Abstract
Sulfation and glucuronidation are the principal metabolic pathways of flavonoids, and extensive phase II metabolism is the main reason for their poor bioavailabilities. The purpose of this study was to compare the similarities and differences in the positional preference of glucuronidation versus sulfation in the mouse liver S9 fraction. The conjugating rates of seven mono-hydroxyflavones (HFs) (i.e., 2’-, 3’-, 4’-, 3-, 5-, 6-, and 7-HF), and five di-hydroxyflavones (diHFs), (i.e., 6,7-, 4’,7-, 3,7-, 5,7-, and 3,4’-diHF) were determined in three separate enzymatic reaction systems: (A) sulfation only, (B) glucuronidation only, or (C) simultaneous sulfation and glucuronidation (i.e., Sult-Ugt co-reaction). In general, glucuronidation rates were much faster than the sulfation rates. Among the HFs, 7-HF was the best substrate for both conjugation reactions, whereas 3-HF was rapidly glucuronidated but was not sulfated. As a result, the rank order of sulfation was very different from that of glucuronidation. Among the diHFs, regiospecific glucuronidation was limited to 7-OH and 3-OH positions, whereas regiospecific sulfation was limited to 7-OH and 4’-OH positions. Other positions (i.e., 6-OH and 5-OH) in diHFs were not conjugated. The positional preferences were essentially maintained in a Sult-Ugt co-reaction system, although sulfation was surprisingly enhanced. Lastly, sulfation and glucuronidation displayed different regiospecific- and substrate-dependent characteristics. In conclusion, glucuronidation and sulfation shared the same preference for 7-OH position (of flavonoids) but displayed unique preference in other positions in that glucuronidation preferred 3-OH position whereas sulfation preferred 4’-OH position.
Introduction
Flavonoids have a variety of “claimed” biological activities, including anti-inflammatory, anti-allergic, anti-viral, anti-cancer, and anti-oxidant (1–3). However, their bioavailabilities are poor due to rapid and extensive first-pass metabolism via the phase II metabolic pathways in the gut and liver. As a result, there are large amounts of sulfates and glucuronides in the plasma following oral administration of flavonoids, flavonoid-rich food or diets (4–7). For example, a significant portion of the absorbed flavonoid aglycones (e.g. fisetin and 7-hydroxyflavone or 7-HF) was rapidly bio-transformed into sulfates or glucuronides in rats (8). Separately, quercetin absorbed from the rat intestine was present in the conjugated forms (glucuronides or sulfates) in the mesenteric blood (9). In humans, following the ingestion of quercetin-rich diets/food, only quercetin metabolites (e.g. sulfate conjugates, glucuronide conjugates, or isorhamnetin conjugates), were found in the plasma (10), and the major conjugates were identified as quercetin-3-glucuronide, 3'-methylquercetin-3-glucuronide and quercetin-3'-sulfate (7). In contrast, 5-hydroxyflavone (5-HF) was exclusively metabolized to glucuronide (8), whereas chrysin (5,7-dihydroxyflavone, 5,7-diHF) and quercetin were both glucuronidated and sulfated (11–13). Similarly, extensive intestinal sulfation and glucuronidation of apigenin revealed that most apigenin were not transported intact across intestinal epithelium (14).
Most of the published studies on flavonoid metabolism were focused on glucuronidation (15–19). These studies have demonstrated that glucuronidation is regiospecific and isoforms-dependent (20, 21). Furthermore, the concentrations of flavonoids used moderately impacted the dominant isoforms for their metabolism, because UDP-glucuronosyltransferases 1As (or UGT1As, especially UGT1A1) may display substrate inhibition kinetics (21). In contrast, much less is known about isoform-dependent regiospecific sulfation of flavones. To our knowledge, no information is available regarding the question as to whether rapidly glucuronidated flavonoids will be similarly sulfated. More importantly, there are no published data showing whether flavone metabolism via sulfation or glucuronidation pathway shares or displays unique structural requirements towards their substrates. The latter is important in order to elucidate if these two conjugation pathways are compensatory (the slower the glucuronidation is, the faster the sulfation is, or vice versa), competitive, or independent of each other.
Therefore, the purpose of this study is to determine if sulfation and glucuronidation pathways share or display unique structural requirements for their flavone substrates. Liver S9 fraction was used here because S9 fraction is routinely used in the metabolism studies, especially for the phase II metabolic pathways involving sulfation. In addition, liver is enriched with both Ugts and sulfotransferases or Sults. Intact cells or organs were not used here since the focus is on the formation of the phase II conjugates, which cannot passively diffuse across the cell membrane.
Materials and Methods
Materials
Seven mono-hydroxyflavones (or HFs) and five di-hydroxyflavones (or diHFs) were purchased from LC Laboratories (Woburn, MA). Uridine diphosphoglucuronic acid (UDPGA), β-glucuronidase, alamethicin, magnesium chloride, and 3'-phosphoadenosine 5'-phosphosulfate (PAPS) were purchased from Sigma-Aldrich (St Louis, MO). All other materials (typically analytical grade or better) were used as received.
Mouse liver S9 fraction preparation
Male FVB mouse (18~22 g) were obtained from Cyagen Biosciences Inc. (Guangzhou, China) and kept in an environmentally controlled room (temperature: 25 ±2°C, humidity: 50±5%, 12 h dark-light cycle) for at least three days before the experiments. The animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health, and the procedures were approved by the Ethical Committee of Southern Medical University (Guangzhou, China). Male mouse liver S9 fraction was prepared using a procedure published previously with minor modification (22, 23). Briefly, ten freshly harvested mouse livers were washed and then perfused with ice-cold saline, weighed and minced. Minced livers were homogenized using a motorized homogenizer (4 strokes) in ice-cold homogenization buffer (50 mM potassium phosphate, 250 mM sucrose, 1 mM EDTA, pH 7.4) and centrifuged at 7,700 × g for 15 min at 4°C. Fat layer was carefully aspirated, and the supernatant was collected with pasture pipettes into microfuge tubes (1 ml each), which were stored at −80 °C until use. The concentration of S9 fraction protein (normally 5–20 mg/ml) was determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA, USA) using the bovine serum albumin as the standard.
Ugt activity measurements
The incubation procedures for measuring mouse Ugt activities in vitro were essentially the same as those published previously (16, 18), and all the studies were conducted in triplicates. Briefly, different concentrations of flavone substrates, mouse liver S9 fraction protein (final concentration: 0.053–0.37 mg/mL), magnesium chloride (0.88 mM), alamethicin (0.022 mg/mL), and diphosphate glucuronic acid (UDPGA) at 3.5 mM (added last) were mixed in a 50 mM potassium phosphate buffer (pH 7.4, containing 0.1% (w/v) vitamin C as the stabilizing agent for flavones). Vitamin C (0.1%) was added to stabilize the substrate and the reaction rates did not change because of its presence (not shown). The mixture (final volume ≈ 400 µL) was incubated at 37°C for predetermined periods of time (5 to 90 min to limit the percent metabolism of substrate to less than 30%, allowing the accurate determination of initial rates) based on results of pilot experiments. Reaction was stopped by the addition of 60 µL 94% acetonitrile plus 6% glacial acetic acid containing 50 µM testosterone as the internal standard (Stop Solution). For glucuronidation, three starting substrate concentrations were used: 5, 10, and 40 µM. We found that for phase II metabolism of flavonoids, 10 µM was considered as a moderate concentration (24–26), whereas 40 µM was considered as the high test concentration since reaction rates at this concentration is generally above reported Vmax values of most flavonoid glucuronidation. A concentration of 5 µM (designated as low concentration in this paper) is achievable after pharmacological dose of flavonoids (e.g., genistein) (27). We did not conduct studies at lower concentrations because of quantification limits of our method was around 1.5 µM (30% of 5 µM) for model flavonoids with the lowest molar extinction coefficients (UV).
We have also tested chemical stability of the tested compounds and found that 2’-, 3’-, 4’-, 3-, 6-, and 7-hydroxyflavones(2’-HF, 3’-HF, 4’-HF, 3-HF, 6-HF and 7-HF), 6,7-, 4’,7-, 3,7-, 5,7-, and 3,4’-dihydroxyflavones(6,7-diHF, 4’,7-diHF, 3,7-diHF, 5,7-diHF and 3,4’-diHF) were stable in 50 mM potassium phosphate buffer (pH 7.4) at 37°C with 0.1% vitamin C for 30 to 90 min. The 5-HF flavone was only stable for 15 min in 50 mM potassium phosphate buffer (pH 7.4) at 37°C with 0.1% vitamin C. We then conducted pilot study to measure the formation of 6-O-glucuronide and 7-O-glucuronide (from 6HF and 7HF) as a function of time (at 0, 15, 30, 45, 60, and 90 min) using three starting concentrations (5, 10, and 40 µM), and the results indicated that formation was approximately linear for up to 90 min (not shown). Formation of 5-O-glucuronide (from 5-HF) was measured at 0, 5, 10, 15, and 20 min using three concentrations, and the results indicated the formation was approximately linear for up to 15 min (not shown). Therefore the incubation time for 5-HF was 5 to 15 min, and for other flavones was from 30 to 90 min.
Sult activity measurements
The S9 fraction (0.053–0.37 mg/mL protein), 0.1 mM adenosine 3'- phosphate 5'-phosphosulfate or PAPS, and 0.1% vitamin C in a 50 mM potassium phosphate buffer (pH 7.4) were mixed with different concentrations of a substrate. The mixture (final volume ≈400 µL) was incubated at 37°C for a predetermined period of time (5 to 90 min) and the reaction was stopped by the addition of 60 µL Stop Solution. For sulfation reactions, three substrate concentrations were used as well: 5, 10, and 40 µM. From pilot studies, the best incubation time for 5-HF was from 5 to 15 min, and for other flavones was from 30 to 90 min.
