Mouse RNase H2 is essential to remove ribonucleotides from the genome to prevent DNA damage.
Abstract
Ribonucleases H (RNases H) are endonucleases which cleave the RNA moiety of RNA/DNA hybrids. Their function in mammalian cells is incompletely understood. RNase H2 mutations cause Aicardi-Goutières syndrome, an inflammatory condition clinically overlapping with lupus erythematosus. We show that RNase H2 is essential in mouse embryonic development. RNase H2–deficient cells proliferated slower than control cells and accumulated in G2/M phase due to chronic activation of a DNA damage response associated with an increased frequency of single-strand breaks, increased histone H2AX phosphorylation, and induction of p53 target genes, most prominently the cyclin-dependent kinase inhibitor 1 encoding cell cycle inhibitor p21. RNase H2–deficient cells featured an increased genomic ribonucleotide load, suggesting that unrepaired ribonucleotides trigger the DNA damage response in these cells. Collectively, we show that RNase H2 is essential to remove ribonucleotides from the mammalian genome to prevent DNA damage.
Because of the reactive 2′ hydroxyl group on the ribose ring, ribonucleotides contained in DNA pose a threat to genome stability (Nick McElhinny et al., 2010). High numbers of ribonucleotides were shown to be misincorporated into yeast DNA during replication because ribonucleotide exclusion of replicative polymerases is not absolute (Nick McElhinny et al., 2010). Other sources of ribonucleotides in DNA are Okazaki fragment RNA primers or RNA/DNA hybrids which arise from base pairing of nascent RNA transcripts with their template (Helmrich et al., 2011; Wahba et al., 2011). Ribonuclease H (RNase H) endonuclease activity is defined by specific cleavage of the RNA component of RNA/DNA hybrids (Cerritelli and Crouch, 2009). RNase H2, which accounts for most of the RNase H activity in mammalian cells, recognizes single ribonucleotides in DNA (Cerritelli and Crouch, 2009), suggesting a role in the repair of misincorporated single ribonucleotides in genomic DNA. A PCNA–RNase H2 complex was proposed to scan the genome for ribonucleotides (Bubeck et al., 2011). In addition, RNase H2 may contribute to the removal of Okazaki fragment RNA primers (Turchi et al., 1994). Yeast deficient for RNase H2 was found to display increased genomic ribonucleotide content (Nick McElhinny et al., 2010). Although this phenotype was not associated with reduced proliferation, phosphorylation of Rad53 checkpoint kinase, an accumulation of mutant cells in S phase, and increased sensitivity to replicative stress were observed (Nick McElhinny et al., 2010; Lazzaro et al., 2012). Concomitant abrogation of RNase H1 and 2 in yeast demonstrated that also RNase H1 contributes to ribonucleotide removal (Lazzaro et al., 2012). RNase H–deficient yeast can replicate their ribonucleotide-containing genomes by means of postreplication repair pathways (Lazzaro et al., 2012). RNase H2 deficiency in yeast was demonstrated to result in a mutator phenotype (Nick McElhinny et al., 2010), suggesting that ribonucleotides in DNA are mutagenic if not repaired by RNase H2. In the absence of RNase H2, topoisomerase I (topo I) processes misincorporated ribonucleotides and is responsible for part of the increased mutation rate (Kim et al., 2011).
Mutations in RNase H2 cause Aicardi-Goutières syndrome (AGS), a pediatric inflammatory disorder resembling intrauterine viral infection (Crow et al., 2006b). The condition shows clinical overlap with systemic lupus erythematosus (SLE) and, like SLE, is characterized by an uncontrolled type I IFN response. AGS is also caused by mutations in TREX1 (three prime repair exonuclease 1; Crow et al., 2006a) and SAMHD1 (Rice et al., 2009) encoding an intracellular 3′-5′ exonuclease and a deoxynucleoside triphosphate triphosphohydrolase, respectively. Mutations of Trex1 are associated with human lupus (Lee-Kirsch et al., 2007), and Trex1 deficiency results in type I IFN–dependent multi-organ inflammation in mice (Morita et al., 2004; Stetson et al., 2008; Gall et al., 2012). This led to a concept of autoimmunity caused by defective degradation of intracellular nucleic acids and their sensing by innate receptors. In this study, we describe that loss of RNase H2 results in increased numbers of ribonucleotides in genomic DNA, spontaneous DNA breaks, and activation of a DNA damage response in mouse cells.
RESULTS AND DISCUSSION
Early embryonic lethality in Rnaseh2c−/− mice
All three subunits of yeast and human RNase H2 are required for enzymatic activity in vitro (Cerritelli and Crouch, 2009) and mutations in any of the three human subunits give rise to AGS. Biallelic null mutations have not been described for any of the three Rnaseh2 genes and presumably result in early lethality. We generated Rnaseh2c−/− mice carrying a deletion of the entire coding region (Fig. 1), likely resulting in complete loss of RNase H2 activity. Homozygous embryos were small at embryonic day 9.5 (E9.5) and were not detected at later time points (not depicted), whereas heterozygous neonates were obtained in expected numbers (Fig. 1). Embryos with presumably complete abrogation of RNase H2 activity were also generated by crossing a conditional Rnaseh2b mouse line (see Materials and methods) to a general Cre-deleter strain (Lallemand et al., 1998) and displayed lethality similar to Rnaseh2c−/− embryos (unpublished data). Thus, RNase H2 activity is essential in mouse embryonic development.
Figure 1.
Rnaseh2c−/− mice die in utero around day E9.5. (A) Targeting strategy for the generation of Rnaseh2c−/− mice. Probe, position of the probe for Southern blot analysis. A, ApaLI cleavage site; HR, homologous recombination; DT, diphtheria toxin; NEO, neomycin/G418 resistance cassette. (B) Southern blot of a targeted ES cell clone. (C) Weaned offspring from heterozygous (Rnaseh2c−/WT) breedings. (D) Embryos from heterozygous (Rnaseh2c−/WT) breedings at E9.5.
