Abstract
The NarI recognition sequence (5′-G1G2CG3CN-3′) is the most vulnerable hot spot for frameshift mutagenesis induced by the carcinogen 2-aminofluorene and its analogs in Escherichia coli. Lesioning of the guanine in the G3 position induces an especially high frequency of -2 deletion mutations; vulnerability to these mutations is modulated by the nature of nucleotide in the N position (C ~ A > G ≫ T). The objective of the present study was to probe the structural basis of this N-mediated influence on the propensity of the G3-lesion to form a slipped mutagenic intermediate (SMI) during translesion synthesis. We studied NarI-based fully paired [(5′-CTCG1G2CG3*CNATC-3′)(5′-GATNCGGCCGAG-3′), N = dC or dT] and -2 deletion [(5′-CTCG1G2CG3*CNATC-3′)(5′-GATNGCCGAG-3′), N = dC or dT] duplexes, in which G* was either AF [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene] or the 19F probe FAF [N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene]. The latter sequences mimic the bulged SMI for -2 deletion mutations. Dynamic 19F NMR, circular dichroism, and UV-melting results indicated that the NarI-dC/-2 deletion duplex adopts exclusively an intercalated conformer, whereas the NarI-dT/-2 deletion duplex exists as multiple conformers. The data support the presence of a putative equilibrium between a carcinogen-intercalated and a carcinogen-exposed SMI for the dT/-2 duplex. A similar dT-induced conformational heterogeneity was observed for the fully-paired duplexes in which all three guanines were individually modified by AF or FAF. The frequency of the carcinogen stacked S-conformation was found to be highest (69~75 %) at the G3 hot spot in NarI-dC duplexes. Taken together, our results support the hypothesis that the conformational stability of the SMI is a critical determinant for the efficacy of -2 frameshift mutagenesis in the NarI sequence. We also provide evidence for AF/FAF conformational compatibility in the NarI sequences.
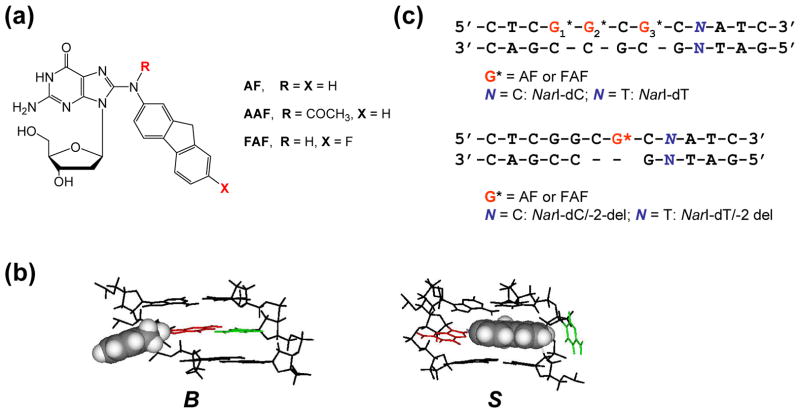
Arylamines are an important group of mutagen/carcinogens that cause human cancers (1). 2-Aminofluorene and its analogs are perhaps the most studied mutagens; they produce two major C8-substituted dG adducts in vivo: AF [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene and AAF [N-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene](Fig. 1a)(2,3). AF and AAF induce distinct mutation and repair activities. Accurate nucleotide incorporation is feasible, albeit at a low frequency, opposite N-deacetylated AFs, but not opposite bulky acetylated AAFs which block nucleotide incorporation (4,5). The AF-adduct has drawn much attention because of its unique ability to adopt different conformations depending on the location of the carcinogen moiety: the major-groove binding external ‘B-type’ (B), the base-displaced ‘stacked’ (S), and the minor-groove binding ‘wedged’ (W) conformations (Fig. 1b)(6–8). When in fully paired DNA duplexes, AF equilibrates between B- and S-conformations (9–14). The W-conformer has been observed in duplexes in which the AF lesion is mismatched with a purine base (6,11). Solution NMR structures of AF-adducts have been studied extensively in various DNA sequence contexts (6–14). AF-induced S/B/W-heterogeneity is modulated not only by base sequence context, but also by primer length (15). As a result, the AF-adduct influences polymerase function through a long-range distortion effect (16). A similar conformational feature has been observed for duplexes containing adducts derived from the heterocyclic amine mutagens, such as PhIP (2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine)(17) and IQ (2-amino-3-methylimidazo[4,5-f]quinoline)(18–20).
Figure 1.
(a) Chemical structures of AF-adduct [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene], AAF-adduct [N-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene], and FAF-adduct [N-(2′-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene]. (b) The major groove views of the central trimer segments of the B- and S-conformers of AF-modified duplex. The modified dG and the complementary dC are shown in red and green, respectively, and the AF-moiety is highlighted with grey CPK. In the B conformer, anti-[AF]dG maintains Watson-Crick hydrogen bonds, thereby placing the AF ring in the major groove. The AF moiety of the S-conformer stacks into the helix with the modified dG in the syn conformation. (c) Sequence contexts of the fully-paired and -2 deletion NarI duplexes.
Local sequence context plays an important role in adduct-induced frameshift mutagenesis (21–26). The most well known sequence-sensitive mutation locus is the NarI recognition sequence (5′-G1G2CG3CC-3′), which represents the most active hot spot for AAF and AF-induced frameshift mutagenesis in Escherichia coli (27). Replication of SOS-induced E. coli pBR322 resulted in -2 deletion mutations when the bulky AAF was incorporated at the G3, but not the G1 and G2 positions, even though the three guanines have similar chemical reactivities (27). AF produces -2 deletion mutations at a frequency of about 10-fold less than AAF (22). Interestingly, both AF and AAF produce point mutations (primarily G → T transversions) when replicated in COS-7 mammalian cells (28,29). Fuchs and coworkers (30,31) have studied AAF-modified NarI sequences (5′-NaGCG*CNb-3′) and found that over 90% of the mutations were targeted -2 deletion mutations, in which a GC dinucleotide was deleted between the Na and Nb bases. They also discovered that varying Nb strongly modulated mutagenesis (C ~ A > G ≫ T), whereas varying Na located on the 5′ end of the sequence had little effect. For example, the mutational efficiency of -2 deletion for the NarI sequence 5′-G1G2CG3*CC-3′ (Na=G; Nb=C) was about 6-fold greater than for the 5′-G1G2CG3*CT-3′ sequence (Na=G; Nb=T).
Propensity for deletion mutation generation depends on the characteristics of the base inserted opposite the modified G*, the 5′-sequence context surrounding the adduct, the rate of translesion DNA synthesis (21–24), and the nature of polymerases (25). The ability of B-conformeric AF at G3 to pair correctly with dC during translesion synthesis facilitates formation of a slippage mutagenic intermediate (SMI) for eventual -1 or -2 deletion mutation. Gill and Romano (32) showed that incorporation of AAF at G3 in the NarI sequence resulted in a -2 deletion in both E. coli pol II exo- and E. coli Klenow fragment exo- (KFexo-). These results implied that there may be a common AAF-induced SMI structure in this sequence. The study also showed that incorporation of an AAF adduct in the NarI sequence produces a very different SMI in the polymerase activation site compared with that produced by a similar modification in a non-NarI sequence. Fuchs and coworkers have shown (25) that DNA pol II is the enzyme responsible for AAF-induced frameshift pathway, while bypass is error free by pol V in E. coli. These results emphasize the complex role of a polymerase which must be taken into consideration. Replication of a NarI-based -2 deletion generated by replication of the food mutagen C8-IQ-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) as well as in E. coli pol II and KFexo- was found to be strongly influenced by the regioisomeric nature of the adduct in addition to the local sequence (33).