Co-reaction system of glucuronidation and sulfation
In the co-reaction system, co-factors for Sult and Ugt were both present at the same time, and their concentration of in the co-reaction system was the same as the single reaction system. Due to chemical stability consideration, incubation of 5-HF was limited to 5 to 15 min, whereas the incubation of other flavones was limited to 30 to 90 min. To minimize variability, experiments for two single reaction systems (Sult and Ugt) and one co-reaction system were conducted simultaneously for each of the flavones.
Elimination half-life of aglycone in single and co-reaction system of glucuronidation and sulfation
Amounts of 6,7-diHF (10 µM) remaining as a function time were determined at 0, 10, 15, 30, 45, 60 and 75 min for single Ugt reaction system and Sult-Ugt co-reaction system or at 0, 10, 20, 40, 60, 80 and 120 min for single Sult reaction system. The elimination half-life of aglycone in single and co-reaction system of glucuronidation and sulfation were determined based on percent of flavones remained when compared the starting concentration.
UPLC analysis of flavones and their glucuronides and sulfates
We analyzed seven HFs, five diHFs, and their respective glucuronides and/or sulfates by using the method same as those that was published (20). Briefly, the system, Waters Acquity UPLC with photodiode array detector and Empower™ software; column, BEH C18, 1.7 µm, 2.1 × 50 mm; mobile phase B, 100% acetonitrile; mobile phase A, 100% aqueous buffer (2.5 mM NH4Ac, pH 7.4); flow rate, 0.45 mL/min; gradient, 0 to 2.0 min, 10–35% B, 2.0 to 3.0 min, 35–70% B, 3.2 to 3.5 min, 70–10% B, 3.5 to 4.0 min, 10% B; injection volume, 10 µL; and detection wavelength, 254 nm for testosterone, and other values were different depending on compounds (actual value listed in Table 1). Linearity was established over the range 1.5625–50 µM for all compounds. The lower limit of quantification (LLOQ) for all compounds was at least 1.5625 µM. The analytical method for each compound was validated for inter-day and intra-day variations using 6 samples at each of three concentrations (50, 12.5, and 1.56 µM). Error for all flavones was <15%.
Table.1.
UV characteristics, LC/MS/MS characteristics, conversion factors (K) of glucuronides and sulfates of selected mono-hydroxylfavones and dihydroxyflavones. The metabolic positions of metabolites were determined by UV characteristics of relevant HFs.
| Flavones | Glucuronides | Sulfates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UV λmax, nm | Position | TW | K | UV λmax, nm | GCI in LC/MS/MS | Position | TW | K | UV λmax, nm | SCI in LC/MS/MS | ||||||
| Band I | Band II | Band I | Band II | [M+H]+ | [M−G+H]+ | Band I | Band II | [M+H]+ | [M−S+H]+ | |||||||
| 2’-HF | 323.9 | 254.9 | 2’-O-G | 254 | 1.26±0.08& | 321.5 | 254.9 | 415 | 239 | 2’-O-S | 254 | 0.72±0.06 | 319.2 | 253.7 | 319 | 239 |
| 3’- HF | 298.9 | 240.7 | 3’-O-G | 254 | 0.93±0.02& | 301.3 | 256.1 | 415 | 239 | 3’-O-S | 254 | 1.40±0.09 | 306.0 | 243.1 | 319 | 239 |
| 4’- HF | 324.2 | 252.5 | 4’-O-G | 254 | 1.16±0.02& | 318.0 | 253.7 | 415 | 239 | 4’-O-S | 254 | 1.21±0.05 | 315.7 | 253.7 | 319 | 239 |
| 3-HF | 343.8 | 239.5 | 3-O-G | 254 | 0.96±0.06& | 309.2 | 247.8 | 415 | 239 | 3-O-S | NA | NA | ||||
| 5- HF | 335.8 | 269.1 | 5-O-G | 254 | 0.84±0.13& | 324.1 | 262.0 | 415 | 239 | 5-O-S | 254 | 1.52±0.03 | 294.1 | 257.3 | 319 | 239 |
| 6- HF | 304.8 | 268.0 | 6-O-G | 254 | 0.99±0.03& | 306.0 | 260.8 | 415 | 239 | 6-O-S | 254 | 1.59±0.04 | 306.0 | 257.5 | 319 | 239 |
| 7- HF | 309.0 | 252.3 | 7-O-G | 254 | 0.96±0.03& | 309.0 | 252.3 | 415 | 239 | 7-O-S | 254 | 0.98±0.04 | 308.4 | 252.5 | 319 | 239 |
| 3,4’- diHF | 356.9 | 232.4 | 3-O-G | 254 | 1.02±0.03& | 334.7 | 232.4 | 415 | 239 | 3-O-S | NA | NA | ||||
| 4’-O-G | 254 | 0.87±0.03& | 352.1 | 246.6 | 431 | 255 | 4’-O-S | 355 | 1.06±0.076 | 348.5 | 246.6 | 335 | 255 | |||
| 3,7- diHF | 343.8 | 252.5 | 3-O-G | 263 | 1.29±0.03& | 314.4 | 251.4 | 431 | 255 | 3-O-S | NA | NA | ||||
| 7-O-G | 254 | 0.86±0.09& | 341.5 | 250.2 | 431 | 255 | 7-O-S | 340 | 0.97±0.08 | 341.5 | 246.6 | 335 | 255 | |||
| 4’,7- diHF | 331.1 | 253.7 | 4’-O-G | NA | NA | 4’-O-S | 254 | 1.01±0.014 | 314.4 | 253.7 | 335 | 255 | ||||
| 7-O-G | 254 | 1.09±0.04 | 331.1 | 252.5 | 431 | 255 | 7-O-S | 254 | 1.18±0.04 | 331.1 | 252.5 | 335 | 255 | |||
| 5,7- diHF | 312.2 | 266.8 | 7-O-G | 254 | 1.16±0.02 | 306.2 | 266.8 | 431 | 255 | 7-O-S | 254 | 1.45±0.10 | 308.1 | 268.0 | 335 | 255 |
| 6,7- diHF | 316.8 | 266.8 | 6-O-G | 309 | 1.61±0.14 | 314.4 | 264.4 | 431 | 255 | 6-O-S | NA | NA | ||||
| 7-O-G | 309 | 1.22±0.11 | 312.2 | 264.4 | 431 | 255 | 7-O-S | 309 | 1.21±0.76 | 312.0 | 266.8 | 335 | 255 | |||
Using published data (20), TW: Test Wavelength (nm), GCI: Glucuronide characteristic ions, SCI: Sulfate characteristic ions, NA: not available due to low intensity of the peak.
LC-MS/MS analysis of flavonoid glucuronides and sulfates
Seven HFs, five diHFs, and their respective glucuronides and sulfates were separated by the same UPLC system. The effluent was introduced into a Premier XE mass spectrometer triple quadrupole mass spectrometer (Waters Corp., Milford, MA, USA). The mass spectrometer was operated in the negative ion mode to perform the analysis of flavones and their metabolites. The main working parameters for the mass spectrum were capillary voltage, 3.0 kV; cone voltage, 35 V; ion source temperature, 110°C; and desolvation temperature, 500°C. Flavone mono-glucuronides and mono-sulfates were identified by MS and MS2 full scan modes (Table 1).
Determination of position of conjugation by UV spectra shift method
The molecular positions of glucuronidation and sulfation on HFs and diHFs were identified by changes in UV spectra using the same λmax shift method published previously (20). Details are shown in Table 1 and described in the results.
Molar extinction coefficient ratio of flavone conjugates
In order to provide more accurate estimation of substrate and metabolite concentrations, and of glucuronidation and sulfation rates, a conversion factor (K) representing the ratio between the molar extinction coefficient of the glucuronide, or sulfate and its aglycone, was determined for each flavone and its respective phase II metabolite. The conversion factors (K) of the glucuronides from the seven mono-hydroxyflavones and two dihydroxyflavones (3,4’-, and 3,7-diHF) were taken from previously published results (20). To calculate K of sulfates, sulfates of flavones were prepared using S9 fraction at a flavone concentration of 10 µΜ and 40 µΜ. The supernatant was then removed and extracted twice with methylene chloride (sample/dichloromethane=1:3, v/v) to remove >95% of flavone aglycones and protein present in the S9 fraction. The resulting sample was then divided into two parts, one of which was analyzed following incubation with water and the other after hydrolysis with sulfatase (30 U/ml) at 37°C for 2 h. The difference in the amount of aglycones found in these two samples was the amount of metabolite formed. The relationship between the spectral peak areas of the metabolites before hydrolysis and the peak areas of aglycones after the hydrolysis was used to establish the conversion factor required to quantify the amounts of flavone conjugates as described previously (28, 29).
Statistical Analysis
One-way ANOVA, factorial design ANOVA (Univariate Analysis of Variance) with Tukey multiple comparison (post-hoc) tests or Student’s t-test were used to evaluate statistical differences. Differences were considered significant when p values were less than 0.05 (or p<0.05).