Perinatal lethality in mice with reduced RNase H2B expression
In addition to Rnaseh2c−/− mice, a mouse line carrying a gene trap cassette inserted into Rnaseh2b intron 4/5 (Rnaseh2b knockout first [KOF] mice) was analyzed. The gene trap cassette is expected to be spliced to exon 4 and terminate translation (Testa et al., 2004). Mice homozygous for the KOF allele were underrepresented at birth (Fig. 2), were significantly smaller compared with littermate controls, and died perinatally (Fig. 2). Already at E14.5, homozygous embryos were small but present at Mendelian ratios (Fig. 2). Histological analysis revealed proportionally small organs. Many tissues were edematous and displayed extended vessels, as well as bleedings, suggesting defects in vessel wall architecture (Fig. 2). FACS analysis of E18.5 liver and thymus revealed a massive reduction of cellularity (unpublished data). Because the gene trap cassette may allow normal mRNA splicing to some degree, we analyzed E14.5 fetal liver (not depicted) or fibroblasts for residual normal transcripts and found them reduced to a small percentage compared with control levels (Fig. 2). Correspondingly, RNase H2 activity was reduced to 6% of normal values in extracts from E14.5 fibroblasts (Fig. 2) and liver (not depicted). Fibroblasts from two additional Rnaseh2bKOF/KOF and control embryos yielded similar results (unpublished data). Thus, the Rnaseh2bKOF mutation represents a hypomorphic allele with low residual enzyme activity.
Figure 2.
Phenotype of Rnaseh2bKOF mice with low levels of residual RNase H2 activity. (A) Control and stillborn homozygous Rnaseh2bKOF/KOF neonate. (B) Neonates from heterozygous (Rnaseh2bKOF/WT) breedings. *, all homozygous neonates were dead upon first inspection. (C) E14.5 embryos from heterozygous (Rnaseh2bKOF/WT) breedings. (D) Histology of E14.5 Rnaseh2bKOF/KOF embryos and littermate controls. Left and middle, H&E-stained section of E14.5 Rnaseh2bKOF/KOF and control skin. Note massive edema in mutant skin. Right, H&E-stained section of E14.5 Rnaseh2bKOF/KOF tongue. Note the extravasated erythrocytes (arrow) in the muscle tissue. Similar bleedings were detected in numerous tissues of most embryos. (E) Quantification of WT RNaseh2b transcripts in fibroblasts from Rnaseh2bKOF/KOF and control E14.5 embryos. Mean ± SD of three pairwise comparisons are displayed. (F) RNase H2 activity in E14.5 fibroblasts. Cleavage of an 18-bp dsDNA substrate containing a single ribonucleotide was measured and activity was calculated from the slope of the graphs (means, represented as percentage of WT).
RNase H2 deficiency impairs proliferation and triggers a DNA damage response
Rnaseh2bKOF/KOF E14.5 fibroblasts proliferated slower than control cultures (Fig. 3) and this defect became more pronounced with increasing passage numbers. Reduced proliferation of these cultures was not a result of enhanced apoptosis because fractions of propidium iodide+ and annexin V+ cells were not increased (unpublished data). DNA content analysis revealed mildly increased numbers of cells in S and G2/M phase in (asynchronous) 1-wk-old mutant versus control cultures (Fig. 3). The fraction of mutant cells in G2/M markedly increased with the duration of the culture. In E18.5 fibroblasts, the accumulation of cells in G2/M was striking already after 1 wk of culture, and 4-wk-old E18.5 fibroblast cultures contained 70–80% of cells in G2/M (Fig. 3). This finding suggested that the problem caused by RNase H2 deficiency exacerbates with increasing numbers of divisions.
Figure 3.
RNase H2–deficient cells show impaired proliferation and accumulate in G2/M. (A) 3H-thymidine incorporation by E14.5 Rnaseh2bKOF/KOF (KOF) versus Rnaseh2bKOF/WT or RnaseH2bWT/WT control (Ctrl) fibroblasts. (B) DNA content analysis. Fibroblasts from E14.5 Rnaseh2bKOF/KOF (n = 10) or control (WT or heterozygous, n = 56) embryos as well as fibroblasts from E18.5 Rnaseh2bKOF/KOF (n = 2) or control (WT or heterozygous, n = 5) embryos were analyzed 1 or 4 wk after initiation of the cultures. One representative result is displayed. (C) Lethally irradiated recipients were transplanted with 4 × 106 fetal liver cells from E14.5 Rnaseh2bKOF/KOF (n = 3) or WT embryos (n = 3). Chimerism was quantified by detection of the congenic CD45.1/2 marker on CD11c+Gr1+ neutrophilic granulocytes. (D) RNase H2 activity in sorted splenic B220+CD19+ B cells from Rnaseh2bFLOX/FLOX CD19-Cre+ mice (n = 2) and controls (WT/FLOX Cre+, n = 1; and WT/FLOX Cre-negative, n = 2). Purity of the sorted B cells was 96.5–97.7%. (E) Numbers of splenic CD19+B220+ B cells in adult Rnaseh2bFLOX/FLOX CD19-Cre+ (KO, n = 6) and controls (Ctrl, FLOX/FLOX Cre− or FLOX/WT CRE+, n = 6). (F) 3H-thymidine incorporation by Rnaseh2bFLOX/FLOX CD19-Cre+ (n = 9) and control (Ctrl, FLOX/FLOX Cre− or FLOX/WT CRE+, n = 9) B cells. Total splenocytes were analyzed after 3 d of culture in the presence of LPS. *, P < 0.05 (two-tailed Student’s t test). (G) DNA content analysis. Splenic B cells from Rnaseh2bFLOX/FLOX CD19-Cre+ (Δ/Δ, n = 6) and control mice (Ctrl, FLOX/FLOX Cre− or FLOX/WT CRE+, n = 6) were analyzed after 3 d in vitro LPS stimulation. One representative result is displayed. Error bars indicate ±SD in all graphs.
