Abstract
Arginine methylation is a common posttranslational modification (PTM). This type of PTM occurs on both nuclear and cytoplasmic proteins, and is particularly abundant on shuttling proteins. In this review, we will focus on one aspect of this PTM: the diverse roles that arginine methylation of the core histone tails play in regulating chromatin function. A family of nine protein arginine methyltransferases (PRMTs) catalyze methylation reactions, and a subset target histones. Importantly, arginine methylation of histone tails can promote or prevent the docking of key transcriptional effector molecules, thus playing a central role in the orchestration of the histone code.
Keywords: arginine methylation, histone code, tudor domain, CARM1, ChIP-seq
Arginine Methylation
The mammalian family of protein arginine methyltransferases (PRMTs) has nine members. These enzymes transfer a methyl group from S-adenosylmethionine (AdoMet) to a guanidino nitrogen of arginine resulting in S-adenosylhomocysteine (AdoHcy) and methylarginine. The PRMT family of AdoMet-dependent methyltransferases harbor a set of four conserved signature amino acid sequence motifs (I, post-I, II, and III), and a THW loop [1]. Motifs I, post-I and the THW loop form part of the AdoMet-binding pocket [2]. There are three main forms of methylated arginine identified in mammalian cells: monomethylarginines (MMA); asymmetric dimethylarginines (ADMA); and symmetric dimethylarginines (SDMA). PRMTs are classified as either type I, type II, or type III, enzymes, which methylate the terminal (or ω) guanidino nitrogen atoms of arginine. Type IV enzyme activity catalyzes the monomethylation of the internal guanidine nitrogen atom and this type of activity has only been described in yeast. Type I and type II enzymes catalyze the formation of an MMA intermediate, then type I PRMTs (PRMT1, 2, 3, 4, 6 and 8) further catalyze the production of ADMA, while type II PRMTs (PRMT5 and 7) catalyze the formation of SDMA (Figure 1A). Certain substrates are only monomethylated by PRMT7, which is referred to as type III enzymatic activity. Histone tails are a prime target for this family of enzymes (Figure 1B).
Figure 1. Types and sites of histone tail arginine methylation.
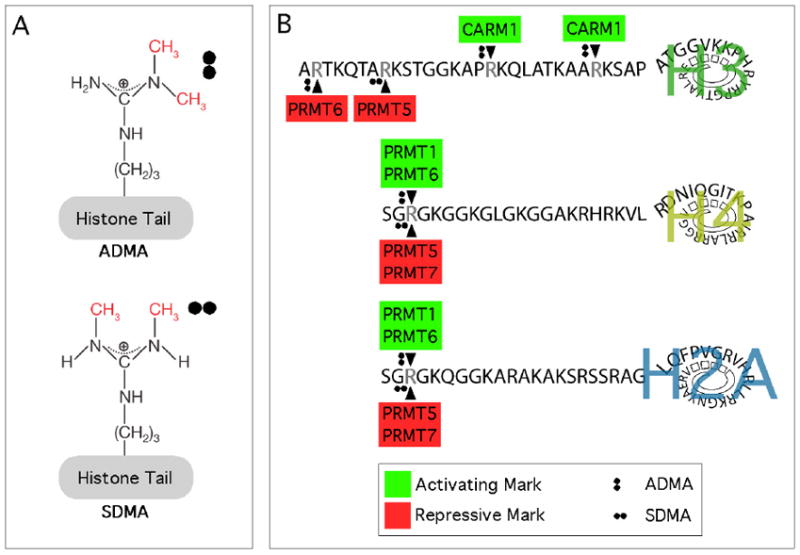
(A) Arginine residues in the tails of histones can be monomethylarginines (MMA), asymmetric dimethylarginines (ADMA), and symmetric dimethylarginines (SDMA). The MMA form of arginine is generally regarded as an intermediate on its way to the dimethylated state and is not depicted here. (B) The known sites of histone H3, H4, and H2A arginine dimethylation are shown. Red denotes transcriptional repressor activity and green denotes transcriptional activator activity.
Mammalian Arginine Methyltransferases
PRMT1
PRMT1 was the first mammalian protein arginine methyltransferase identified [3], and it is also responsible for the bulk (about 85%) of total protein arginine methylation activity [4]. The central role that PRMT1 plays as a diverse regulator of protein function is revealed by the disruption of this enzyme in mice, which die shortly after implantation [5]. PRMT1 displays wide substrate specificity, with a preference for methylating arginine residues that are flanked by one or more glycine residues – motifs often referred to as GAR sequences (Glycine & Arginine Rich) [6,7]. The three-dimensional structure of PRMT1 reveals that it is active as a homodimer [8]. PRMT1 also methylates histone H4 at arginine 3, generating the H4R3me2a mark [9], and thus contributing to the histone code. This modification on histone H4 functions as a transcriptional activation mark, which can recruit methyl-binding proteins and influence the deposition of other posttranslational marks in the vicinity. As a transcriptional coactivator, PRMT1 is recruited to promoters by a number of different transcription factors [10].