Results
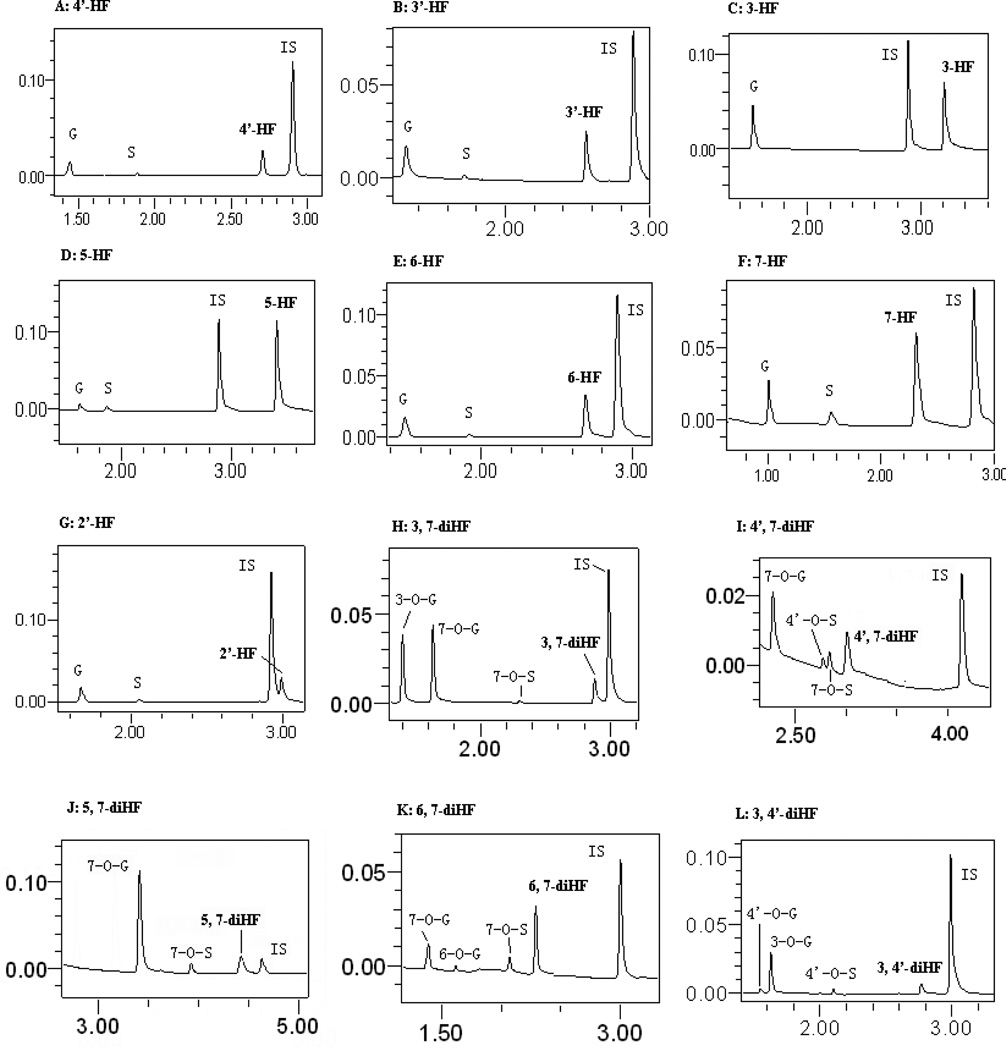
Confirmation of flavone conjugate formation by LC-MS/MS
We conducted LC-MS/MS studies of the metabolites of all flavones (Figure 1) used in the present study to show that all glucuronides and sulfates formed using FVB mice S9 fraction were mono-glucuronides and mono-sulfates (Table 1). The UPLC chromatograms and MS data of these flavones has been published in our earlier publication (20). All the HFs formed had one glucuronide and one sulfate moiety except for 3-HF, which formed only one glucuronide but no sulfate. There were no di-glucuronides or di-sulfates of any diHFs. Conjugating reaction of 3,4’-, 3,7-, and 6,7-diHF all produced two mono-glucuronide isomers but only one mono-sulfate. In contrast, conjugation of 4’,7-diHF resulted in two sulfate isomers but only one glucuronide, whereas conjugation of 5,7-diHF resulted in only one glucuronide and one sulfate.
Figure 1. Structures of model flavones.
Structures of aglycone forms of tested flavones used in the present study. Seven monohydroxyflavones and five dihydroxyflavones were chosen for these experiments. Available conjugating –OH positions include 3, 5, 6, 7, 2’, 3’, and 4’.
The positions of conjugation of dihydroxyflavones
There are no commercial standards for flavone glucuronides and sulfates, and therefore it was necessary to determine the likely molecular positions of mono-glucuronides and mono-sulfates using the UV spectra and relative reaction rates. The site of glucuronidation of tested flavones was tentatively assigned using the UV peak (λmax) shift method developed previously in our lab (31), and actual position could be further confirmed by NMR. In general, the UV spectra of flavones have two λmax values at around 300 nm (band I), and at around 240 nm (band II). Glucuronidation at the 3-hydroxyls and 4'-hydroxyls resulted in band I hypsochromic shifts (or blue shift) of 13–30 and 5–10 nm, respectively. Glucuronidation of the 5-hydroxyl group caused a band II hypsochromic shift of 5–10 nm. In contrast, glucuronidation of the 7-hydroxyl group did not cause any change in band I or II (32). Because we only have 5 diHFs with limited substitution pattern (5 possible positions at 3, 5, 6, 7, and 4’), this method of positional identification is very accurate and reliable for every compound except for 6,7-diHF, whose positional identification required additional factors (see below) (20). Therefore, the 3-O-G and 7-O-G of 3,7-diHF, the 3-O-G and 4’-O-G of 3,4’-diHF were tentatively assigned according to the published UV method (Table 1) (20). For 4’,7-diHF, the unique glucuronide showed no shift in band I, which indicated that the metabolite was likely the 7-O-glucuronide of 4’,7-diHF (Table.1). For 5,7-diHF, the unique glucuronide showed no shift in band I or band II, which indicated that this metabolite was likely the 7-O-glucuronide of 5,7-diHF (Table 1). For 6,7-diHF, the unique glucuronide was presumed to be 7-O-glucuronide because the glucuronide production from 7-HF was much greater than that from 6-HF, indicating that 7-OH was much more active than 6-OH. This is consistent with our previous observation that the more slowly metabolized phenolic group in a diHF was always more similar to that of the more slowly metabolized HF than the more rapidly metabolized HF (20).
Similarly, we also tentatively assigned the position of mono-sulfates from diHFs using the same UV λmax shift method, which could be further confirmed by NMR.. 3-HF could not be sulfated by FVB mice liver S9 fraction. For 4’-HF, sulfation at the 4’-OH position severely diminished the peak at 321.5 nm, and caused a band I hypsochromic shift of 5.8 nm. For 7-HF, there was nearly no band I hypsochromic shift. Sulfation of the 5-hydroxyl group of 5-HF resulted in a major 41.7 nm hypsochromic shift in Band I, suggesting sulfation at this site significantly altered electron conjugation of the flavone ring structure. Identification of the sulfate position using the shift in band I and band II values are shown in Table 1, and the extinction coefficient ratios of all HFs and diHFs are also listed in Table 1.
For diHFs, a similar approach was used to tentatively assigned the position of sulfation, which was based on the UV peak (λmax) shift in band I and band II regions. Using a similar methodology for 4’,7-diHF, the sulfate 1 had a shorter retention time and a major hypsochromic shift of 16.7 nm of the band I region. It was assigned as 4’-O-sulfate, whereas sulfate 2 showed nearly no change at either band I or band II and was assigned as 7-O-glucuronide (Table 1). For 3, 4’-diHF, the unique sulfate produced a band I induced a λmax hypsochromic shift of 8.4 nm, so it was assigned as 4’ -O-sulfate. For 3,7-diHF and 5,7-diHF, both had only one sulfate, and each sulfate produced nearly no change in the UV spectrua at either band I or band II regions compared to aglycones, and so it was identified as a 7-O-sulfate. This identification of sulfate position is constructive and necessary because it provided the basis for determining the molar extinction coefficient ratios as shown in Table 1. On the other hand, both 3,7-diHF and 3,4’-diHF had only one sulfate, and we assigned the sulfates of 3,7-diHF and 3,4’-diHF as 7-O-sulfate and 4’-O-sulfate, respectively, since 3-HF was not sulfated. The sulfation reaction results for mono-hydroxyflavones demonstrated that 7-OH was more active than 6-OH and the amount of sulfate 1 of 6,7-diHF was greater than sulfate 2. As shown in our previous studies, the more slowly metabolized phenolic group in a diHF was always similar to that of the more slowly metabolized HF than the more rapidly metabolized HF (20). So we presumed that sulfate 1 of 6,7-diHF with shorter retention time was 7-O-sulfate and the other rare sulfate was 6-O-sulfate.
Sulfation rates of mono-hydroxyflavones
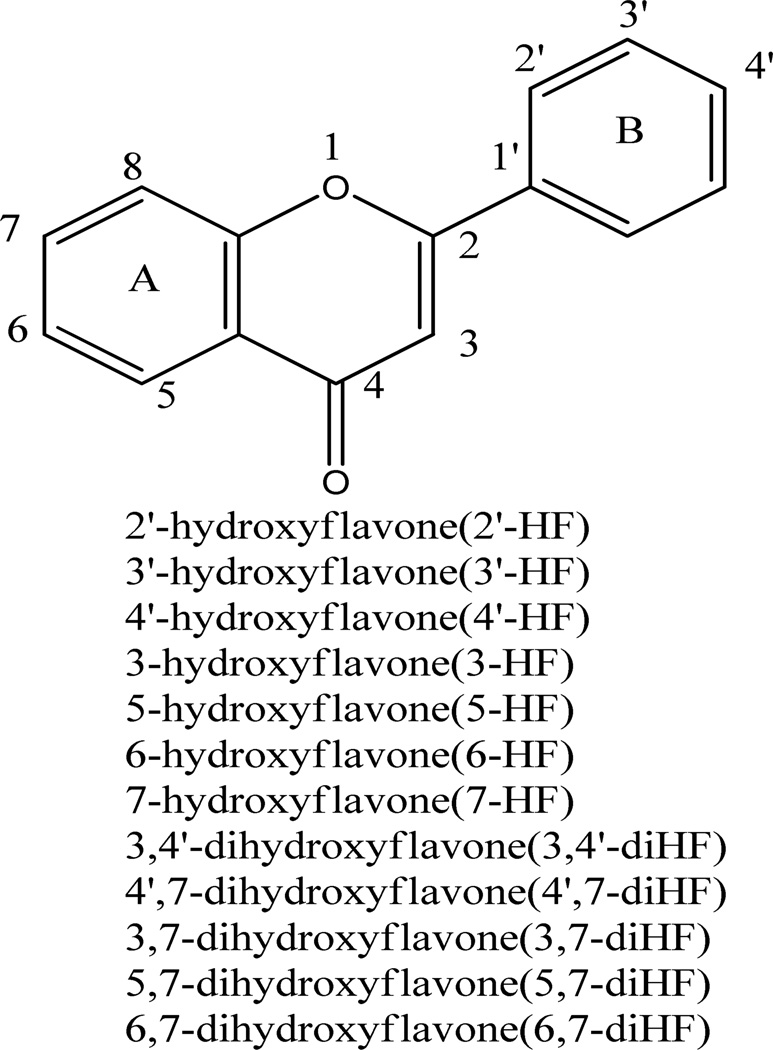
The rates of sulfation of seven mono-hydroxyflavones by FVB mice liver S9 fraction in Sult single reaction were determined at 5, 10, and 40 µM (Figure 3A). The results showed that the sulfation rate of 7-HF was always the fastest of all mono-hydroxyflavones at all substrate concentrations, while 3-HF was not sulfated by FVB mice liver S9 fraction at all. At 10 µM substrate concentration, the rank order of the sulfation rates (in nmol per min per mg of protein) was 7-HF (0.0998 ± 0.0061) > 4’-HF (0.0793 ± 0.0031) > 2’-HF (0.0719 ± 0.0030) > 5-HF (0.0524 ± 0.0019) > 3’-HF (0.0312 ± 0.0095) > 6-HF (0.0211 ± 0.0002) > 3-HF (0.00 ± 0.00). At 5 µM substrate, the rank order was the same, while the rank order in the middle changed slightly at 40 µM. The results clearly indicated that there were significant differences in the sulfation rates with respect to different positions of a hydroxyl group (p<0.05, one way ANOVA) (Fig. 3A).
