Abstract
The heterogeneity in the healthy human immune system, and the immunological changes that portend various diseases, have been only partially described. Their comprehensive elucidation has been termed the ‘Human Immunology Project’. The accurate measurement of variations in the human immune system requires precise and standardized assays to distinguish true biological changes from technical artefacts. Thus, to be successful, the Human Immunology Project will require standardized assays for immunophenotyping humans in health and disease. A major tool in this effort is flow cytometry, which remains highly variable with regard to sample handling, reagents, instrument setup and data analysis. In this Review, we outline the current state of standardization of flow cytometry assays and summarize the steps that are required to enable the Human Immunology Project.
Flow cytometry has increasingly become a tool of choice for the analysis of cellular phenotype and function in the immune system (BOX 1). It is arguably the most powerful technology available for probing human immune phenotypes, because it measures multiple parameters on many individual cells. Flow cytometry thus allows for the characterization of many subsets of cells, including rare subsets, in a complex mixture such as blood. And because of the wide array of antibody reagents and protocols available, flow cytometry can be used to assess not only the expression of cell-surface proteins, but also that of intracellular phosphoproteins1 and cytokines2, as well as other functional readouts3–6.
Box 1 | Flow cytometry.
The study of cells moving in suspension through an image plane — flow cytometry — is a potent tool for immunology research and immune monitoring. Its main advantages are that it makes multiparameter measurements and that it does so on a single-cell basis. The result is that this technique can dissect the phenotypes and functions of cell subsets in ways that are not possible using bulk assays, such as Western blots, microarrays or enzyme-linked immunosorbent assays (ELISAs). Nowhere has this proven more useful than in a mixed suspension of immune cells, such as the blood. Newer instrumentation allows for the analysis of eight or more parameters at flow rates of thousands of cells per second; the resulting rich data sets are unparalleled for the knowledge of immune function that they have contributed. Flow cytometry will thus be used in this Review as the prime example of an immune-monitoring technology that would benefit from standardization.
This diversity of reagents and applications has led to a large variety of ways in which flow cytometry is being used to monitor the immune systems of humans and model organisms (mostly mice). These uses include the identification of antigen-specific T cells using tetramers7 or intracellular cytokine staining8,9; the measurement of T cell proliferation using dyes such as 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE)3; and the use of immunophenotyping assays to identify lymphocyte, monocyte and/or granulocyte subsets. The information to be gained from such assays is immense and varied. Here, we cite just a few examples for human disease. First, a high proliferative capacity10 and the production of multiple cytokines11 by HIV-specific CD8+ T cells have been linked to non-progression in patients with HIV/AIDS, whereas declining CD4+ T cell counts12, inverted ratios of CD4+ to CD8+ T cells13 and increased numbers of CD38+CD8+ T cells14 have been shown to signify disease progression. Second, the diagnosis and monitoring of leukaemia and lymphoma have been aided by the immunophenotyping of blood and bone marrow15. Third, ageing has been associated with an increase in the number of CD28− late-stage effector T cells16 and an increase in the number of oligoclonal cytomegalovirus-specific T cells17.
Animal studies tend to be relatively small and self-contained, as the mice used are inbred (genetically identical) and their environment and treatments are carefully controlled. Human studies, by contrast, are often larger, to account for genetic and environmental variables. They may require longitudinal assays over a considerable length of time, owing to the difficulty of recruiting suitable subjects and/or the need to follow those subjects over time. There may also be a need to aggregate data from multiple sites, or even across multiple studies, to achieve sufficient sample sizes for statistical analysis or to compare different treatments. In these situations, the standardization of reagents and protocols becomes crucial.
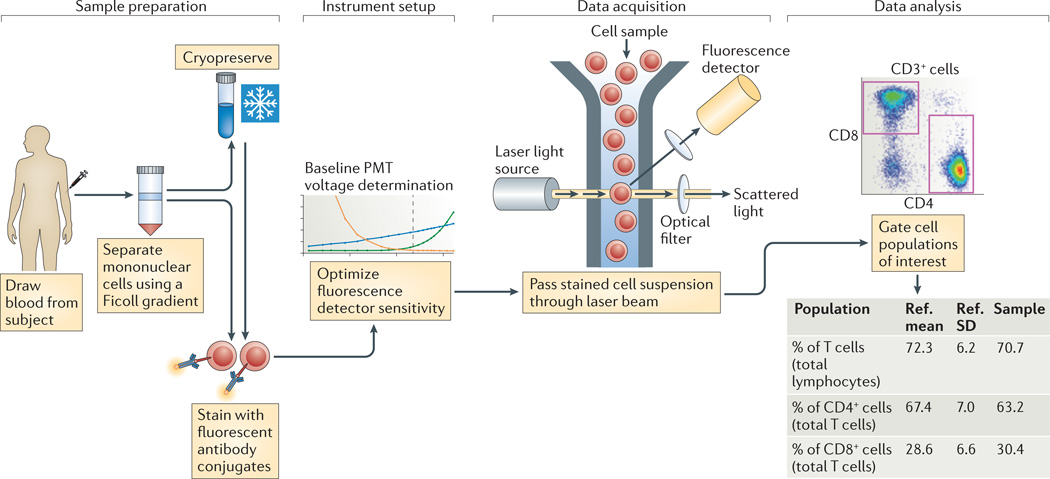
The steps in a typical flow cytometry experiment are shown in FIG. 1, and they present several variables that need to be controlled for effective standardization. These variables involve the general areas of reagents, sample handling, instrument setup and data analysis (TABLE 1). The effects of changes in these variables are largely known18–22. For example, the stabilization and control of staining reagents through the use of preconfigured lyophilized-reagent plates, and centralized data analysis, have both been shown to decrease variability in a multi-site study20. However, the widespread adoption of standards for controlling such variables has not taken place. This is in contrast to other technologies, such as gene expression microarrays, which have achieved a reasonable degree of standardization in recent years23–29. Led by consortia such as the Microarray Quality Control (MAQC) consortium28,29, the External RNA Controls Consortium (ERCC)24 and the EMERALD project23, this standardization was facilitated by the move to a few standardized vendor platforms for microarrays. Of course, microarray data are less complex than flow cytometry data, which are based on many hierarchical gates. Still, a reasonable degree of standardization of flow cytometry assays should be possible to achieve.
Figure 1. A typical flow cytometry experiment.