PRMT2
PRMT2 harbors a methyltransferase domain and a SH3 domain [11]. The SH3 domain binds the PRMT8 N-terminal domain and may also target it to substrates [12]. Initially, it was not believed to possess methyltransferase activity. However, recently very weak Type I activity was eked out of this enzyme [13]. PRMT2 can methylate histone H4, although the site of methylation has not been mapped. PRMT2 is a coactivator of gene expression much like PRMT1 and CARM1, and this activity appears to rely upon the integrity of the methyltransferase domain. PRMT2 is a coactivator of both the androgen receptor and the estrogen receptor alpha [14,15]. In addition, PRMT2 can promote apoptosis and inhibit NF-kappaB function by blocking IkappaB-alpha nuclear export [16]. PRMT2 null mice are viable and grossly normal [17].
PRMT3
PRMT3 harbors a zinc-finger domain at its N-terminus, which is the substrate recognition module of this enzyme [18,19]. The 40S ribosomal protein S2 (rpS2) is a zinc-finger dependent substrate of mammalian PRMT3 [20]. Importantly, in fission yeast PRMT3 (called Rmt3) also methylates rpS2 [21], emphasizing the conserved nature of this enzyme/substrate pairing. In yeast, loss of PRMT3 results in a ribosomal subunit imbalance, which can be restored with the expression of a catalytic inactive mutant [22]. Mouse embryos with a targeted disruption of PRMT3 are small in size, but survive after birth and attain a normal size in adulthood. In these null mice, rpS2 is hypomethylated, demonstrating that it is an in vivo PRMT3 substrate [23]. PRMT3 seems to be located exclusively in the cytosol, thus it may not directly impact epigenetic pathways.
CARM1 (PRMT4)
CARM1, which is also referred to as PRMT4, was identified as a steroid receptor coactivator and provided the first evidence that arginine methylation regulates transcription [24]. The recruitment of CARM1 to transcriptional promoters results in the methylation of histone H3 (H3R17me2a & H3R26me2a) and transcriptional regulators [10]. CARM1 is not only a steroid receptor coactivator, but it also reinforces other transcription factor pathways [10]. In addition, CARM1 methylates splicing factors and regulates the coupling of transcription and splicing [25]. CARM1 and PRMT1 do not share substrates (Lee and Bedford, 2002). CARM1 null mice die just after birth and are smaller than their wild-type littermates [26]. Further analysis of these null mice have revealed key in vivo roles for CARM1 in early T cell development [27], in adipocyte differentiation [28], in chondrocyte proliferation [29], and in the proliferation and differentiation of pulmonary epithelial cells [30]. CARM1 requires its enzymatic activity for all of its known in vivo functions [31].
PRMT5
PRMT5 is the predominant Type II arginine methyltransferase in mammals and is generally regarded as a strong transcriptional repressor [32]. It was first identified as a Jak2-binding protein and shown to methylate histones H2A, H3 and H4 [33,34]. In the nucleus, PRMT5 binds to COPR5 (cooperator of PRMT5), which appears to be responsible for its transcriptional corepressor activities. The COPR5 interaction alters the specificity of PRMT5, causing it to preferentially methylate H4R3 over H3R8 [35]. PRMT5 is recruited by numerous transcription factors and repressor complexes, including Snail [36], ZNF224 [37], Ski [38], and at the globin locus [39]. In the cytoplasm, PRMT5 is involved in snRNP biogenesis through its ability to methylate a number of Sm proteins [40,41], and it also methylates Piwi proteins, which regulate a class of small noncoding RNAs [42]. It should be noted that αFLAG M2-agarose enriches for PRMT5 activity [43], thus many affinity purified FLAG-tagged complexes are “contaminated” with PRMT5, confounding the field.
PRMT6
PRMT6 is predominantly localized to the nucleus and like PRMT1 methylates GAR motifs [44]. It is the primary enzyme responsible for H3R2 methylation in mammalian cells, [45-47]. H3R2 methylation, by PRMT6, counteracts the H3K4me3 activation mark, making it a transcriptional repressor. Thrombospondin-1 (TSP-1) is the first characterized transcriptionally repressed target of PRMT6 [48]. The general assumption that PRMT6 functions as a transcriptional repressor is not clear cut, as a recent report demonstrates that it functions as a coactivator with nuclear receptors [49]. The ability of PRMT6 to function as a coactivator may be linked to its capacity to also methylate the R3 position of the N-terminal tails of histones H4 and H2A (the first 5 residues of these histones are the same) [46]. Thus, in certain contexts, PRMT6 may methylate H3R2 and repress transcription, and in other contexts, it may methylate H4R3/H2AR3 and activate transcription – this hypothesis needs to be confirmed.
PRMT7
PRMT7 is one of two PRMTs that harbor two putative AdoMet-binding motifs [50]. Indirect evidence correlates PRMT7 activity with either resistance or sensitivity to DNA damaging agents [51-53], and sensitivity of the kidney to damage caused by certain antibiotics [54]. The in vitro enzymatic activity of PRMT7 is not particularly robust, but it has been described as having type III activity towards some substrates [55] and type II activity towards others [56]. It is thus possible that distinct substrates are methylated in different fashions by this enzyme. PRMT7 plays a role in male germline imprinted gene methylation through its interaction with CTCFL and subsequent symmetrical methylation of H4R3 [57]. PRMT7 might also play a role in embryonic stem cell (ESC) pluripotency, as its expression is lost when ESCs differentiate [58].
