Abstract
The glmS ribozyme is the first known example of a natural ribozyme that has evolved to require binding of an exogenous small molecule for activity. In Gram-positive bacteria, this RNA domain is part of the mRNA encoding the essential enzyme that synthesizes glucosamine-6-phosphate (GlcN6P). When present at physiologic concentration, this small molecule binds to the glmS ribozyme and uncovers a latent self-cleavage activity that ultimately leads to degradation of the mRNA. Biochemical and structural studies reveal that the RNA adopts a rigid fold stabilized by three pseudoknots and the packing of a peripheral domain against the ribozyme core. GlcN6P binding to this pre-organized RNA does not induce conformational changes; rather, the small molecule functions as a co-enzyme, providing a catalytically essential amine group to the active site. The ribozyme is not a passive player, however. Active site functional groups are essential for activity, even in the presence of GlcN6P. In addition to being a superb experimental system with which to analyze how RNA catalysts can exploit small molecule coenzymes to broaden their chemical versatility, the presence of the glmS ribozyme in numerous pathogenic bacteria make this RNA an attractive target for the development of new antibiotics and antibacterial strategies.
1. Introduction
A dichotomy developed in the first quarter-century of ribozyme studies. On one hand a number of larger ribozymes were found to have broad phylogenetic distribution. The ribosome is universal, ribonuclease (RNase) P is nearly so (Randau et al., 2008), and self-splicing introns are widely distributed in bacteria and organelles (Koonin, 2009; Raghavan & Minnick, 2009). On the other hand, the smaller self-cleaving catalytic RNAs, namely the hammerhead, hairpin, hepatitis delta virus (HDV) and Varkud satellite (VS) ribozymes, appeared to be highly restricted in phylogeny (Fedor, 2009; Ferré-D’Amaré & Scott, 2010; Lilley, 2003; Wilson & Lilley, 2009). These ribozymes, which catalyze the same overall chemical transformation, were only known from a few satellite RNAs and other similar parasitic RNAs, but not from free-living organisms. This was surprising because such small ribozymes could be found readily in random RNA sequences by in vitro selection methods, implying that they are not uncommon in RNA sequence space (Salehi-Ashtiani & Szostak, 2001).
Despite their seemingly limited distribution, the small self-cleaving ribozymes were subjected to intensive studies, both because their relatively small sizes (60–200 nucleotides, nt) make them amenable to experimentation and because they represent four structurally distinct, evolutionarily independent solutions to the same biochemical problem. These studies led to a number of breakthroughs in our understanding of the biochemical versatility of RNA. They provided the first high-resolution glimpses of ribozyme architecture (Pley et al., 1994; Scott et al., 1995), the first view of a preorganized ribozyme active site (Ferré-D’Amaré et al., 1998), and yielded the most detailed structural descriptions of RNA-catalyzed chemical reactions (Chi et al., 2008; Klein et al., 2007a; Rupert et al., 2002). These studies also led to a paradigm shift from the view that all ribozymes depend on metal ion cofactors (Pyle, 1993) to one in which RNA functional groups play a direct role in chemical catalysis (Bevilacqua & Yajima, 2006; Murray et al., 1998; Wilson & Lilley, 2009).
The advent of whole-genome sequencing led to the discovery of riboswitches, which are broadly distributed phylogenetically (Dambach & Winkler, 2009; Edwards et al., 2007; Henkin, 2008; Serganov, 2009). It was during the search for riboswitches that the glmS ribozyme was discovered. This ribozyme, which is distributed across Gram-positive bacteria, was found to catalyze the same overall reaction as that catalyzed by the four previously known small self-cleaving ribozymes, but to require binding of a small molecule for activity (Barrick et al., 2004; Winkler et al., 2004). Structural and biochemical characterization revealed that this ribozyme employs its cognate small molecule as a coenzyme, making it the first natural catalytic RNA known to harness the chemical properties of exogenous small molecules to achieve chemical catalysis (Klein & Ferré-D’Amaré, 2006; McCarthy et al., 2005). We are witnessing an accelerating pace of discovery of the broad distribution of small catalytic RNAs, fuelled by the explosion in available sequence data and improvements in sequence analysis (Martick et al., 2008; Salehi-Ashtiani & Szostak, 2001; Webb et al., 2009; Winkler et al., 2004). Thus, self-cleaving RNAs are probably abundant in genomes, and the dichotomy in the extent of phylogenetic distribution of large and small ribozymes spurious. Moreover, given that convergent evolution is common, it is likely that other natural ribozymes that employ small molecule coenzymes to broaden the chemical versatility of RNA await discovery.
2. Riboswitches and discovery of the glmS ribozyme
2.1 Riboswitches: gene regulation by small molecule-binding RNAs
Riboswitches are gene-regulatory mRNA domains that recognize small molecules without employing proteins, and modulate gene expression in cis. Binding to their specific ligands stabilizes a conformation of the ligand-binding domain (the ‘aptamer domain’) of the riboswitch that in turn affects the conformation of an RNA segment (the ‘expression platform’) that interfaces with the transcription, translation or RNA processing machinery that ultimately leads to modulation of gene expression. Although the proposal that gene-regulatory RNAs may themselves be directly responsible for sensing the intracellular concentration of small molecules goes at least as far back as the idea of the ‘repressor’ (Jacob & Monod, 1961), and characterization of the T-box system demonstrated that gene regulatory RNAs can sense the aminoacylation state of tRNA (i.e. a small-molecule modification of an RNA) (Grundy & Henkin, 1993), formal demonstration of the existence of riboswitches did not occur until the XXI century (Mironov et al., 2002; Winkler et al., 2002). At present, riboswitches have been discovered that respond specifically to more than a dozen different metabolites and one bacterial second messenger. Their structural and biochemical characterization has progressed apace (reviewed in Baird et al., 2010; Edwards et al., 2007; Henkin, 2008; Serganov, 2009).
2.2 Catalytic activation of the glmS ribozyme by GlcN6P
Most riboswitches known to date have been discovered by bioinformatic approaches in which phylogenetically conserved elements are sought by aligning non-coding RNA sequences. This approach has been particularly successful in the discovery of bacterial riboswitches, whose 5′-UTRs are short compared to UTRs and introns of typical eukaryotic transcripts. Once a candidate riboswitch is discovered, its cognate ligand has to be established. Often, the biochemical function of the structural genes that are adjacent in the genome, and therefore potentially under control of the candidate riboswitch, offers useful clues. By employing this heuristic, Barrick et al. (2004) discovered a conserved element that is widespread in Gram-positive bacteria upstream of the glmS gene. This gene encodes the protein glucosamine-6-phosphate synthetase (GlmS), an enzyme that catalyzes the conversion of fructose-6-phosphate and glutamine to glucosamine-6-phosphate (GlcN6P) and glutamate (reviewed in Milewski, 2002), the first committed step in the metabolic pathway that leads to synthesis of the bacterial cell wall. As expected from its function, glmS is an essential gene in the model organism Bacillus subtilis (Kobayashi et al., 2003). Chemical probing of the glmS 5′ UTR RNA in the presence of GlcN6P did not reveal protection patterns diagnostic of RNA folding in the presence of the metabolite. Instead, a dramatic increase in the rate of scission of one particular phosphodiester bond of the presumed riboswitch domain was detected. Further characterization established that physiologic concentrations of GlcN6P activate a latent self-cleavage activity of the RNA domain that leads to specific scission of one internucleotide phosphodiester bond, with production of products bearing 2′,3′-cyclic phosphate and 5′-OH termini, analogous to those produce by the previously well characterized hammerhead, hairpin, HDV and VS ribozymes. GlcN6P was not consumed in the reaction. Winkler et al. (2004) showed that, in vitro, it is possible to convert the glmS 5′-UTR into a GlcN6P-dependent multiple-turnover catalyst, that is, a ribozyme.
2.3 Regulation of gene expression by the glmS ribozyme
How does the GlcN6P-induced self-cleavage activity of the glmS ribozyme lead to the modulation of gene expression? In Gram-positive bacteria, intact mRNAs are capped by a 5′ triphosphate. Activation of the glmS ribozyme by high levels of GlcN6P releases a short (~25 nt) leader sequence that bears the triphosphate, and exposes a new 5′-OH group in the ribozyme domain. This hydroxyl group is recognized by RNase J1, a ribonuclease conserved among bacteria, which degrades the mRNA. Because the GlmS protein is unstable, degradation of its mRNA results in down-regulation of the enzymatic activity, thus completing a negative-feedback loop (Figure 1). The physiologic importance of the glmS ribozyme has been demonstrated by employing ribozyme-riboswitch mutants that lack GlcN6P-induced self-cleavage activity. B. subtilis strains harboring glmS genes downstream of such catalytically defective mutant RNA domains fail to sporulate under conditions that lead to sporulation of wild-type bacteria. Presumably, the differentiation program leading to sporulation requires precise regulation of GlcN6P levels (Collins et al., 2007). The ribozyme regulation of the GlmS activity appears to be restricted to Gram-positive bacteria, whose GlcN6P synthetase is not an allosterically regulated enzyme. The paralogous enzymes of Gram-negative bacteria and eukaryotes are allosterically regulated by GlcN6P; thus the activity of the enzyme is controlled at the protein level (Milewski, 2002). In addition, it was recently found that the model Gram-negative bacterium Escherichia coli regulates expression of its glmS gene employing trans-acting non-coding RNAs that do not bind GlcN6P and are not catalytic (i.e. are neither riboswitches nor ribozymes) (Görke & Vogel, 2008; Reichenbach et al., 2008).
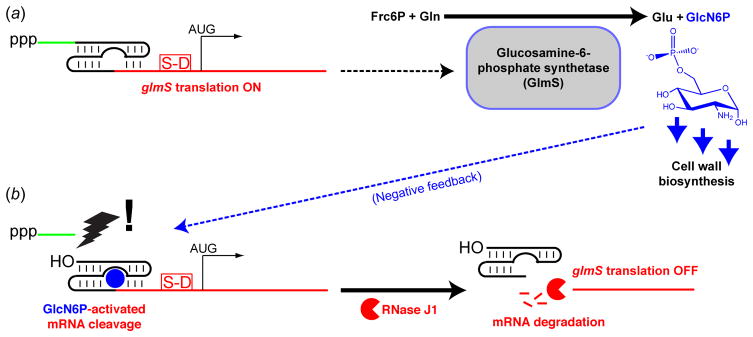
Figure 1.