Sample preparation from blood often involves Ficoll gradient separation of mononuclear cells, and sometimes cryopreservation, before staining with fluorescent antibody conjugates. Each of these steps can introduce variability in the assay results. Instrument setup involves setting voltage gains for the photomultiplier tubes (PMTs) so as to achieve optimal sensitivity. To the extent that this is not standardized, it becomes a source of variability as well. Data acquisition involves passing the stained cells through a laser beam and recording the fluorescence emission from all of the bound antibody conjugates. Here, the main variable is the type of instrument, including the lasers and optical filters used. This is followed by data analysis, in which cell populations of interest are defined and reported on, which is another significant source of variation. Ref., reference; SD, standard deviation.
Table 1.
Technical variables in immunophenotyping and approaches to minimize them
| Variable | Approaches to minimize effects |
|---|---|
| Reagents |
|
| Sample handling |
|
| Instrument setup |
|
| Data analysis |
|
PBMC, peripheral blood mononuclear cell.
Beyond facilitating large, longitudinal and multi-site studies, the global standardization of flow cytometry-based immunophenotyping would enable an even larger goal: the Human Immunology Project30. By analogy to the Human Genome Project, the Human Immunology Project would attempt to characterize the detailed immune phenotypes of healthy individuals, for comparison with various disease states or to elucidate the changes that occur in response to defined perturbations, such as vaccination. In this context, an ‘immune phenotype’ would encompass not only the proportions of major immune cell subsets, but also, for example, their activation state, responsiveness to stimuli, and T cell receptor or B cell receptor repertoires, among other parameters. The Human Immunology Project would create a repository of immunological data that could be mined for biomarkers of immunological ageing, of disease diagnosis and prognosis, and of responses to therapy.
But such a comprehensive enterprise can only be successful with informative and well-standardized assays. Whereas the information content of immunophenotyping assays has grown exponentially in recent years, their level of standardization has not. Therefore, we outline here the steps that we feel need to be taken for assay standardization to enable a successful Human Immunology Project, using flow cytometry of peripheral blood mononuclear cells (PBMCs) as the prime example from which to start the difficult process of standardization. Similar principles might then be applied to cells from target organs and/or lymphoid tissue and to more complex functional assays and rare cell analyses (BOX 2).
Box 2 | Extending standardization to other assays.
We recognize that eight-colour phenotyping of peripheral blood mononuclear cells (PBMCs), although useful, is unlikely in the long term to be the most important assay in our planned immunophenotyping armamentarium. Instead, it is a relatively simple place to start, paving the way for the standardization of more complex assays, such as:
Functional flow cytometry assays, including intracellular cytokine staining, proliferation and phosphoepitope flow cytometry;
Whole blood analysis of rare cells, such as circulating tumour cells, stem cells or dendritic cell subsets;
Analysis of other tissue types, such as tissue biopsies, or cerebrospinal or synovial fluid.
These types of assay represent new frontiers for standardization that are more challenging than the surface-marker phenotyping of PBMCs because they involve the enumeration of rare cells, in vitro activation or tissue processing. However, the rewards for global standardization of such assays are likely to be high, in terms of our understanding of immune mechanisms and eventual clinical utility.
Current state of flow cytometry standardization
The definition of particular subsets of immune cells using cell-surface markers continues to evolve, particularly for cell types that are the subject of intense current research. These include regulatory T (TReg) cells, interleukin-17 (IL-17)-secreting T helper (TH17) cells, dendritic cells (DCs) and natural killer (NK) cells. However, even well-characterized subsets, such as naive and memory T cells, are defined differently in various studies. For example, the classical T cell subsets of naive, central memory, effector memory and effector T cells were first defined on the basis of their expression of CC-chemokine receptor 7 (CCR7; also known as CD197) and CD45RA31 (where naive T cells are CCR7+CD45RA+, central memory T cells are CCR7+CD45RA−, effector memory T cells are CCR7−CD45RA− and effector T cells are CCR7−CD45RA+). Other investigators have since used different markers, such as CD62L or CD27 in place of CCR7 (REFS 32,33), and CD45RO in place of CD45RA. Although these different combinations of markers generally define similar cell subsets, they introduce an unknown amount of heterogeneity that makes comparisons between studies difficult.
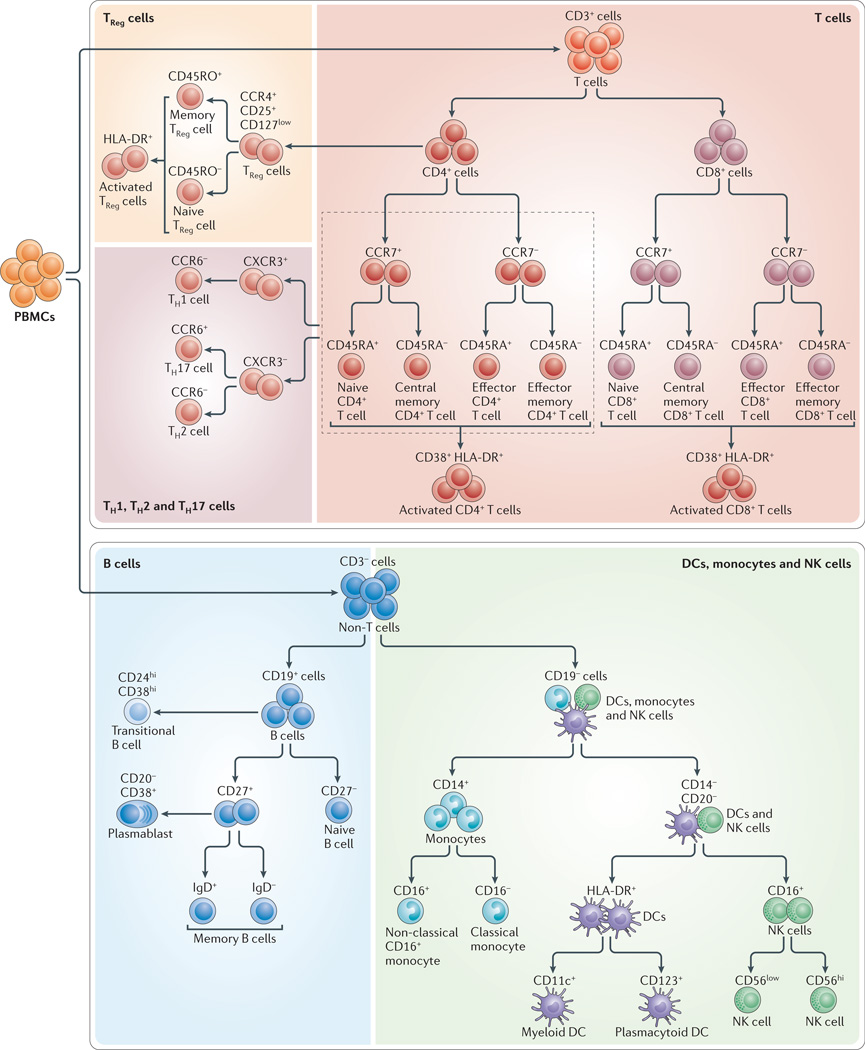
It is reasonable to think that the discovery of new markers and new cell subsets will continue for some time in the future. However, we propose that there is sufficient maturity in the field of cellular immunology to reach consensus working definitions for most of the commonly studied subsets of immune cells. At a high level, these definitions are unlikely to change very much as new discoveries are made, although there are likely to be new subsets of these cell types described over time. In other words, it should be possible to define a stable set of markers that delineate the major classes of B, T and NK cells, monocytes and DCs. Towards this end, we review here the literature that indicates what the key differentiation markers for these cell types might be; these markers could then, in our opinion, form the basis of a standard working definition. We further discuss how consensus is being reached in the immunological community regarding these markers and definitions, leading to a standardized immunophenotyping panel for human immune monitoring. Although there may be disagreements about the choice and utility of certain markers, and the names of the subsets so defined, we propose that the markers and subsets shown in FIG. 2, and discussed below, can form a working definition of cell types that will help to drive standardization in the future.