To test the proliferative capacity of mutant hematopoietic cells, we transferred E14.5 Rnaseh2bKOF/KOF liver cells into lethally irradiated recipients. Transfer of a standard radioprotective number of liver cells did not result in stable chimerism (unpublished data), whereas transfer of 4 × 106 Rnaseh2bKOF/KOF cells (20× radioprotective dose) resulted in long-term rescue with chimerism close to 100% (Fig. 3). In these chimeric animals, leukocyte numbers tended to be lower compared with recipients of control cells but were in the normal range (unpublished data). Red blood cell counts were not different between the groups (unpublished data). Thus, reduced RNase H2 activity impairs cell cycle progression but does not preclude substantial proliferative expansion of hematopoietic cells.
To bypass lethality of complete RNase H2 deficiency, we generated Rnaseh2bFLOX mice by Flpe-mediated deletion of the gene trap cassette of the Rnaseh2bKOF allele to create conditional Rnaseh2b knockout mice. Rnaseh2bFLOX mice were crossed to CD19-Cre mice (Rickert et al., 1997) for B cell–specific gene inactivation. (Cre-mediated deletion of exon5 introduces a frame shift.) B cell numbers of Rnaseh2bFLOX/FLOXCD19-Cre+ mice were normal in spleen (Fig. 3). Overgrowth of the B cell compartment by cells that had escaped Cre-mediated gene inactivation was excluded by analysis of protein extracts from sorted B cells for RNase H2 activity (Fig. 3). Rnaseh2bFLOX/FLOX CD19-Cre+ B cells featured a reduction of activity to 5.6 ± 2.1% (n = 2). Post-sort reanalysis determined the purity of the sorted B cells to range from 96.5–97.7% (unpublished data), indicating that about half of the residual RNase H2 activity was accounted for by contaminating non–B cells. Thus, most Rnaseh2bFLOX/FLOXCD19-Cre+ B cells were completely devoid of RNase H2 activity. CD19-Cre–driven recombination commences in B cell development and is ongoing at the pre–B cell stage (Rickert et al., 1997). Our findings indicate that complete deficiency for RNase H2 in precursor B cells does not preclude a limited number of divisions required to fill up peripheral B cell compartments. Proliferation of LPS-stimulated Rnaseh2bFLOX/FLOXCD19-Cre+ B cell cultures was reduced to 80% compared with controls (Fig. 3) with a moderate accumulation of mutant B cells in G2/M (Fig. 3). Collectively, RNase H2 deficiency impairs proliferation but does not preclude considerable proliferative expansion in vivo.
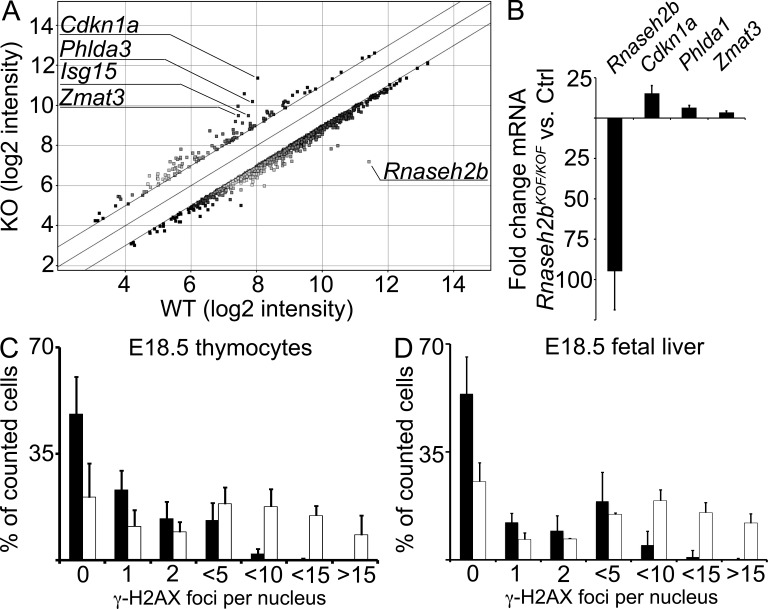
Microarray-based gene expression profiling of Rnaseh2bKOF/KOF versus WT E14.5 liver cells revealed a striking up-regulation of p53-inducible genes in Rnaseh2bKOF/KOF versus control E14.5 liver cells (GEO database accession no. GSE36687). The top seven genes up-regulated in mutant versus control included four p53 target genes, with Cdkn1a, encoding cyclin-dependent kinase inhibitor 1 (p21), being the gene with the most prominent up-regulation of all differentially regulated genes (Fig. 4). Enhanced transcription of these genes was validated by quantitative (q) RT-PCR (Fig. 4). A large number of genes involved in mitosis showed reduced transcript levels in the mutant samples (unpublished data). Our results suggested that RNase H2–deficient cells feature cell cycle arrest mediated via p53-induced p21 expression. To detect DNA damage that triggered p53 activation, we stained E18.5 Rnaseh2bKOF/KOF thymocytes and liver cells for phosphorylated histone H2AX (γ-H2AX), which is induced in chromatin flanking double-strand breaks, and found significantly increased numbers of γ-H2AX foci per nucleus in mutant cells (Fig. 4). In addition to enhanced H2AX phosphorylation, we detected increased numbers of spontaneous single-strand breaks in DNA from embryos with complete RNase H2 deficiency (see next section). Collectively, our data indicate that RNase H2–deficient cells spontaneously activate a DNA damage response.