PRMT8
PRMT8 displays a high degree of sequence identity with PRMT1 [59]. It has a unique N-terminal end that harbors a functional myristoylation motif that facilitates its association with the plasma membrane. This myristoylation motif is conserved in the puffer fish orthologue of PRMT8 (fuguL3) [60]. The alternate use of initiator methionine residues may affect the localization of the endogenous protein [61]. Furthermore, PRMT8 expression is largely restricted to the brain, and more specifically in neurons. The in vitro activity of the full-length recombinant enzyme is low; however, removal of the N-terminal domain by truncation or proteolysis results in increased activity [12]. The N-terminal region contains two proline-rich sequences that can bind a number of SH3 domains, including that of PRMT2.
PRMT9 (4q31)
PRMT9 (4q31) was first identified at the same time as PRMT8 was described [59]. PRMT9, like PRMT7, harbors two putative AdoMet-binding motifs. In addition, at its N-terminal end, PRMT9 has two tetratricopeptide repeats (TPR), which often mediate protein-protein interactions [62]. We refer to this protein with the human chromosomal location, because the PRMT9 designation has also been used for the product of the human FBXO11 gene on chromosome 2p16 [63], although FBXO11 is unlikely to be a bona fide PRMT [64]. PRMT9 (4q31) has yet to be biochemically characterized.
Arginine demethylation
Arginine methylation is a very stable mark, and it is unclear if the modification can be enzymatically reversed. This is a very active field of investigation.
JMJD6
The first putative arginine demethylase was recently identified [65]. This enzyme, JMJD6, a Jumonji domain-containing protein, was reported to demethylate H3R2me2 and H4R3me2. Furthermore, it could demethylate both, asymmetrically and symmetrically, dimethylated substrates. Recently, these findings were brought into question by studies performed by Webby et al., who showed that JMJD6 is actually a lysine-hydroxylase [66]. They were unable to detect demethylase activity on either H3R2me2 or H4R3me2 peptides. In addition, the structural analysis of JMJD6 suggests that it is not an arginine demethylase [67,68]. Thus, the issue of whether an arginine demethylase really exists remains unanswered.
PADIs
Arginine residues are not only substrates for methylation, they can also be converted to citrulline by deimination [69]. The conversion of arginine to citrulline is catalyzed by a family of enzymes called peptidylarginine deiminases (PADIs). The core histones H2A, H3 and H4 comprise a major group of deiminated proteins [70]. PADI4 is a nuclear protein that targets the same arginine residues on histones H3 and H4 as the PRMTs methylate [71,72]. A recent study showed that PADI4 is recruited to the pS2 promoter region just prior to H3R17me2a loss, suggesting that it is responsible for removing this methyl-mark [73]. However, PADIs catalyze the deimination of arginine, but not methylated arginine residues [74,75]. Furthermore, no enzyme has been identified than can convert citrulline back to arginine. Thus, PADIs can block methylation on an arginine residue by converting it to citrulline, but are not “true” demethylases.
Sites of arginine methylation on histone tails
Methyl-specific antibodies have been raised to many of the arginine methylation sites on histone tails. These antibodies have been used in a variety of studies, including traditional ChIP, ChIP-chip, and ChIP-seq experiments. Here we will summarize these findings, and attempt to consolidate them into unified rules that will associate a specific mark with a specific chromatin state. In some cases, these attempts fall short because of conflicting data in the literature.
H3R17me2a
The H3R17me2a mark is generated by CARM1 [76], which was the first PRMT to be identified as a transcriptional regulator [24]. CARM1 has been definitively characterized as a coactivator, and it functions as such with nuclear receptors and a variety of additional transcription factors including p53, NF-κB, PPARγ and c-Fos [77]. Indeed, ChIP analysis has identified elevated levels of H3R17me2a at the promoters of pS2 [78,79], E2F1 [80], CCNE1 [81], aP2 [28], Oct 4 and Sox2 [82], CITED2 [83], and Scn3b [30]. It should be noted here that the H3R17me2a antibody [UpState], used for the vast majority of these ChIP experiments, recognizes a number of different CARM1 substrates, including AIB1 and a cohort of splicing factors [25,26,84]. Thus, this antibody is a great reporter for CARM1 activity, but not a specific readout for H3R17me2a methylation per se.