Figure 2. Identification of immune cell subsets by eight-colour antibody staining.
The figure shows the cell populations that can be identified using the markers targeted by each of the five antibody cocktails of the Human Immunophenotyping Consortium (HIPC) phenotyping panel shown in TABLE 2. CCR, CC-chemokine receptor; CXCR3, CXC-chemokine receptor 3; DC, dendritic cell; NK, natural killer; PBMC, peripheral blood mononuclear cell; TH, T helper; TReg, regulatory T.
T cells
The major subsets of T cells can be defined by the expression of CD4 and CD8, together with, for example, CCR7 and CD45RA31. CCR7 has been a difficult marker for use in flow cytometry because of its low expression levels; however, a new CCR7-specific antibody clone (150503; commercially available from multiple vendors) that provides brighter staining has greatly improved this situation. Furthermore, other substitute markers for CCR7 are more problematic. For example, the expression level of CD62L is highly affected by density gradient separation34 or cryopreservation35 of PBMCs. As PBMC cryopreservation is integral to the workflow of many human studies, owing to the need to batch samples and store cells for future assays, this limits the broad use of CD62L for immunophenotyping. Another possible CCR7 replacement, CD27, is expressed not only by naive and CCR7+ central memory T cells, but also by a population of effector memory T cells that lack expression of CCR7 (REF. 36), meaning that CD27 is not a full substitute for CCR7. So, a panel containing CD3 (to define T cells), CD4, CD8, CD45RA and CCR7 seems most applicable for distinguishing naive, central memory, effector memory and effector CD4+ and CD8+ T cells. With the addition of activation markers, such as CD38 and HLA-DR, it is possible to define activated subsets of each of these cell types as well (FIG. 2; TABLE 2).
Table 2.
Eight-colour antibody panels proposed by the Human Immunophenotyping Consortium
| Fluorochrome | Marker | |||||
|---|---|---|---|---|---|---|
| T cells | TReg cells | TH1, TH2 and TH17 cells |
B cells | DCs, monocytes and NK cells |
||
| FITC | Live or dead | Live or dead | Live or dead | Live or dead | Live or dead | |
| PE | CCR7 | CD25 | CXCR3 | CD24 | CD56 | |
| PerCP-Cy5.5 | CD4 | CD4 | CD4 | CD19 | CD123 | |
| PE-Cy7 | CD45RA | CCR4 | CCR6 | CD27 | CD11c | |
| APC | CD38 | CD127 | CD38 | CD38 | CD16 | |
| APC-H7 | CD8 | CD45RO | CD8 | CD20 | CD3, CD19 and CD20 | |
| V450 | CD3 | CD3 | CD3 | CD3 | CD14 | |
| V500 | HLA-DR | HLA-DR | HLA-DR | IgD | HLA-DR | |
APC, allophycocyanin; APC-H7, allophycocyanin–cyanine H7 tandem; CCR, CC-chemokine receptor; CXCR3, CXC-chemokine receptor 3; DC, dendritic cell; FITC, fluorescein isothiocyanate; NK, natural killer; PE, phycoerythrin; PE-Cy7, phycoerythrin–cyanine 7 tandem; PerCP-Cy5.5, peridinin chlorophyll protein–cyanine 5.5 tandem; TH, T helper; TReg, regulatory T; V450, violet 450; V500, violet 500.
B cells
Tonsillar B cells were first classified into five groups, termed Bm1–Bm5, according to their expression of IgD and CD38 together with that of CD23 and CD77 (REF. 37). Some versions of all of these cell types can also be found in peripheral blood38. Bm1 and Bm2 cells are CD38−IgD+, and include mostly resting (CD23−) naive B cells and some activated (CD23+) cells. Bm3 and Bm4 cells are CD38+IgD− germinal centre (or germinal centre precursor) cells, which can be further subdivided by their expression of CD77, a marker of germinal centre location (Bm3 cells are CD77−; Bm4 cells are CD77+). Bm5 cells are CD38−IgD− memory B cells. CD27 can also be used to define naive (CD27−) and memory (CD27+) B cells39. CD24 has been shown to be useful in defining immature ‘transitional’ B cells (which are CD24hiCD38hi)40. Recently, circulating plasmablasts were also identified in peripheral blood, using either CD38 and CD138 (REF. 41) or CD38 and CD27 (REF. 42). Notably, these plasmablasts are low or negative for CD20 expression and, because of the use of CD20-specific antibody therapy in various diseases, it is prudent to also include CD19 as a marker to define the parental B cell population. Although it is not possible to include all of the potentially relevant B cell markers in a single eight-colour antibody cocktail, a highly useful panel can be constructed using antibodies specific for CD19 and CD20 (to define B cells), CD38 (to identify plasmablasts as well as transitional B cells), CD24 (for transitional B cells), and IgD and CD27 (for naive and memory B cell populations) (FIG. 2).
NK cells
NK cells have been shown in recent years to be much more heterogeneous than previously thought, with a large variety of activating and inhibitory receptors expressed by overlapping subsets of cells that vary widely between individuals43. However, at a more basic level, NK cells can be subdivided into two major categories based only on the markers CD16 and CD56. The vast majority of peripheral blood NK cells are CD16+CD56low, whereas a smaller subset is CD16+CD56hi. These populations differ in terms of function and tissue localization, and the latter subset has been shown to be an intermediate phenotype that can give rise to the former subset44.