Schematic representation of the mechanism of gene regulation by the glmS ribozyme. (a) The glmS mRNA comprises a triphosphate cap, a leader sequence (green) preceding the ribozyme domain (black) and a conventional Shine-Delgarno sequence (S-D) followed by the initiation codon for the ORF encoding the protein Glucosmine-6-phosphate synthetase (GlmS). This enzyme catalyzes conversion of fructose-6-phosphate and glutamine into glutamate and glucosamine-6-phosphate (GlcN6P). The latter serves as the starting point for the synthesis of the bacterial cell wall. (b) When GlcN6P accumulates cytoplasmically, it binds to the glmS ribozyme domain, activating a latent self-cleavage activity. This releases the leader sequence, exposing a new 5′-OH terminus in the cleaved mRNA. RNase J1 recognizes the 5′-OH, and degrades the glmS mRNA. (Collins et al., 2007).
3. Structure of the glmS ribozyme
3.1 A triply pseudoknotted ribozyme fold
From the biochemical standpoint, the most remarkable feature of the glmS ribozyme-riboswitch is its GlcN6P-induced catalytic activation. Depending on the precise experimental conditions employed, the change in the rate of cleavage between the inactive and fully activated ribozyme can approach 107 (Brooks & Hampel, 2009; Klein et al., 2007b). Deletion analyses demonstrated that RNAs comprising sequences spanning from one nucleotide upstream of the scissile phosphate, denoted as residue (−1) to approximately 75 nucleotides downstream exhibit GlcN6P dependent catalytic activity. However, RNAs comprising residues from (−1) to ~ 145 nt 3′ of the cleavage site exhibited maximal activity. Based on comparative analysis of glmS ribozyme homologs from a number of bacteria, Barrick et al. (2004) and Winkler et al. (2004) proposed a secondary structure for the minimal core (residues −1 to 75) comprised of three conserved stem-loops, denoted as paired regions P1 through P3. These authors demonstrated biochemically that the nucleotides that form the loop (L1) that closes helix P1, which are not phylogenetically conserved, are not functionally important. Moreover, the glmS ribozyme can be split at L1, and the resulting bimolecular RNA constructs perform multiple-turnover catalysis in a GlcN6P-dependent manner. The additional sequences between residues ~ 75 and ~ 150, required for maximal activity, were proposed to fold into a long helix (P4) interrupted by a central bulge (Barrick et al., 2004; Winkler et al., 2004). Subsequent sequence and biochemical analyses led Wilkinson and Been (Wilkinson & Been, 2005) to suggest formation of a canonical H-type pseudoknot between nucleotides near the 3′ terminus of the glmS ribozyme and those in the loop capping helix P3.
In order to elucidate the mechanism of GlcN6P binding and ligand-dependent catalysis by the glmS ribozyme, we set out to determine its three-dimensional structure. As is customary in crystallographic structure determination, homologs from a number of different species were subjected to crystallization experiments. A ribozyme construct spanning nucleotides −1 through 145 of the glmS ribozyme from the thermophilic Gram-positive bacterium Thermoanaerobacter tengcongensis (Xue et al., 2001) produced highly ordered crystals (Klein & Ferré-D’Amaré, 2009). This crystallization construct is comprised of two RNA chains: a synthetic oligonucleotide that corresponds to residues (−1) to the tip of loop L1, and a longer strand (made by in vitro transcription) that includes residues from L1 to the end of the conserved ribozyme domain. Assembly of the crystallization employing a synthetic oligonucleotide that spans the scissile phosphate between residues (−1) and (+1) allowed facile introduction of chemical modifications nearby to inhibit cleavage, analogous to previous studies on the hammerhead (Scott et al., 1995) and hairpin (Rupert & Ferré-D’Amaré, 2001) ribozymes. The structure of the ribozyme was solved by multiple isomorphous replacement (Klein & Ferré-D’Amaré, 2006) and refined, ultimately against diffraction data extending to 1.7 Å resolution (Klein et al., 2007b) (Figure 2). Subsequently, the structure of the glmS ribozyme from B. anthracis (identical in sequence that from B. cereus, which has been employed for some biochemical studies) was determined at ~ 2.5 Å resolution (Cochrane et al., 2007). As expected from the high degree of sequence conservation, the glmS ribozymes from T. tengcongensis and B. anthracis adopt virtually identical structures. Since the precision of the atomic coordinates of the T. tengongensis structure is markedly higher, this structure will be discussed hereafter and its numbering scheme employed throughout.
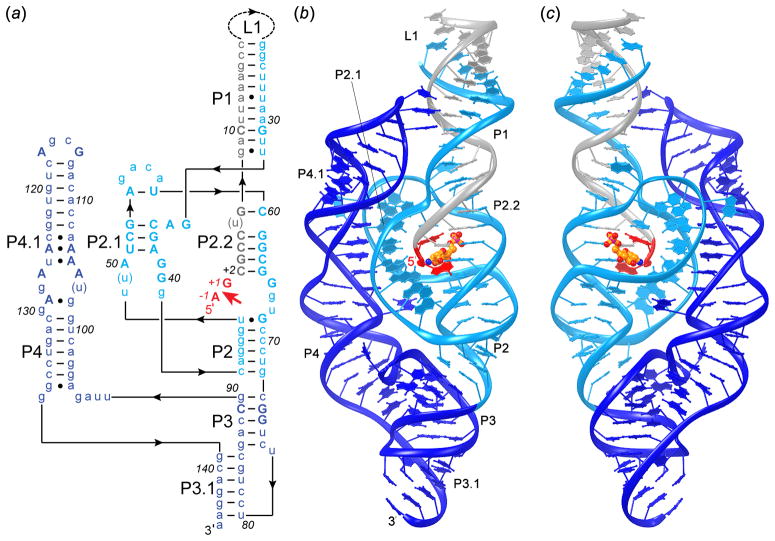
Figure 2.
Overall structure of the glmS ribozyme. (a) Schematic secondary structure of the glmS ribozyme from Thermoanaerobacter tengcongensis based on its crystal structure. Nucleotides that flank the scissile phosphate are colored red. Helices (or paired regions) are numbered according to convention. In the RNA construct employed for crystallization, there is a covalent break in loop L1. This construct comprises two RNA chains: the substrate (red and gray) and the ribozyme (blue). The core of the ribozyme is colored in cyan; the peripheral P4–P4.1 region in dark blue. Nucleotides that are >90% conserved throughout phylogeny are in upper case letters. Lines with embedded arrowheads denote connectivity. (b) Ribbon representation of the crystal structure, colored as in (a); the bound GlcN6P is yellow (Klein & Ferré-D’Amaré, 2006). (c) View from the side opposite (b). Structure figures prepared with Ribbons (Carson, 1997).
The crystal structure revealed that the glmS ribozyme is comprised not of four stem-loops and a pseudoknot, but of three parallel helical stacks (Figure 2) comprising a total of three pseudoknots (Figure 3). Parallel packing of the three helical stacks results in a molecule with approximate dimensions of 100 × 50 × 20 Å3 (the smallest dimension being that of the width of an RNA double helix). The minimal functional core of the ribozyme [red, gray and cyan in Figure 2, residues (−1) to 73] is comprised of four helices P1, P2, P2.1 and P2.2. Three of these (P1, P2 and P2.2) stack coaxially, and this stack packs side-by-side with the short helix P2.1, thus forming a compact, contiguous structure. The four helices are connected by four strand crossovers that define two nested pseudoknots (Figure 3a). The scissile phosphate and the bound GlcN6P lie at the center of this core structure, with the metabolite binding site in the major groove of P2.2. Consistent with this, the most highly conserved nucleotides of the glmS ribozyme are in P2.1, P2.2, and the non-helical segments connecting these two helices (Klein & Ferré-D’Amaré, 2006).
Figure 3.
Three pseudoknots form the core of the glmS ribozyme (Klein & Ferré-D’Amaré, 2006). (a) Ribbon representation of the core of the ribozyme shown from the same direction as Figure 2c. Nucleobases that form Watson-Crick pairs are depicted as short cylinders. Nucleotides not forming such pairs are omitted for clarity. Arrows denote direction of the polynucleotide chain. M and m denote the major and minor grooves, respectively. (b) Canonical H-type pseudoknot formed by helices P3 and P3.1. Dashed arrows indicate that the minor groove crossover is comprised of the entirety of helices P4 and P4.1
3.2 Stabilization of the structure by a peripheral element
The peripheral domain comprised by the coaxial stack of helices P4 and P4.1 (dark blue in Figure 2) is connected covalently to the ribozyme core through the predicted H-type pseudoknot formed by helices P3 and P3.1 (Figures 2 and 3b), and two sets of tertiary interactions: packing of the oblique A-minor stack against P2.1 (see below), and a class-I A-minor interaction (Nissen et al., 2001) of the GNRA tetraloop capping the peripheral domain with the conserved C10-G31 base pair in P1. Association of the peripheral and core domains of the ribozyme buries a total of ~2500 Å2 of solvent accessible surface area, and generates the most solvent-inaccessible interface in the RNA. The pseudoknotted helices P3 and P3.1 stack coaxially with P1, P2, and P2.2 to form the longest helical stack (~ 100Å) of the ribozyme. An H-type pseudoknot is defined by two helices and three connecting loops (Aalberts & Hodas, 2005). In the glmS ribozyme peripheral domain, the first loop crosses the major groove of P3.1 and is comprised of a single nucleotide (U79). As in the majority of H-type pseudoknots (Klein et al., 2009), the P3/P3.1 pseudoknot has a second loop of length zero. The third loop crosses the minor groove of P3 and is unusually long, being in effect comprised of the entire 48 nt P4/P4.1 coaxial stack (Figure 3) (Klein & Ferré-D’Amaré, 2006).