Figure 3. Glucuronidation and sulfation of seven mono-hydroxyflavones in single Sult or Ugt reaction system.
Three different concentrations (5, 10, and 40 µM) were used in the experiments done in triplicates (n=3). The rates of glucuronidation and sulfation were calculated in nmol/min/mg protein. The error bar is the SD of three determinations. The data were analyzed by one-way ANOVA with Tukey post-hoc test. The asterisk (*) indicates a statistically significant difference for the glucuronidation (Figure 3A) and sulfation (Figure 3B) rates among seven mono-hydroxyflavones (p< 0.05, one-way ANOVA). Significant differences between three concentrations for each flavone are marked as follows: ●P<0.05 for 7-HF, ▼P<0.05 for 4’-HF, ▲P<0.05 for 2’-HF, □P<0.05 for 5-HF, ○P<0.05 for 3’-HF, ■P<0.05 for 6-HF, □P<0.05 for 3-HF.
Glucuronidation rates of mono-hydroxyflavones
The rates of glucuronidation of seven mono-hydroxyflavones by the FVB mouse liver S9 fraction were again determined at 5, 10, and 40 µM substrate concentrations (Figure 3B). The glucuronidation rates of 7-HF were still the fastest of all mono-hydroxyflavones at all three substrate concentrations, and 3-HF was glucuronidated the second fastest at 5 and 10 µM. However, when the concentration was 40 µM, 4’-HF and 3’-HF jumped ahead of 3-HF, whereas 5-HF was always glucuronidated the slowest. This were consistent with our previous study showing that 5-HF was the least reactive among the seven mono-hydroxyflavones when the reaction was catalyzed by human recombinant UGT1A1 or UGT1A10, and in human liver and intestinal microsomes at concentrations of 2.5, 10, and 35 µM (20). Also, the rank orders of glucuronidation were the same at concentrations of 5 µM and 10 µM: 7-HF > 3-HF > 3’-HF > 4’-HF > 2’-HF > 6-HF > 5-HF. At the 40 µM substrate concentration, the rank order in the middle changed except for 7-HF and 5-HF. The results clearly indicated that the glucuronidation of the seven hydroxyflavones were structure-dependent (p<0.05, one way ANOVA) (Figure. 3B). However, the rank order for glucuronidation was very different from that of the sulfation (Fig.3), especially for 3-HF, which was rapidly glucuronidated but not sulfated at all.
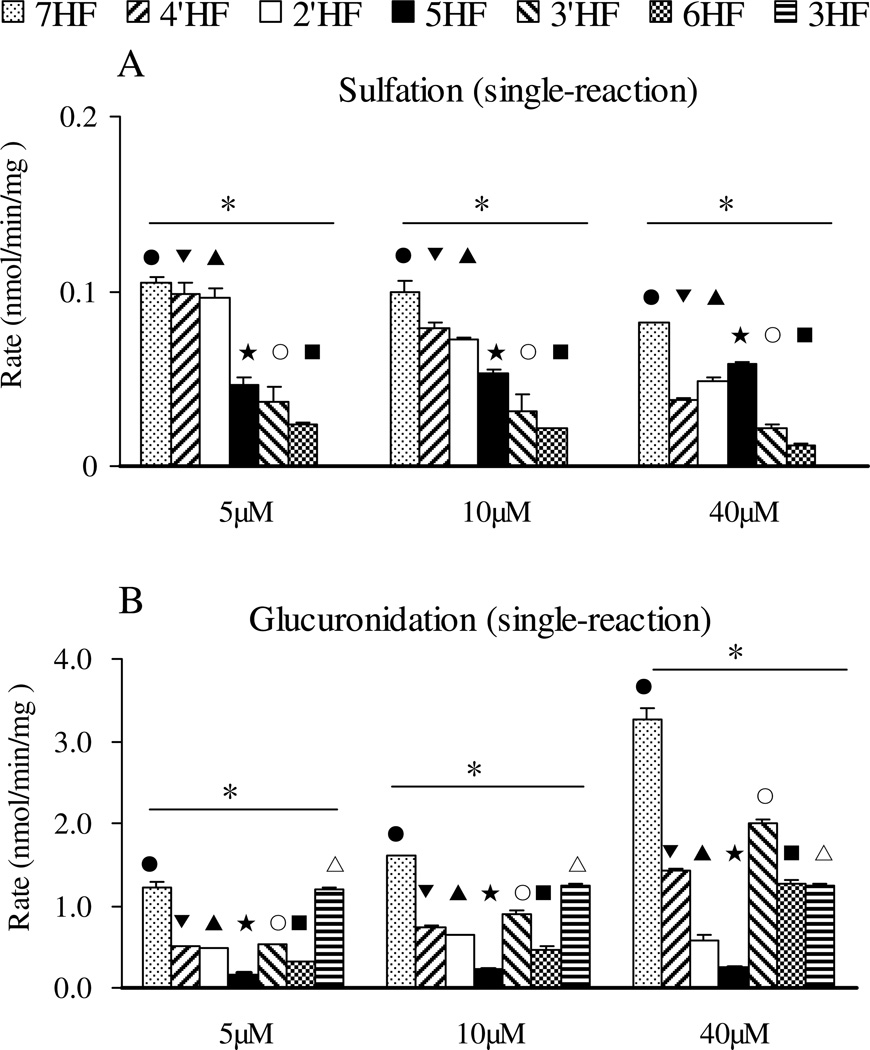
Sulfation rates of di-hydroxyflavones
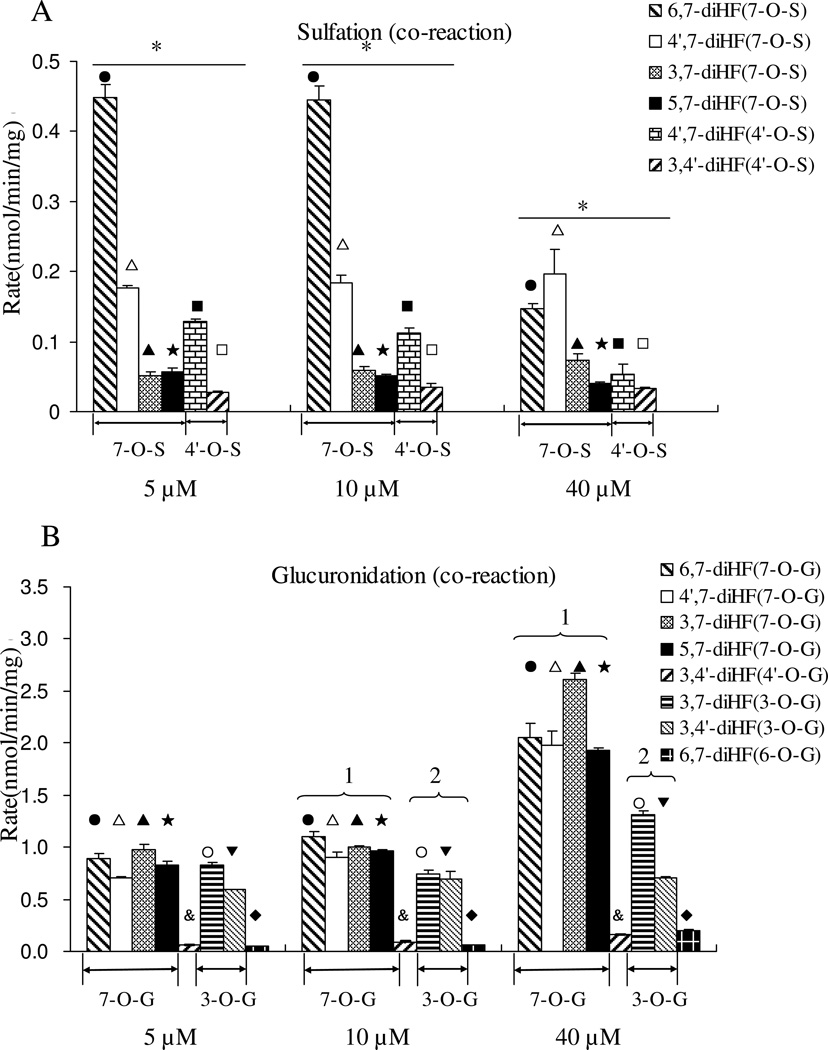
The rates of sulfation of the five di-hydroxyflavones were determined at 5, 10, and 40 µM (Figure 4A). All of the diHFs formed only one mono-sulfate, each of which was determined to be 7-O-sulfate, except for 4’,7-diHF which formed two mono-sulfates. The formation rates of 7-O-sulfate from 4’,7-diHF was always higher than formation rates at the 4’-OH position at all three concentrations, consistent with reaction rates of mono-hydroxyflavones from 4’-HF and 7-HF.