The objective of the present work was to probe the structural/conformational basis of the ‘hotness’ of the arylamine lesion at G3 for -2 frameshift mutagenesis in the NarIsequence (5′-CTCG1G2CG3*CNATC-3′, N = C or T). The underlined portion of this 12-mer sequence is identical to that used in the afore-mentioned mutation study by Fuchs and coworkers (30,31). To this end, we prepared several model duplexes that were site-specifically modified by AF or FAF (Figure 1) and then examined their conformational profiles by dynamic 19F NMR, UV-melting, and circular dichroism (CD) spectroscopy. We hypothesized that the ease with which the SMI can form and the SMI’s conformational stability are important factors in determining the propensity of a sequence for frameshift mutagenesis. Specifically, we investigated the contribution of the identity of nucleotide N on the conformational stability of the lesion at G3 and its propensity to slip during replication. We further examined the nature of the conformational heterogeneity at G3 in the -2 deletion duplex and compared it to the well-established S/B-equilibrium observed in the fully paired duplex.
EXPERIMENTAL PROCEDURES
Caution:2-Aminofluorene derivatives are mutagens and suspected human carcinogens and therefore must be handled with caution.
Crude oligodeoxynucleotides in 10 ~15 μmol scales in desalted form were obtained from Sigma-Genosys (The Woodlands, TX). All HPLC solvents were purchased from Fisher Inc. (Pittsburgh, PA).
The model sequence systems
The NarI sequences used in the present study and their respective designations are listed in Figure 1. The three guanines in the 12-mer sequence (5′-CTCG1G2CG3CNATC-3′, N = C or T) were site-specifically modified by AF or FAF. The modified sequences were annealed with complementary 12-mer sequences to form fully paired 12/12-mer NarI-dC and NarI-dT duplexes, designated by the identity of the nucleotide N (Figure 1c). G3-modified sequences were also annealed with complementary 10-mer strands (5′-GATNGCCGAC-3′, N = G or A) to produce 12/10-mer -2 deletion duplexes (NarI-dC/-2 del and NarI-dT/-2 del)(Figure 1c). These 12/10-mer bulge duplexes mimic SMIs for -2 deletion mutations. The high-resolution 1H NMR solution structures of AF-modified NarI-dC (34) and NarI-dC/-2 (35) duplexes were reported previously.
Preparation and characterization of modified oligodeoxynucleotides
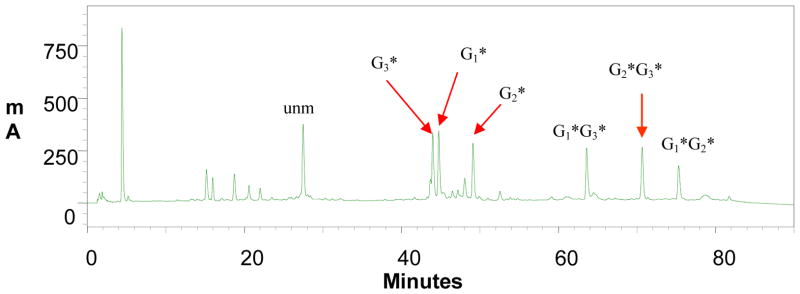
AF- or FAF-modified oligodeoxynucleotides were prepared using the general procedures described previously (9,12,14). Briefly, an unmodified oligodeoxynucleotide was treated with either activated carcinogens in a pH 6.0 sodium citrate buffer and placed in a shaker for 18 – 24 hrs at 37 °C: N-acetoxy-N-trifluoroacetyl-2-aminofluorene (12) and N-acetoxy-N-trifluoroacetyl-7-fluoro-2-aminofluorene (9,14), respectively for AF and FAF adducts. Figure 2 shows a typical reverse phase HPLC of a mixture derived from treatment of an unmodified NarI-dC 12-mer (5′-CTCGGCGCCATC-3′) with the activated 7-fluoro-2-aminofluorene. The oligomer FAF-adducts appeared in the 40–80 min range were separated and purified up to > 97 % purity by repeated injections. The HPLC system consisted of a Hitachi EZChrom Elite HPLC unit with a L2450 diode array detector and a Waters XTerra MS C18 column (10 × 50 mm, 2.5 μm). A 60-min gradient system involving 3 to 15% acetonitrile in pH 7.0 ammonium acetate buffer (0.10 M) with a flow rate of 2.0 mL/min was used. The purified sequences were characterized by analyses of their UV and electrospray ionization time-of-flight mass spectrometry (ESI-TOF-MS) characteristics. The mass analyses were carried out using a Waters ESI-mass spectrometer in the negative ion mode on the basis of the exonuclease (3′-5′ or 5′-3′) strategies described previously (12).
Figure 2.
HPLC chromatogram of a mixture from reaction between NarI-dC 12-mer sequence and an activated FAF (N-trifluoroacetyl-N-acetoxy-7-fluoro-2-aminofluorene). The mono- (G3*, G1*, G1*) and di- (G1*G3*, G2*, G3*, G1*G2*) FAF adducts eluted in the 40–50 and 60–80 min ranges were purified by reverse HPLC and characterized by LC-TOF-MS. The HPLC system consisted of Waters Xterra 5 m colum with flow rate of 2 mL/min. A gradient condition was 3–15% acetonitrile in triethylammonium acetate buffer (pH 7.0, 100 mM) for 60 min, followed by 15%–20% for 20 minutes and back to 3% acetonitrile in 10 min, flow rate 2 ml/min.
UV-Melting Experiments
UV-melting data were obtained using a Beckman DU 800 UV/VIS spectrophotometer equipped with a 6-chamber, 1-cm path length Tm cell. Sample cell temperatures were controlled by a Peltier temperature controller. Duplexes with a total concentration in the range of 0.2 – 14 μM were prepared in solutions containing 0.2 M NaCl, 10 mM sodium phosphate, and 0.2 mM EDTA at pH 7.0. Thermomelting curves were constructed by varying the temperature of the sample cell (1°C/min) and monitoring the absorbance of the sample at 260 nm. A typical melting experiment consisted of forward/reverse scans and was repeated three times. Thermodynamic parameters were calculated using the program MELTWIN® version 3.5 as described previously (12).
Circular Dichroism (CD) Spectra
CD measurements were conducted on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller. Typically, 2 ODS of each strand were annealed with an equimolar amount of a complementary sequence. The samples were dissolved in 400 μL of a neutral buffer (0.2 M NaCl, 10 mM sodium phosphate, 0.2 mM EDTA) and placed in a 1-mm path-length cell. The samples were heated at 85°C for 5 min and then cooled to 15°C over a 10 min period to ensure complete duplex formation. Spectra were acquired every 0.2 nm with a 2-s response time from 200 to 400 nm at a rate of 50 nm/min and were the averages of 10 accumulations and smoothed using 17 point adaptive smoothing algorithms provided by Jasco. For salt experiments, CD spectra were recorded at 15 °C with increasing amount of NaCl: 0.2, 0.5, 1.0, 2.0, 3.0 and 5.0 M.
NMR Experiments
Approximately 50 ~ 80 ODS of a pure modified oligonucleotide was annealed with an equimolar amount of a complementary sequence to produce the corresponding duplex. The samples were centrifugated using a Pall Microsep MF centrifugal device (Yellow, MW cutoff = 1,000). The samples were then dissolved in 300 μL of a pH 7.0 buffer (10% D2O/90% H2O, 100 mM NaCl, 10 mM sodium phosphate, and 100 μM tetrasodium EDTA) and filtered into in a Shigemi tube through a 0.2 μm membrane filter for NMR measurements.