3.3 The oblique A-minor motif
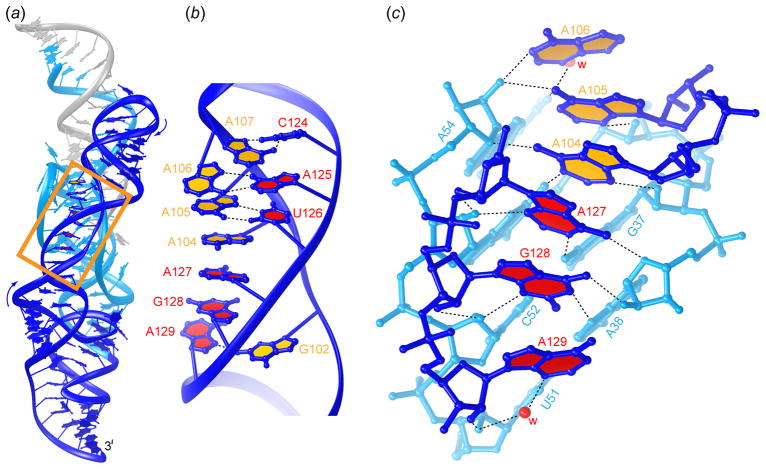
The central portion of the P4/P4.1 stack, which includes five highly conserved adenosine residues, folds into an overwound helix that packs at an oblique angle against P2.1 (Figure 4a). The conformation of this portion of the peripheral domain results in the purine nucleobases of three residues from each strand stacking on each other and projecting into the minor groove (Figure 4b). Oblique (~70°) docking of this structure against the minor groove of P2.1 differs from a stack of canonical A-minor interactions (Nissen et al., 2001) in which each adenosine contacts base and sugar atoms from a single base pair. Instead, A105 and G128 of the glmS ribozyme contact two, and A104 and A127 contact three consecutive base pairs of P2.1 (Figure 4c). Although stacks of inclined adenosines that pack against the minor groove of a helix have been observed previously [e.g. in the TPP riboswitch (Serganov et al. 2006)], the stack in the glmS ribozyme bridges both strands of the helix against which it packs. This oblique A-minor motif, which was first described in the glmS ribozyme (Klein & Ferré-D’Amaré, 2006), has since also been observed in the crystal structure of the Class-II S-adenosylmethionine riboswitch, in which loop nucleotides of an H-type pseudoknot contact the minor groove of a helix in a similar manner (Gilbert et al., 2008). The P4/P4.1 peripheral domain of glmS riboswitches is quite variable across phylogeny (Roth et al., 2006), and it is possible that other structural solutions to the stabilization of the core domain exist in ribozymes from organisms distantly related to T. tengcongensis and B. anthracis.
Figure 4.
The oblique A-minor motif forms the interface between the core and the peripheral domains (Klein & Ferré-D’Amaré, 2006). (a) Representation of the entire ribozyme, seen from the direction of P4 and P4.1. In this figure, all the nucleotides in the substrate strand are colored gray. Nucleobases of the ascending and descending strands of the oblique A-minor motif are filled in yellow and red, respectively. Note oblique angle of the region boxed in gold (dark blue) with respect to the core of the ribozyme (cyan). (b) Detailed view of the boxed region in panel (a). Black dashed lines denote hydrogen bonds between nucleobases. (c) Ball-and-stick representation of the oblique A-minor interaction formed between the peripheral domain nucleotides 104–106 and 127–129, and the core domain. Red spheres labeled “w” depict water molecules.
4. Catalysis of phosphodiester cleavage by proteins and RNA
4.1 RNase A: a canonical protein nuclease
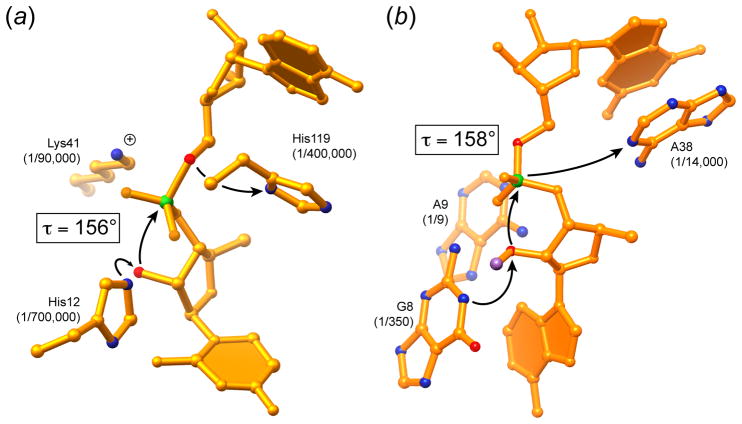
In discussing the catalytic mechanism of the glmS ribozyme, the mechanism of a protein nuclease, and those of the other well-characterized natural self-cleaving ribozymes provide useful frames of reference [reviewed in (Fedor, 2009; Ferré-D’Amaré & Scott, 2010; Lilley & Eckstein, 2008; Raines, 1998)]. RNase A is a protein nuclease that catalyzes RNA cleavage through the same overall chemical mechanism as the glmS ribozyme and other small self-cleaving RNAs (unlike the ribozymes, the protein enzyme hydrolyzes the cyclic phosphate in a subsequent step.) Although RNase A exhibits limited sequence specificity, it achieves a rate enhancement of ~1011 over the uncatalyzed reaction. The principal factors responsible are schematized in Figure 5a (Raines, 1998). Like all enzymes, RNase A overcomes the entropic and steric penalties of bringing the reactants into the active site in a productive conformation through binding energy. The transesterification reaction proceeds through a concerted SN2 mechanism which requires that the 2′ ribose oxygen, the phosphorus, and the 5′ ribose oxygen be aligned. Richards et al. (1971) solved a crystal structure of the enzyme bound to a dinucleotide mimic inhibitor (made non-cleavable by replacing the 5′-oxo leaving group with a methylene) and discovered that RNase A splays apart the nucleotides flanking the scissile phosphate to achieve a nearly linear arrangement of the three reactive atoms. [This angle, denoted τ (Soukup & Breaker, 1999), is typically ~150°, not 180°, in inhibitors bound to crystal structures, which represent the ground state –greater τ angles require that atoms be brought closer than their van der Waals radii.] This structure revealed the location of two catalytic histidines and one lysine. His12 functions as a general base catalyst, deprotonating the 2′-OH, and His114 functions as a general acid, protonating the 5′-oxo leaving group. The reaction proceeds through a trigonal bipyramidal oxyphosphorane transition-state whose excess negative charge is stabilized by the ammonium group of Lys41. As predicted by this mechanism, mutating these catalytic amino acids greatly impairs RNase A (Figure 5a).
Figure 5.
Comparison of the active sites and mechanisms of RNase A and the hairpin ribozyme. (a) Active site of RNase A bound to the dinucleotide mimic inhibitor UpcA (Richards et al., 1971). In this compound, the atom corresponding to the 5′-oxo leaving group has been replaced with a methylene group. The degree of impairment of the enzyme when the catalytic histidines are replaced with alanines, or the catalytic lysine with cysteine (Raines, 1998) is indicated. τ is the angle formed between the nucleophile, electrophile and leaving group of the transesterification reaction. (b) Active site of the hairpin ribozyme (Rupert et al., 2002). The cleavage reaction was inhibited by replacing the hydrogen atom of the nucleophile 2′-OH with a methyl group (magenta). The degree of impairment of the ribozyme when the catalytic nucleotides are replaced with abasic substitutions (Kuzmin et al., 2005; Lebruska et al., 2002) is indicated.
4.2 The small, self-cleaving ribozymes
The hammerhead, hairpin, HDV and VS ribozymes all adopt different three-dimensional structures. Crystal structures have been determined for the first three (Ferré-D’Amaré et al., 1998; Pley et al., 1994; Rupert & Ferré-D’Amaré, 2001; Scott et al., 1995), while molecular models for the VS ribozyme have been developed based on biochemical characterization, FRET and SAXS data (Lilley, 2004). The hammerhead, hairpin and VS ribozymes are organized around multi-helical junctions (Lilley, 1999), and differ in this fundamental manner from the glmS ribozyme. The hammerhead ribozyme folds around a three-helix junction, and its overall Y-shaped architecture (de la Peña et al., 2009) is stabilized by loop-loop interactions distal to the junction (Martick & Scott, 2006). The active site of the hairpin ribozyme results from precise juxtaposition of the minor grooves of two irregular helical stems (Rupert & Ferré-D’Amaré, 2001). Association of the two helices is facilitated by a four helix junction from which they radiate (Murchie et al., 1998). The VS ribozyme is larger than the hammerhead or hairpin ribozymes (being ~150 nt long) and its structure comprises two separate three-helix junctions (Lipfert et al., 2008). All three ribozymes bind their substrate through canonical Watson-Crick base-pairing to nucleotides both 5′ and 3′ to the cleavage site (although not to the nucleotides immediately adjacent to the scissile phosphate). This base-pairing specifies the cleavage site, and has the consequence that the substrate RNA is presented to the ribozyme in the context of an RNA duplex. Because of this, both products of RNA cleavage may remain associated with the ribozyme through base pairing, allowing the reverse ligation reaction to be catalyzed by the hammerhead, hairpin and VS ribozymes. The efficiency of the cleavage and ligation reactions (i.e. the internal equilibrium) depends on the specifics of the RNA constructs and solution conditions. The HDV ribozyme differs from the hammerhead, hairpin and VS ribozymes in that its structure is organized not around a helical junction, but by a double pseudoknot. As in the case of the core of the glmS ribozyme, the four helical crossovers that define its fold allow close parallel packing of two helical stacks, with the active site lying in the interhelical interface. Nonetheless, the glmS and HDV ribozyme folds are completely different: the scissile phosphate faces the major groove of P2.1 in the glmS ribozyme and the minor groove of P3 in the HDV ribozyme. Unlike the ribozymes organized around helical junctions, the HDV and glmS ribozymes only base pair with their substrates 3′ to the site of cleavage. As a result, the 5′ product of the cleavage reaction (which bears a 2′,3′ cyclic phosphate) readily dissociates from the ribozymes after scission, and neither ribozyme catalyzes the ligation reaction [reviewed in (Ferré-D’Amaré & Scott, 2010)].
4.3 Nucleobase participation in ribozyme catalysis
In large measure, the catalytic power of RNase A derives from its use of histidine side-chains, with pKa’s close to neutrality, and a lysine side-chain, whose side-chain amino group is positively charged at physiologic pH (Figure 5a). RNA has no functional groups with analogous ionization properties. The groups with pKa’s closest to neutrality are the N1 nitrogens of purines (3.5 and 9.2 for A and G, respectively) and the N3 nitrogens of pyrimidines (4.2 and 9.2 for C and U, respectively). As a polyanion, however, RNA binds cations, and in principle, these can function either as electrostatic catalysts, as Lewis acids that perturb the pKa of RNA functional groups bound to them, or as Brønsted-Lowry acids that lower the pKa of the cation-coordinated waters. Thus, RNA-bound cations might assist in catalysis by providing a localized positive charge, or by providing hydroxide or hydronium ions to function as reactants or specific acid or base catalysts. Because the Group I intron has been demonstrated to employ tightly-bound Mg2+ ions for electrostatic transition-state stabilization and as Lewis acids, it was widely believed that all catalytic RNAs would employ bound metal ions as cofactors (Pyle, 1993). It is now known that this is not the case for the small self-cleaving ribozymes.