Kinetic ChIP analysis of the estrogen-responsive pS2 promoter reveals that CARM1 is recruited in a cyclic manner, occurring at 40 minute intervals after E2-treatment [85]. This recruitment of CARM1 correlates with an increase in the H3R17me2 mark, which arises at the same time that CBP/p300 is recruited. The concomitant recruitment of CARM1 with CBP/p300 is important for H3R17me2 methylation, because acetylation of H3K18 makes the histone H3 tail a better substrate for CARM1 [86]. Crosstalk between CARM1 and p300 also occurs at the GADD45 gene, which is regulated by p53 [87]. A detailed analysis of this H3K18ac priming for CARM1 methylation reveals that the acetylated peptide does not enhance its affinity for CARM1, but instead this priming increases the rate of the methyltransferase reaction itself [88]. It is proposed that by neutralizing the positive charge, acetylation of H3K18 would facilitate the nucleophilic attack on the sulphur-methyl bond of SAM, thus explaining why CARM1 has five-fold higher activity towards the H3K18ac-peptide than the unmodified peptide. Furthermore, the methylation of the H3R17 mark may be regulated in a cell cycle-dependent manner. Histones isolated from M-phase cells harbor the highest level of H3R17 methylation, and this peak in signal correlates well with the H3S10p [89].
Recently, the first genome wide localization of CARM1 activity (the H3R17me2a mark) was performed using a ChIP-chip approach [90]. In MCF7 breast cancer cells, CARM1 activity was localized to distinct classes of ERα binding sites that have been termed enhancer-rich clusters Ec1 and Ec3. Surprisingly, CARM1 activity localizes to enhancer regions that are a great distance away from the transcription start sites (TSS) of the genes that undergo E2-stimulated gene expression. Thus CARM1 activity maps to ERα-specific enhancers rather than promoter regions, although a number of groups have ChIPed CARM1 activity at the estrogen-responsive pS2 promoter [78,79,85]. The pS2 promoter may represent an exception, because ERα is recruited to both its promoter and enhancer regions [91]. ChIP-seq experiments for CARM1 or CARM1 activity have not yet been reported.
H3R26me2a
The H3R26me2a mark is also generated by CARM1 [76], although this modification has not been studied much. This mark has been identified at the CCNE1 promoter using a ChIP approach [81]. An interesting feature of the H3R26 site is its proximity to H3K27, which when methylated is a major repressive mark. If the H3R26me2a mark antagonizes the Polycomb repressive function (by either preventing EZH2 methylation of H3K27, or blocking the docking of the Polycomb family of chromodomains and/or the EED WD40 domain to H3K27me3 [92,93]), then profound effects on gene expression would be expected. These studies have yet to be performed.
H3R2me2a
The H3R2me2a mark was initially thought to be generated by CARM1 [76], but recent studies have identified it as a major mark deposited by PRMT6 [45-47]. Methylation of the H3R2 site essentially prevents the MLL1 complex from methylating H3K4 [46]. However, PRMT6 can strongly methylate H3K4me1 and H3K4me2 peptides, and weakly methylate a H3K4me3 peptide, so dually modified histone tails likely exist [47]. H3K4me3 marks the TSS of actively transcribed genes. Many effectors that bind H3K4me3 are blocked from docking if H3R2 is also methylated [47]. Thus, PRMT6 functions as a transcriptional repressor by blocking the recruitment of transcriptional activators to the methylated H3K4 mark. ChIP analysis at 185 human promoters supports this hypothesis, demonstrating a counter-correlation between H3K4me3 and H3R2me2a levels [45]. This study also shows a counter-correlation between H3R2me2a and gene expression levels. Further ChIP analysis has revealed PRMT6 activity at the promoters of the HoxA2 gene [46] and the TSP-1 gene [48], which correlate with transcriptional repression.
The first ChIP-seq survey to be performed on histone methyl-arginine marks found no enrichment of H3R2me1 or H3R2me2a at either active or silenced gene promoters [94]. However, more recent ChIP-seq studies have revealed enrichment of the H3R2me2a mark at pericentromeric regions, and H3R2me1 mark at subtelomeric regions [95]. This same study also shows that both the H3R2me1 and H3R2me2a marks are associated with highly expressed genes, which is rather unexpected considering the ChIP experiment described above and the ability of the H3R2me2a mark to block H3K4me3 effector molecules. However, this report is consistent with the finding that PRMT6 functions as a coactivator of a number of nuclear receptors [49]. Of course, the capacity of PRMT6 to function as a coactivator may be due to its ability to also methylate H4R3me2a, which is associated with active chromatin. The interaction of PRMT6 with different regulators may alter its substrate specificity (H3R2me2a or H4R3me2a), thus allowing this PRMT to toggle between an activator and a repressor.
H4R3me2a and H2AR3me2a
The first five residues of histones H4 and H2A are identical, and it is thus likely that most of the activities reported for H4R3 will also hold true for H2AR3. Importantly, antibodies raised against H4R3 methylaion marks will also recognize H2AR3. We will thus refer to these two sites jointly as the “R3 motif”. The fact that this motif has been duplicated, provides an efficient means of amplifying the signaling that are mediated by these two histone tails.
The asymmetric dimethylation of the R3 motif is catalyzed by PRMT1, PRMT6 and PRMT8 [9,46,47], and is associated with actively transcribed promoters. PRMT1 functions as a coactivator with ER, YY1, p53 and RUNX1 [77]. ChIP analysis has identified elevated levels of R3 motif methylation at the promoters of pS2 [73], CYP3A4 [96], CITED2 [83], and the β-globin locus [97].