All 1H and 19F NMR results were recorded using a dedicated 5-mm 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 400.0 and 376.5 MHz, respectively. Imino proton spectra at 5°C were obtained using a phase sensitive jump-return sequence and referenced relative to DSS. 19F NMR spectra were acquired in the 1H-decoupled mode and referenced to CFCl3 by assigning external C6F6 in C6D6 at -164.90 ppm. One-dimensional 19F NMR spectra were measured between 5°C and 60°C with increment of 5 ~ 10 °C. Temperatures were maintained by a BRUKER-VT unit with the aid of controlled boiling liquid N2 into the probe. Each spectrum was obtained by collecting 65,536 points using a 37,664-Hz sweep width and a recycle delay of 1.0 s. A total of 1,600 scans were acquired for each dynamic NMR spectrum. All free induction decays were processed by zero-filling and exponential multiplication using a 20 Hz line broadening factor before Fourier transformation. NOESY/exchange 19F NMR spectra were obtained in the phase-sensitive mode using the following parameters: sweep width 4529 Hz, number of complex data points in t2 1024, number of complex FIDs in t1 256, number of scans 96, dummy scans 16, recycle delays 1.0 s, and mixing time 400 ms. The data were apodized with SSB function using 2 Hz line broadening in both dimensions and Fourier transformed with the 1024 × 256 data matrix.
Complete Line Shape Analysis
Complete line-shape analysis was carried out using the simulation program WINDNMR-Pro (version 7.1.6, J. Chem. Educ. Software Series; Reich, H. J., University of Wisconsin, Madison, WI) to determine rate constants (k), free energy difference ΔG° (−RTlnKeq), and the S/B interconversion energy barrier ΔG≠ (36). The values of frequencies and the S/B-population ratios were determined at the slow exchange limit (5 °C) by area integration of the base-line corrected spectra using TOPSPIN software (Bruker, Billerica, MA). Subsequently, spectra were recorded at various temperatures between 5 and 60 °C. Additional spectra were obtained to clarify coalescence of two slowly exchanging signals. The samples were then cooled back to the slow exchange limit to insure that no irreversible processes have occurred at the higher temperatures.
RESULTS
Synthesis and Characterization
Figure 2 shows the HPLC profile of a reaction mixture derived from treatment of an unmodified NarI sequence with an activated 7-fluoro-2-aminofluorene. The photodiode array UV of the peaks eluting after 41 min exhibited shoulders in the 300–350 nm UV range (data not shown). These UV shoulders represent the aminofluorene chromophore attached at the C8 of dG and their intensities correlate with aminofluorene moiety quantity (28,37). The relative intensities of the three late eluting peaks at 63.6, 70.2, and 74.6 min were double those of the early eluting peaks at 42.8, 44.2 and 48.6 min. Hence, the late eluting peaks were designated as di-FAF adducts and the early eluting ones as mono-FAF adducts. Adduct position assignments were made on the basis of the exonuclease-digest/ESI-MS procedure as described previously (12); the details of the position assignment work will be published elsewhere. The molecular mass ions of all the modified sequences used herein are listed in Supporting Information Table S1.
CD Experiments
Fully paired NarI-dC and NarI-dT duplexes
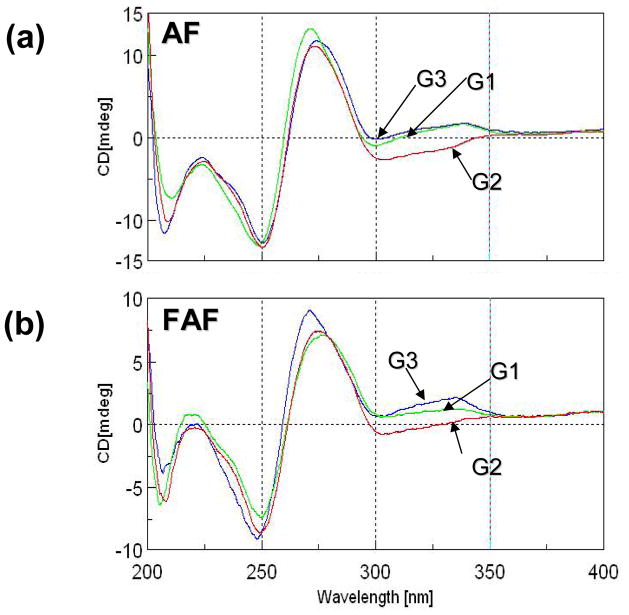
The CD data of the NarI-dC duplexes in which the three guanines in the sequence were individually modified by either AF or FAF were compared (Figure 3). The highly positive CD at around 275 nm represents a typical B-DNA helical conformation. We have shown previously that induced circular dichroism in the 290–360 nm range (ICD290–360nm) is a sensitive conformational marker for AF-induced S/B-heterogeneity (11–15). The fully-paired modified duplexes exhibited sequence-dependent ICD290–360nm. The control duplexes however showed no such ICD (Supporting Information Fig. S1). The ICD290–360nm exhibited by AF-adducts (Figure 3a) and by FAF-adducts (Figure 3b) was strikingly similar. The data confirm that incorporation of a fluorine atom at the remote C7 position of AF did not change the local conformation in the repetitive NarI sequences (7,9,14). In both AF- and FAF-modified sequences, G1- and G3-adducts showed a positive ICD290–360nm, while the G2-adduct exhibited a strong negative ICD290–360nm. These results are in good agreement with previous 1H NMR data on the same sequence contexts, which showed that AF at G1, G2, and G3 existed in 70/30, 90/10, and 50/50 ratios of B/S-conformer populations, respectively (34). A similar general trend in ICD290–350nm was observed for the NarI-dT duplexes (Supporting Information Fig. S2), except that G1-adducts exhibited negative CD.
Figure 3.
CD spectral overlay of fully paired NarI-dC duplexes with (a) AF or (b) FAF modification at G1 (green), G2 (red), and G3 (blue), recorded at 15 °C.
NarI-dC and NarI-dT -2 deletion duplexes
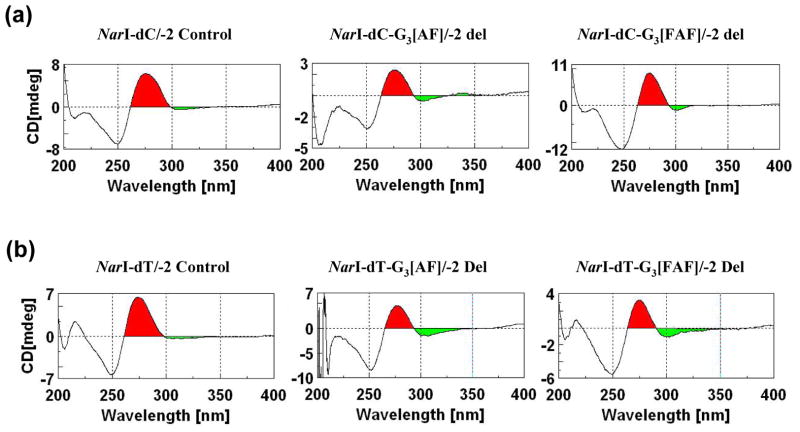
Figure 4 shows the CD spectra of -2 deletion duplexes relative to the unmodified control duplexes. The CD profiles of the control NarI-dC/-2 and NarI-dT/-2 del duplexes were similar; both exhibited a weak negative ICD290–360nm. The fully paired control duplexes exhibited no such ICD290–360nm (Supporting Information Fig. S1). The negative ICD290–360nm associated with the deletion duplexes could arise from bending of the DNA helix as would be expected for bulging structures (6,8,35,38,39). An observed negative intensification of ICD290–360nm upon adduct formation indicated that the bent structures provide binding pockets for the hydrophobic carcinogen and are maintained upon adduct formation (Figure 4). AF-modified -1 and -2 deletion duplexes in different sequence contexts share the common structural feature of base-displacement intercalation with an increase in Tm and significant bending of the DNA helix around the adduct site (6,8,33,38).
Figure 4.