The hammerhead, hairpin and VS ribozymes are fully active in the presence of high concentrations of monovalent ions alone, or if Mg2+ is replaced with cobalt (III) hexammine. The latter is an isoster of hexahydrated Mg2+. However, unlike the waters of hydration of Mg2+, the amine ligands of cobalt hexammine do not dissociate, and therefore this complex ion cannot function as a Lewis acid or perturb the pKa of hydration waters (Cowan, 1993). Thus, activity of the hammerhead, hairpin and VS ribozymes implies that RNA functional groups are catalytic. The structure of the hairpin ribozyme (Figure 5b) shows that the bound substrate is distorted to τ ~ 158°, and that three conserved nucleobases are positioned in a manner reminiscent of catalytic amino acid residues in the active site of RNase A. Thus, G8 and A38 may function as general base and acid catalysts, respectively, in the cleavage reaction (their roles would be reversed for the ligation reaction), and A9 may provide electrostatic stabilization of the transition state. Because the total rate enhancement achieved by the hairpin ribozyme (~ 106) is modest compared to that of RNase A, individual deletion of these nucleobases, while detrimental to catalysis, does not have effects of nearly the same magnitude as individual mutation of RNase A active site residues (Figure 5a). Nonetheless, the effect of deletions is approximately additive, as in the case of the protein enzyme (Figure 5b).
The HDV ribozyme, which can achieve rate enhancements of nearly 1010, supplements a catalytic nucleobase with a catalytic metal ion (Nakano et al., 2000). The highly negatively charged environment of the active site of the HDV ribozyme perturbs the pKa of the N3 group of the catalytic C75 residue of the RNA, raising it from 4.2 to ~ 7. Experiments in which the stability of the leaving group of the cleavage reaction was altered demonstrate that C75 functions as a general acid catalyst (Das & Piccirilli, 2005). While this nucleobase provides the bulk of the rate enhancement achieved by the HDV ribozyme, the ribozyme activity decreases in the absence of divalent cations. This is because a hydrated cation in the active site facilitates cleavage by delivering a water with a perturbed pKa that functions as a specific base catalyst.
5. GlcN6P is a coenzyme of the glmS ribozyme
5.1 Allosteric activator or coenzyme function for GlcN6P?
In principle, activation of the glmS ribozyme by GlcN6P could occur through two different mechanisms. Binding of GlcN6P to the RNA could drive a conformational change of the RNA; that is, GlcN6P might function as an allosteric activator. Alternatively, GlcN6P may bind in the active site of the ribozyme, and provide a catalytic functional group, that is, function as a coenzyme. Although the two mechanisms are not mutually exclusive, the available biochemical and structural evidence indicates that GlcN6P is a coenzyme of the glmS ribozyme (Klein & Ferré-D’Amaré, 2006; McCarthy et al., 2005).
5.1.1 Importance of the amine group of GlcN6P
Characterization of the ligand specificity of the glmS ribozyme indicates that several small molecules other than GlcN6P can activate the RNA, although to a lesser extent than the cognate activator. Thus, glucosamine-6-sulphate (GlcN6S), glucosamine (GlcN), Tris (2-amino-2-hydroxymethyl-propane-1,3-diol), L-serine, serinol (2-amino-propane-1,3-diol), and even ethanolamine, can activate cleavage of the glmS ribozyme if present at elevated concentrations. Of these compounds, GlcN6S and GlcN were the most potent activators, which is not surprising given their structural similarity to GlcN6P. Remarkably, glucose-6-phosphate (Glc6P) which differs from GlcN6P only in having a hydroxyl group rather than an amino group at position 2 of the pyranose ring, was found not to be an activator of the ribozyme. Instead, Glc6P can function as an antagonist, inhibiting GlcN6P-activaed cleavage of the glmS ribozyme when added in high concentrations. This observation suggests a critical function for the amine group of GlcN6P in glmS ribozyme activation (McCarthy et al., 2005; Winkler et al., 2004).
5.1.2 Similarity of crystal structures in multiple states
We determined crystal structures of the glmS ribozyme in multiple functional states by employing either a substrate or inhibitor oligonucleotide (§ 3.1), and varying the small molecule employed for cocrystallization. This allowed us to compare three pairs of states that bear on the question of allostery vs. coenzyme function for GlcN6P. First, by replacing the 2′-OH nucleophile of residue (−1) with either a hydrogen or an amine group it was possible to inhibit RNA cleavage in the presence of saturating concentrations of GlcN6P. This led to structures of the ribozyme bound to GlcN6P. Because the ribozyme is inactive in the absence of GlcN6P, a structure of the ribozyme bound to an intact all-ribose (i.e. cleavable) substrate oligonucleotide was solved free of activator. Remarkably, both structures superimpose on each other with root-mean-square (r.m.s.) differences comparable to the precision of their atomic coordinates (0.5 Å or less); that is, the structures are experimentally identical, indicating that GlcN6P binding does not induce measurable conformational changes in the RNA structure (Klein & Ferré-D’Amaré, 2006; Klein et al., 2007b). Second, since Glc6P does not activate the ribozyme, a structure of the ribozyme bound to an all-ribose substrate oligonucleotide and to Glc6P was also determined. Comparison of the Glc6P-bound and GlcN6P-bound structures also revealed precise superposition (Klein & Ferré-D’Amaré, 2006; Klein et al., 2007b). This suggests that inhibition of ribozyme activity by Glc6P is not a result of an inhibitory conformational change in the ribozyme active site, and, conversely, that GlcN6P activation of the ribozyme results from its chemical differences from Glc6P. Third, the structure of the product state of the ribozyme, that is, bound to an oligonucleotide whose sequence starts at residue (+1), was also determined. Except for residue (−1), which is absent in this structure, the product complex structure also superimposed precisely on the previous structures (Klein & Ferré-D’Amaré, 2006). Comparison of the active sites of precursor and product states demonstrated that even fine details, such as the positions of tightly coordinated water molecules, remained invariant between the inhibitor and product complex structures. Together, these analyses support the hypothesis that GlcN6P does not function as an allosteric activator of the ribozyme, and that the glmS ribozyme does not undergo large conformational changes as it is activated and carries out the cleavage reaction. However, a caveat of these experiments is that the comparison is between the structures of ribozymes in crystals. Thus, it is possible that the lack of measurable structural differences simply indicates that crystal contacts are forcing the RNA to adopt the same conformation in all examined states.
5.1.3 Activation and catalysis in the crystalline state
In order to evaluate whether the conformation of the glmS ribozyme observed in the crystal structures is capable of binding the activator productively and carrying out chemical catalysis, we performed a series of post-crystallization soaking experiments (Klein & Ferré-D’Amaré, 2006). As previously, we crystallized the glmS ribozyme bound to an all-ribose substrate RNA strand, in the absence of Glc6P or GlcN6P. These crystals were then soaked briefly (~ 5 minutes) in solutions containing saturating concentrations of Glc6P before flash-cooling and diffraction data collection. The resulting structure showed an RNA in identical conformation to those of structures obtained previously, with Glc6P bound also in the same location. This experiment demonstrated that Glc6P can diffuse into crystals of the glmS ribozyme and bind to the RNA. Therefore, the conformation of the RNA in the crystalline state is competent for small molecule binding, and small molecule binding does not require that the RNA undergo large-scale conformational changes, since such excursions would not be compatible with crystalline packing. We then briefly soaked crystals from the same crystallization experiment in solutions containing saturating concentrations of GlcN6P before flash-cooling and diffraction data collection. The resulting structure showed an RNA with unchanged conformation, but lacking any electron density for residue (−1) or the scissile phosphate. This indicates that the cleavage reaction has proceeded to completion, and that the 5′ RNA cleavage product has left the active site of the ribozyme, consistent with the inability of the glmS ribozyme to carry out the ligation reaction. This experiment indicates that the crystalline glmS ribozyme can be activated for cleavage by GlcN6P, and that the cleavage reaction can take place without the RNA undergoing large conformational changes. Together, these crystallographic experiments indicate that GlcN6P binding, ribozyme activation, and catalysis do not require that the RNA undergo measurable conformational changes (Klein & Ferré-D’Amaré, 2006).
Three additional lines of evidence support the conclusion that neither GlcN6P binding nor RNA cleavage by the glmS ribozyme is accompanied by large conformational changes. First, structures have been determined of the glmS ribozyme from B. anthracis in the precursor state, free and bound to GlcN6P and Glc6P, and in the product state (Cochrane et al., 2009; Cochrane et al., 2007). All these structures also superimpose closely on each other. Since the B. anthracis RNA differs in sequence from the T. tengcongensis RNA, and crystallizes in a different unit cell under different crystallization conditions, this similarity reinforces the conclusion that the specific crystalline environment of either RNA is not suppressing functionally important conformational changes. Second, biochemical analyses of the B. subtilis glmS ribozyme in different functional states employing hydroxyradical footprinting and UV crosslinking (Hampel & Tinsley, 2006), as well as terbium and RNase V1 footprinting (Tinsley et al., 2007), did not reveal any conformational changes. Third, FRET analysis of trans-acting forms of the B. subtilis ribozymes in the pre- and post-cleavage states also indicated that the ribozyme is folded prior to catalysis, and does not change conformation as a result of the cleavage reaction (Tinsley et al., 2007).
5.2 Active site structure
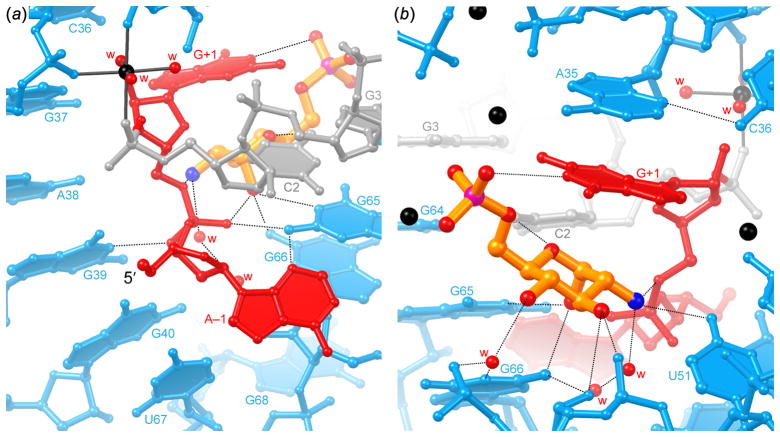
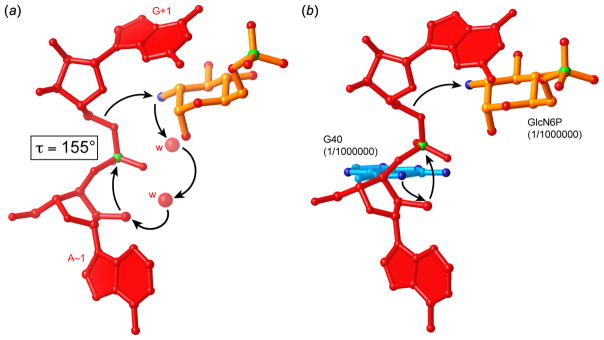
What is the organization of the glmS ribozyme active site, and if GlcN6P is a coenzyme, how does it complement it? The nucleotides flanking the scissile phosphate, (A–1) and (G+1) traverse the thin dimension of the core of the ribozyme in a conformation that brings the reactive groups of the substrate into a near-in-line conformation (τ = 155° for the 2′-aminoribose structure). The nucleobase of (A–1) makes a hydrogen bond with that of G65. An adenosine at the (−1) position is conserved among glmS ribozymes. While the single hydrogen bond would not specify an A at this position, replacing a G into this position would result in steric clash with G65 (Figure 6a). The scissile phosphate is held in place by hydrogen bonds from G39 and G65. Nucleotide (G+1) projects onto the other face of the ribozyme, where its nucleobase forms the roof of the GlcN6P binding pocket (Figure 6b). Although the nucleobase of (G+1) does not make any hydrogen bonds with the rest of the RNA, its position is defined by its stacking underneath the base of A35, and the sharp bend in the phosphate-ribose backbone following it. This bend results from the formation of canonical Watson-Crick pairs in P2.2 starting with residue C2, and appears to be stabilized by a tightly-bound magnesium ion that coordinates the phosphates of C2, C36 and G37 (Klein & Ferré-D’Amaré, 2006).