Dendritic cells
Circulating blood DCs have long been identified as HLA-DRhi cells that are negative for various lineage-specific markers, including CD3 (which defines T cells), CD14 (which is monocyte specific), and CD19 and CD20 (which define B cells)45. DCs can be divided into cells of the myeloid lineage (CD11c+ cells) and the plasmacytoid lineage (CD123+ cells). Myeloid DCs can be further subdivided by their expression of CD16, CD1c and CD141 (REFS 46,47). Although these myeloid DC subsets may have functional significance, their enumeration in cryopreserved PBMC populations is too problematic to warrant the inclusion of these markers in a general phenotyping panel. Thus, one can construct a basic DC panel based on a lineage cocktail, and antibodies specific for HLA-DR, CD11c and CD123.
Monocytes
With further study, monocytes might also reveal substantial heterogeneity, but currently two major categories are widely recognized: classical monocytes (which are CD14hiCD16−) and non-classical monocytes (which are CD14lowCD16hi)48. Because monocyte, NK cell and DC phenotyping can all include CD16, it is possible to imagine a phenotyping cocktail for all three cell types that includes a lineage cocktail, together with antibodies specific for HLA-DR, CD11c, CD14, CD16, CD56 and CD123.
Activation markers
Activation markers for these cell types are of course of equal interest to the differentiation markers discussed above, as the activated subsets can provide clues to an individual’s response to infection, vaccination, cancer or autoimmune disease. For this purpose, one can assess intracellular markers of recent cell division (such as Ki67) or surface markers of activation that have varying kinetics of expression (for example, CD69 is a transient marker, whereas HLA-DR, CD38 and CD71 are expressed for longer periods). The choice of marker(s) will certainly vary depending on the experimental question, and the expression of these markers may not be perfectly correlated with proliferation in vivo49. However, the combination of CD38 and HLA-DR has proven to be useful for identifying activated T cells in various contexts, including HIV infection14. It can be hypothesized that the frequencies of activated T cells of various phenotypes could become biomarkers of several conditions or outcomes, including: vaccine-induced protection (at defined time point(s) post-vaccination); disease activity (for T cell-mediated autoimmune or infectious diseases); and chronic inflammation (as has been shown in HIV infection). The similar use of HLA-DR to track activated NK cells is a possibility that has been less well explored, but that might be equally fruitful. Plasmablasts — the activated (CD38+) subset of antibody-secreting B cells — have been shown to increase in number post-vaccination50 and might also be used as biomarkers of disease status and/or vaccine efficacy.
From the above discussion, it should be clear that it is possible, using current reagents and instrumentation, to carry out immunophenotyping in a far greater level of detail than is currently achieved clinically. Furthermore, there are some clear choices that we think could form the basis of standard working definitions for different immune cell subsets using eight colours of fluorescence or less (FIG. 2; TABLE 2). However, no current clinical test includes all, or even most, of the subsets and cell types mentioned above. As a result, clinical immunophenotyping is carried out only at the level of lymphocytes, monocytes and granulocytes, or using limited four- to six-colour staining for major cell types in diseases such as HIV/AIDS and leukaemia or lymphoma. All other immunophenotyping is carried out in the research laboratory, with essentially no standardization. A clinician who wishes to know the status of a patient’s immune system cannot yet order a ‘complete immunophenotype’ in the way that we currently order a ‘complete blood count’.
How can this situation be improved?
Recognizing the need for the standardization of immunophenotyping, a consortium of laboratories was formed in 2009, led by the National Institutes of Health (NIH) in the United States and the Federation of Clinical Immunology Societies (FOCIS). In 2010, the consortium convened an international group of laboratories, representing FOCIS Centers of Excellence (FCEs), recipients of NIH grants in the Human Immunophenotyping Consortium (HIPC), and key academic and industrial leaders in flow cytometry. About 50 participants from these groups met at a workshop in January 2011 called the Flow Immunophenotyping Technical Meeting at NIH (FITMaN; see the spring 2011 FOCIS newsletter). The main outcome of the meeting was the proposal of a set of desired standard markers for the phenotyping of B, T and NK cells, monocytes and DCs. These evolved into a set of defined eight-colour antibody cocktails, which could be expanded by the addition of more marker-specific antibodies but which, by themselves, could enumerate the basic subsets of the above cell types and could determine their activation status (TABLE 2).
To see where this effort may lead, and to define the remaining obstacles to the standardization of flow immunophenotyping, let us examine in more detail the major areas of flow cytometry variability and how these can be addressed through this and similar community efforts.
Reagents
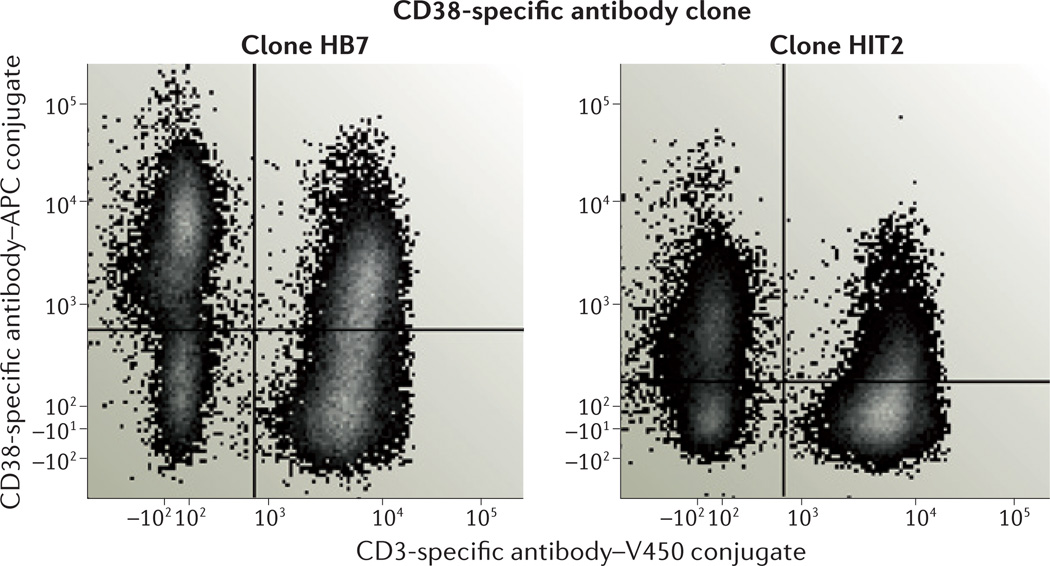
How important is it to define the actual antibody cocktails used for immunophenotyping? As described above, the choice of markers is clearly important, but different antibody clones and fluorochromes can also greatly influence results. As shown in FIG. 3, two clones of CD38-specific antibody give widely disparate staining results for PBMCs from the same healthy donor. Moreover, the fluorescence separation of positive and negative populations for a particular marker can vary by about 20-fold between the brightest commercially available fluorochromes (such as phycoerythrin and allophycocyanin) and the dimmest ones (such as AmCyan and Pacific Blue)51. This difference affects whether a population that is positive for a particular marker can be reliably separated from the negative population and may even influence the apparent percentage of positive cells (as dim positive cells become mixed with negative cells when the separation is poor). Needless to say, the antibody titre also has an important role in this variability.