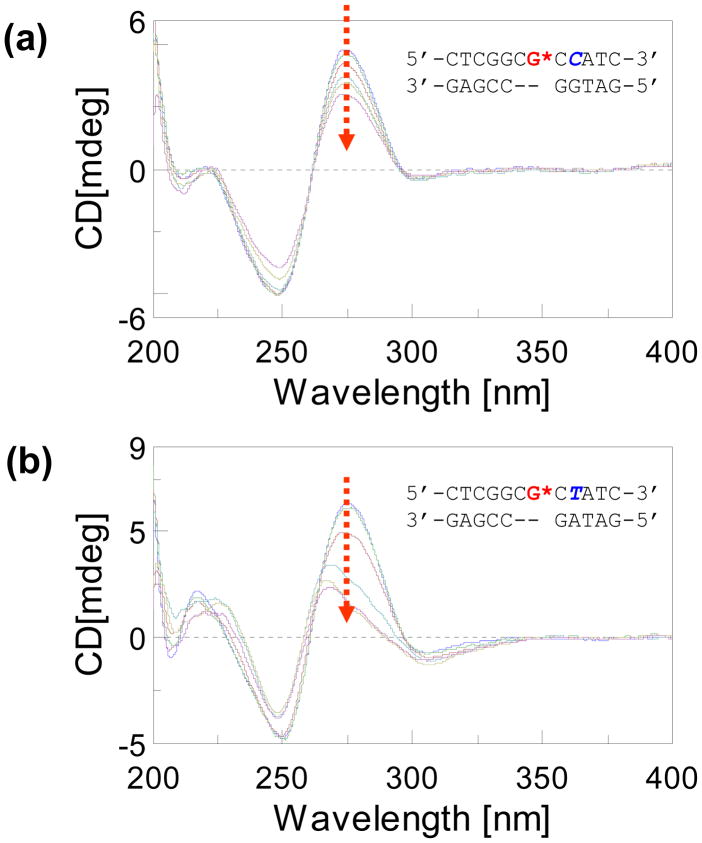
CD spectra of -2 deletion duplexes (control, AF- and FAF-modified at G3*) recorded at 15 °C for the (a) NarI-dC/-2 and (b) NarI-dT/-2 del series (see Figure 1 for structure and sequences). The carcinogen-induced ellipticities in the 290–360 nm range (ICD290–360nm) are colored with green.
Salt effects on NarI/G3-FAF -2 deletion duplexes
Salt experiments were performed to assess conformational flexibility at the lesion site (37). Figure 5 shows overlays of CD spectra of G3-FAF dC/and dT/-2 deletion duplexes at various salt concentrations (0.2 ~ 5.0 M NaCl). The ellipticity at 275 nm of the dC/-2 del duplex decreased incrementally with increasing salt concentration, indicating a gradual structural collapse of the helices. In contrast, the dT/-2 del duplex exhibited dramatic spectral changes under the identical experimental conditions. This modest salt effect with the dC/-2 del duplex is consistent with previous 1H NMR findings with the same AF-modified sequence that showed an exclusive presence of the AF-intercalative conformer structure. The dramatic salt effect with the dT/-2 del duplex provides a robust demonstration of the conformational flexibility at the lesion site.
Figure 5.
CD spectral overlays of G3-FAF-modified (a) NarI-dC/-2 and (b) NarI-dT/-2 deletion duplexes at 15°C. Dotted arrows denote increasing NaCl concentration: 0.2, 0.5, 1.0, 2.0, 3.0 and 5.0 M.
NMR
Fully paired NarI-dC duplexes
S/B-Conformational Heterogeneity
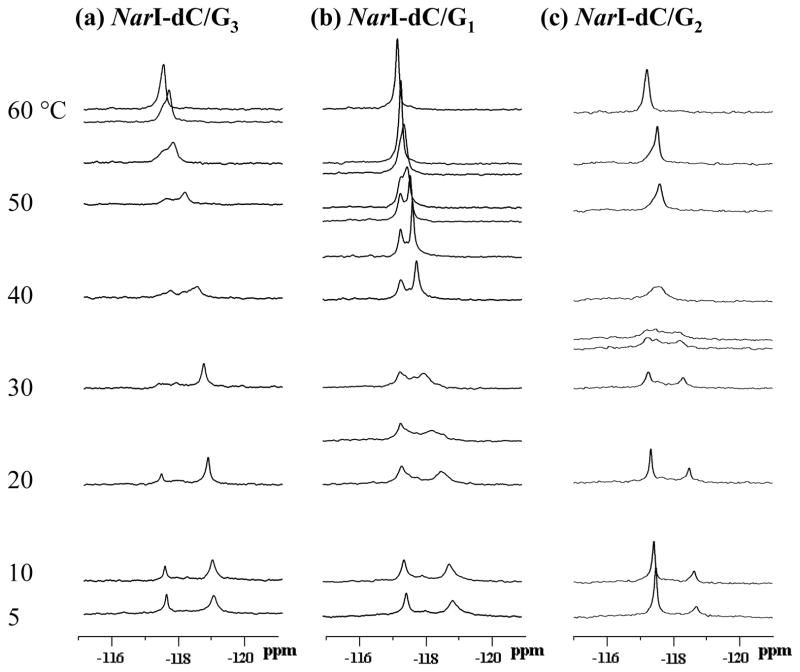
Figure 6 shows dynamic 19F NMR spectra of three fully paired duplexes with FAF-modifications at G1, G2 and G3 (NarI-dC/G1, NarI-dC/G2, and NarI-dC/G3, respectively). These duplexes exhibited two major 19F signals, each representing a unique conformation. The 19F signal assignments were based primarily on the deuterium isotope and NOESY results as described previously (9,13–15). NOESY spectra (Supporting Information Fig. S3) showed that two prominent signals in each of the duplexes slowly interconvert between S- and B-conformers. The downfield B-conformer peaks consistently exhibited a larger D2O effect (0.19 – 0.28 ppm) than the upfield S-conformer peaks (0.07 – 0.18 ppm)(40). The H/D isotope effect details are summarized in Supporting Information Table S2.
Figure 6.
Dynamic 19F NMR spectra of fully-paired NarI-dC duplexes with FAF-modification at (a) G3, (b) G1, and (c) G2. Spectra were obtained at seven standard temperatures (5, 10, 20, 30, 40, 50, and 60 °C). Additional temperatures were used to clarify coalescent exchange process (see text).
The base sequence context of the fully-paired FAF-modified NarI-dC duplexes investigated in the present study was identical to that used in a previous 1H NMR study by Mao et al (34), except that herein the hydrogen at the C7 position of the aminofluorene moiety was replaced with fluorine. Not surprisingly, our imino proton spectra are nearly identical to those of Mao et al (34)(Supporting Information Fig. S4). They reported the relative S-conformer population at 1°C for the G1-, G2-, and G3-AF duplexes to be 30%, 10%, and 50%, respectively. The present 19F NMR data (Figure 6) indicated that the FAF-duplexes followed a similar %S population trend at a similar temperature (2 °C): 56%, 29%, and 69%, respectively. The % S at 20 °C was 75%, 35%, and 44%, respectively (Table 1). A collectively greater S-conformer population was observed for the FAF- than for the AF-modified duplexes (see below).
Table 1.
Dynamic NMR and thermodynamic parameters for the FAF-modified 12-mer NarI-dC duplexes
| NarI-dC/G3 | NarI-dC/G1 | NarI-dC/G2 | |
|---|---|---|---|
| B:S ratioa @ 20 °C | 25%:75% | 56%:44% | 65%:35% |
| B:S ratioa @ 2 °C | 31%:69% | 46%:56% | 71%:29% |
| kc @ 30 °C (s−1)b | 163 | 378 | 378 |
| τ @ 30 °C (ms) c | 6.14 | 3.00 | 3.00 |
| ΔG0 (cal/mol)d | −639.6 | 140.4 | 360.3 |
| ΔG≠ @ 30 °C (kcal/mol)e | 14.7 | 14.2 | 14.2 |
Percent population of B and S conformers based on integration of 19F NMR signals.