Kinetic ChIP analysis of the estrogen-responsive pS2 promoter reveals that PRMT1 is recruited in cyclic manner, and is the first PRMT to be recruited after an E2 pulse [85]. PRMT1 recruitment and its subsequent methylation of the R3 motif occurs during the first transcriptionally nonproductive cycle, and defines the transcriptional competency of the pS2 promoter. This is consistent with the fact that R3 motif methylation is required for the subsequent acetylation of histone H3 and H4 [97,98]. Interestingly, while the H3R17me2a marks every transcriptionally productive cycle (at 40 min intervals), methylation of the R3 motif covers two of these cycles before it is lost and then re-established again. This phenomenon has been observed in at least two independent studies [73,85] and remains unexplained.
Using a Top Down mass spectrometric approach, the possible combinations of different histone H4 modifications were quantified [99]. Less than 2% of total histone H4 is mono- or dimethylated at the H4R3 site. This study did not distinguish between symmetric and asymmetric dimethylation at H4R3. However, they did report that the H4R3me species is often also lysine acetylated, which is consistent with transcriptional activation. A second mass spectrometric study also found H4R3 methylation in combination with extensive acetylation, and always with the H4K20me2 mark [100].
H4R3me2s and H2AR3me2s
PRMT5 symmetrically methylates both H3R8 and the R3 motif [101], but its specificity is tilted towards the R3 motif by one of its regulatory binding proteins, COPR5 [35]. There is evidence that PRMT7 can also methylate these sites [57]. H4R3me2s motif ChIP experiments for PRMT5 target genes have been performed, localizing this mark to repressed promoter regions [102], and the silenced fetal globin gene [39]. Chip experiments have also identified the R3me2s motif mark at sites within the H19 imprinting control region [57]. ChIP-seq experiments have been performed for the H4R3me2s mark by Keji Zhao’s group [94]. Unexpectedly, they did not find any correlation between this mark and transcriptionally repressed loci. However, reevaluation of this data, by a different group, indicated that the H4R3me2s mark is strongly associated with repressed gene expression [103].
As mentioned above, this R3 motif is also a site for asymmetric methylation (an activation mark) by a number of enzymes. Thus, at genes that are actively maintained in an “on” or “off” state there is constant pressure on this bifunctional site to be either asymmetrically or symmetrically dimethylated.
H3R8me2s
The H3R8 site is symmetrically methylated by PRMT5 [101]. This methylation event is linked to transcriptional repression and is tightly associated with H4R3me2s methylation, which is also generated by PRMT5. PRMT5 is recruited by numerous transcriptional repressors, including Snail, ZNF224 and Ski [36-38]. H3R8me2s ChIP experiments for a number of PRMT5 target genes have been performed, localizing this mark to repressed promoter regions [102].
The prior acetylation of H3K9 or H3K14 prevents the methylation of H3R8 by PRMT5 [101]. The reverse has not been tested, so it is not clear if the H3R8me2s modification has an impact on H3K9 acetylation. However, G9a methylation of H3K9 is blocked by the prior existence of the H3R8me2s modification [104]. Also, the recognition of the guanidinium amino group of H3R8 contributes to sequence specificity of H3S10ph recognition by 14-3-3ζ [105]. The effect of H3R8 methylation on H3K9me3 recognition by chromodomain-containing proteins (HP1, MPP8 or CDYs) has not yet been established. Thus, the mechanism by which H3R8 methylation links to transcriptional repression remains unclear, but a number of intriguing possibilities exist. Genome-wide chromatin profiles of PRMT5 or H3R8me2s have not yet been reported.
The consequences of histone tail arginine methylation
The different arginine methylation events that occur on histone tails likely impact the generation of other histone code marks and/or influence the binding of effector molecules. Here we will highlight a few examples of these different means of regulation.
Blocking the docking of effector molecules
It is clear that there is cross-talk between lysine methylation and serine phosphorylation on histone tails [106]. In a similar fashion, it has been proposed that cross-talk between arginine and lysine methylation is also a wide-spread phenomenon, and has been termed an “arginine/lysine-methyl/methyl switch” [107]. A number of arginine/lysine pairs exist, i.e H3R2/H3K4; H3R8/H3K9; H3R26/H3K27, where both residues are methylated. Furthermore, there are many more of these potential switches on the histone tails.
One of these sites of cross-talk that has been well-defined is the H3R2/H3K4 switch (Figure 2A). PRMT6 is the primary enzyme responsible for the methylation of H3R2 in mammalian cells [45-47]. In mammalian cells, these two marks may coexist, because PRMT6 can strongly methylate H3K4me1 and H3K4me2 peptides, and weakly methylate a H3K4me3 peptide [47]. However, prior methylation of the H3R2 site essentially prevents the MLL1 complex from methylating H3K4 [46]. It is not clear whether other H3K4 methyltransferases (there are at least nine of them) are active on a H3R2me2a substrate. Methylation of H3K4 is found at the 5’ end of active genes and is responsible for recruiting chromatin-remodeling enzymes to establish and maintain a transcriptionally active state. The effectors that bind H3K4me3 harbor methyl-specific binding domains, including PHD, Chromo, Tudor and WD40 domains. Methylation at the H3R2 site blocks the ability of most of these domains to bind the N-terminal tail of histone H3, in in vitro assays [47]. In cells, the chromatin binding of both WDR5 and ING2 is sensitive to the level of PRMT6 expression [45-47]. Thus, PRMT6 functions as a transcriptional repressor by blocking the recruitment of transcriptional activators to the methylated H3K4 mark.
