular Biology 25: 251–255 [Google Scholar]
- Guerber JC, Liu B, Correll JC, Johnston PR. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872–895 [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98 [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA , et al. 2011. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. 2001. MrBayes: Bayesian inference of phylogenetic trees. Biometrics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Abd-Elsalam K, Cai L. 2010. Morphology: still essential in a molecular world. Mycotaxon 114: 439–451 [Google Scholar]
- Jin J. 2011. Conidial morphology changes in four Phyllosticta species. Mycotaxon 115: 401–406 [Google Scholar]
- Kobayashi T. 2007. Index of fungi inhabiting woody plants in Japan. Host, distribution and literature. Zenkoku-Noson-KyoikuKyokai Publishing Co., Ltd. [Google Scholar]
- Motohashi K, Araki I, Nakashima C. 2008. Four new species of Phyllosticta, one new species of Pseudocercospora, and one new combination in Passalora from Japan. Mycoscience 49: 138–146 [Google Scholar]
- Motohashi K, Inaba S, Anzai K, Takamatsu S, Nakashima C. 2009. Phylogenetic analyses of Japanese species of Phyllosticta sensu stricto. Mycoscience 50: 291–302 [Google Scholar]
- Myllys L, Stenroos S, Thell A. 2002. New genes for phylogenetic studies of lichenized fungi: glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist 34: 237–246 [Google Scholar]
- Nylander JAA. 2004. MrModeltest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University [Google Scholar]
- Okane I, Lumyong S, Nakagiri A, Ito T. 2003. Extensive host range of an endophytic fungus, Guignardia endophyllicola (anamorph: Phyllosticta capitalensis). Mycoscience 44: 353–363 [Google Scholar]
- Page RDM. 1996. TreeView: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Persoon CH. 1818. Traité sur les Champignons Comestibles, Contenantl’ Indication des Espèces Nuisibles Précédéd’une Introduction à l’Histoire des Champignons. Paris, Belin-Leprieur [Google Scholar]
- Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311 [DOI] [PubMed] [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Common wealth Mycological Institute; Kew, Surrey, UK [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116 [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, USA [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala P, Ravaz L. 1892. Sur la dénomination botanique (Guignardia bidwellii) du black-rot. Bulletin de la Société Mycologique de France 8: 63 [Google Scholar]
- Wang XH, Chen GQ, Huang F, Zhang JZ, Hyde KD, Li HY. 2012. Phyllosticta species associated with citrus diseases in China. Fungal Diversity 52: 209–224 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR Protocols: A guide to methods and applications: 315–322. Academic Press, San Diego, USA [Google Scholar]
- Wikee S, Udayanga D, Crous PW, Chukeatirote K, McKenzie EHC , et al. 2011. Phyllosticta – an overview of current status of species recognition. Fungal Diversity 51: 43–61 [Google Scholar]
- Wingfield MJ, Beer ZW de, Slippers B, Wingfield BD, Groenewald JZ , et al. 2012. One fungus, one name promotes progressive plant pathology. Molecular Plant Pathology doi: 10.1111/J.1364-3703.2011.00768.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulandari NF, To-anun C, Hyde KD, Duong LM, Gruyter J de , et al. 2009. Phyllosticta citriasiana sp. nov., the cause of Citrus tan spot of Citrus maxima in Asia. Fungal Diversity 34: 23–39 [Google Scholar]
- Yip HY. 1989. New species of Phyllosticta. Mycological Research 93: 489– 496 [Google Scholar]
- Zhang TY. 1999. Taxonomic studies of Alternaria from China VIII. New species and new records on Gramineae, Iridiaceae, and Liliaceae. Mycotaxon 72: 443–453 [Google Scholar]
- Zhang TY. 2006. Flora Fungorum Sinicorum. Vol. 26. Botrytis, Ramularia. Science Press, Beijing [Google Scholar]
- Zhaxybayeva O, Gogarten JP. 2002. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. Genomics 3: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]