Figure 4.
Enhanced transcription of p53-inducible genes and DNA damage in RNase H2B–deficient cells. (A) Comparison of gene expression profiles in liver cells from Rnaseh2bKOF/KOF and WT control E14.5 embryos (n = 3 both groups) by microarray analysis. The graph shows genes with more than twofold difference in transcript levels between the groups. (B) Validation of mRNA levels in E14.5 fetal liver by qRT-PCR for the Rnaseh2b and three p53-inducible genes. Results of three pairwise comparisons of Rnaseh2bKOF/KOF and WT control samples are displayed. (C and D) Rnaseh2bKOF/KOF (n = 6, white bars) or control (WT/WT or KOF/WT, n = 11, black bars) E18.5 thymocytes (C) and Rnaseh2bKOF/KOF (n = 4, white bars) or control (WT/WT or KOF/WT, n = 9, black bars) E18.5 liver cells (D) were immunostained for phosphorylated histone H2AX (γ-H2AX). 400 nuclei per individual thymus or liver were evaluated in a blinded fashion. Error bars indicate ±SD in all graphs.
Similar to our findings in RNase H2–deficient cells, compromised proliferation, spontaneous DNA damage, and chronic checkpoint activation were also reported for a Trex1−/− mouse embryonic fibroblast line and fibroblasts from AGS patients with Trex1 mutations by Yang et al. (2007). Trex1−/− cells featured p21 expression, a defective G2/M cell cycle checkpoint, and a block in G1/S transition (Yang et al., 2007). These findings, however, were not reproduced by Stetson et al. (2008), who did not find impaired proliferation or evidence of a DNA damage response in primary embryonic fibroblasts from Trex1−/− mice. In our primary RNase H2–deficient cells, the G2/M checkpoint seemed to be intact and may be responsible for the observed p21-mediated arrest. Although p21 is well known to block G1/S transition, p21 was also shown to arrest cells in G2 by inhibition of CDK1 (Bunz et al., 1998).
RNase H2 deficiency results in an increased genomic ribonucleotide load
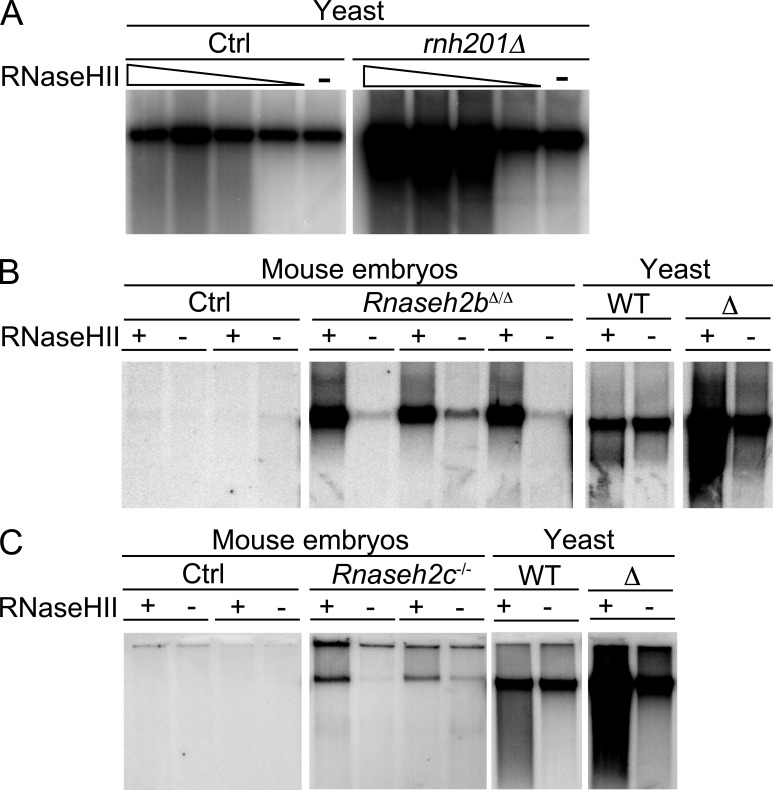
DNA from RNase H2–deficient yeast displays enhanced sensitivity to alkaline hydrolysis, indicating an increased genomic ribonucleotide content (Nick McElhinny et al., 2010). To test whether RNase H2 deficiency in mammalian cells is associated with increased numbers of ribonucleotides in DNA, we used bacterial RNase HII to introduce nicks into chromosomal DNA at positions of ribonucleotides. DNA was purified using standard procedures and contains few double-strand breaks as a result of shear stress, resulting in large fragments in the range of several hundred kilobases. Nicks were detected by DNA polymerase I–dependent nick translation incorporating label into the DNA, which was then run on an agarose gel and visualized by autoradiography. Assuming equal mean length of the single strand polymerized de novo by pol I in mutant versus WT DNA and loading of equal amounts of DNA per lane (determined photometrically for each sample), incorporation of label correlates with numbers of nicks present in the genomic DNA. We found massive RNase HII–dependent incorporation of label into DNA from RNase H2–deficient (rnh201Δ; Nick McElhinny et al., 2010) but not WT (Fig. 5) yeast, confirming that the increased sensitivity of mutant yeast DNA to alkaline hydrolysis (Nick McElhinny et al., 2010) was caused by the presence of increased numbers of ribonucleotides. Likewise, DNA from E9.5 Rnaseh2bΔ/Δ (Fig. 5) and Rnaseh2c−/− (Fig. 5) embryos displayed RNase HII–dependent incorporation of label, indicating increased numbers of ribonucleotides. Similar results were obtained for two additional Rnaseh2bΔ/Δ and four additional Rnaseh2c−/− embryos (unpublished data). Importantly, for all 11 mutant embryos tested, the assay consistently revealed increased incorporation of label compared with DNA from control embryos also in those samples without pretreatment with bacterial RNase HII (Fig. 5 and not depicted), demonstrating that DNA from RNase H2–deficient cells contains higher numbers of spontaneous single-strand breaks as compared with control DNA. The increased genomic ribonucleotide load in RNase H2–deficient mouse cells demonstrates that, like the yeast enzyme, mammalian RNase H2 also functions to remove ribonucleotides from genomic DNA. Our assay did not discriminate whether the accumulating ribonucleotides in RNase H2–deficient DNA represented single nucleotides misincorporated during replication, incompletely removed Okazaki primers, or R-loop structures. Because RNase H1—which cannot cleave at single ribonucleotides but requires at least four subsequent ribonucleotides (Cerritelli and Crouch, 2009)—did not compensate for the loss of RNase H2, it appears likely that the accumulating ribonucleotides in RNase H2–deficient mouse cells are single nucleotides misincorporated by DNA polymerases.