Rate constant. (STD ±25%). The data were obtained by complete line shape analysis of temperature dependent 19F NMR results using the WINDNMR-Pro Shareware.
Exchange time (1/k) indicates the amount of time the adduct spends in one conformation before jumping into another conformation.
The energy difference between the two conformers: ΔG° = −RT ln Keq [Keq=S/B].
The S/B interconversion energy barrier @ 30 °C.
The S/B interconversion energy barrier @ 30 °C obtained from Eyring equation (36): ΔG≠C=4.58 TC (10.32 + log TC/kC) cal/mol. SD ± 0.2 kcal/mol.
Dynamic 19F NMR
The three fully paired NarI-dC duplexes (Figure 6) exhibited a typical two-site chemical exchange. Thus, the S/B 19F signals broadened with increasing temperature; the broadening signals gradually moved closer to one another and eventually coalesced into single sharp signals in the 50 ~ 60°C range, which represented the denatured FAF-modified single strands. The dynamic system is comprised of two well-defined equilibriums: the S/B-equilibrium below 10°C and the melting equilibrium between the duplex and separated single strands above 20°C (14). Therefore, the signals in the intermediate temperatures (20 ~ 50°C) are a mixture of the two equilibriums. Additional signals observed in this intermediate range are probably due to transit conformers (B* or S*).
The results of complete line-shape analyses performed to calculate kinetic and thermodynamic parameters are summarized in Table 1. An example of the computer simulation is shown in Supporting Information Figure S5. The S/B interconversion energy barriers (ΔG≠) were found to be in a narrow 14–15 kcal/mol range. The energy differences (ΔG° = −RTlnKeq) of the two conformers at 20 °C were small (140 ~ 639 cal/mol). We were unable to determine exact coalescence temperatures (TC) due to lack of sufficient numbers of recorded dynamic spectra. However, a close inspection of emerging signals indicated that TC decreases in the order of NarI-dC/G3 (-CG*G-) ~ NarI-dC/G2 (-GG*C-) > NarI-dC/G1 (-CG*G-). The Tm values were within the narrow range of 59 ~ 62°C, probably because the FAF-lesions were flanked by G:C or C:G pairs.
The exchange rate constants at TC (kC=20.5 π*Δν, Δν = the separation in frequency in Hz between the two conformer signals at 5°C, when dynamic exchange is minimal) were determined to be in the range of 903 ~ 1170s−1 (Table 1). The correlation times (τ =1/κ), which signify the amount of time the lesion spends in one conformation before changing to another conformation, were in the narrow range of 2.2–5.2 ms at 20 °C (14).
Fully paired NarI-dT duplexes
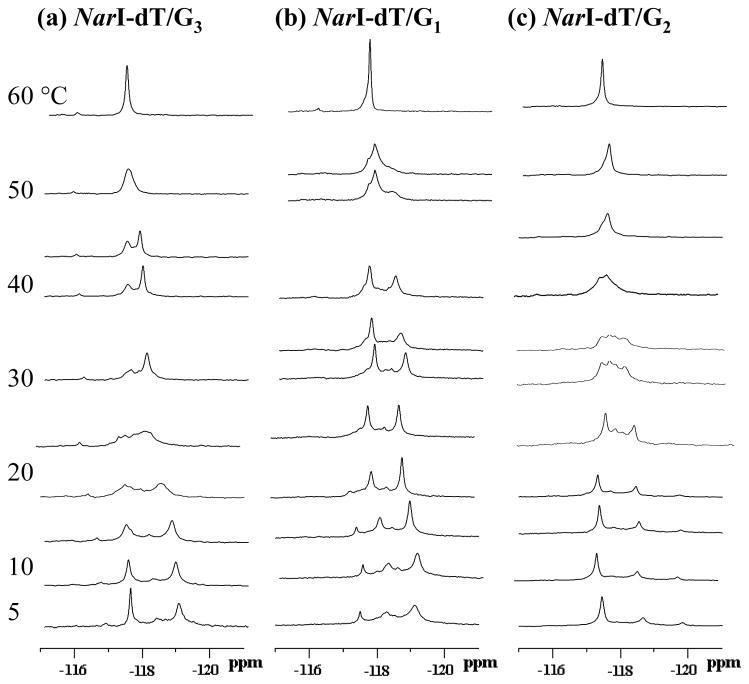
Figure 7 shows dynamic 19F NMR spectra of three fully paired duplexes with FAF-modification at G1, G2 and G3 (NarI-dT/G1, NarI-dT/G2, and NarI-dT/G3, respectively). At low temperatures (5 – 10 °C), both G3- and G2-duplexes exhibited dynamics and signal patterns similar to those of dC duplexes discussed above. The two signals were slowly exchangeable as evidenced by NOESY (Supporting Information Fig. S3). The downfield signals showed greater (0.19 – 0.30 ppm) H/D effects than the upfield ones (0.07 – 0.14 ppm)(Supporting Information Table S2). These results confirm the presence of a typical S/B equilibrium (14,15) It should be noted that the dT/G1 duplex displayed at least five major conformations in the 5 – 10°C range. In the 20 – 35°C range, all three NarI-dT duplexes exhibited significantly greater conformational heterogeneity than their NarI-dC counterparts. This is probably due to the presence of additional intermediate conformers (B*, S*). The heterogeneity persisted at high temperatures (40–60 °C) and was complicated by a melting equilibrium. Conformeric complexities in the intermediate temperature range precluded execution of line shape analyses.
Figure 7.
Dynamic 19F NMR spectra of fully-paired NarI-dT duplexes with FAF-modification at (a) G3, (b) G1, and (c) G2. Spectra were obtained at seven standard temperatures (5, 10, 20, 30, 40, 50, and 60 °C. Additional temperatures were used to clarify coalescent exchange process (see text).
It should be noted that the only difference between the fully paired NarI-dC and NarI-dT duplex series is the base (dC or dT) at the N position (5′-CTCG1G2CG3CNATC-3′). It appears that the primary effect of the dC/dT swap is to increase conformational complexities. Conformational disturbance was the greatest for FAF-modification at G1, a remarkable long-range conformational effect. These data indicated that the thermodynamic properties depend not only flanking sequences, but also on sequences next to surrounding the lesion site.
NarI-dC/-2 and NarI-dT/-2 deletion duplexes
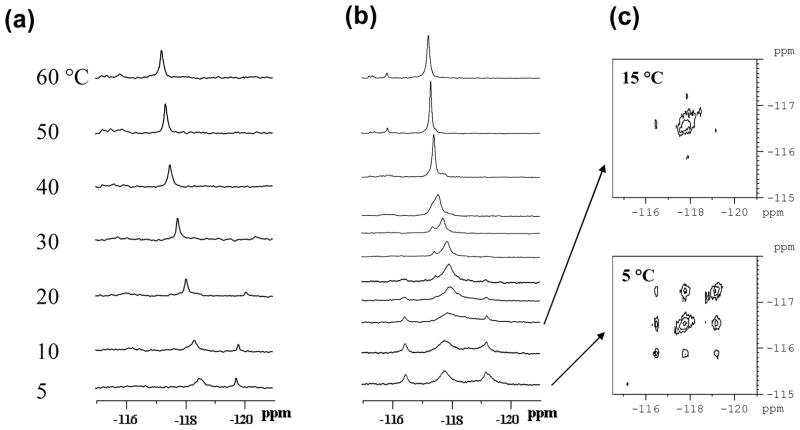
Dynamic 19F NMR spectra were recorded for the NarIdC/-2 and NarIdT/-2 del duplexes, in which G3 was modified by FAF. Using 1H NMR, Mao et al found an AF-intercalated conformer with -CG*-unpartnered (Figure 1c) with the modified G* and its 5′-C displaced into the major groove (35). The modified G* adopted a syn glycosidyl conformation and the methylene edge of the aminofluorene ring was directed toward the minor groove. The imino spectrum of the dC/-2 deletion (del) duplex showed ten well-resolved signals, indicating a single predominant conformation (Supporting Information Fig. S6). Consistent with this observation, the dC/-2 del duplex exhibited a single exclusive 19F signal throughout the entire temperature range tested (5 ~ 60 °C)(Figure 8a). The small signal at −120 ppm was determined to be an impurity when it shifted upfield at increasing temperatures and disappeared after membrane filtering. The signal at −118.8 ppm at 20 °C displayed a very little H/D isotope effect (+0.02 ppm), indicative of a buried 19F signal as predicted for the intercalated conformer (9).