Figure 3. The importance of antibody choice.
The staining patterns of two commercially available clones of human CD38-specific antibody are very different, despite the fact that both antibodies were conjugated to allophycocyanin (APC) by the same vendor, and were used to stain peripheral blood mononuclear cells (PBMCs) from the same healthy subject under identical conditions. V450, violet 450. Data courtesy of Angelique Biancotto, National Heart, Lung and Blood Institute, USA.
The use of preformatted lyophilized-reagent plates (Lyoplates, BD Biosciences) can help, by ‘locking in’ the above reagent variables, once optimal antibody clones, fluorochromes and titres have been established. The use of such lyophilized-reagent plates has been shown to decrease staining variability compared with using individual liquid reagents in multiple studies20,52. The decrease in variability can be attributed to the stabilization of reagents against degradation, as well as to the elimination of pipetting variation and the avoidance of errors in reagent addition. Lyophilized-reagent plates have also been used as a means of standardizing stimulation reagents for functional assays53,54.
Of course, standardization does not end with defining the reagents (that is, the antibody specificities, clones and titres, and the fluorochromes) in a staining cocktail. Sample handling, instrument setup and data analysis are crucial variables as well.
Sample handling
The presumed ideal sample-handling protocol (staining fresh whole blood or PBMCs immediately after blood draw) is not always practical in large clinical trials, and it may not even be desirable with regard to comparing samples longitudinally. The cryopreservation of PBMCs is an attractive alternative for the purpose of batching samples over time, and for banking samples for future use. But it has the disadvantage of depleting certain interesting cell types (such as plasmablasts and DCs), and generally decreasing cell yield and function. Cryopreservation is also technically demanding, such that not all clinical trial sites are equipped and competent to carry it out. This leads to the compromise of shipping whole blood or PBMCs to a central laboratory for cryopreservation, which can further decrease cell viability and function and can introduce additional variability owing to shipping time and conditions.
A case has been made for empowering individual laboratories to carry out their own processing, including cryopreservation and possibly even flow cytometry, in large multicentre studies18. Done correctly, this could significantly decrease the compromises associated with shipping and/or cryopreservation of samples. However, it requires an infrastructure commitment in terms of equipment, training and the maintenance of standardization between a set of disparate laboratories to minimize variability. In many cases though, such empowerment is becoming a requirement for collaboration with scientists in countries such as India and Japan, as the primary samples are not usually allowed to leave these countries.
Another solution is to use some type of automation to carry out either the staining or, for functional assays, the activation of whole blood, followed by fixation and freezing, at individual laboratories where samples are drawn. The fixed and frozen samples can then be batch-shipped to a central laboratory for further processing and analysis, or this could be carried out at the local site in a batch manner. A commercial instrument to automate the activation and fixation of whole blood is available (from Smart Tube, Palo Alto, California), as are instruments to stain and fix whole blood samples (such as Sample Prep Assistant, BD Biosciences; and Biomek, Beckman Coulter).
Instrument setup
In addition to optimizing certain settings, such as laser time delays, instrument setup is mostly concerned with the setting of the voltage gains applied to each fluorescence detector, which influences the sensitivity of the detector to dim versus bright signals. This used to be entirely a matter of user preference, but some guiding principles have now emerged55 based on measuring the variance of a population of dim particles over a series of voltage gains. Equally important, software to automatically achieve a predetermined setup has become available (such as BD Cytometer Setup and Tracking, BD Biosciences; and Beckman Coulter’s MXP software). There are still, however, marked differences in performance associated with instruments of different configurations and from different manufacturers. In fact, a high proportion of multicolour flow cytometers are customized by the manufacturer and/or by individual laboratories with a variety of laser and filter options. This makes standardization between sites (or even between two different instruments at the same site) difficult. The only method currently available to overcome these variations is to establish target values across the cytometers using standardized control particles. By setting voltage gains to place the control particles at a particular target channel, a relatively similar instrument setup is achieved. The best particles in this regard are ones that contain the actual fluorochromes to be used experimentally, such as stained antibody capture beads56, so that the performance using actual samples is most closely mimicked.
Data analysis
Data analysis is one of the largest variables in flow cytometry, as shown by studies analysing data at multiple sites versus centrally20. It is also one of the easiest variables to address, as re-analysis of existing data is always possible, whereas choices made about sample handling and instrument setup are irrevocable. Central analysis is simple in concept, but it can be overwhelming in terms of the demands placed on one or a few coordinated personnel who must review all of the data to achieve consistent gating. Fortunately, automated gating algorithms have proliferated and improved, such that they now compete favourably with expert manual gating57. A group of interested experts (FlowCAP), with support from the NIH, has completed two workshops aimed at comparing and improving the use of automated gating algorithms for flow cytometry analyses. Their continued work in this area should lead to standard software packages that can carry out easy and unbiased analyses of flow cytometry files.
From the above, it is clear that individual solutions are available in each of the areas that lead to flow cytometry variability. What is missing is: guidance for those designing clinical trials on the integrated use of these solutions; resources to enable the implementation of all appropriate solutions; and some standardized approaches to the most common flow cytometry immunophenotyping challenges. With regard to such standardized approaches, the preformatted antibody cocktails being produced by the HIPC might provide a good start. By predetermining the reagent choices and simplifying sample handling, they should alleviate much of the variability in these areas. Because they represent a single set of reagent cocktails, they are also amenable to standardized instrument setup and data analysis routines, which should further decrease variability. Their use by the HIPC and by other interested laboratories should produce an example body of data that could demonstrate to the field the benefits of a standardized approach to flow cytometry, and to immunological assays in general.
What needs to happen next?