Figure 5.
Increased numbers of ribonucleotides in genomic DNA of RNase H2–deficient embryos. Large chromosomal fragments obtained by standard DNA purification were nicked by bacterial RNase HII at ribonucleotide positions, followed by DNA polymerase I–dependent nick translation in the presence of 32P-dCTP. Labeled DNA was run on a 1% agarose gel (focusing the large chromosomal fragments [several hundred kilobases] in a distinct band) and visualized by autoradiography. Control samples were not digested with RNase HII to detect background single strand breaks. (A) DNA from RNase H2–deficient (rnh201Δ) or control yeast (Ctrl) was digested with increasing amounts (1–10 mU/µl) of RNase HII or subjected to nick translation without RNase HII pretreatment. (B) 200 ng DNA from WT (Ctrl) and Rnaseh2bΔ/Δ embryos (obtained by crossing Rnaseh2bFLOX mice with PGK-Cre mice) was digested with 10 mU/µl RNase HII before nick translation. Negative control samples from each embryo were not pretreated with RNase HII. DNA from RNase H2–proficient (WT) and –deficient (Δ) yeast served as controls. (C) Experiment as in B performed with DNA from Rnaseh2c−/− and WT control embryos. All panels in A, B, and C originate from the same exposure of one single gel each.
Our data strongly suggest that defective ribonucleotide repair triggers the spontaneous damage response of mutant cells. Breaks in RNase H2–deficient DNA may in part reflect lower chemical stability of the ribonucleotide-containing genome. In addition, topoisomerase-mediated cleavage of ribonucleotide-containing DNA could explain the higher frequency of strand breaks in RNase H2–deficient cells. Topo I was reported to cleave double-stranded DNA substrates containing single ribonucleotides resulting in a single-strand break with one 2′,3′-cyclic phosphate end (Sekiguchi and Shuman., 1997; Kim et al., 2011). The damage response could also be a result of collisions of widespread topo I cleavage complexes with replication forks resulting in replication-mediated double-strand breaks (Furuta et al., 2003) or to compromised resolution of RNA/DNA hybrids arising from spontaneous errors of RNA biogenesis. Such R-loop structures represent substrates for RNase H activity in yeast and their persistence was demonstrated to predispose to double-strand break formation (Wahba et al., 2011; Stirling et al., 2012).
The relevance of the spontaneous damage response we found in RNase H2–deficient mouse cells for the pathogenesis of AGS is unclear. Genome instability and increased incidence of neoplastic disease were not reported in RNase H2–associated AGS. Embryonic lethality, spontaneous DNA damage response, and increased genomic ribonucleotide load were also described in another study of RNase H2 mutant mice (Reijns et al., 2012).
Lethal phenotype of RNase H2–deficient embryos is independent of type I IFN
Deficiency for Trex1 causes AGS and/or Lupus, both of which are inflammatory conditions characterized by a chronic type I IFN response (Crow, 2011). Trex1−/− mice develop type I IFN and lymphocyte-dependent autoimmune multi-organ inflammation (Morita et al., 2004; Stetson et al., 2008; Gall et al., 2012). The IFN response initiated in utero before E14 (Gall et al., 2012). Similar to Trex1 mutations, defects in RNase H2 also cause AGS. We therefore expected spontaneous IFN-mediated pathology in our RNase H2–deficient mice. However, crossing the RNaseh2bKOF strain to type I IFN receptor-deficient (IFNAR1−/−) mice (Müller et al., 1994) did not result in a rescue of perinatal lethality (unpublished data). Both RNaseh2bKOF/KOF IFNAR1−/− E17.5 embryos obtained so far featured similar growth retardation as RNaseh2bKOF/KOF animals with intact type I IFN system and living RNaseh2bKOF/KOF IFNAR1−/− neonates were not found. In accordance with this finding, gene expression profiling of RNaseh2bKOF/KOF E14.5 liver cells (Fig. 4) did not reveal a clear signature of up-regulated IFN-responsive genes (unpublished data). Few IFN-inducible genes were found up-regulated; however, several of these are also responsive to p53 and may not reflect type I IFN effects in our mice. Inflammatory changes were not found upon detailed histological analysis of serial sections of whole mount E14.5, E18.5, or stillborn RNaseh2bKOF/KOF embryos (unpublished data). A cross to RAG1−/− mice did not rescue the pathology of RNaseh2bKOF/KOF animals (unpublished data), indicating that lymphocytes have no role in the lethal phenotype. We speculate that IFN-mediated autoimmune pathology might commence at later stages of ontogeny in mouse models of less severe or conditional RNase H2 deficiency which bypass early lethality. A milder reduction of RNase H2 activity in AGS patients compared with our mouse models may explain that a DNA damage–related phenotype dominates in the latter but not in RNase H2–associated AGS. Whether a low-level chronic DNA damage response contributes to the pathogenesis of AGS and related human disorders remains an open question.