Figure 2. Mechanisms by which histone tail arginine methylation regulates transcription.
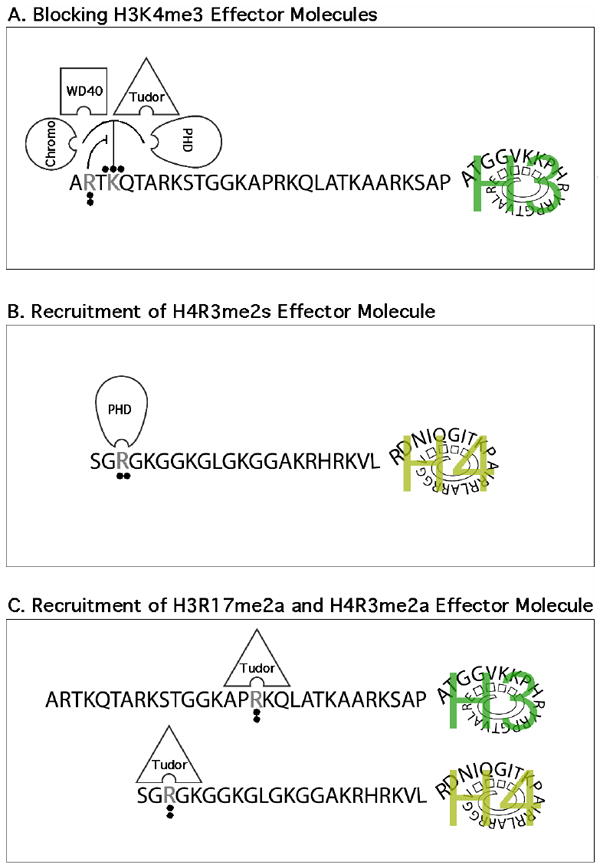
(A) The symmetrical dimethylation of H3R2 blocks the binding of H3K4me3 effector molecules. In addition, the H3R2me2a mark prevents methylation of H3K4me3 by MLL1. (B) The H4R3me2s mark may function as a docking site for the PHD finger of the de novo DNA methyltransferase DNMT3A, thereby linking PRMT5 activity to stable and heritable DNA methylation. (C) The activation marks deposed by PRMT1 (H4R3me2a) and CARM1 (H3R17me2a) can be “read” by the tudor domain of TDRD3. TDRD3 itself functions as a coactivator, in some manner relaying the intent of these two PRMTs.
Recruitment of repressive effector molecules
Do readers of symmetrically methylated arginine motif exist? If there are SDMA binders, these effector molecules should possess repressor functions that would link PRMT5 (and PRMT7) to its clearly defined role as an attenuator of transcription. Recently, the PHD domain of the de novo DNA methyltransferase DNMT3A was identified as a binder of the H4R3me2s mark [108]. A repressive mechanism can now be envisioned where PRMT5 is recruited to a promoter, generating a patch of H4R3me2s that in turn will facilitate the binding of DNMT3A, thereby promote DNA methylation and prolonged gene silencing (Figure 2B). This sequence of events has been challenged by Otani et al., who confirmed previous reports that the PHD domain (also called the ADD domain) of DNMT3A binds H3K4me0 [109], but were not able to reproduce the interaction between this PHD and a H4R3me2s peptide [110].
Recruitment of effector molecules with activator functions
Do readers of asymmetrically methylated arginine motif exist? If there are ADMA binders, these effector molecules should possess activator functions that would link PRMT1 and CARM1 to their clearly defined roles as transcriptional coactivators. Using a protein domain microarray approach, one such effector molecule was recently identified [111]. TDRD3 contains a tudor domain that binds both the H3R17me2a and H3R4me2a activator marks (Figure 2C). TDRD3 itself functions as a coactivator in ERE-luciferase assays. Furthermore, endogenous TDRD3 ChIPs at the pS2 promoter in a estrogen-dependent manner, and global ChIP-seq analysis reveals that TDRD3 is highly enriched at the promoters of actively transcribed genes. Although TDRD3 is recruited to estrogen-responsive promoters in an estrogen-dependent manner, it remains unclear how it promotes transcriptional activation. Indeed, TDRD3 harbors no enzymatic activity. It is thus likely that TDRD3 recruits a protein complex that assists in opening and activating chromatin in the vicinity of H3R17me2a and H3R4me2a activator marks.