Figure 8.
Dynamic 19F NMR spectra of G3-FAF-modified (a) NarI-dC/-2 and (b) NarI-dT/-2 deletion duplexes. Spectra were obtained at seven standard temperatures (5, 10, 20, 30, 40, 50, and 60 °C. Additional temperatures (15, 22, 25, 35 °C) were used for NarI-dT/-2 to clarify coalescent exchange process (see text). (c) 19F NMR NOESY/exchange spectra of NarI-dT/-2 recorded at 5 and 15 °C.
A similar set of dynamic NMR spectra were obtained for the FAF-modified NarI-dT/-2 del duplex. The imino proton spectrum exhibited complex proton signals with varying degrees of intensity, in contrasted to the dC/-2 counterpart, which showed well-resolved signals (Fig. S6, Supporting Information). The 19F NMR spectrum for the FAF-modified NarI-dT/-2 del duplex (Figure 8B) showed pronounced conformational heterogeneity with at least three signals at −116.8, −117.9 and −119.0 ppm at 5 °C; each of these signals represents a distinct conformer with a population ratio of 18%, 47%, and 35%, respectively. Only the major signal at −117.9 ppm exhibited an H/D isotope effect (+0.15 ppm). These results suggest that the intense signal is probably due to an external B-type conformer, while the minor signals likely represent the FAF-intercalated S-type conformers. As the temperature was raised from 5°C to 22 °C, the main signal broadened and ultimately merged with the two minor signals. Over this low temperature range, the population intensities of the minor signals were reduced significantly. Off-diagonal contour peaks in the NOESY spectra were intense at 5 °C, but disappeared at 15 °C (Figure 8, insets). This strong temperature dependence indicates that the conformers were in slow exchange. Upon raising the temperature further from 20–40 °C the major peak −117.9 ppm narrowed as a new downfield signal emerged at −117.5 ppm. Thus the exchange dynamic characteristics of the dT/-2 del duplex were distinct from the S/B-equilibrium observed with the corresponding fully paired dT duplex described above.
Thermodynamic comparison between the NarI-dC and NarI-dT series
Table 2 summarizes the van’t Hoff parameters obtained for the FAF-modified NarI-dC and -dT of both fully-paired and -2 deletion sequences relative to their controls. The fully paired NarI-dC duplex was thermally (ΔTm = +2.2 °C) and thermodynamically (ΔΔG = +4.5 kcal/mol) more stable than the NarI-dT duplex. FAF-modification resulted in the expected thermal (ΔTm = 6.8 ~ 8.3 °C) and thermodynamic (ΔΔG = 4.2 ~ 4.6 kcal/mol) destabilization. No significant differences were observed among the isomeric NarI-dC duplexes. Thus, the position of the adduct and the polarity (G:C vs. C:G) of base pairs surrounding the lesion did not seem to significantly influence duplex stability. Relative to the fully paired dC-duplexes, the fully-paired dT-duplexes exhibited a similar extent of thermal destabilization (7.0 ~ 13.7 °C) but less thermodynamic destabilization (ΔΔG = −0.5 ~ 0.7 kcal/mol).
Table 2.
Effects of FAF-modification on the thermal and thermodynamic stability of NarI-based DNA duplexesa,b
| entry | −ΔG°c (kcal/mol) | −ΔH°c (kcal/mol) | Tmc,d (°C) | ΔΔG°c,e (kcal/mol) | ΔΔH°c,f (kcal/mol) | ΔTmc,g (°C) |
|---|---|---|---|---|---|---|
| NarI-dC Control | 16.2 | 107.9 | 68.5 | |||
| NarI-dC/G3 | 12.0 | 65.9 | 60.2 | 4.2 | 42 | −8.3 |
| NarI-dC/G1 | 11.7 | 76.7 | 59.1 | 4.5 | 31.2 | −9.4 |
| NarI-dC/G2 | 11.6 | 70.7 | 61.7 | 4.6 | 37.2 | −6.8 |
| NarI-dT Control | 11.7 | 77.5 | 66.3 | |||
| NarI-dT/G3 | 11.0 | 81.9 | 56.3 | 0.7 | 4.4 | −10.0 |
| NarI-dT/G1 | 12.3 | 85.7 | 59.3 | −0.6 | 8.2 | −7.00 |
| NarI-dT/G2 | 12.2 | 121.4 | 52.6 | −0.5 | 43.9 | −13.7 |
see Figure 1 for full structure and sequence details (G* = FAF-adduct).
The results of curve fit and Tm−lnCt dependence were within ± 15% of each other and therefore these numbers are average of the two methods.
The average standard deviations for −ΔG°, −ΔH°, and Tm are ± 0.22, ± 6.33, and ± 0.4 respectively.
Tm values at 14 μM taken from the 1/Tm − lnCt/4 Meltwin plots
ΔΔG = ΔG° (FAF-modified duplex) − ΔG° (control duplex).
ΔΔH = ΔH° (FAF-modified duplex) − ΔH° (control duplex).
ΔTm = Tm (FAF-modified duplex) − Tm (control duplex).
FAF modification of the dT/-2 del duplex produced only a small increase in free energy (ΔΔG = −0.5 kcal) and a small decrease in Tm (ΔTm = −1.0 °C). On the other hand, FAF-modification of the dC/-2 duplex resulted in significant thermal (ΔTm = 3.3 °C) and thermodynamic (ΔΔG = −3.0 kcal/mol) stabilization. These data are consistent with the 19F NMR results described above in which single and multiple signals for NarI-dC/-2 and -dT/-2 del duplexes, respectively, were observed. In other words, the dC/-2 del duplex is homogeneous, and thus conformationally stable, whereas the multi-conformeric dT/-2 del duplex is prone to destabilization.
DISCUSSION
The focus of the present work is the examination of the sequence-specific conformation-function-relationships. Here we used a combination of 19F NMR, UV-melting, and CD spectroscopy to probe the hotness of the FAF lesion at G3* and N in the NarI sequence (5′-CTCG1G2CG3*CNATC-3′, N = dC and dT). The present experiments demonstrated that the AF-induced conformational heterogeneity at G3* is strongly dependent on the nature of the base at N in both the fully-paired and -2 deletion duplexes. While the dC/-2 deletion duplex (N=C) adopted exclusively a single intercalated S-type conformer, the corresponding dT/-2 del duplex (N=T) existed as multiple conformers. A similar dT-induced conformational heterogeneity was observed for the fully-paired NarI duplexes. The results support a model that the conformational and thermodynamic stabilities of the induced SMI are critical determining factors for the efficacy of -2 frameshift mutagenesis in the NarI sequence.
dT-induced conformational heterogeneity
The NarI-dC duplexes exhibited typical S/B-equilibrium characteristics as evidenced by two-site exchanges in their dynamic 19F NMR spectra (Figure 6, Table 1). The corresponding NarI-dT duplexes exhibited marked conformational heterogeneity (Figure 7), probably due to the considerable presence of various intermediate B*- and S*-conformers. We refer this phenomenon as “dT-induced conformational heterogeneity.” This effect was particularly dramatic for the G1 duplex (Figure 7B), which is surprising given that G1 is located five bases away from the N position. The reason for this unusual long-range effect is unknown.