Figure 4. Glucuronidation and sulfation of five dihydroxyflavones by FVB mice liver S9 in a single Sult (A) and Ugt (B) reaction system.
Concentrations used, number of experiments and calculation of the rates of glucuronidation and sulfation are the same as in the legend to Figure 3. The data were analyzed by one-way ANOVA with Tukey post-hoc test (Figure 4A) or Univariate ANOVA with Tukey test (Figure 4B). The asterisk (*) in Figure 4A indicates a statistically significant difference in the sulfation rates among five diHFs (p < 0.05, one way ANOVA). The number “1” in Figure 4B, shows that 7-OH position is the group of reaction with the fastest glucuronidation rates, and the number “2” shows the –OH group with the second fastest glucuronidation rates at 3-OH. Significant differences between three concentrations for each flavone are marked as follows: ●P<0.05 for 6, 7-diHF (7-O-S or 7-O-G), □P<0.05 for 4’, 7-diHF(7-O-S or 7-O-G), ▲P<0.05 for 3, 7-diHF (7-O-G), □P<0.05 for 5, 7-diHF (7-O-S or 7-O-G), ■P<0.05 for 4’, 7-diHF (4’-O-S), □P<0.05 for 3, 4’-diHF (4’-O-S), &P<0.05 for 3, 4’-diHF (4’-O-G), ○P<0.05 for 3, 7-diHF (3-O-G), ▼P<0.05 for 3, 4’-diHF (3-O-G)6-HF, ♦P<0.05 for 6, 7-diHF (6-O-G)
At a 5 µM substrate concentration, the fastest formation rate (in nmol per min per mg of protein) was for formation of 7-O-sulfate of 6,7-diHF (0.2217 ± 0.0102), followed closely behind by 7-O-sulfate of 4’,7-diHF (0.1826 ± 0.0033), then 7-O-sulfate of 4’,7-diHF (0.1036 ± 0.0051), 7-O-sulfate of 3,7-diHF (0.0486 ± 0.0083), 7-O-sulfate of 5,7-diHF (0.0387 ± 0.0025), and 4’-O-sulfate of 3,4’-diHF (0.0210 ± 0.0022). At 10 µM substrate concentration, the rank order was the same as the order observed at 5 µM. The fast sulfation rate occurred at 7-OH for 4’,7-diHF with 40 µM substrate. These results clearly indicated that the sulfation tendency at 7-OH position was much greater than those at the other position (p<0.05, one way ANOVA), and rank order of positional preferences in diHF followed that of HFs.
Glucuronidation rates of di-hydroxyflavones
The glucuronidation rates of five diHFs were determined to rank order the activity of different hydroxyl groups. Among the diHFs, there are four flavones containing 7-OH, two containing 3-OH, two containing 4’-OH, one containing 5-OH and one containing 6-OH group. The glucuronidation rate at 7-OH was always faster than that at other –OH groups for all di-hydroxyflavones at all three substrate concentrations. The glucuronidation rates at 3-OH were again the second fastest among the other –OH groups. The glucuronidation activity of –OH groups in diHFs were 7-OH > 3-OH > 4’ -OH (p>0.05) ≈ 6-OH > 5-OH (Univariate Analysis of Variance, p<0.05). For example, at 10 µM, the glucuronidation rate rank order (nmol per min per mg of protein) was 1.1059 ± 0.1311 (7-O-G of 6,7-diHF) ≈ 1.0763 ± 0.0399 (7-O-G of 5,7-diHF) ≈ 1.0621 ± 0.0413 (7-O-G of 4’,7-diHF) ≈ 1.0201 ± 0.0288 (7-O-G of 3,7-diHF) > 0.7564 ± 0.0018 > (3-O-G of 3,7-diHF) > 0.0642 ± 0.0010 (4’-O-G of 4’,7-diHF) ≈ 0.0644 ± 0.0078 (6-O-G of 6,7-diHF) > 0.00 ± 0.00 (5-O-G of 5,7-diHF). This was also consistent with the previous results that the glucuronidation rate of 7-HF was always the highest, followed by that of 3-HF, 4’-HF, and 6-HF. Also, the activity rank orders of glucuronidation were the same at 5 µM and at 10 µM (Figure 4B).
Taken together, for diHFs, 7-OH was a preferred position for both sulfation and glucuronidation, whereas 3-OH was a preferred position for glucuronidation but a non-reactive position for sulfation. In contrast, 4’-OH was a preferred position for sulfation, but not for glucuronidation.
Conjugation of HFs in the Sult-Ugt co-reaction system
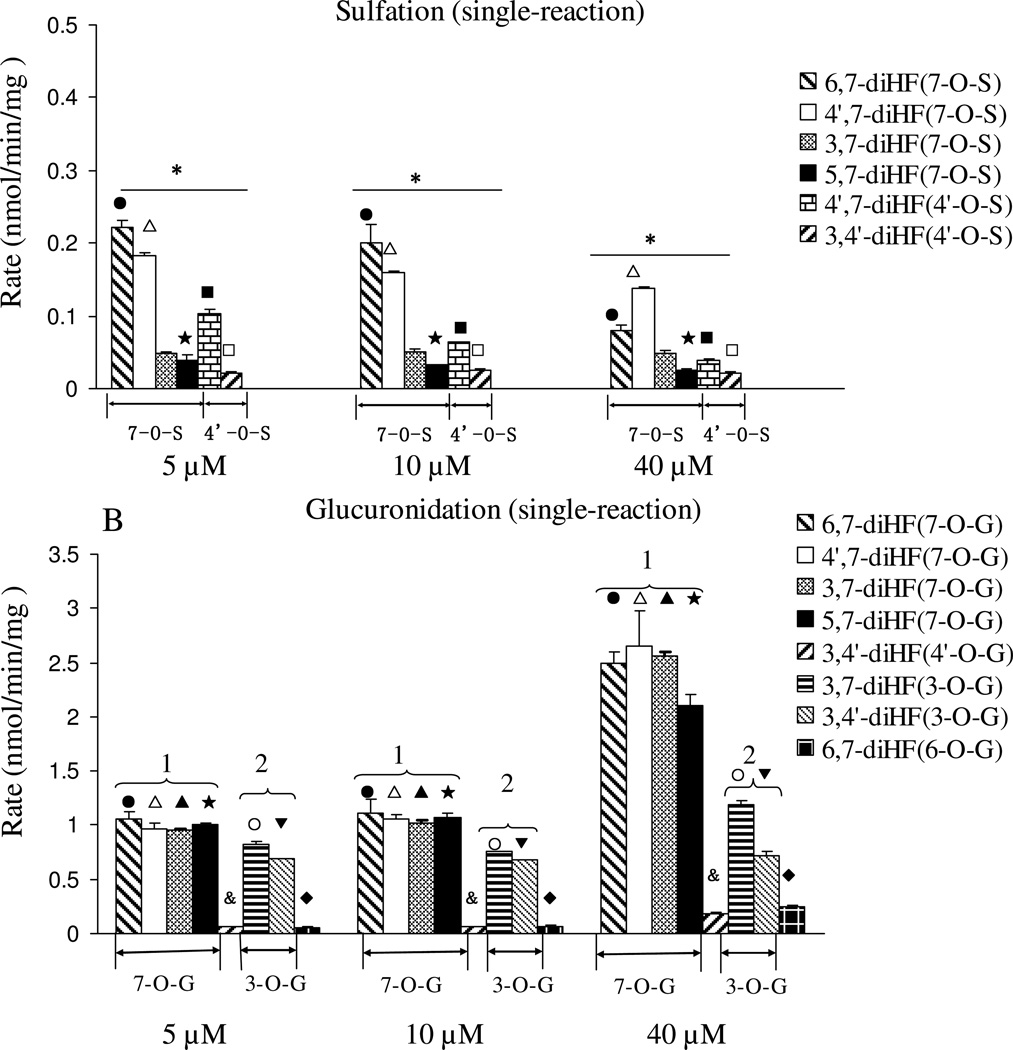
We investigated the sulfation and glucuronidation in a Sult-Ugt co-reaction system in order to determine if effects of structural changes observed in a single reaction system seen above will be affected in a Sult-Ugt co-reaction system, which somewhat better mimic cellular conditions where both sulfation (in cytosol) and glucuronidation (at endothelium reticulum) occurs albeit at different cellular compartments. The experiments were conducted in the Sult-Ugt co-reaction system using 5, 10, and 40 µM substrate concentrations. We did not see significant changes with respect to the rank order of sulfation or glucuronidation in a co-reaction system (Figure 5), when compared to the single reaction systems (Fig.3), although there was some minor shift in one or two of the compounds at one concentration.
Figure 5. Glucuronidation and sulfation of seven mono-hydroxyflavones in Sult-Ugt co-reaction system.
Concentrations used, number of experiments, calculation of the rates of glucuronidation and sulfation, and data analysis, are the same as in the legend to Figure 3. The asterisk (*) indicates a statistically significant difference for the glucuronidation and sulfation rates among seven mono-hydroxyflavones (p< 0.05, one way ANOVA). Significant differences between three concentrations for each flavone are the same as in the legend to Figure 3.