Exciting prospects for the near future
It is likely that we have identified all the arginine methyltransferases within this particular PRMT family. It is possible that convergent evolution has generated other classes of arginine methyltransferases, as it has for lysine methyltransferases and demethylases. The search and identification of additional effector molecules that “read” ADMA and SDMA motifs will be critical to help us understand the mechanisms of action of this posttranslational modification. Clearly, a comprehensive ChIP-seq analysis of all the arginine methylated histone code marks is required for us to better understand the roles of each mark. New arginine methylation sites will likely be identified on histone tails in the coming years. The novel marks may well exist in the context of “arginine/lysine-methyl/methyl switch”. These “switches” have not attracted much attention, and the importance of the H3R8/H3K9 and H3R26/H3K27 nodes have yet to be determined. The holy grail in the field remains the elusive arginine demethylases.
Acknowledgments
Mark Bedford is supported by NIH grant number DK62248.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katz JE, Dlakic M, Clarke S. Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics. 2003;2:525–40. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. Embo J. 2000;19:3509–19. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–44. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 4.Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–30. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 5.Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–69. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–73. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Cheng X. Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides. Structure (Camb) 2003;11:509–20. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–7. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 10.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, Henry MF. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2) Genomics. 1998;48:330–40. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 12.Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J Biol Chem. 2007;282:36444–53. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- 13.Lakowski TM, Frankel A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl- and asymmetric dimethyl-arginine residues on histone H4. Biochem J. 2009;421:253–61. doi: 10.1042/BJ20090268. [DOI] [PubMed] [Google Scholar]
- 14.Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107:1–14. doi: 10.1016/j.jsbmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–30. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 16.Ganesh L, Yoshimoto T, Moorthy NC, Akahata W, Boehm M, Nabel EG, Nabel GJ. Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis. Mol Cell Biol. 2006;26:3864–74. doi: 10.1128/MCB.26.10.3864-3874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshimoto T, Boehm M, Olive M, Crook MF, San H, Langenickel T, Nabel EG. The arginine methyltransferase PRMT2 binds RB and regulates E2F function. Exp Cell Res. 2006;312:2040–53. doi: 10.1016/j.yexcr.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. CONFERRAL OF SUBSTRATE SPECIFICITY BY A ZINC-FINGER DOMAIN [In Process Citation] J Biol Chem. 2000;275:32974–82. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- 19.Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–45. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 20.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. Embo J. 2004;23:2641–50. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perreault A, Gascon S, D’Amours A, Aletta JM, Bachand F. A methyltransferase-independent function for Rmt3 in ribosomal subunit homeostasis. J Biol Chem. 2009;284:15026–37. doi: 10.1074/jbc.M109.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swiercz R, Cheng D, Kim D, Bedford MT. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J Biol Chem. 2007;282:16917–23. doi: 10.1074/jbc.M609778200. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 25.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464–8. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Lee J, Yadav N, Wu Q, Carter C, Richard S, Richie E, Bedford MT. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279:25339–44. doi: 10.1074/jbc.M402544200. [DOI] [PubMed] [Google Scholar]
- 28.Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008;9:193–8. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito T, et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9:47. doi: 10.1186/1471-213X-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien KB, et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137:2147–56. doi: 10.1242/dev.037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285:1147–52. doi: 10.1074/jbc.M109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabbrizio E, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 2002;3:641–5. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–42. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 34.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. Prmt5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–6. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 35.Lacroix M, Messaoudi SE, Rodier G, Le Cam A, Sardet C, Fabbrizio E. The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 2008;9:452–8. doi: 10.1038/embor.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, Rauscher FJ., 3rd The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cesaro E, De Cegli R, Medugno L, Florio F, Grosso M, Lupo A, Izzo P, Costanzo P. The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene. J Biol Chem. 2009;284:32321–30. doi: 10.1074/jbc.M109.043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tabata T, Kokura K, Ten Dijke P, Ishii S. Ski co-repressor complexes maintain the basal repressed state of the TGF-beta target gene, SMAD7, via HDAC3 and PRMT5. Genes Cells. 2009;14:17–28. doi: 10.1111/j.1365-2443.2008.01246.x. [DOI] [PubMed] [Google Scholar]
- 39.Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010 doi: 10.1182/blood-2009-10-251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishioka K, Reinberg D. Methods and tips for the purification of human histone methyltransferases. Methods. 2003;31:49–58. doi: 10.1016/s1046-2023(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 44.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–43. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 45.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–7. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 46.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–80. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iberg AN, Espejo A, Cheng D, Kim D, Michaud-Levesque J, Richard S, Bedford MT. Arginine methylation of the histone h3 tail impedes effector binding. J Biol Chem. 2008;283:3006–10. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 48.Michaud-Levesque J, Richard S. Thrombospondin-1 is a transcriptional repression target of PRMT6. J Biol Chem. 2009;284:21338–46. doi: 10.1074/jbc.M109.005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison MJ, Tang YH, Dowhan DH. Protein arginine methyltransferase 6 regulates multiple aspects of gene expression. Nucleic Acids Res. 2010;38:2201–16. doi: 10.1093/nar/gkp1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004 doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 51.Bleibel WK, Duan S, Huang RS, Kistner EO, Shukla SJ, Wu X, Badner JA, Dolan ME. Identification of genomic regions contributing to etoposide-induced cytotoxicity. Hum Genet. 2009;125:173–80. doi: 10.1007/s00439-008-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gros L, et al. Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 2003;63:164–71. [PubMed] [Google Scholar]