Figure 6.
Active site of the glmS ribozyme. (a) View from the direction of Figure 2b. A partially dehydrated hexacoordinate magnesium ion is depicted as a black sphere, linked to its coordinating atoms with thin black lines. Note position of the 5′-OH of nucleotide (−1). Nucleotides upstream of the ribozyme domain in the natural transcript would approach the active site from this direction. For this crystal structure, the ribozyme was prevented from cleaving the substrate by replacing the ribose of nucleotide (−1) with a 2′-deoxyribose. (b) View from the direction of Figure 2c. The GlcN6P binding pocket includes several well-ordered magnesium ions (black spheres), and is extensively hydrated. Only selected water molecules are shown for clarity.
5.2.1 GlcN6P recognition
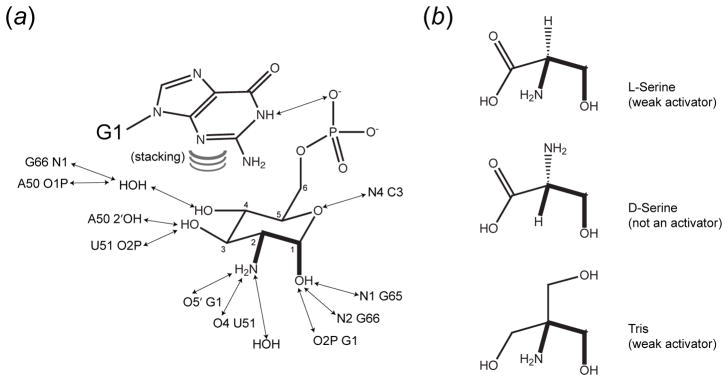
The activator, GlcN6P, binds to an open, hydrated binding site that is lined with phylogenetically conserved residues, burying ~80% of its solvent-accessible surface area (Klein & Ferré-D’Amaré, 2006). Nucleotides that line the ligand-binding pocket and active site are highly conserved phylogenetically. In addition to (A–1) and (G+1), nucleotides conserved in more than 90% of glmS ribozyme sequences include residues 35–40 and 64–65 on the side opposite the GlcN6P binding pocket, and residues 2, 35–36, 50–53, and 59–65 in the GlcN6P pocket (Klein & Ferré-D’Amaré, 2006). Although GlcN6P exists as a mixture of the α and β anomers in solution, in a ratio of ~ 40:60, with less than 1% in the acyclic conformation (Lim et al., 2006), it binds to the RNA exclusively in the α-axial anomeric conformation. The interactions made by GlcN6P are summarized in Figure 7a. Studies of the ability of non-cognate small molecules to activate the glmS ribozyme point to the importance of the stereochemical arrangement of the anomeric OH and the amine at position 2. Thus, L-serine, whose side-chain OH and amine groups are in the same stereochemical relationship as the anomeric OH and amine groups of GlcN6P (Figure 7b) is an activator of the glmS ribozyme, but D-serine is not (McCarthy et al., 2005). Non-chiral activators of the ribozyme all have vicinal amine and hydroxyl groups. Thus, Tris (Figure 7b) and ethanolamine are activators, but neither methylamine nor ethanol are (McCarthy et al., 2005).
Figure 7.
Recognition of glucosamine-6-phosphate and analogs by the glmS ribozyme (Klein & Ferré-D’Amaré, 2006). (a) Schematic depiction of interactions responsible for recognition of GlcN6P. Hydrogen bonds are shown as two-headed arrows. Curved lines depict stacking of the nucleobase of G1 on the sugar. Numbering scheme for the sugar is indicated. (b) L-serine and Tris function as weak activators of the glmS ribozyme in vitro; D-serine does not (McCarthy et al., 2005). The three molecules are drawn to emphasize their similarity to pyranose ring positions 1 and 2 of GlcN6P.
The functionally critical anomeric OH and amine groups of GlcN6P each make three hydrogen bonds. The anomeric OH hydrogen bonds to the N1 and N2 groups of G65 and G66, respectively, and to a non-bridging oxygen of the scissile phosphate (Figures 6a, 7a). The amine hydrogen bonds to the 5′ oxygen of (G+1) (the leaving group of the cleavage reaction), to O4 of U51, and to a well-ordered water molecule seen in all high-resolution structures (Figures 6b,7a). The OH group in position 3 of GlcN6P hydrogen bonds to both the 2′-OH of A50 and to a non-bridging phosphate oxygen of A50. The OH group at position 4 of GlcN6P only makes water-mediated contacts to the RNA, and the pyranose ring oxygen accepts a hydrogen bond from the exocyclic amine of C3. The N1 imine of residue (G+1) of the glmS ribozyme donates a hydrogen bond to one of the non-bridging oxygens of the phosphate of GlcN6P. In addition, the nucleobase of (G+1) stacks on the sugar (Figure 7a) (Klein & Ferré-D’Amaré, 2006; Klein et al., 2007b). Most of these interactions between the ribozyme and GlcN6P are also observed in the crystal structure of the B. anthracis RNA (except for the water-mediated interactions, not resolved due to the low resolution of that structure) (Cochrane et al., 2007).
The importance of the interaction between the N1 imine of (G+1) and the phosphate of GlcN6P for ribozyme function was explored by substituting this RNA residue with a series of natural and unnatural purine nucleotides and comparing the rates of cleavage of the variant RNAs when activated by either GlcN6P or GlcN (Klein et al., 2007b). With the wild-type ribozyme bearing a guanine at position (+1), GlcN6P is the stronger activator. This preference for the phosphate-bearing activator is maintained when (G+1) is replaced with inosine, another purine whose N1 position is protonated at neutral pH. However, when (G+1) is replaced with a purine whose N1 position is not protonated at neutral pH, such as A, 2-aminopurine, or 6-dimethyladenine, the selectivity of the ribozyme switches. These ribozymes are more active in the presence of GlcN than in the presence of GlcN6P. These observations were interpreted to mean that if the N1 imine of the purine residue at position (+1) of the glmS ribozyme is not protonated, then steric clash with the phosphate of GlcN6P is sufficiently detrimental to ribozyme activity that GlcN, not having the phosphate, is a better activator. In these studies, the variant ribozymes with mutations or non-natural purines at position (+1) were all less active than wild-type, regardless of whether GlcN or GlcN6P was employed for activation. This indicates that aspects other than the hydrogen bond between the N1 imine and the phosphate of GlcN6P (or the relief of steric clash with the phosphate) are important for ribozyme function (Klein et al., 2007b).
5.2.2 Role of metal ions in glmS ribozyme function
Like other complex RNAs (reviewed in Draper, 2004), the glmS ribozyme requires divalent cations to adopt a native structure under physiological concentrations (Cayley et al., 1991) of monovalent cations (Brooks & Hampel, 2009). Biochemical analyses (Roth et al., 2006; Winkler et al., 2004) have shown that the glmS ribozyme is fully active not only in the presence of physiologic (Romani & Scarpa, 1992) concentrations of Mg2+, but also under similar concentrations of Mn2+ or Ca2+, or indeed, of cobalt hexammine (§ 4.3). Monovalent ions at molar concentrations support glmS catalysis at rates ~ 1% of those supported by Mg2+. These results indicate that the ribozyme is employing the cations for folding, not catalysis, and that the higher charge density of divalent (Mg2+, Mn2+, Ca2+,) or trivalent (cobalt hexammine) cations compared to monovalent cations is needed for native folding. The crystal structures reveal a number of Mg2+ binding sites, the best ordered or which is formed by the sharp turn of the RNA backbone at the 5′-end of P2.2 (Figure 6a). Although this Mg2+ ion makes three inner-sphere coordinations, full biochemical activity in cobalt hexammine implies that an outer-sphere coordinated cobalt hexammine can functionally occupy this binding site. This structural plasticity is reminiscent of the cation binding sites that are important for function of other RNAs, for instance, the hairpin ribozyme (Ferré-D’Amaré, 2004) or the FMN riboswitch (Serganov et al., 2009).
The crystal structures show no metal ions bound in the immediate vicinity of the scissile phosphate (Klein & Ferré-D’Amaré, 2006). This is consistent both with the metal ion substitution experiments summarized above, and with the results of experiments in which the scissile phosphate was replaced with a phosphorothioate. Sulfur is both a larger and a softer (more polarizable) atom than oxygen. Thus, replacement of a phosphate with a phosphorothioate at the cleavage site of a ribozyme can be employed to probe for the effect of the larger sulfur atom or, in conjunction with replacement of hard metal ions such as Mg2+ with soft ones such as Mn2+, to probe for coordination of one or both non-bridging phosphate oxygens by a hard metal ion in the wild-type (Jaffe & Cohn, 1979). If a diasteromeric mixture (Burgers & Eckstein, 1979) of chemically synthesized phosphorothiaoates is introduced at the cleavage site of a ribozyme, and one of the non-bridging phosphate oxygens is engaged in a functionally essential coordination of a hard cation, then half the substrate should be rendered uncleavable. This inhibition should be rescued in the presence of a soft cation. In the case of the glmS ribzyme, the rate of cleavage of a substrate containing a diasteromeric phosphorothioate mixture at the cleavage site is reduced to ~1/3 of the wild-type substrate. This inhibition is not rescued in the presence of soft cations. Moreover, at long time-points, the phosphorothioate-containing substrate is cleaved to equal extents in either hard or soft cations (Roth et al., 2006). These results are consistent with the absence of cations near the scissile phosphate in the crystal structure. The inhibition of the ribozyme by phosphorothioate substrates is also consistent with the close apposition of ribozyme and GlcN6P functional groups with the scissile phosphate (Figure 6), that is, it probably arises from steric clashes.