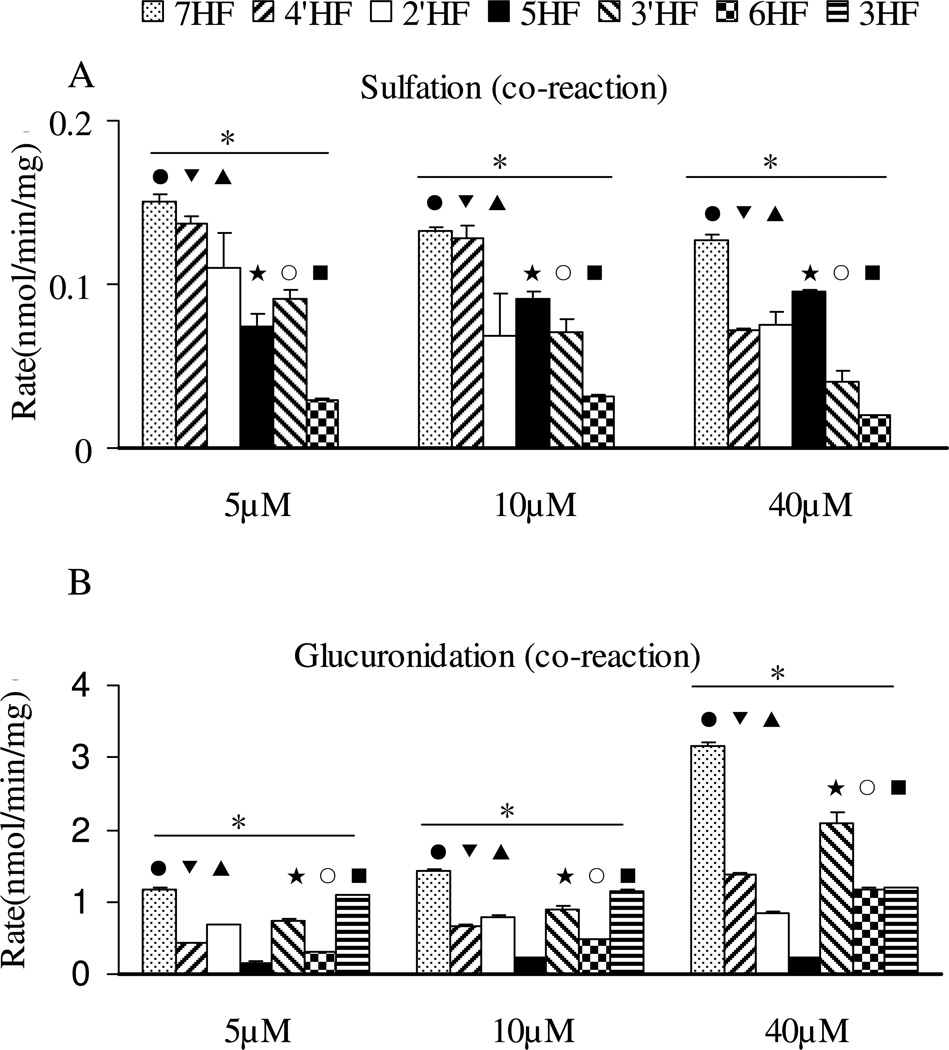
Conjugation of diHFs in the Sult-Ugt co-reaction system
The results indicated that effects of structural changes on a sulfation and glucuronidation were essentially maintained in the co-reaction system (Figure 6), when compared to the single reaction systems (Fig.4). Specifically, 7-OH sulfation was once again preferred over 4’-OH sulfation (Fig.6), whereas other groups were not sulfated. The results again showed that the sulfation rate was unexpectedly enhanced (p<0.05) in the presence of an Ugt reaction system (Figure 6A) (p<0.05), whereas the formation rates of glucuronides were usually unchanged or slightly decreased in the presence of a Sult reaction system.
Figure 6. Glucuronidation and sulfation of five dihydroxyflavones by FVB mice liver S9 in a Sult-Ugt co-reaction system.
Concentrations used, number of experiments, and calculation of the rates of glucuronidation and sulfation are the same as in the legend to Figure 3. The data were analyzed by one-way ANOVA with Tukey post-hoc test (Figure 6A) or Univariate ANOVA with Tukey test (Figure 6B). The asterisk (*) in Figure 6A indicates a statistically significant difference for the sulfation rates among five dihydroxyflavones (p < 0.05). Number “1” and the number “2” are the same as in the legend to Figure 4. Significant differences between three concentrations for each flavone are the same as in the legend to Figure 4.
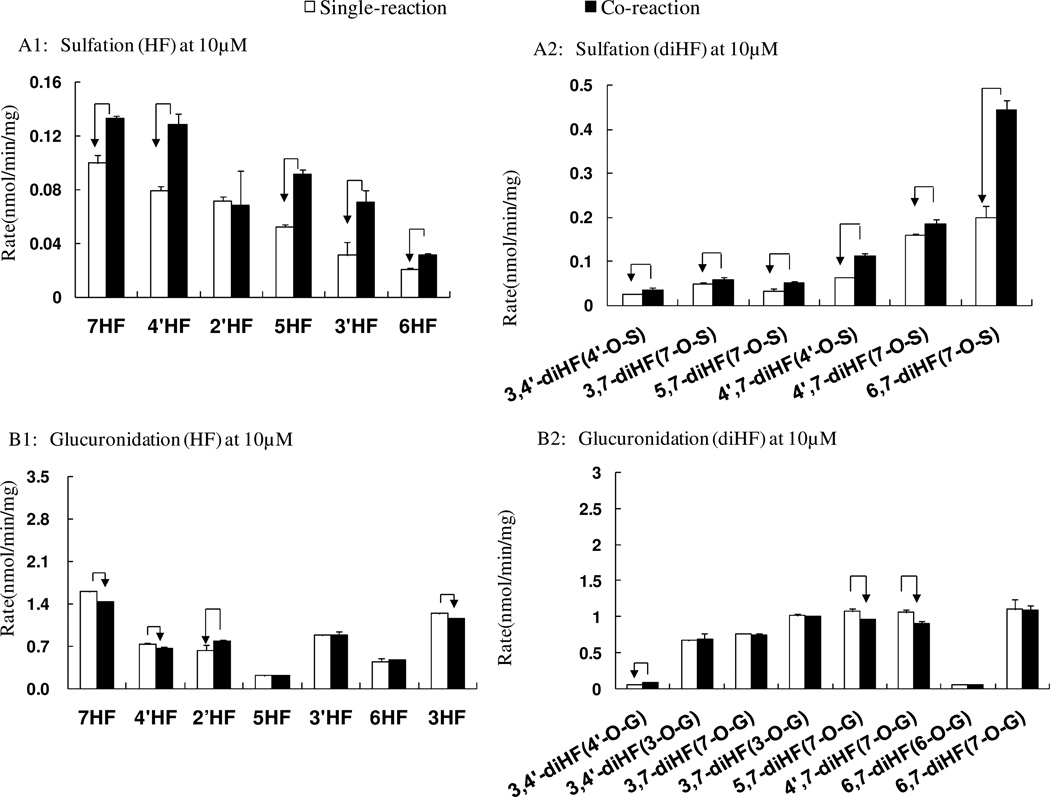
Interactions of conjugation of HFs in the SULT-UGT co-reaction system
In order to determine whether there was an expected competitive interaction between glucuronidation and sulfation for these flavones, we made a direct comparison plot for sulfation or glucuronidation in Sult/Ugt single and Sult-Ugt co-reaction systems at 10 µM (Figure 7). Unexpectedly, the sulfation of HFs and diHFs were enhanced significantly in the Ugt-Sult co-reaction system (p<0.05, Student’s t-test). We found that the higher the substrate concentration, the higher the percentages increase in sulfation rates (with the exemption of 2’-HF) in a co-reaction system. In contrast to sulfation reaction, the formation rates of glucuronides from HFs and diHFs were usually unchanged or only mildly impacted in the co-reaction system. The phenomenon was almost the same at the other two concentrations (i.e., 5 and 40 µM).
Figure 7. Interaction between sulfation and glucuronidation for hydroxyflavones in a Sult and Ugt co-reaction system.
Concentration of 10 µM was shown. Sulfation in the Sult single reaction system and the Sult-Ugt co-reaction system were compared (A1, A2). Glucuronidation in the single Ugt reaction system and the Sult-Ugt co-reaction system were also compared (B1–B2). The data were analyzed by Student’s t-test. The arrow indicates a statistically significant difference (p < 0.05).
Disappearance half-life of 6,7-diHF in single and co-reaction system of glucuronidation and sulfation
In order to determine if increase of the sulfation in the co-reaction system would enhance the elimination, we compared the elimination half life (t1/2) values of 6,7-diHF in the single system vs. the co-reaction system. The reason why we chose 6,7-diHF was because the formation rate of its 7-O-sulfate was enhanced significantly in the Sult-Ugt co-reaction system. The t1/2 values of 6,7-diHF were 244.31 ± 36.81, 26.78 ±1.32 and 21.18 ± 2.03 min in Sult reaction, Ugt reaction, and Sult-Ugt co-reaction system, respectively. The results indicated that the increase of sulfation in co-reaction system only lead to a moderate albeit significant change on rate of the elimination of the flavones by the mouse liver S9 fraction.
Concentration-dependent conjugation in the single and co-reaction systems
The data from above plots were reorganized to display concentration-dependence of various conjugating reactions. Both in the Sult single-reaction and co-reaction system, sulfation rates for all the selected mono-hydroxflavones (besides 5-HF) decreased as substrate concentrations increased (substrate inhibition) (Figure 3A, 5A). This was also observed for diHFs in single and co-reaction systems (Figure 4A, 6A), although in the co-reaction system, the rank order of sulfation at 7-hydroxyl position of 3,7- and 4’,7-diHF were also slightly altered as the result of concentration change. Therefore, the data appear to suggest that most of the selected flavones cause substrate inhibition at high concentrations. In contrast, the glucuronidation rate for 11 hydroxyflavones (besides 2’-HF) increased with increasing substrate concentration, regardless if the Sult reaction system was present or not (Figure 3B, 4B, 5B, and 6B).