MATERIALS AND METHODS
Mice.
Rnaseh2c−/− mice were generated by homologous recombination in JM8.F6 (C57BL/6) cells. For targeted deletion of all four exons of the Rnaseh2c gene, upstream and downstream homology regions (1,800 and 900 bp, respectively) were amplified from C57BL/6 genomic DNA (primers: 5′HR up, 5′-GGACTTCTAGATGGTACTTTGCAAGCAGCCCCTGGAGGAGA-3′; 5′HR down, 5′-CGTACCGGATCCGTGATGAGGATTGGACTCAGGGCCTTGGAC-3′; 3′HR up, 5′-TATGGTGTCCTAGGCTTTAGCTAGTCGCAGTCATTAAGCTTCTTTGGTCCACGTGTTCAGAGCGGACCTGG-3′; and 3′HR down, 5′-CCTAATGAATTGGCCTGTAGCTGGTAGTCTATCCATCTCGAGACTCCGCCTCCTGTGCTCTATTGCTTTAGG-3′) and cloned into the vector pBluescript (using restriction sites contained in the primers) to flank a PGK-gb2-neomycin resistance cassette, which had been inserted into the multiple cloning site. For negative selection against random integration of the targeting vector, a diphtheria toxin cassette was inserted downstream of the 3′ homology region. The linearized vector was transfected into embryonic stem (ES) cells and recombinant clones were detected by Southern blot analysis of ApaLI-digested DNA. The probe was amplified from BAC RP23-156A18 (BACPAC Resources Center; primers: H2C probe up, 5′-CTGAAACGAGATGGAGTGACCATGAGGAC-3′; and H2C probe down, 5′-GTCCTAATAGGGACAGGAAATTGCTCTG-3′) and labeled fragments of 9.3 and 5.7 kb in WT and targeted locus, respectively. Positive clones were injected into albino C57BL/6 foster mothers. Chimeric animals were crossed to C57BL/6 mice and offspring was genotyped by PCR (primers: H2c wt forward, 5′-CACAGATCCCTTCTCACCACTTGGA-3′; H2c neo forward, 5′-CTGCTTGCCGAATATCATGGTGGAA-3′; and H2c reverse, 5′-CCGAAGAACAGACCCAAGAGGAGAA-3′) to detect germline transmission of the mutation.
Heterozygous Rnaseh2bKOF mice (C57BL/6) were purchased from the International Knockout Mouse consortium and were genotyped by PCR as recommended by the vendor. For generation of the conditional Rnaseh2bFLOX mouse line, Rnaseh2bKOF mice were bred to the Flpe-deleter mouse strain (Rodríguez et al., 2000) and successful deletion of the frt-flanked gene trap cassette was demonstrated by PCR. All mouse strains were on the C57BL/6 background. Mice were housed at the Experimental Centre at University of Technology Dresden, Medical Faculty Carl-Gustav Carus, under specific pathogen-free conditions. All procedures were in accordance with institutional guidelines on animal welfare and were approved by the Landesdirektion Dresden.
Cell culture.
Upon cesarean section, E14.5 or 18.5 mouse embryos were decapitated and internal organs were removed. Tissue was minced, digested with 1× trypsin (2.5%; Invitrogen) for 15 min at 37°C, and disaggregated by pipetting. The cell suspension was seeded into 10-cm Petri dishes in DME (Gibco) containing 10% fetal bovine serum (Biochrom), 100 U/ml penicillin and 100 µg/ml streptomycin (Biochrom), 1× nonessential amino acids (Biochrom), and 100 µM β-mercaptoethanol (Sigma-Aldrich).
Cell cycle analysis.
Embryonic fibroblasts were seeded into a 6-well plate (5 × 105 cells/well) and cultivated for 3 d. Cells were harvested, washed once with PBS, fixed in 70% ice-cold ethanol, and chilled for at least 12 h at −20°C, followed by washing in PBS and resuspension and 10-min incubation at room temperature in 0.5 µg/ml RNase A (Sigma-Aldrich) and 2.5 µg/ml propidium iodide (Sigma-Aldrich). Cells were analyzed on a MACSQuant flow cytometer (Miltenyi Biotec).
3H-thymidine incorporation.
Embryonic fibroblasts were seeded into flat-bottom 96-well plates (5 × 103 cells/well) and cultivated for 48 h in complete DME (see Cell culture). 3.7 × 104 Bq 3H-thymidine (HARTMANN ANALYTICS) was added to each well, and after a further 48 h of culture, cells were harvested and washed. Incorporation of 3H-thymidine was determined using a 1450 MicroBeta TriLux scintillation counter (Perkin Elmer).
B cell in vitro stimulation.
Total splenocytes were cultivated in 6-well plates at a density of 3 × 105 cells/ml in RPMI 1640 supplemented with 10% fetal bovine serum (Biochrom) and 100 U/ml penicillin and 100 µg/ml streptomycin, 2 mM l-alanyl-l-glutamine (Biochrom), 10 µM β-mercaptoethanol, and 25 µg/ml lipopolysaccharide from Salmonella minnesota Re 595 (Sigma-Aldrich). For DNA content analysis, cells were harvested after 3 d. For proliferation assays, 3.7 × 104 Bq 3H-thymidine per well was added after 48 h and the cells were incubated for an additional 24 h, followed by harvesting and quantification of 3H-thymidine incorporation.
Quantitative PCR.