Crystal structures reveal that natural and artificial riboswitches that bind to anionic ligands recognize them as chelates of divalent cations (Serganov et al., 2006; Serganov et al., 2009; Xiao et al., 2008a). Although several Mg2+ ions are observed in the vicinity of the phosphate moiety of GlcN6P bound to the glmS ribozyme in the 1.7 Å resolution structure (Figure 6b), none appear to be tightly coordinated based on two criteria. First, the distances separating the Mg2+ ions and the phosphate oxygens of GlcN6P are all greater than 2.5 Å, indicating that none of the cations make inner-sphere (direct) coordination with the phosphate. Second, the GlcN6P binding site of the glmS ribozyme is heavily hydrated, and, consequently, the Mg2+ ions occupying it are surrounded by well-ordered water molecules. Yet, none of the Mg2+ ions in the GlcN6P binding pocket have ligands that complete their octahedral coordination shell with good geometry (Klein et al., 2007b). This differs from the highly ordered Mg2+ ion that coordinates to the phosphates of C2, C36 and G37 (Figure 6a), and indicates that the Mg2+ ions and associated waters in the GlcN6P-binding site do not have one largely preferred arrangement, i.e., they are statically disordered. This contrasts, for instance, with the partially hydrated Mg2+ ions chelating the pyrophosphate moiety of the thiamine pyrophosphate bound to its cognate riboswitch. In that case, the cations are highly ordered, and even at 2.5 Å resolution, water molecules forming the first hydration shell of the metal ions could be readily discerned in difference Fourier electron density maps (Edwards & Ferré-D’Amaré, 2006). Thus, in the case of the glmS ribozyme, divalent cations play a non-specific electrostatic role in facilitating binding of GlcN6P, rather than participating directly in small molecule recognition.
5.3 A role for GlcN6P in proton transfer
The fundamental biochemical phenomenon that any proposed catalytic mechanism for the glmS ribozyme must explain is the complete lack of measurable activity of the ribozyme in the absence of GlcN6P. Given the compelling biochemical and structural evidence for a pre-organized active site in which the substrate is distorted into a near-inline conformation prior to GlcN6P binding, the most parsimonious model is that the small molecule provides an essential catalytic functional group. The location of GlcN6P in the active site of the glmS ribozyme is reminiscent of those of Lys 41 and His119 in the active site of RNase A (Figures 5a, 8a). In solution, the pKa of the amine of GlcN6P is 8.2 (the pKa of the phosphate is ~6.2) (McCarthy et al., 2005); therefore, free GlcN6P will mostly exist in its ammonium form under physiologic conditions. Since the ribozyme is strongly negative, the ammonium form of GlcN6P would appear to be a better ligand for the RNA than the amine form. Binding of the ammonium form of GlcN6P to a pre-organized glmS ribozyme would place a positive charge in van der Waals contact with the scissile phosphate, analogous to the ammonium group of the side-chain of RNase A. Thus, GlcN6P could function as an electrostatic catalyst. Moreover, the ammonium group would also hydrogen bond to the 5′-oxo leaving group of the cleavage reaction, and may be able to stabilize it by donating a proton, that is, by carrying out general acid catalysis, analogous to His199 of RNase A (Klein & Ferre-D’Amare, 2006).
Figure 8.
Hypothetical catalytic mechanisms of the glmS ribozyme (Klein & Ferré-D’Amaré, 2006). (a) GlcN6P deprotonates the 2′-OH nucleophile through a proton relay employing two tightly bound water molecules, functioning as a general base catalyst. The resulting ammonium form of GlcN6P functions to stabilize the transition state electrostatically and also donates a proton to the 5′-oxo leaving group, functioning as a general acid catalyst. (b) The N1 imine of G40 functions as a general base catalyst. GlcN6P, in its ammonium form, functions as an electrostatic and general-acid catalyst. The ribozyme is inactive in the absence of GlcN6P or if a G40A mutation is made. The degree of impairment of the ribozyme indicated in parentheses is therefore an estimate based on the in vitro cleavage rate.
The analogy to RNase A can be extended further if it is postulated that the amine form of GlcN6P binds to the ribozyme. The active site of the ribozyme is heavily hydrated and two well-ordered water molecules are seen in all high-resolution structures (Figure 6) that might form a proton relay connecting the amine of GlcN6P to the 2′-OH of (A–1). Thus, the amine of GlcN6P might be able to deprotonate the 2′-OH indirectly, functioning as a general base catalyst (Klein & Ferré-D’Amaré, 2006). Once this has happened, GlcN6P would exist in the ammonium form, and would be able to carry out electrostatic and general acid catalysis as proposed above (Figure 8a). While it might appear fanciful, this proposed mechanism has two attractive features. First, it is consistent with the observation that the RNA itself, even though pre-organized, is completely inactive in the absence of GlcN6P. Therefore, a mechanism in which GlcN6P is responsible for all the catalytic chemistry is parsimonious. Second, mechanisms involving a proton relay through water molecules have been proposed for a number of protein enzymes, for instance, some DNA polymerases (Wang et al., 2007; Wang & Schlick, 2008), the F1-ATPase (Dittrich et al., 2008), and some β-lactamases (Damblon et al., 1996).
Analysis of the activity of the glmS ribozyme as a function of pH also supports a role in proton transfer for GlcN6P. The rate of the ribozyme was found to increase linearly with pH and plateau above pH ~ 8 (Winkler et al., 2004), with a pKa of 7.8 (McCarthy et al., 2005). If the pKa of the reaction corresponds to that of the amine of GlcN6P, this indicates that binding to the ribozyme lowers the pKa of the coenzyme by ~ 0.5 pH units, potentially making it a better general acid-base catalyst. These authors also compared the apparent pKa of the cleavage reaction when the ribozyme is activated by either GlcN or serinol, and found them to be 7.9 and 8.7, respectively. Free in solution, GlcN and serinol have pKa’s of 7.8 and 8.8 respectively. That the reaction pKa tracks closely that of the activator employed is evidence that an ionization event involving the amine group of the activator is part of the rate-limiting step of the reaction. A similar argument has been made in support of the participation of a nucleobase in general acid-base catalysis in the HDV ribozyme, where mutation of the proposed catalytic residue C75 to an A changed the pKa of the reaction from 6.1 to 5.7. Since the pKa’s of the unperturbed N3 of C and N1 of A are 4.2 and 3.5, respectively, this suggests that C75 is a general acid-base catalyst, and that the local environment of C75 in the ribozyme active site raises its pKa by ~ 2 pH units (Nakano et al., 2000; Perrotta et al., 1999).
5.4 A role for G40 in catalysis
G40 is highly conserved in all glmS ribozymes, and crystal structures show that its nucleobase is adjacent to the ribose of (A–1), with the N1 imine of the guanine ~ 3.2 Å from the nucleophilic 2′-OH (Klein & Ferré-D’Amaré, 2006). G40, therefore, could be postulated to function as a general base, deprotonating the nucleophile, analogous to His12 in RNase A and G8 in the hairpin ribozyme (Figure 5). Then, GlcN6P could function as a electrostatic catalyst, a general acid, or both (Figure 8b). A fundamental problem with such a model is that in the absence of GlcN6P, the glmS ribozyme is completely inactive, even though the substrate is in a reactive conformation and G40 is in position. This contrasts greatly from the behavior of other catalysts. Mutation of either of the catalytic histidines of RNase A lowers the activity of the enzyme several hundred thousand-fold, but the mutant enzymes are still active (Figure 5a). Likewise, deletion of either of the general acid-base catalysts of the hairpin ribozyme lowers the activity of the ribozyme several hundred to several thousand-fold, but the ribozymes with the abasic substitutions also retain activity (Figure 5b).
The importance of G40 was evaluated by mutagenesis. A G40A mutant of the glmS ribozyme was found to be completely inactive in the presence of saturating concentrations of GlcN6P. Indeed, the rate of cleavage measured for this mutant was indistinguishable from that of the spontaneous degradation of RNA under the experimental conditions. In principle, this result could simply indicate that the mutant RNA cannot fold correctly, or bind GlcN6P productively. However, the mutant RNA could be crystallized readily. Its structure was solved bound to an all-ribose (cleavable) substrate in the presence of GlcN6P. The crystal structure revealed that the substrate is uncleaved, and that GlcN6P is bound in precisely the same location as in the wild-type ribozyme. Residue 40 is nearly in the same position in the mutant as in wild-type: the distance between the N1 imine of A and the 2′-OH of (A–1) is 4.1 Å instead of 3.2 Å as observed in the wild-type (Klein et al., 2007a).
Because the N1 of G40 is near the nucleophile (the 2′-OH of A–1), it might hydrogen bond to it in the transition state, helping to orient it. Previously, vanadate has been employed to mimic the transition state of the hairpin ribozyme (Rupert et al., 2002). Crystal structures showed that that ribozyme makes more hydrogen bonds to the transition state than to either the precursor or product state. Presumably because the glmS ribozyme does not bind tightly to the 5′ cleavage product (i.e., A–1) it could not be successfully crystallized with vanadate serving as a transition state mimic. Instead, structures were solved of the wild-type and G40A T. tengcongensis glmS ribozymes bound to a an RNA strand in which (A–1) and (G+1) are linked through a 2′-5′ phosphodiester linkage. This type of linkage has been proposed to mimic the transition state of the hairpin ribozyme (Torelli et al., 2007). The structures of the glmS ribozyme bound to such 2′-5′ linked transition-state mimics did not show any additional interactions between G40 (or A40) and the substrate RNA (Klein et al., 2007a). Thus, the available structural information does not support a role for G40 in preferential hydrogen bonding to the transition state.