- 53.Verbiest V, et al. Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 2008;582:1483–9. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- 54.Zheng Z, et al. A Mendelian locus on chromosome 16 determines susceptibility to doxorubicin nephropathy in the mouse. Proc Natl Acad Sci U S A. 2005;102:2502–7. doi: 10.1073/pnas.0409786102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miranda TB, Miranda M, Frankel A, Clarke S. PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J Biol Chem. 2004;279:22902–7. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, et al. PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J Biol Chem. 2005;280:3656–64. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- 57.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buhr N, Carapito C, Schaeffer C, Kieffer E, Van Dorsselaer A, Viville S. Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis. 2008;29:2381–90. doi: 10.1002/elps.200700738. [DOI] [PubMed] [Google Scholar]
- 59.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–6. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 60.Hung CM, Li C. Identification and phylogenetic analyses of the protein arginine methyltransferase gene family in fish and ascidians. Gene. 2004;340:179–87. doi: 10.1016/j.gene.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 61.Kousaka A, Mori Y, Koyama Y, Taneda T, Miyata S, Tohyama M. The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience. 2009;163:1146–57. doi: 10.1016/j.neuroscience.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 62.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001;134:117–31. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 63.Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S. FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun. 2006;342:472–81. doi: 10.1016/j.bbrc.2006.01.167. [DOI] [PubMed] [Google Scholar]
- 64.Fielenbach N, et al. DRE-1: an evolutionarily conserved F box protein that regulates C. elegans developmental age. Dev Cell. 2007;12:443–55. doi: 10.1016/j.devcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 65.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 66.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–3. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 67.Hong X, et al. Interaction of JMJD6 with single-stranded RNA. Proc Natl Acad Sci U S A. 2010;107:14568–72. doi: 10.1073/pnas.1008832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mantri M, et al. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J Mol Biol. 2010;401:211–22. [PubMed] [Google Scholar]
- 69.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–41. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 70.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277:49562–8. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 71.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 73.Denis H, Deplus R, Putmans P, Yamada M, Metivier R, Fuks F. Functional connection between deimination and deacetylation of histones. Mol Cell Biol. 2009;29:4982–93. doi: 10.1128/MCB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hidaka Y, Hagiwara T, Yamada M. Methylation of the guanidino group of arginine residues prevents citrullination by peptidylarginine deiminase IV. FEBS Lett. 2005;579:4088–92. doi: 10.1016/j.febslet.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 75.Raijmakers R, et al. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–29. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 76.Schurter BT, et al. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–56. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 77.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carascossa S, Dudek P, Cenni B, Briand PA, Picard D. CARM1 mediates the ligand-independent and tamoxifen-resistant activation of the estrogen receptor alpha by cAMP. Genes Dev. 2010;24:708–19. doi: 10.1101/gad.568410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–6. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- 81.Messaoudi El, et al. Coactivator-associated arginine methyltransferase 1 (CARM1) is a positive regulator of the Cyclin E1 gene. Proc Natl Acad Sci U S A. 2006;103:13351–6. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, Langford CF, Glover DM, Zernicka-Goetz M. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27:2637–45. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–13. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naeem H, Cheng D, Zhao Q, Underhill C, Tini M, Bedford MT, Torchia J. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–34. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 86.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–7. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 87.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–48. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 88.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. Embo J. 2007;26:4402–12. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010 doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29:3413–23. doi: 10.1128/MCB.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 95.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xie Y, Ke S, Ouyang N, He J, Xie W, Bedford MT, Tian Y. Epigenetic regulation of transcriptional activity of pregnane X receptor by protein arginine methyltransferase 1. J Biol Chem. 2009;284:9199–205. doi: 10.1074/jbc.M806193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–93. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X, et al. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood. 2010;115:2028–37. doi: 10.1182/blood-2009-07-236059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–37. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phanstiel D, et al. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4093–8. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang L, Pal S, Sif S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol Cell Biol. 2008;28:6262–77. doi: 10.1128/MCB.00923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu X, Hoang S, Mayo MW, Bekiranov S. Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinformatics. 2010;11:396. doi: 10.1186/1471-2105-11-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rathert P, et al. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–6. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–9. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 107.Migliori V, Phalke S, Bezzi M, Guccione E. Arginine/lysine-methyl/methyl switches: biochemical role of histone arginine methylation in transcriptional regulation. Epigenomics. 2010;2:119. doi: 10.2217/epi.09.39. [DOI] [PubMed] [Google Scholar]
- 108.Zhao Q, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–11. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–41. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang Y, Lu Y, Espejo A, Wu J, Xu X, Liang S, Bedford MT. TDRD3 is an Effector Molecule for Arginine Methylated Histone Marks. Molecular Cell In Press. 2010 doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