Discussion
This is the first systematic study determining the positional preference of Sult-catalyzed sulfation of flavonoids. We also compared this preference with that of Ugt-catalyzed glucuronidation. The results reveal that structure activity relationship (or SAR) with respect to sulfation is quite different from that for the glucuronidation when using the mouse liver S9 fraction. The rank order of reaction was shown to be very different between sulfation and glucuronidation, although sulfation and glucuronidation at 7-OH position was favored among the 12 flavonoids studied here. Moreover, the Sult-Ugt co-reaction system unexpectedly enhanced the sulfation of flavonoids, suggesting that the two metabolic pathways are unlikely to compete with each other for substrates as long as the substrates are not exhausted.
The distinctive SAR displayed by Sult suggests that sulfation pathway will not compensate for the glucuronidation pathway or vice versa in limiting the bioavailability of flavonoids. In other words, there is no relationship between a flavonoid’s susceptibility to sulfation vs. glucuronidation, and that independent SARs are needed if the goal is to predict sulfation vs. glucuronidation. This new finding suggest that we will need to consider a flavonoid’s potential for sulfation and glucuronidation separately, if we were to improve its bioavailability.
The rank order of the sulfation between 7 individual HFs and 5 individual diHFs suggests that a more slowly sulfated phenolic group in a dihydroxylflavones was always predicable using results derived from the monohydroxylflavones (positional preference for HF and diHF were similar), which was also shown previously for glucuronidation (20). For example, the sulfation reaction results for all HFs had shown that 7-OH was more active than 4’-OH and therefore, the formation of 7-O-sulfate from 4’, 7-diHF was greater than the 4’-O-sulfate (Figure.3A, 4A). As expected, sulfation at 5-OH position of diHFs was below the detection limit (Fig.3 and Fig.4). On the other hand, the glucuronidation relationship shown previously for a different group of compound holds true for the current group of 12 flavonoids. For example, the hydroxyl group activity of diHFs for glucuronidation had the same order as HFs (7-OH > 3-OH > 4’-OH >= 6-OH). As expected, more glucuronide was produced from 7-HF than from 5-HF, indicating that 7-OH was more reactive. For 5,7-diHF, the unique glucuronide was determined to be 7-O-glucuronide (Figure.3B, 4B), and 5-O-glucuronide was not found. Taken together, 5-OH is not usually glucuronidated as long as another –OH position was available.
The fact that Sult could not metabolize 3HF is consistent with the small size of reported active binding cavity of the sulfotransferases revealed in crystallography studies. For example, it has been reported that human SULT1A3 selectively sulfates only the D-dopa (not L-Dopa) and prefer 3-OH position over 4-OH position of D-Dopa (33). Our own study showed that human SULT1A3 only metabolizes flavonoids with 7-OH group (not shown). The small size of the active pocket means that sterically hindered 3-OH group is not available for sulfation. In contrast, human UGT1A9 have broad substrate specificity against flavonoids, and almost all flavonoids chosen are its substrates, and no hydroxyl position is off the limit for UGTs, although some positions (e.g., 7-OH and 3-OH) are clearly preferred over other positions (e.g., 5-OH) (34). An analysis of the literature suggested that the 3-OH group was the most reactive for glucuronidation, even in the more complex flavones with 2 phenolic groups. In addition, the 7-OH group was also highly reactive (7, 20, 35). The later is consistent with published data showing that 7-HF was sulfated while 3-HF was not by SULTs in human liver and duodenum S9 fractions (36). This analysis provided the structural basis why Sults and Ugts will have different SARs.
We found strong evidence that mouse Sults and Ugts would not compete for the same substrate as would have been expected, suggesting that the binding of the substrate to the enzyme is not the rate-limiting step in the conjugation process. We were surprised that sulfation rates were enhanced in the Sult-Ugt co-reaction system since we thought the more rapid metabolism via the glucuronidation pathway would have taken the substrate away from sulfation, and thereby decreasing its sulfation. The fact that sulfation profiles of most flavonoids displayed substrate inhibition (Fig.7) provided a plausible explanation for this surprised finding, but more studies are needed to identify the actual causes.
This surprising discovery suggests that Ugt and Sult conjugation pathways could display kinetic interplay capable of changing the kinetics of conjugation reaction and profiles. For example, the elimination half life (t1/2) values of 6,7-diHF in the co-reaction was moderately shorter than the single reaction system. It is not entirely clear what the basis of this interplay are and whether this will happen in an intact cells where flavone aglycones are to encounter Sult (cytosol) first, then Ugt (endoplasmic reticulum). If there are enough Sults to bind and metabolize aglycones, Ugt will never encounter the substrate. On the other hand, if there is an excess amount of flavones, they will interfere with Sult functions by substrate inhibition. It appears that Ugts are capable of facilitating the sulfation reaction by reacting with excess flavone aglycone. In any rate, the interplay is consistent with previous observation that excretion rates of genistein, daidzein and glycitein sulfates in intact cells where both metabolic pathways are present were all significantly higher than predicted based on their formation rates in the Caco-2 cell lysates using a single reaction system (15). Additionally, higher percentages of apigenin conjugates are present as sulfates at lower loading concentration than at higher loading concentration (of apigenin) in the Caco-2 cells (14).
The S9 fraction was employed for the present study because the focus was on the metabolite formation processes. The whole cells were not used for this purpose since phase II metabolites of flavonoids (i.e., glucuronides and sulfates) are too hydrophilic to passively diffuse across a cell membrane. In other words, excretion rates from whole cell or organ model systems would not have represented the formation rates of hydrophilic phase II metabolites. Therefore, the imperfect S9 fraction model is the best model available for the determination of the formation rates. This model is not perfect since cellular production of sulfates is likely to be more favored in a whole cell system than the S9 fraction used in the present study, perhaps because in mammalian cells flavonoids encounter Sults (in cytosolic domain) first, and then UDP-glucuronosyltransferase or Ugt (at the endoplasmic reticulum). In this liver S9 fraction model, a substrate will have equal access to Sults and Ugts, and hence glucuronidation could have been overestimated. Therefore, it was highly unexpected that the presence of glucuronidation reaction system in the mixed reaction system actually increased the sulfonation rates for some flavonoids.
In conclusion, Ugts and Sults have different and independent SARs, and there appears to be limited kinetic interplay between the sulfation and glucuronidation pathways in the mouse liver S9 fraction. Both sulfation and glucuronidation pathways showed stronger preference for the 7–OH group. Besides 7-OH group, glucuronidation also showed similar preference for 3-OH, whereas sulfation also showed preference for the 4’-OH position. Unexpected enhancement of the sulfation reaction in the co-reaction system is likely the result of lowering of the substrate concentrations, since most sulfation displayed substrate inhibition kinetic profiles. The latter deserves more serious investigation to confirm our preliminary finding here since the S9 fraction used in the present study does not have cellular structures, which separate Sults and Ugts into different compartments inside cells.
Figure 2. UPLC chromatograms of HFs and di-HFs, and their phase II metabolites in Sult- and Ugt-associated reactions.
The UPLC chromatograms were used to quantify the flavones and their respective metabolites, which were obtained after incubation of flavones with the FVB mouse liver S9 fraction.
Acknowledgment
This work was mainly supported by the National Basic Research Program of China (973 Program, 2009CB5228008), grant of the Key Project of National Natural Science Foundation of China (U0832002). MH was also supported by NIH grant GM070737.