The inactivity of the G40A mutant is surprising for three reasons. First, if GlcN6P is responsible for all catalysis in the wild-type, its presence in the active site of the mutant structure implies that the mutation should not abrogate all (or any) ribozyme activity. Second, if the N1 nitrogen of the purine at position 40 is functioning as a general base catalyst, then A, whose unperturbed pKa is 3.2 (and is therefore unprotonated at physiologic pH) would be better suited than a G, whose unperturbed pKa is 9.2 (and is protonated at physiologic pH). Third, although the 0.9 Å difference in distance between the N1 imine of G40A and wild-type to the 2′-OH nucleophile could well lead to a decrease in activity, it would be surprising if it led to complete inactivation of the catalyst. Thus, this experiment appears to have uncovered an interdependency in the catalytic activity of GlcN6P and G40: neither is functional in the absence of the other. This is even more surprising in light of the structure of the active site (Figure 6): the closest approach of GlcN6P and residue 40 is 7Å, measured from the amine of GlcN6P to the N1 imine of G40. These two functional groups which appear to interact functionally in the transition state are, in fact, on opposite sides of the scissile phosphate. How it is that the active site of the glmS ribozyme modulates the ability of GlcN6P to function as a catalyst (and vice versa) is a question that remains unanswered.
5.5 pH-dependence of GlcN6P binding
Another unexplained feature of the glmS ribozyme is that the affinity of the ribozyme for GlcN6P increases with pH. This increase in affinity is not sufficient entirely to explain the increase in rate with pH (Cochrane et al., 2007; McCarthy et al., 2005), but is nonetheless very apparent, even in crystallization experiments. Thus, if the glmS ribozyme is crystallized at pH 5.5, GlcN6P binding is not observable crystallographically (even though the ribozyme is active in the crystal at that pH) (Klein & Ferré-D’Amaré, 2006), but at pH 8.5 electron density for the bound coenzyme is readily apparent (Klein et al., 2007b). The phenomenon is likely not due to titration of the phosphate of GlcN6P (which could lead to weaker binding to Mg2+ ions, and decrease the through-cation binding of the phosphate of GlcN6P to the ribozyme) as suggested by Cochrane et al., (2009), because Glc6P binds to the ribozyme comparably at low or high pH. Therefore, it appears that the glmS ribozyme has a “selectivity filter” that allows it preferentially to bind to the amine form of GlcN6P over the ammonium form.
6. Concluding remarks
The results of biochemical and structural experiments make a compelling case for the function of GlcN6P as a coenzyme of the glmS ribozyme. However, the lack of activity of the G40A mutant ribozyme, which appears to be correctly folded suggests that the glmS ribozyme is not a passive scaffold. The result of this mutation suggests that GlcN6P and the ribozyme active site mutually modulate each other (§ 5.4). Although the physical-chemical nature of this interaction is unknown, the precedent of the HDV ribozyme suggests that a ribozyme can tune the pKa of functional groups in its active site. Thus, it is possible that the pKa’s of GlcN6P and G40 (and their ability to function as general acid-base catalysts) depend on each other. This, however, is unlikely to be the whole explanation. It has been argued for the hairpin ribozyme that even if the pKa’s of active-site functional groups are not perfectly tuned, some level of catalysis should obtain (Bevilacqua, 2003). Thus far, all structural information for the glmS ribozyme has been obtained by X-ray crystallography. At the resolutions common for RNA, this technique reveals only the time-averaged location of non-hydrogen atoms. While it is clear that unlike some ribozymes (Xiao et al., 2008b) and conventional riboswitches (Baird & Ferré-D’Amaré, 2010; Kulshina et al., 2009), the glmS ribozyme does not undergo large conformational changes concomitant with ligand binding, subtle changes in structure, and potentially more importantly, in the dynamics of specific ribozyme moieties, cannot be excluded. Thus, the dynamics of the glmS ribozyme, the perturbation of these by binding of GlcN6P, and the effect of active site dynamics on catalysis need to be explored.
Because the glmS ribozyme regulates an essential bacterial gene, and because it has evolved to depend on small-molecule binding for activity, this RNA is in principle an attractive target for the development of new anti-bacterial compounds. Collins et al., (2007) have demonstrated that disruption of the activity of the glmS ribozyme in living B. subtilis is deleterious for fitness. Thus, the stage is set for a discovery of unnatural activators or inhibitors of the ribozyme, and methodology suitable for high-throughput screening of compound libraries against the ribozyme has been described (Blount et al., 2006; Mayer & Famulok, 2006). In this regard, it is interesting that the apparent Kd of the ribozyme for GlcN6P is only of the order of 100 μM (Cochrane et al., 2007; McCarthy et al., 2005). Because the glmS ribozyme regulates the intracellular concentration of an abundant metabolite, this low affinity may simply reflect the physiologic need of the ribozyme to remain inactive until high GlcN6P levels have been achieved. Nonetheless, since even synthetic riboswitches can achieve sub-nanomolar dissociation constants for their cognate ligands (Xiao et al., 2008a), compounds that bind and activate the glmS ribozyme at very low concentrations may exist. Binding of their ligands to conventional riboswitches that couple folding to binding is often enthalpically driven (Baird & Ferré-D’Amaré, 2010; Batey et al., 2004; Kulshina et al., 2010). The thermodynamics of natural and unnatural ligand binding by the much more rigid and pre-organized glmS ribozyme remain to be studied.
The demonstration that a natural catalytic RNA can employ a small molecule to broaden its chemical repertory expands the potential role of this nucleic acid both in present-day organisms and in the primordial RNA world. In particular, the ability of the glmS ribozyme to employ a non-covalently bound coenzyme suggests that early ribozymes could have employed chemistry unavailable to ‘plain’ RNA even before the advent of enzymes capable of modifying RNA post-transcriptionally (Ferré-D’Amaré, 2003). An argument has been made that some modern coenzymes (of protein enzymes) that have nucleotide-like moieties may be molecular fossils of the RNA world (White, 1976). Discovery of the role of GlcN6P in activating the glmS ribozyme suggests that even before primitive translation systems evolved (Xiao et al., 2008b), free amino acids may have been recruited by ribozymes as coenzymes; that is, that amino acids could be, in this sense, fossils of the RNA world.
Acknowledgments
The author thanks past and current members of the Ferré-D’Amaré laboratory for their many contributions, and in particular D. Klein for his trail-blazing work on the glmS ribozyme. Studies on the glmS ribozyme in the author’s laboratory have been funded by the Bill and Melinda Gates Foundation, the Damon Runyon Cancer Research Foundation, the Howard Hughes Medical Institute (HHMI), the National Institutes of Health (GM63576 and RR15943), and the W.M. Keck Foundation. The author is an Investigator of the HHMI.
References
- AALBERTS DP, HODAS NO. Asymmetry in RNA pseudoknots: observation and theory. Nucleic Acids Res. 2005;33:2210–2214. doi: 10.1093/nar/gki508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIRD NJ, FERRÉ-D’AMARÉ AR. Idiosyncratically tuned switching behavior of riboswitch aptamer domains revealed by comparative small-angle X-ray scattering analysis. RNA. 2010;16:598–609. doi: 10.1261/rna.1852310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAIRD NJ, KULSHINA N, FERRÉ-D’AMARÉ AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010 doi: 10.4161/rna.7.3.11932. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARRICK JE, CORBINO KA, WINKLER WC, NAHVI A, MANDAL M, COLLINS J, LEE M, ROTH A, SUDARSAN N, JONA I, WICKISER JK, BREAKER RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATEY RT, GILBERT SD, MONTANGE RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- BEVILACQUA PC. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- BEVILACQUA PC, YAJIMA R. Nucleobase catalysis in ribozyme mechanism. Current Op Chem Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- BLOUNT K, PUSKARZ I, PENCHOVSKY R, BREAKER R. Development and application of a high-throughput assay for glmS riboswitch activators. RNA Biol. 2006;3:77–81. doi: 10.4161/rna.3.2.3102. [DOI] [PubMed] [Google Scholar]
- BROOKS KM, HAMPEL KJ. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry. 2009;48:5669–5678. doi: 10.1021/bi900183r. [DOI] [PubMed] [Google Scholar]
- BURGERS PMJ, ECKSTEIN F. Diasteromers of 5′-O-adenosyl 3′-O-uridyl phosphorothioate: chemical synthesis and enzymatic properties. Biochemistry. 1979;18:592–596. doi: 10.1021/bi00571a007. [DOI] [PubMed] [Google Scholar]
- CARSON M. Ribbons. Meth Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- CAYLEY S, LEWIS BA, GUTTMAN HJ, RECORD MT. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- CHI YI, MARTICK M, LARES M, KIM R, SCOTT WG, KIM SH, JOYCE GF. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCHRANE JC, LIPCHOCK SV, SMITH KD, STROBEL SA. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COCHRANE JC, LIPCHOCK SV, STROBEL SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COLLINS JA, IRNOV I, BAKER S, WINKLER WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COWAN JA. Metallobiochemistry of RNA. Co(NH3)63+ as a probe for Mg2+ (aq) binding sites. J Inorg Biochem. 1993;49:171–175. doi: 10.1016/0162-0134(93)80002-q. [DOI] [PubMed] [Google Scholar]
- DAMBACH MD, WINKLER WC. Expanding roles for metabolite-sensing regulatory RNAs. Curr Opin Microbiol. 2009;12:161–169. doi: 10.1016/j.mib.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAMBLON C, RAQUET X, LIAN LY, LAMOTTE-BRASSEUR J, FONZE E, CHARLIER P, ROBERTS GC, FRÈRE JM. The catalytic mechanism of beta-lactamases: NMR titration of an active-site lysine residue of the TEM-1 enzyme. Proc Natl Acad Sci USA. 1996;93:1747–1752. doi: 10.1073/pnas.93.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAS SR, PICCIRILLI JA. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- DE LA PEÑA M, DUFOUR D, GALLEGO J. Three-way RNA junctions with remote tertiary contacts: A recurrent and highly versatile fold. RNA. 2009;15(11):1949–1964. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DITTRICH M, HAYASHI S, SCHULTEN K. ATP Hydrolysis in the βTP and βDP Catalytic Sites of F1-ATPase. Biophysical Journal. 2008;87:2954–2967. doi: 10.1529/biophysj.104.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRAPER DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDWARDS TE, FERRÉ-D’AMARÉ AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- EDWARDS TE, KLEIN DJ, FERRÉ-D’AMARÉ AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Op Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- FEDOR MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- FERRÉ-D’AMARÉ AR. RNA-modifying enzymes. Curr Opin Struct Biol. 2003;13:49–55. doi: 10.1016/s0959-440x(02)00002-7. [DOI] [PubMed] [Google Scholar]