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. For qRT-PCR, RNA was digested with DNaseI (New England Biolabs, Inc.) and purified using the RNeasy Mini kit (QIAGEN). 200 ng–1 µg RNA was reverse transcribed using a poly(dT) primer of the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations. 1 µl of the cDNA product served as template in the subsequent qRT-PCR using the Maxima SYBR green/ROX qPCR Master Mix (Thermo Fisher Scientific) using 0.6 µM target specific primers, initial denaturation at 95°C for 15 min, 40 cycles of denaturation (15 s at 95°C), annealing (30 s at 58°C), and elongation (30 s at 72°C). Incorporation of SYBR green was analyzed using the Mx3005P QPCR system (Agilent Technologies) and the MxPro QPCR Software. RT-PCR primers were as follows: p21 up, 5′-GCAATCCACAGCGATATCC-3′; p21 down, 5′-CAACTGCTCACTGTCCACGG-3′ (Kanatsu-Shinohara et al., 2010); Zmat3 up, 5′-GAGTCACTCATTCTCGGACTCCGC-3′; Zmat3 down, 5′-GAGCGGGCATTGAAGTAAGGGC-3′; Phlda3 up, 5′-TTCGCCCGCATCAAAGCCGT-3′; Phlda3 down, 5′-AGGGGGCAGCGGAAGTCGAT-3′ (Brady et al., 2011); Rnaseh2b up, 5′-AGGTTTCCAGGGACAAGGAAGAGGA-3′; Rnaseh2b down, 5′-GTCAATGAAGCTGGAGGTTCTGGAAG-3′; β-actin up, 5′-TGACCCAGATCATGTTTGAGACCTTCA-3′; and β-actin down, 5′-GGAGTCCATCACAATGCCTGTGG-3′. Measured transcript levels were normalized to β-actin. All samples were run in triplicate.
Microarray analysis of fetal liver of RNaseH2b KOF mice.
Fetal livers of E14.5 embryos were homogenized using the gentleMACS Dissociator (Miltenyi Biotec). RNA was isolated using the acid guanidinium thiocyanate-phenol-chloroform method (Chomczynski and Sacchi, 2006). RNA quality was determined on a Bioanalyzer instrument (Agilent Technologies). The RNA was reverse transcribed, amplified, and biotin labeled using the Message Amp II-Biotin Enhanced kit (Ambion). Hybridization, staining, and scanning of the GeneChip Mouse Genome 430 2.0 microarrays were performed according to the GeneChip Expression Analysis Technical Manual P/N 702232 Rev. 3 (Affymetrix). Microarray data were analyzed using GeneSpring GX software. Deregulated genes of interest were validated by qRT-PCR. All microarray data were deposited in the GEO database (accession no. GSE36687).
Nick translation assay for ribonucleotide content of genomic DNA.
200 ng mouse or yeast genomic DNA (quantified using an ND-1000; Nanodrop Technologies) was digested with 0.5 U E. coli RNase HII (New England Biolabs, Inc.) in a total volume of 50 µl and the recommended buffer at 37°C for 2.5 h, precipitated in 0.3 M sodium acetate, pH 5.2, and ethanol. DNA was dissolved in water and nick translation reagents were added to final concentrations of 1× buffer 2 (New England Biolabs, Inc.) and 20 µM unlabeled dATP, dGTP, and dTTP (Thermo Fisher Scientific). 5 U E. coli DNA polymerase I (New England Biolabs, Inc.) and 3.7 × 105 Bq 32P-dCTP (Hartmann Analytics) were added, and the reaction volume was adjusted to 20 µl. The reaction was incubated at 16°C for 35 min. Labeled high molecular weight DNA was separated from unincorporated nucleotides by electrophoresis on a 1% TAE agarose gel and visualized by autoradiography (PhosphorImager SI; Molecular Dynamics) of dried gels (GD 2000 Vacuum Gel Dryer system; GE Healthcare).
Generation of chimeric mice.
Fetal liver cells were isolated from E14.5 embryos and homogenized by repeated aspiration through an 18G needle followed by aspiration through a 20G needle. Cell numbers were determined without lysis of red blood cells. Lethally irradiated (7 Gy) CD45.1/2 congenic recipient mice were reconstituted with 2.5 × 105 or 4 × 106 total fetal liver cells (1.25 or 20× radioprotective dose, respectively) by retro-orbital intravenous injection. Chimerism was assessed in CD11b+Gr1+ neutrophilic granulocytes by flow cytometry FACSCalibur (BD) at various time points starting at week 4 after transfer.
Staining for phosphorylated histone H2AX (γ-H2AX).
Cytospins of 105 E18.5 fetal liver or E18.5 fetal thymocytes were washed in Tris-buffered saline (TBS). Cells were fixed in 4% formaldehyde for 30 min at RT and permeabilized in TBS containing 0.25% Triton X-100 for an additional 10 min at room temperature, followed by blocking with 3% bovine serum albumin (Serva) in TBS for at least 1 h at room temperature, incubation with a 1:500 dilution of the anti–γ-H2AX antibody (clone JBW301; Millipore) in blocking buffer, washing, and detection using a 1:1,000 dilution of Alexa Fluor 488 goat anti–mouse IgG (Life Technologies) in blocking buffer. Nuclei were counterstained with DAPI.
Assay for RNase H2 activity.
Cell lysates were prepared and assayed for specific cleavage of an 18-bp double-stranded DNA substrate containing a single ribonucleotide in one strand as previously described (Reijns et al., 2011). RNase H2–specific activity was determined by subtracting the cellular activity against a sequence-matched DNA duplex without ribonucleotides. Cell lysate protein concentration was determined and lysates were added to the reaction mix at a final protein concentration of 100 ng/µl for fetal liver and fibroblast lysates. Lysates of B cells contained less protein and were added to reactions to final concentration of 5 ng/µl.
Statistical analysis.
Data are shown as means ± SD. Statistical analysis was performed with the Student’s t test.