A previous 1H NMR study on the NarI-dC/-2 deletion (del) sequence with AF at G3 indicated an exclusive S-type intercalated SMI structure (38). As depicted in Figure 9a, in this structure the modified G and its 5′-C residues are displaced into the major groove. A similar intercalated SMI structure was reported for a non-NarI-based d(CG*AC).d(G-G) duplex; however, the modified G3 was found to stack with its 3′ neighbor A bulging into the major groove (37). These intercalated structures have shown unusual thermal stability independent of sequence context. Our dynamic 19F NMR data showed a single signal for the FAF-modified dC/-2 del duplex throughout the entire melting process (Figure 8a), indicating conformational homogeneity. The 19F signal at 5°C was significantly shielded, indicating that the fluorine atom was buried within the helix as occurs in the S-type conformation. This conformational homogeneity was further evidenced by the presence of well-resolved signals in the imino proton NMR spectrum (Supporting Information Fig. S6).
Figure 9.
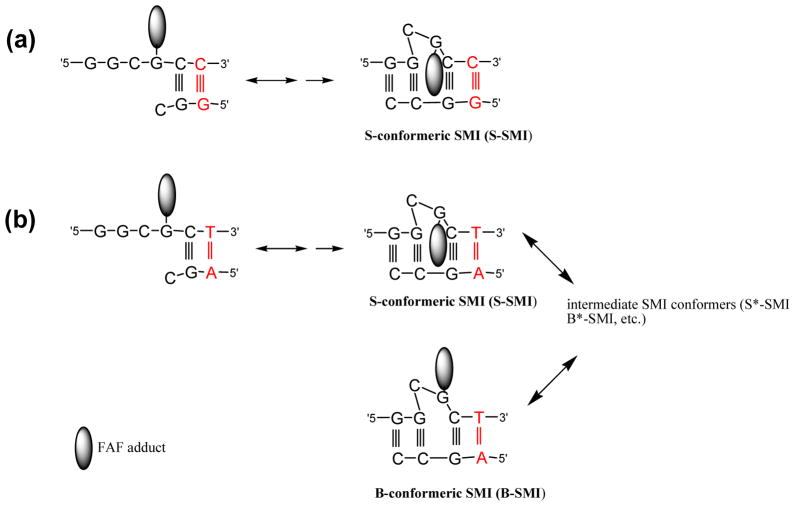
Carton representation oftranslesion synthesis of NarI-induced -2 deletion mutations. Incorporation of correct dC opposite the AF and subsequent nucleotide incorporation (e.g. dC) can lead formation of stable S- and B-conformeric slipped mutagenic intermediates (S-SMI and B-SMI, respectively). (a) NarI-dC/-2 del resulted in an exclusive formation of S-SMI. (b) In contrast, the dT/-2 del produced a complex conformational heterogeneity involving not only S- and B-SMI, but also various intermediate conformers (*). The conformational stability of SMI is strongly modulated by the nature (Watson-Crick, orientation) of the base pairs at N position (red colored)(see text).
Unlike the dC/-2 del duplex, the dT/-2 del duplex displayed severe conformational heterogeneity in both 19F and 1H NMR spectra (Figure 8b, Supporting Information Fig. S6). We observed at least three major conformers exchanging slowly at low temperatures (<15 °C). Consistent with van’t Hoff parameters, our 19F NMR results showed that the dT/-2 del duplex exhibited significantly smaller increases in thermal and thermodynamic stabilities than the dC/-2 duplex (Table 3). Furthermore, the dT/-2 del duplex exhibited a dramatic salt dependent CD effect, indicating the conformational flexibility at the lesion site. Thus it appears that the so-called “dT-induced heterogeneity” also plays a key role in deletion duplexes.
Table 3.
Effects of FAF-modification on the thermal and thermodynamic stability of NarI/-2 deletion duplexesa,b
| entry | Sequence | −ΔG° c (kcal/mol) | −ΔH°c (kcal/mol) | Tmc,d (°C) | ΔΔG°c,e (kcal/mol) | ΔΔH°c,f (kcal/mol) | ΔTmc,g (°C) |
|---|---|---|---|---|---|---|---|
| NarI-dC/-2 Control | -GGCG*CC- -CC-- GG- |
8.2 | 30.5 | 54.5 | |||
| NarI-dC/G3*/-2 del | -GGCG*CC- -CC-- GG- |
11.2 | 91.3 | 57.8 | −3.0 | −60.8 | 3.3 |
| NarI-dT/-2 Control | -GGCG*CT- -CC-- GA- |
8.5 | 30.5 | 45.7 | |||
| NarI-dT/G3*/-2 del | -GGCG*CT- -CC-- GA- |
9.0 | 58.5 | 44.7 | +0.5 | −28.0 | −1.0 |
see Figure 1 for full structure and sequence details (G* = FAF-adduct).
The results of curve fit and Tm−lnCt dependence were within ± 15% of each other and therefore these numbers are average of the two methods.
The average standard deviations for −Δ G°, −ΔH°, and Tm are ± 0.22, ± 6.33, and ± 0.4 respectively.
Tm values at 14 μM taken from the 1/Tm − lnCt/4 Meltwin plots
ΔΔG = ΔG° (FAF-modified duplex) − ΔG° (control duplex).
ΔΔH = ΔH° (FAF-modified duplex) − ΔH° (control duplex).
ΔTm = Tm (FAF-modified duplex) − Tm (control duplex).
The dT/-2-induced heterogeneity can be envisioned as a putative equilibrium between the B- and S-conformeric SMI as depicted in Figure 9b. The signals observed in the 19F NMR spectra of the dT/-2 del duplex (Figure 8b) are probably due to an equilibrium between B- and S-conformeric SMIs and other intermediate conformers (S*-SMI, B*-SMI, etc). Theoretical studies suggest that the syn glycosidic S-conformeric SMI is much more stable than the anti B-conformeric SMI (ΔE >30 kcal/mol), and that it can be stabilized by many favorable interactions between a carcinogen and flanking base pairs inside the bulging pocket (39). In the anti B-SMI structure, however, the carcinogen is situated in the major groove with both faces completely exposed to solvent and virtually no interaction with DNA. As shown in Figure 9b, the carcinogen in the S-conformeric dC/-2 duplex is well-accommodated within the bulge pocket with significant stacking with the 3′ immediate neighbor G:C base pair. The SMI structure of the dT/-2 del duplex has not yet been elucidated by 1H NMR. However, our 19F NMR results indicate clearly that the presence of dT at position N causes pronounced conformational flexibility of the FAF-adduct at G3 within the SMI pocket.
The “dT-effect” in the NarI sequences described above applies to both fully paired and -2 deletion duplexes. It should be noted that in both cases the intercalated carcinogen moiety stacks with the 3′ flanking C:G base pair rather than with N. This type of long-range conformational effect is novel and has not been reported previously. However, an immediate flanking dT-effect has been observed previously. For example, Geacintov and coworkers (41,42) have shown that trans-anti-BaP-dG flanked by dT (-TG*T-) existed in multiple conformations, whereas the same adduct in the -CG*C- context adopted a single conformation. Multi-conformeric TG*T sequences have been shown to be more susceptible to NER than a single conformeric-CG*C sequence (43). Similar sequence dependent NER results have also been obtained for various AF- and AAF-modified sequences (13,14,44). The results indicated greater NER susceptibility for intercalated S-conformers than Watson-Crick paired B-type conformers and hence reinforce the complexity of a multipartite model for NER (45).