The efforts of the HIPC with regard to eight-colour immunophenotyping of the major PBMC subsets are of course only a start to the process of standardization. For a glimpse at where this process could lead, one needs only to consider another immunophenotyping assay that has already been standardized — namely, CD4+ T cell counting for the staging of HIV/AIDS. As HIV directly infects and depletes CD4+ T cells, the enumeration of these cells is crucial for determining disease progression and for tracking the efficacy of therapeutic intervention. Because this assay requires multiple markers for accurate quantitation (generally CD3 and CD4 at a minimum), it is well-suited to a flow cytometry platform. And because of the large global burden of HIV infection, manufacturers have responded with multiple systems-level solutions. As a result, the counting of CD4+ T cells has been standardized to achieve an inter-laboratory coefficient of variation of <10%58. It can be carried out around the world on one of several standardized platforms, at least one of which takes advantage of lyophilized reagents (BD FACSCount, BD Biosciences), and several of which use automated analysis software (TetraCXP, Beckman Coulter; BD Multiset, BD Biosciences; Guava EasyCD4, Millipore). Sample-handling integrity is ensured by simple protocols and the mandatory proficiency testing that is prescribed by government agencies overseeing clinical laboratories.
Why not apply these principles to a broader set of immunophenotyping assays (BOX 2)? With the availability of standard antibody panels (such as those described above), specific instrument-setup and staining protocols, and centralized and/or automated data analysis, the potential exists to create highly comparable data. But will such systems be widely adopted? And how will the participating laboratories be monitored? As long as the assays are strictly research based, there is little or no enforcement power, other than that of the funding agencies of grant recipients. Beyond that, we believe that it is only in the power of example that the adoption of standardized panels and practices can be encouraged. Still, this can be a powerful force in itself; after all, the microarray community quickly adopted standard commercial platforms once they became available and their quality became apparent. Eventually, when the clinical utility of immunophenotyping has been demonstrated for specific disease indications, quality assurance can be achieved through the same regulatory means as for other clinical assays.
Extending to other assays
There is no reason to believe that similar standardized panels cannot be developed for functional assays, such as intracellular cytokine staining and phosphoepitope flow cytometry. Efforts are already underway in Europe to standardize functional assays that are used in clinical diagnostics, through the European Network for Translational Immunology Research and Education (ENTIRE). And an NIH-supported cooperative research programme, Clinical Trials in Organ Transplantation (CTOT), is engaged in cross-validating cellular function assays as well as immunophenotyping panels. Their findings should help to inform reagent choices for standardized functional assay panels, including those used in non-flow cytometric assays such as enzyme-linked immunosorbent spot (ELISPOT) assays and immunohistochemistry.
Adopting new technologies
To some degree, the need for standardization and the effort required to achieve it will be reduced by new technologies. One area of technology growth is in systems that can both process samples and analyse them by flow cytometry, all in a highly automated manner (such as the Load & Go systems from Blue Ocean Biomedical). This type of system works from a blood draw tube, processing and staining cells, acquiring data and carrying out routine analysis, all in a pre-programmed manner. The proliferation and continued development of such systems could eliminate, to a large degree, the need to ship blood to central laboratories for flow cytometry assays. In the process, the quality and reproducibility of data should also be improved.
Another new technology, mass cytometry (CyTOF, DVS Sciences), is poised to revolutionize flow cytometry by allowing many more antibody probes to be combined in a single assay, while simultaneously eliminating the issue of optical spillover59–61. If reagents for this platform become standardized, the design of antibody panels would be much easier, because the inclusion of certain reagents would not influence the performance of other reagents (as it does for conventional flow cytometry). This would allow for standardized immunophenotyping panels to be extended as desired for particular investigations, without compromising the core reagents or the quality of data.
Data centralization and mining
To realize the full benefit of standardized assays, data need to be centrally collected and mined. In this way, normal ranges can be determined across the variables of age, gender and ethnicity, for cell subsets and functions about which little is currently known. Disease cohorts can be compared with matched healthy controls. Patients treated in various ways can be compared with each other and with controls. Data can be used and reused, beyond the intention of the original research, if the same standardized assays have been applied, and if the data are centrally aggregated in a way that allows this type of exploration.
To this end, we have recently collaborated with CytoAnalytics to develop an online system for data integration and exploration. By modifying existing open-source tools in the business intelligence field (such as online analytical processing (OLAP)), we were able to create a data-mining environment in which laboratory results are integrated with clinical and demographic data (‘metadata’), filtered using any desired variables, and displayed with automated aggregate statistics (such as the mean, median and sample number). Similar systems could be used for the aggregation of flow cytometry phenotyping (and functional) data in general, and they should be made publicly available as quickly as possible to allow other groups to integrate their own data sets.
All of the above elements should help to facilitate the pursuit of the Human Immunology Project, the value of which is directly dependent on the ability to compare the results of immunological assays across studies and across laboratories.
Conclusions
With the major exception of CD4+ T cell counting for immune monitoring in patients with HIV and the immunophenotyping of leukaemias and lymphomas, flow cytometry is mostly carried out in a research setting. Particular assays may be internally validated for a given clinical study, but comprehensive standardization is rare to non-existent.
However, there is a clear and growing interest in the standardization of human immunophenotyping. This is evidenced by standardization efforts, such as those of the European ENTIRE and United States CTOC networks. The NIH have also invested in a set of awards focused on human immunophenotyping, and both FOCIS and the NIH are working to create standard immunophenotyping panels.
The application of attention and resources to this area should produce tangible results. Previous studies have identified crucial variables in flow cytometry in the areas of sample handling, reagents, instrument setup and data analysis. Control of these variables can markedly decrease variability in clinical studies. What remains to be achieved is the widespread adoption of standard reagents and protocols for the collection of human immunophenotyping data. In addition, an infrastructure for aggregating and mining results will be needed to achieve the vision of the Human Immunology Project. With such standardization and accompanying infrastructure, it should be possible to rapidly mine the data for metrics of immunological health and disease, much as we can currently mine genetic data using common databases and tools. The result will be the more rapid discovery of biomarkers that can aid in the diagnosis, prognosis and therapeutic monitoring of immunologically related diseases.
Acknowledgements
The authors thank all the contributors to the HIPC/FITMaN immunophenotyping panel, in particular S. Heck, F. Nestle, A. Biancotto, S. Gupta, M. Malipatlolla, L. Devine, R. Montgomery and D. Hafler. The mention of any commercial products in this manuscript does not imply endorsement by the US Government or the US National Institutes of Health.
Glossary
- Immune phenotypes
Measurable aspects of the immune system, such as the proportions of various cell subsets or measures of cellular immune function.