Acknowledgments
The authors thank Martin Reijns and Andrew Jackson for performing enzyme activity assays and for sharing unpublished data. We are grateful to Thomas Kunkel for providing RNase H2–deficient yeast strains, Julia Jarrells and Britta Schilling for microarray analyses, Dora Schreier for ES cell transfection and morula injections, Min-Ae Lee-Kirsch, Werner Müller, Michael Bachmann, Rolf Jessberger, Frank Buchholz, Dragomir Krastev, and Alexander Gerbaulet for advice and discussions, and Christina Hiller and Tobias Häring for expert technical assistance.
This work was supported by the German Research Foundation (Klinische Forschergruppe KFO 249, project TP7, Ro2133/6-1) to A. Roers and “MeDDrive” grants 60.230 and 60.294 of the Medical Faculty Carl Gustav Carus, TU-Dresden, to R. Behrendt.
The authors have no competing financial interest.
Footnotes
Abbreviations used:
- AGS
- Aicardi-Goutières syndrome
- ES
- embryonic stem
- KOF
- knockout first
- RNase H
- ribonuclease H
- topo I
- topoisomerase I
- TREX1
- three prime repair exonuclease 1
References
- Brady C.A., Jiang D., Mello S.S., Johnson T.M., Jarvis L.A., Kozak M.M., Kenzelmann Broz D., Basak S., Park E.J., McLaughlin M.E., et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 145:571–583 10.1016/j.cell.2011.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D., Reijns M.A., Graham S.C., Astell K.R., Jones E.Y., Jackson A.P. 2011. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 39:3652–3666 10.1093/nar/gkq980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 282:1497–1501 10.1126/science.282.5393.1497 [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., Crouch R.J. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494–1505 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 1:581–585 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- Crow Y.J. 2011. Type I interferonopathies: a novel set of inborn errors of immunity. Ann. N.Y. Acad. Sci. 1238:91–98 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Hayward B.E., Parmar R., Robins P., Leitch A., Ali M., Black D.N., van Bokhoven H., Brunner H.G., Hamel B.C., et al. 2006a. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat. Genet. 38:917–920 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. 2006b. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38:910–916 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- Furuta T., Takemura H., Liao Z.Y., Aune G.J., Redon C., Sedelnikova O.A., Pilch D.R., Rogakou E.P., Celeste A., Chen H.T., et al. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303–20312 10.1074/jbc.M300198200 [DOI] [PubMed] [Google Scholar]
- Gall A., Treuting P., Elkon K.B., Loo Y.M., Gale M., Jr, Barber G.N., Stetson D.B. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 36:120–131 10.1016/j.immuni.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., Tora L. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 44:966–977 10.1016/j.molcel.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Takashima S., Shinohara T. 2010. Transmission distortion by loss of p21 or p27 cyclin-dependent kinase inhibitors following competitive spermatogonial transplantation. Proc. Natl. Acad. Sci. USA. 107:6210–6215 10.1073/pnas.0914448107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Huang S.N., Williams J.S., Li Y.C., Clark A.B., Cho J.E., Kunkel T.A., Pommier Y., Jinks-Robertson S. 2011. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 332:1561–1564 10.1126/science.1205016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand Y., Luria V., Haffner-Krausz R., Lonai P. 1998. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7:105–112 10.1023/A:1008868325009 [DOI] [PubMed] [Google Scholar]
- Lazzaro F., Novarina D., Amara F., Watt D.L., Stone J.E., Costanzo V., Burgers P.M., Kunkel T.A., Plevani P., Muzi-Falconi M. 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 45:99–110 10.1016/j.molcel.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Kirsch M.A., Gong M., Chowdhury D., Senenko L., Engel K., Lee Y.A., de Silva U., Bailey S.L., Witte T., Vyse T.J., et al. 2007. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 39:1065–1067 10.1038/ng2091 [DOI] [PubMed] [Google Scholar]
- Morita M., Stamp G., Robins P., Dulic A., Rosewell I., Hrivnak G., Daly G., Lindahl T., Barnes D.E. 2004. Gene-targeted mice lacking the Trex1 (DNase III) 3′—>5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 24:6719–6727 10.1128/MCB.24.15.6719-6727.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Steinhoff U., Reis L.F., Hemmi S., Pavlovic J., Zinkernagel R.M., Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science. 264:1918–1921 10.1126/science.8009221 [DOI] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Kumar D., Clark A.B., Watt D.L., Watts B.E., Lundström E.B., Johansson E., Chabes A., Kunkel T.A. 2010. Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6:774–781 10.1038/nchembio.424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., Bubeck D., Gibson L.C., Graham S.C., Baillie G.S., Jones E.Y., Jackson A.P. 2011. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J. Biol. Chem. 286:10530–10539 10.1074/jbc.M110.177394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F., Devenney P.S., Sexton D., Grimes G., Holt I.J., Hill R.E., Taylor M.S., Lowson K.A., Dorin J.R., Jackson A.P. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 149:1008–1022 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G.I., Bond J., Asipu A., Brunette R.L., Manfield I.W., Carr I.M., Fuller J.C., Jackson R.M., Lamb T., Briggs T.A., et al. 2009. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41:829–832 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., Rajewsky K. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Shuman S. 1997. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1:89–97 10.1016/S1097-2765(00)80010-6 [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., Hieter P. 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26:163–175 10.1101/gad.179721.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G., Schaft J., van der Hoeven F., Glaser S., Anastassiadis K., Zhang Y., Hermann T., Stremmel W., Stewart A.F. 2004. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 38:151–158 10.1002/gene.20012 [DOI] [PubMed] [Google Scholar]
- Turchi J.J., Huang L., Murante R.S., Kim Y., Bambara R.A. 1994. Enzymatic completion of mammalian lagging-strand DNA replication. Proc. Natl. Acad. Sci. USA. 91:9803–9807 10.1073/pnas.91.21.9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Amon J.D., Koshland D., Vuica-Ross M. 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 44:978–988 10.1016/j.molcel.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., Barnes D.E. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 131:873–886 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]