Abbreviations used
- 3-HF
3-hydroxyflavone
- 5-HF
5-hydroxyflavone
- 6-HF
6-hydroxyflavone
- 7-HF
7-hydroxyflavone
- 2’-HF
2’-hydroxyflavone
- 3’-HF
3’-hydroxyflavone
- 4’-HF
4’-hydroxyflavone
- 3,7-diHF
3,7-dihydroxyflavone
- 3,4’-diHF
3,4’-dihydroxyflavone
- 5,7-diHF
5,7-dihydroxyflavone
- 6,7-diHF
6,7-dihydroxyflavone
- 4’,7-diHF
4’,7-dihydroxyflavone
- KPI
potassiµM phosphate buffer (pH7.4)
- UDPGA
uridine diphosphoglucuronic acid
- PAPS
3'-phosphoadenosine 5'-phosphosulfate
- UPLC
ultra performance liquid chromatography
- Ugt
UDP-Glucuronosyltransferase
- Sult
Sulfotransferase
Literature cited
- 1.Thomasset SC, Berry DP, Garcea G, Marczylo T, Steward WP, Gescher AJ. Dietary polyphenolic phytochemicals--promising cancer chemopreventive agents in humans? A review of their clinical properties. Int J Cancer. 2007;120(3):451–458. doi: 10.1002/ijc.22419. [DOI] [PubMed] [Google Scholar]
- 2.Benavente-Garcia O, Castillo J. Update on uses and properties of citrus flavonoids: new findings in anticancer, cardiovascular, and anti-inflammatory activity. J Agric Food Chem. 2008;56(15):6185–6205. doi: 10.1021/jf8006568. [DOI] [PubMed] [Google Scholar]
- 3.Birt DF, Hendrich S, Wang W. Dietary agents in cancer prevention: flavonoids and isoflavonoids. Pharmacol Ther. 2001;90(2–3):157–177. doi: 10.1016/s0163-7258(01)00137-1. [DOI] [PubMed] [Google Scholar]
- 4.Moon JH, Nakata R, Oshima S, Inakuma T, Terao J. Accumulation of quercetin conjugates in blood plasma after the short-term ingestion of onion by women. Am J Physiol Regul Integr Comp Physiol. 2000;279(2):R461–R467. doi: 10.1152/ajpregu.2000.279.2.R461. [DOI] [PubMed] [Google Scholar]
- 5.de Vries JH, Hollman PC, Meyboom S, Buysman MN, Zock PL, van Staveren WA, Katan MB. Plasma concentrations and urinary excretion of the antioxidant flavonols quercetin and kaempferol as biomarkers for dietary intake. Am J Clin Nutr. 1998;68(1):60–65. doi: 10.1093/ajcn/68.1.60. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Lin G, Zuo Z. Involvement of UDP-glucuronosyltransferases in the extensive liver and intestinal first-pass metabolism of flavonoid baicalein. Pharm Res. 2007;24(1):81–89. doi: 10.1007/s11095-006-9126-y. [DOI] [PubMed] [Google Scholar]
- 7.Day AJ, Mellon F, Barron D, Sarrazin G, Morgan MR, Williamson G. Human metabolism of dietary flavonoids: identification of plasma metabolites of quercetin. Free Radic Res. 2001;35(6):941–952. doi: 10.1080/10715760100301441. [DOI] [PubMed] [Google Scholar]
- 8.Shia CS, Tsai SY, Kuo SC, Hou YC, Chao PD. Metabolism and pharmacokinetics of 3,3',4',7-tetrahydroxyflavone (fisetin), 5-hydroxyflavone, and 7-hydroxyflavone and antihemolysis effects of fisetin and its serum metabolites. J Agric Food Chem. 2009;57(1):83–89. doi: 10.1021/jf802378q. [DOI] [PubMed] [Google Scholar]
- 9.Crespy V, Morand C, Besson C, Manach C, Demigne C, Remesy C. Comparison of the intestinal absorption of quercetin, phloretin and their glucosides in rats. J Nutr. 2001;131(8):2109–2114. doi: 10.1093/jn/131.8.2109. [DOI] [PubMed] [Google Scholar]
- 10.Azuma K, Ippoushi K, Ito H, Horie H, Terao J. Enhancing effect of lipids and emulsifiers on the accumulation of quercetin metabolites in blood plasma after the short-term ingestion of onion by rats. Biosci Biotechnol Biochem. 2003;67(12):2548–2555. doi: 10.1271/bbb.67.2548. [DOI] [PubMed] [Google Scholar]
- 11.De Santi C, Pietrabissa A, Spisni R, Mosca F, Pacifici GM. Sulphation of resveratrol, a natural product present in grapes and wine, in the human liver and duodenum. Xenobiotica. 2000;30(6):609–617. doi: 10.1080/004982500406435. [DOI] [PubMed] [Google Scholar]
- 12.Walle UK, Galijatovic A, Walle T. Transport of the flavonoid chrysin and its conjugated metabolites by the human intestinal cell line Caco-2. Biochem Pharmacol. 1999;58(3):431–438. doi: 10.1016/s0006-2952(99)00133-1. [DOI] [PubMed] [Google Scholar]
- 13.Galijatovic A, Walle UK, Walle T. Induction of UDP-glucuronosyltransferase by the flavonoids chrysin and quercetin in Caco-2 cells. Pharm Res. 2000;17(1):21–26. doi: 10.1023/a:1007506222436. [DOI] [PubMed] [Google Scholar]
- 14.Hu M, Chen J, Lin H. Metabolism of flavonoids via enteric recycling: mechanistic studies of disposition of apigenin in the Caco-2 cell culture model. J Pharmacol Exp Ther. 2003;307(1):314–321. doi: 10.1124/jpet.103.053496. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Lin H, Hu M. Absorption and metabolism of genistein and its five isoflavone analogs in the human intestinal Caco-2 model. Cancer Chemother Pharmacol. 2005;55(2):159–169. doi: 10.1007/s00280-004-0842-x. [DOI] [PubMed] [Google Scholar]
- 16.Joseph TB, Wang SW, Liu X, Kulkarni KH, Wang J, Xu H, Hu M. Disposition of flavonoids via enteric recycling: enzyme stability affects characterization of prunetin glucuronidation across species, organs, and UGT isoforms. Mol Pharm. 2007;4(6):883–894. doi: 10.1021/mp700135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang SW, Kulkarni KH, Tang L, Wang JR, Yin T, Daidoji T, Yokota H, Hu M. Disposition of flavonoids via enteric recycling: UDP-glucuronosyltransferase (UGT) 1As deficiency in Gunn rats is compensated by increases in UGT2Bs activities. J Pharmacol Exp Ther. 2009;329(3):1023–1031. doi: 10.1124/jpet.108.147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Tam VH, Hu M. Disposition of flavonoids via enteric recycling: determination of the UDP-glucuronosyltransferase isoforms responsible for the metabolism of flavonoids in intact Caco-2 TC7 cells using siRNA. Mol Pharm. 2007;4(6):873–882. doi: 10.1021/mp0601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong EJ, Jia X, Hu M. Disposition of formononetin via enteric recycling: metabolism and excretion in mouse intestinal perfusion and Caco-2 cell models. Mol Pharm. 2005;2(4):319–328. doi: 10.1021/mp0498852. [DOI] [PubMed] [Google Scholar]
- 20.Tang L, Ye L, Singh R, Wu B, Lv C, Zhao J, Liu Z, Hu M. Use of glucuronidation fingerprinting to describe and predict mono- and dihydroxyflavone metabolism by recombinant UGT isoforms and human intestinal and liver microsomes. Mol Pharm. 7(3):664–679. doi: 10.1021/mp900223c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang L, Singh R, Liu Z, Hu M. Structure and concentration changes affect characterization of UGT isoform-specific metabolism of isoflavones. Mol Pharm. 2009;6(5):1466–1482. doi: 10.1021/mp8002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okita JR, Castle PJ, Okita RT. Characterization of cytochromes P450 in liver and kidney of rats treated with di-(2-ethylhexyl)phthalate. J Biochem Toxicol. 1993;8(3):135–144. doi: 10.1002/jbt.2570080305. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Halls SC, Alfaro JF, Zhou Z, Hu M. Potential beneficial metabolic interactions between tamoxifen and isoflavones via cytochrome P450-mediated pathways in female rat liver microsomes. Pharm Res. 2004;21(11):2095–2104. doi: 10.1023/b:pham.0000048202.92930.61. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Zheng Z, Xia B, Tang L, Lv C, Liu W, Liu Z, Hu M. Use of isoform-specific UGT metabolism to determine and describe rates and profiles of glucuronidation of wogonin and oroxylin A by human liver and intestinal microsomes. Pharm Res. 2010;27(8):1568–1583. doi: 10.1007/s11095-010-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang L, Ye L, Singh R, Wu B, Lv C, Zhao J, Liu Z, Hu M. Use of glucuronidation fingerprinting to describe and predict mono- and dihydroxyflavone metabolism by recombinant UGT isoforms and human intestinal and liver microsomes. Mol Pharm. 2010;7(3):664–679. doi: 10.1021/mp900223c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong EJ, Lin H, Hu M. Disposition mechanisms of raloxifene in the human intestinal Caco-2 model. J Pharmacol Exp Ther. 2004;310(1):376–385. doi: 10.1124/jpet.103.063925. [DOI] [PubMed] [Google Scholar]
- 27.Busby MG, Jeffcoat AR, Bloedon LT, Koch MA, Black T, Dix KJ, Heizer WD, Thomas BF, Hill JM, Crowell JA, Zeisel SH. Clinical characteristics and pharmacokinetics of purified soy isoflavones: single-dose administration to healthy men. Am J Clin Nutr. 2002;75(1):126–136. doi: 10.1093/ajcn/75.1.126. [DOI] [PubMed] [Google Scholar]
- 28.Wang SW, Chen J, Jia X, Tam VH, Hu M. Disposition of flavonoids via enteric recycling: structural effects and lack of correlations between in vitro and in situ metabolic properties. Drug Metab Dispos. 2006;34(11):1837–1848. doi: 10.1124/dmd.106.009910. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Hu M. Absorption and metabolism of flavonoids in the caco-2 cell culture model and a perused rat intestinal model. Drug Metab Dispos. 2002;30(4):370–377. doi: 10.1124/dmd.30.4.370. [DOI] [PubMed] [Google Scholar]
- 30.Tang L, Singh R, Liu Z, Hu M. Structure and Concentration Changes Affect Characterization of UGT Isoform-Specific Metabolism of Isoflavones. Mol Pharm. 2009 doi: 10.1021/mp8002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson DT, Hensley M, Yoshioka H, Mabry TJ. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970;9(7):1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- 32.Singh R, Wu B, Tang L, Liu Z, Hu M. Identification of the position of mono-O-glucuronide of flavones and flavonols by analyzing shift in online UV spectrum (lambdamax) generated from an online diode array detector. J Agric Food Chem. 58(17):9384–9395. doi: 10.1021/jf904561e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itaaho K, Alakurtti S, Yli-Kauhaluoma J, Taskinen J, Coughtrie MW, Kostiainen R. Regioselective sulfonation of dopamine by SULT1A3 in vitro provides a molecular explanation for the preponderance of dopamine-3-O-sulfate in human blood circulation. Biochem Pharmacol. 2007;74(3):504–510. doi: 10.1016/j.bcp.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Wu B, Morrow JK, Singh R, Zhang S, Hu M. Three-dimensional quantitative structure-activity relationship studies on UGT1A9-mediated 3-O-glucuronidation of natural flavonols using a pharmacophore-based comparative molecular field analysis model. J Pharmacol Exp Ther. 2011;336(2):403–413. doi: 10.1124/jpet.110.175356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Pascual-Teresa S, Johnston KL, DuPont MS, O'Leary KA, Needs PW, Morgan LM, Clifford MN, Bao Y, Williamson G. Quercetin metabolites downregulate cyclooxygenase-2 transcription in human lymphocytes ex vivo but not in vivo. J Nutr. 2004;134(3):552–557. doi: 10.1093/jn/134.3.552. [DOI] [PubMed] [Google Scholar]
- 36.Vietri M, Pietrabissa A, Spisni R, Mosca F, Pacifici GM. 7-OH-flavone is sulfated in the human liver and duodenum, whereas 5-OH-flavone and 3-OH-flavone are potent inhibitors of SULT1A1 activity and 7-OH-flavone sulfation rate. Xenobiotica. 2002;32(7):563–571. doi: 10.1080/00498250210130582. [DOI] [PubMed] [Google Scholar]