- Gates
Sequential filters that are applied to a set of flow cytometry data to focus the analysis on particular cell subsets of interest.
- Human Immunology Project
The comprehensive mapping of immune phenotypes in healthy and diseased human populations.
- Density gradient separation
The isolation of mononuclear cells from blood or other sources by centrifugation over a density gradient, which usually consists of a carbohydrate polymer solution (Ficoll).
- Cryopreservation
The processing and storage of cells at sub-zero temperatures under conditions that preserve their viability for later assays.
- Lineage cocktail
A mixture of antibodies specific for various lineage-specific markers, such as CD3 (for T cells), CD14 (for monocytes) and CD19 and CD20 (for B cells).
- Antibody capture beads
Microparticles conjugated with immunoglobulin-specific antibodies. These beads can be stained with fluorescent antibodies to create single-colour controls for flow cytometry instrument setup and compensation.
- Phosphoepitope flow cytometry
(Also known as ‘phospho-flow’). A technique that uses antibodies specific for phosphorylated versions of proteins to analyse cell signalling by flow cytometry.
- Mass cytometry
Flow cytometry using antibodies tagged with heavy metal ions, which are detected by mass spectrometry (as opposed to classical flow cytometry, which uses antibodies tagged with fluorophores and optical detection).
- Optical spillover
The presence of signals from fluorescent antibody staining in multiple detectors of a cytometer, resulting in a loss of resolution sensitivity.
Footnotes
Competing interests statement
The authors declare competing financial interests: see web version for details.
FURTHER INFORMATION
Holden T. Maecker’s homepage: http://himc.stanford.edu
Clinical Trials in Organ Transplantation: http://www.ctotstudies.org
European Network for Translational Immunology Research and Education: http://entire-net.eu
FlowCAP: http://flowcap.flowsite.org
Spring 2011 FOCIS newsletter: http://www.focisnet.org/FOCIS/newsletter/201104.htm
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Perez OD, et al. Multiparameter analysis of intracellular phosphoepitopes in immunophenotyped cell populations by flow cytometry. Curr. Protoc. Cytom. 2005;32:6.20.1–6.20.22. doi: 10.1002/0471142956.cy0620s32. [DOI] [PubMed] [Google Scholar]
- 2.Maecker HT. In: Flow Cytometry Protocols. Hawley TS, Hawley RG, editors. Totowa, New Jersey: Humana Press; 2004. pp. 95–107. [Google Scholar]
- 3.Parish CR, Glidden MH, Quah BJ, Warren HS. Use of the intracellular fluorescent dye CFSE to monitor lymphocyte migration and proliferation. Curr. Protoc. Immunol. 2009;84:4.9.1–4.9.13. doi: 10.1002/0471142735.im0409s84. [DOI] [PubMed] [Google Scholar]
- 4.Betts MR, Koup RA. Detection of T-cell degranulation: CD107a and b. Methods Cell Biol. 2004;75:497–512. doi: 10.1016/s0091-679x(04)75020-7. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nature Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 6.Frentsch M, et al. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nature Med. 2005;11:1118–1124. doi: 10.1038/nm1292. [DOI] [PubMed] [Google Scholar]
- 7.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 8.Prussin C, Metcalfe DD. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J. Immunol. Methods. 1995;188:117–128. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 9.Picker LJ, et al. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood. 1995;86:1408–1419. [PubMed] [Google Scholar]
- 10.Migueles SA, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature Immunol. 2002;3:1061–1068. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 11.Betts MR, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T-cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahey JL, et al. Immune pathogenesis of AIDS and related syndromes. Ann. Inst. Pasteur Immunol. 1987;138:245–252. doi: 10.1016/s0769-2625(87)80075-2. [DOI] [PubMed] [Google Scholar]
- 13.Lifson JD, Finch SL, Sasaki DT, Engleman EG. Variables affecting T-lymphocyte subsets in a volunteer blood donor population. Clin. Immunol. Immunopathol. 1985;36:151–160. doi: 10.1016/0090-1229(85)90116-3. [DOI] [PubMed] [Google Scholar]
- 14.Kestens L, et al. Expression of activation antigens, HLA-DR and CD38, on CD8 lymphocytes during HIV-1 infection. AIDS. 1992;6:793–797. doi: 10.1097/00002030-199208000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Duque RE. Flow cytometric analysis of lymphomas and acute leukemias. Ann. NY Acad. Sci. 1993;677:309–325. doi: 10.1111/j.1749-6632.1993.tb38786.x. [DOI] [PubMed] [Google Scholar]
- 16.Posnett DN, Edinger JW, Manavalan JS, Irwin C, Marodon G. Differentiation of human CD8 T cells: implications for in vivo persistence of CD8+ CD28− cytotoxic effector clones. Int. Immunol. 1999;11:229–241. doi: 10.1093/intimm/11.2.229. [DOI] [PubMed] [Google Scholar]
- 17.Khan N, et al. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J. Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 18. Maecker HT, et al. A model for harmonizing flow cytometry in clinical trials. Nature Immunol. 2010;11:975–978. doi: 10.1038/ni1110-975. This perspective details the ways in which flow cytometry phenotyping assays can vary in the clinical trial setting, and advocates ways to standardize them.
- 19.Suni MA, et al. Performance of plate-based cytokine flow cytometry with automated data analysis. BMC Immunol. 2003;4:9. doi: 10.1186/1471-2172-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maecker HT, et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. Using multi-site data on shared samples, this paper shows the benefits of centralized and/or automated gating and preconfigured lyophilized reagents.
- 21.Dunne JF, Maecker HT. Automation of cytokine flow cytometry assays. JALA. 2004;9:5–9. [Google Scholar]
- 22.Gratama JW, et al. Analysis of variation in results of flow cytometric lymphocyte immunophenotyping in a multicenter study. Cytometry. 1997;30:166–177. doi: 10.1002/(sici)1097-0320(19970815)30:4<166::aid-cyto2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Beisvag V, et al. Contributions of the EMERALD project to assessing and improving microarray data quality. Biotechniques. 2011;50:27–31. doi: 10.2144/000113591. [DOI] [PubMed] [Google Scholar]
- 24.Devonshire AS, Elaswarapu R, Foy CA. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics. 2010;11:662. doi: 10.1186/1471-2164-11-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calza S, Pawitan Y. Normalization of gene-expression microarray data. Methods Mol. Biol. 2010;673:37–52. doi: 10.1007/978-1-60761-842-3_3. [DOI] [PubMed] [Google Scholar]
- 26.Rogers S, Cambrosio A. Making a new technology work: the standardization and regulation of microarrays. Yale J. Biol. Med. 2007;80:165–178. [PMC free article] [PubMed] [Google Scholar]
- 27.Ball CA, et al. Standards for microarray data. Science. 2002;298:539. doi: 10.1126/science.298.5593.539b. [DOI] [PubMed] [Google Scholar]
- 28.Patterson TA, et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nature Biotech. 2006;24:1140–1150. doi: 10.1038/nbt1242. [DOI] [PubMed] [Google Scholar]
- 29. Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nature Biotech. 2006;24:1151–1161. doi: 10.1038/nbt1239. A seminal paper demonstrating that the major commercial microarray platforms can generate similar and reproducible data.