- FERRÉ-D’AMARÉ AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- FERRÉ-D’AMARÉ AR, SCOTT WG. Small self-cleaving ribozymes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 4. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRÉ-D’AMARÉ AR, ZHOU K, DOUDNA JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- GILBERT SD, RAMBO RP, VAN TYNE D, BATEY RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- GÖRKE B, VOGEL J. Noncoding RNA control of the making and breaking of sugars. Genes Dev. 2008;22:2914–2929. doi: 10.1101/gad.1717808. [DOI] [PubMed] [Google Scholar]
- GRUNDY FJ, HENKIN TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- HAMPEL KJ, TINSLEY MM. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- HENKIN T. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F, MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- JAFFE EK, COHN M. Diasteromers of the nucleoside phosphorothioates as probes of the structure of the metal nucleotide substrates and of the nucleotide binding site of yeast hexokinase. J Biol Chem. 1979;254:10839–10845. [PubMed] [Google Scholar]
- KLEIN D, EDWARDS T, FERRÉ-D’AMARÉ A. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLEIN DJ, BEEN MD, FERRÉ-D’AMARÉ AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc. 2007a;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- KLEIN DJ, FERRÉ-D’AMARÉ AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- KLEIN DJ, FERRÉ-D’AMARÉ AR. Crystallization of the glmS ribozyme-riboswitch. Methods Mol Biol. 2009;540:129–139. doi: 10.1007/978-1-59745-558-9_10. [DOI] [PubMed] [Google Scholar]
- KLEIN DJ, WILKINSON SR, BEEN MD, FERRÉ-D’AMARÉ AR. Requirement of helix P2.2 and nucleotide G1 for positioning of the cleavage site and cofactor of the glmS ribozyme. J Mol Biol. 2007b;373:178–189. doi: 10.1016/j.jmb.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOBAYASHI K, EHRLICH SD, ALBERTINI A, AMATI G, ANDERSEN KK, ARNAUD M, ASAI K, ASHIKAGA S, AYMERICH S, BESSIERES P, BOLAND F, BRIGNELL SC, BRON S, BUNAI K, CHAPUIS J, CHRISTIANSEN LC, DANCHIN A, DEBARBOUILLE M, DERVYN E, DEUERLING E, DEVINE K, DEVINE SK, DREESEN O, ERRINGTON J, FILLINGER S, FOSTER SJ, FUJITA Y, GALIZZI A, GARDAN R, ESCHEVINS C, FUKUSHIMA T, HAGA K, HARWOOD CR, HECKER M, HOSOYA D, HULLO MF, KAKESHITA H, KARAMATA D, KASAHARA Y, KAWAMURA F, KOGA K, KOSKI P, KUWANA R, IMAMURA D, ISHIMARU M, ISHIKAWA S, ISHIO I, LE COQ D, MASSON A, MAUEL C, MEIMA R, MELLADO RP, MOIR A, MORIYA S, NAGAKAWA E, NANAMIYA H, NAKAI S, NYGAARD P, OGURA M, OHANAN T, O’REILLY M, O’ROURKE M, PRAGAI Z, POOLEY HM, RAPOPORT G, RAWLINS JP, RIVAS LA, RIVOLTA C, SADAIE A, SADAIE Y, SARVAS M, SATO T, SAXILD HH, SCANLAN E, SCHUMANN W, SEEGERS JF, SEKIGUCHI J, SEKOWSKA A, SEROR SJ, SIMON M, STRAGIER P, STUDER R, TAKAMATSU H, TANAKA T, TAKEUCHI M, THOMAIDES HB, VAGNER V, VAN DIJL JM, WATABE K, WIPAT A, YAMAMOTO H, YAMAMOTO M, YAMAMOTO Y, YAMANE K, YATA K, YOSHIDA K, YOSHIKAWA H, ZUBER U, OGASAWARA N. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOONIN EV. Intron-Dominated Genomes of Early Ancestors of Eukaryotes. Journal of Heredity. 2009;100(5):618–623. doi: 10.1093/jhered/esp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KULSHINA N, BAIRD NJ, FERRÉ-D’AMARÉ AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16(12):1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KULSHINA N, EDWARDS TE, FERRE-D’AMARE AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUZMIN YI, DA COSTA CP, COTTRELL JW, FEDOR MJ. Role of an active site adenine in hairpin ribozyme catalysis. J Mol Biol. 2005;349(5):989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- LEBRUSKA LL, KUZMINE II, FEDOR MJ. Rescue of an abasic hairpin ribozyme by cationic nucleobases. Evidence for a novel mechanism of RNA catalysis. Chem Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- LILLEY DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LILLEY DMJ. Folding of branched RNA species. Biopolymers. 1999;48:101–112. doi: 10.1002/(SICI)1097-0282(1998)48:2<101::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- LILLEY DMJ. The origins of RNA catalysis in ribozymes. Trends Biochem Sci. 2003;28(9):495–501. doi: 10.1016/S0968-0004(03)00191-9. [DOI] [PubMed] [Google Scholar]
- LILLEY DMJ, ECKSTEIN F. Ribozymes and RNA catalysis: introduction and primer. In: Lilley DMJ, Eckstein F, editors. Ribozymes and RNA catalysis. Cambridge: The Royal Society of Chemistry; 2008. pp. 1–10. [Google Scholar]
- LIM J, GROVE BC, ROTH A, BREAKER RR. Characteristics of ligand recognition by a glmS self-cleaving ribozyme. Angew Chem Int Ed Engl. 2006;45:6689–6693. doi: 10.1002/anie.200602534. [DOI] [PubMed] [Google Scholar]
- LIPFERT J, OUELLET J, NORMAN D, DONIACH S, LILLEY D. The complete VS ribozyme in solution studied by small-sngle X-ray scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTICK M, HORAN LH, NOLLER HF, SCOTT WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTICK M, SCOTT WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAYER G, FAMULOK M. High-throughput-compatible assay for glmS riboswitch metabolite dependence. ChemBioChem. 2006;7:602–604. doi: 10.1002/cbic.200500490. [DOI] [PubMed] [Google Scholar]
- MCCARTHY TJ, PLOG MA, FLOY SA, JANSEN JA, SOUKUP JK, SOUKUP GA. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- MILEWSKI S. Glucosamine-6-phosphate synthase–the multi-facets enzymne. Biochem Biophys Acta. 2002;1597:173–192. doi: 10.1016/s0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- MIRONOV AS, GUSAROV I, RAFIKOV R, LOPEZ LE, SHATALIN K, KRENEVA RA, PERUMOV DA, NUDLER E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- MURCHIE AIH, THOMSON JB, WALTER F, LILLEY DMJ. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell. 1998;1:873–881. doi: 10.1016/s1097-2765(00)80086-6. [DOI] [PubMed] [Google Scholar]
- MURRAY JB, SEYHAN AA, WALTER NG, BURKE JM, SCOTT WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- NAKANO SI, CHADALAVADA DM, BEVILACQUA PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- NISSEN P, IPPOLITO JA, BAN N, MOORE PB, STEITZ TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERROTTA AT, SHIH I, BEEN MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- PLEY HW, FLAHERTY KM, MCKAY DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- PYLE AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- RAGHAVAN R, MINNICK MF. Group I Introns and Inteins: Disparate Origins but Convergent Parasitic Strategies. Journal of Bacteriology. 2009;191:6193–6202. doi: 10.1128/JB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAINES RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- RANDAU L, SCHRÖDER I, SÖLL D. Life without RNase P. Nature. 2008;453:120–123. doi: 10.1038/nature06833. [DOI] [PubMed] [Google Scholar]
- REICHENBACH B, MAES A, KALAMORZ F, HAJNSDORF E, GÖRKE B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHARDS FM, WYCKOFF HW, CARLSON WD, ALLEWELL NM, LEE B, MITSUI Y. Protein structure, ribonuclease-S, and nucleotide interactions. Cold Spring Harbor Symp Quant Biol. 1971;36:35–43. doi: 10.1101/sqb.1972.036.01.008. [DOI] [PubMed] [Google Scholar]
- ROMANI A, SCARPA A. Regulation of cell magnesium. Arch Biochem Biophys. 1992;298:1–12. doi: 10.1016/0003-9861(92)90086-c. [DOI] [PubMed] [Google Scholar]
- ROTH A, NAHVI A, LEE M, JONA I, BREAKER RR. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUPERT PB, FERRÉ-D’AMARÉ AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- RUPERT PB, MASSEY AP, SIGURDSSON ST, FERRÉ-D’AMARÉ AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- SALEHI-ASHTIANI K, SZOSTAK JW. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature. 2001;414:82–84. doi: 10.1038/35102081. [DOI] [PubMed] [Google Scholar]
- SCOTT WG, FINCH JT, KLUG A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- SERGANOV A. The long and the short of riboswitches. Curr Op Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERGANOV A, HUANG L, PATEL D. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;457:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERGANOV A, POLONSKAIA A, PHAN AT, BREAKER RR, PATEL DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOUKUP GA, BREAKER RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TINSLEY RA, FURCHAK JR, WALTER NG. Trans-acting glmS catalytic riboswitch: locked and loaded. RNA. 2007;13:468–477. doi: 10.1261/rna.341807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TORELLI AT, KRUCINSKA J, WEDEKIND JE. A comparison of vanadate to a 2′-5′ linkage at the active site of a small ribozyme suggests a role for water in transition-state stabilization. RNA. 2007;13:1052–1070. doi: 10.1261/rna.510807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG L, YU X, HU P, BROYDE S, ZHANG Y. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Y, SCHLICK T. Quantum mechanics/molecular mechanics investigation of the chemical reaction in Dpo4 reveals water-dependent pathways and requirements for active site reorganization. J Am Chem Soc. 2008;130(40):13240–13250. doi: 10.1021/ja802215c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBB CHT, RICCITELLI NJ, RUMINSKI DJ, LUPTÁK A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953–953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE HB. Coenzymes as fossils of an earlier metabolic state. J Mol Evol. 1976;7:101–104. doi: 10.1007/BF01732468. [DOI] [PubMed] [Google Scholar]
- WILKINSON SR, BEEN MD. A pseudoknot in the 3′ non-core region of the glmS ribozyme enhances self-cleavage activity. RNA. 2005;11:1788–1794. doi: 10.1261/rna.2203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILSON TJ, LILLEY DMJ. Biochemistry. The evolution of ribozyme chemistry. Science. 2009;323:1436–1438. doi: 10.1126/science.1169231. [DOI] [PubMed] [Google Scholar]
- WINKLER W, NAHVI A, BREAKER RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- WINKLER WC, NAHVI A, ROTH A, COLLINS JA, BREAKER RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- XIAO H, EDWARDS TE, FERRÉ-D’AMARÉ AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008a;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIAO H, MURAKAMI H, SUGA H, FERRÉ-D’AMARÉ AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008b;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- XUE Y, XU Y, LIU Y, MA Y, ZHOU P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int J Syst Evol Microbiol. 2001;51:1335–1341. doi: 10.1099/00207713-51-4-1335. [DOI] [PubMed] [Google Scholar]