- 30. Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. In this article, Mark Davis lays the foundation for why we need a ‘Human Immunology Project’.
- 31.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 32.Tussey L, Speller S, Gallimore A, Vessey R. Functionally distinct CD8+ memory T cell subsets in persistent EBV infection are differentiated by migratory receptor expression. Eur. J. Immunol. 2000;30:1823–1829. doi: 10.1002/1521-4141(200007)30:7<1823::AID-IMMU1823>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Appay V, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nature Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 34.Lin SJ, Chao HC, Yan DC, Huang YJ. Expression of adhesion molecules on T lymphocytes in young children and infants — a comparative study using whole blood lysis or density gradient separation. Clin. Lab. Haematol. 2002;24:353–359. doi: 10.1046/j.1365-2257.2002.00462.x. [DOI] [PubMed] [Google Scholar]
- 35. Weinberg A, et al. Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin. Vaccine Immunol. 2009;16:1176–1186. doi: 10.1128/CVI.00342-08. One of the best articles on the cryopreservation of PBMCs, its optimization and the caveats for use with T cell assays.
- 36.Fritsch RD, et al. Stepwise differentiation of CD4 memory T cells defined by expression of CCR7 and CD27. J. Immunol. 2005;175:6489–6497. doi: 10.4049/jimmunol.175.10.6489. [DOI] [PubMed] [Google Scholar]
- 37.Pascual V, et al. Analysis of somatic mutation in five B cell subsets of human tonsil. J. Exp. Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohnhorst JO, Bjorgan MB, Thoen JE, Natvig JB, Thompson KM. Bm1–Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjogren’s syndrome. J. Immunol. 2001;167:3610–3618. doi: 10.4049/jimmunol.167.7.3610. [DOI] [PubMed] [Google Scholar]
- 39.Agematsu K, et al. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur. J. Immunol. 1997;27:2073–2079. doi: 10.1002/eji.1830270835. [DOI] [PubMed] [Google Scholar]
- 40.Cuss AK, et al. Expansion of functionally immature transitional B cells is associated with human-immunodeficient states characterized by impaired humoral immunity. J. Immunol. 2006;176:1506–1516. doi: 10.4049/jimmunol.176.3.1506. [DOI] [PubMed] [Google Scholar]
- 41.Rawstron AC. Immunophenotyping of plasma cells. Curr. Protoc. Cytom. 2006;36:6.23.1–6.23.14. doi: 10.1002/0471142956.cy0623s36. [DOI] [PubMed] [Google Scholar]
- 42.Avery DT, et al. Increased expression of CD27 on activated human memory B cells correlates with their commitment to the plasma cell lineage. J. Immunol. 2005;174:4034–4042. doi: 10.4049/jimmunol.174.7.4034. [DOI] [PubMed] [Google Scholar]
- 43.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr. Opin. Immunol. 2005;17:29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Beziat V, et al. CD56brightCD16+ NK cells: a functional intermediate stage of NK cell differentiation. J. Immunol. 2011;186:6753–6761. doi: 10.4049/jimmunol.1100330. [DOI] [PubMed] [Google Scholar]
- 45.Willmann K, Dunne JF. A flow cytometric immune function assay for human peripheral blood dendritic cells. J. Leukoc. Biol. 2000;67:536–544. [PubMed] [Google Scholar]
- 46.Ju X, Clark G, Hart DN. Review of human DC subtypes. Methods Mol. Biol. 2010;595:3–20. doi: 10.1007/978-1-60761-421-0_1. [DOI] [PubMed] [Google Scholar]
- 47.Dzionek A, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 48.Ziegler-Heitbrock HW, et al. Small (CD14+/CD16+) monocytes and regular monocytes in human blood. Pathobiology. 1991;59:127–130. doi: 10.1159/000163629. [DOI] [PubMed] [Google Scholar]
- 49.Allers K, et al. Migration patterns of nonspecifically activated versus nonactivated nonhuman primate T lymphocytes: preferential homing of activated autologous CD8+ T cells in the rectal mucosa. J. Immunother. 2008;31:334–344. doi: 10.1097/CJI.0b013e3181635e7f. [DOI] [PubMed] [Google Scholar]
- 50.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62:169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 52.Nomura L, Maino VC, Maecker HT. Standardization and optimization of multiparameter intracellular cytokine staining. Cytometry A. 2008;73:984–991. doi: 10.1002/cyto.a.20602. [DOI] [PubMed] [Google Scholar]
- 53.Belouski SS, et al. Utility of lyophilized PMA and ionomycin to stimulate lymphocytes in whole blood for immunological assays. Cytometry B Clin. Cytom. 2009;78:59–64. doi: 10.1002/cyto.b.20492. [DOI] [PubMed] [Google Scholar]
- 54.Inokuma M, et al. Functional T cell responses to tumor antigens in breast cancer patients have a distinct phenotype and cytokine signature. J. Immunol. 2007;179:2627–2633. doi: 10.4049/jimmunol.179.4.2627. [DOI] [PubMed] [Google Scholar]
- 55.Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A. 2006;69:1037–1042. doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- 56.Jaimes MC, et al. Quality assurance of intracellular cytokine staining assays: Analysis of multiple rounds of proficiency testing. J. Immunol. Methods. 2010;363:143–157. doi: 10.1016/j.jim.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aghaeepour N, et al. Early immunologic correlates of HIV protection can be identified from computational analysis of complex multivariate T-cell flow cytometry assays. Bioinformatics. doi: 10.1093/bioinformatics/bts082. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hultin LE, et al. Comparison of interlaboratory variation in absolute T-cell counts by single-platform and optimized dual-platform methods. Cytometry B Clin. Cytom. 2010;78:194–200. doi: 10.1002/cyto.b.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 60.Ornatsky O, et al. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 61. Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. An impressive demonstration of the power of mass cytometry for highly multidimensional analyses of haematopoietic cells.