Biological Implications
Translesion DNA synthesis and subsequent elongation are most likely achieved by recruitment of specialized lesion-bypass polymerases (25). We have shown that the AF-lesion at the primer terminus exists in a flexible S/B-equilibrium (15). The AF lesion in repetitive sequences may result in polymerase pause in the S-conformer, which may lead to DNA synthesis blockage or slippage. However, the correct dCTP may be incorporated opposite the modified dG in the B-conformer, followed by elongation producing error-free replication or rearrangement into a stable SMI (21–24)(Figure 9). Deletions are frequent under conditions where the newly inserted nucleotide opposite the lesion can pair with a complementary base 5′- to the lesion. For example, when there is a dG 5′ of the lesion, the newly inserted dC can pair with this base to produce a -1 deletion (21,22). Similarly, -2 deletions can occur when there is a complementary base two positions 5′ of the lesion. The relative frequencies of base insertions opposite the adduct were dCMP > dAMP > dGMP > dTMP (24). Mutations did not occur readily in 5′-CG* and 5′-AG* sequences because neither the S- or B-conformer directs incorporation of dG or dT, which is required for the slippage to occur in these sequences. The formation of mispaired intermediates that lead to base substitutions can compete with the formation of bulge intermediates that produce deletion mutations. Also, repair enzymes may stabilize helical forms of DNA and prevent bulge formation, or vice versa, depending on the nature of adduct and local base sequences.
Our working hypothesis is that the conformational stability of adduct-induced SMIs is a key determining factor for the propensity to form -2 deletion mutations. Fuchs and coworkers have suggested that the intrinsic tendency of the carcinogen to slip during translesion synthesis determines the hotness of N in the NarI 5′-GGCG*CN-3′ sequence (31). In principle, there are two possible SMI structures to account for -2 mutations in the NarI sequence: CG*- or G*C-unpartnered. Fuchs et al have used the latter scenario, albeit without structural proof, to argue that greater stacking of the carcinogen with the C:G base pair over T:A at N is responsible for its significantly higher (>6X) deletion frequency (31). Theoretical calculations indicated that NarI-based SMIs are more stable than their normally extended counterparts and that carcinogen-stacking is more important than the number of correct base pairs around the primer-template junction (38). However, high-resolution 1H NMR studies by Mao et al have shown unequivocally the formation of CG*-unpartnered in the exclusive presence of the S-conformeric SMI for the AF-modified dC/-2 del duplex (35) (Figure 9a). Our 19F NMR data showed that the dC/-2 del duplex adopts a single stable S-type conformer, while the corresponding dT/-2 duplex produces complex conformational heterogeneities involving the putative equilibrium between S- and B-SMIs (Figure 9b). Taken together, it can be concluded that the conformational stability of the SMI is an essential determinant for inducing -2 frameshift mutations.
As mentioned above, the intercalated carcinogen moiety in the S-conformeric SMI stacks with the 3′ immediate flanking C:G base pair, not the nucleotide at position N. Interestingly, however, equally high mutational frequencies have been reported for dC and dA at N (30,31). Whether and to what extent base pair polarity (T:A vs. A:T) at N affects SMI conformational stability is not known. There are no structural data of any kind yet available for the NarI-dA/-2 deletion duplex. If our hypothesis is correct, then the dA/-2 duplex should form a conformationally stable SMI. Our findings indicate that SMI conformational stability is determined not only by the nature of Watson-Crick base pairs (A:T vs. G:C), but also by base pair polarity (A:T vs. T:A or G:C vs. C:G).
AF/FAF Conformational Compatibility
We demonstrated previously the utility of the fluorine probe FAF for studying structure/function of 4-aminobiphenyl (9), AF- (9,11–15) and AAF- (46) adducts. The H/F swap on the aminofluorene moiety generally had little effect on the spectroscopic and repair characteristics in various sequence contexts. In the present work, we provide evidence for AF/FAF-compatibility with repetitive NarI sequences.
The CD profiles of AF- and FAF-adducts are strikingly similar at various positions of the fully-paired NarI sequences (Figures 3, 4 and Supporting Information Fig. S1). We demonstrated previously that AF- and FAF-induced CD in the 290–350 nm range (ICD290–360nm) arises from interactions between the non-chiral carcinogen and the DNA and is a useful marker for sequence dependent S/B-conformational heterogeneity (11,12,14,15). The G2-modified duplexes showed negative ICD290–360 nm, indicative of B-conformation, while the G1 and G3-duplexes exhibited positive ICD290–360 nm, indicative of S-conformation. In concordance with these CD data, our 19F NMR data indicated that S-conformer populations were 56%, 29%, and 69%, respectively, for the G1-, G2-, and G3-modified fully paired NarI-dC duplexes. This convergence of findings supports the compatibility of CD and 19F NMR. Although a similar trend was detected, the present % S-conformer population values derived from 19F NMR were greater than those previously revealed by 1H NMR (30, 10, and 50%, respectively)(34). While data are limited, it is not likely that the AF/FAF swap contributes to the greater S-population observed for FAF. For example, we have shown previously that FAF- or FABP-modified 12-mer duplexes with the TG*A- context result in S/B population ratios similar to the ratios seen with AF-modified duplexes (9). Furthermore, a similar AF/FAF compatibility was obtained for the -2 deletion duplexes.
Concluding remarks
In conclusion, the nature of the base pair and base pair polarity configuration at N in the FAF-modified NarI sequence (5′-CTCGGCG*CNATC) have a substantial influence on conformational and thermodynamic stabilities in both fully-paired and -2 deletion duplexes. Dynamic 19F NMR results reveal the presence of an unusual long-range “dT-induced conformational heterogeneity.” Experimental evidence suggests the existence of a putative equilibrium between the S- and B-conformer SMIs in the bulged dT/-2 deletion duplex. The yield of -2 deletion mutations at G3 in the NarI sequence is determined to a considerable extent by the conformational stability of the S-conformer SMI. In addition, we have provided CD and 19F NMR evidence for AF/FAF compatibility in NarI sequences, demonstrating the utility FAF as a useful conformational marker in investigating AF-adduct structures.
Supplementary Material
Abbreviations
- AF
N-(2′-deoxyguanosin-8-yl)-2-aminofluorene
- AAF
N-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene
- CD
circular dichroism
- 19F NMR
fluorine nuclear magnetic resonance spectroscopy
- ICD290–360 nm
induced circular dichroism at 290–360 nm
- SMI
slipped mutagenic intermediates
- TLS
translesion synthesis
Footnotes
We thank Dr. Paul Chiarelli for providing ESI-TOF-MS spectra of the AF- and FAF-modified 12-mer templates. We are grateful to the NIH (R01CA098296) for their financial support of this work. This research was also made possible in part by the use of the Research Core Facility supported by the NCRR/NIH (P20 RR016457).
Supporting Information Available: CD spectra of NarI-dC (Figure S1) and NarI-dT (Figure S2) duplexes; 19F NOESY (Figure S3) and imino proton (Figure S4) spectra of NarI-dC and NarI-dT duplexes; examples of lineshape dynamic analysis (Figure S5); imino proton spectra of NarI-dC and NarI-dT -2 deletion duplexes (Figure S6); calculated and measured molecular mass ions (Table S1); summary of H/D isotope effects (Table S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Luch A. Nature and nature-lessons from chemical carcinogenesis. Nature Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Heflich RH, Neft RE. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 3.Beland FA, Kadlubar FF. In: Handbook of Experimental Pharmacology. Cooper CS, Grover PL, editors. Springer-Verlag and Heidelberg; 1990. pp. 267–325. [Google Scholar]
- 4.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RPP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 5.Dutta S, Li Y, Johnson D, Dzantiev J, Richardson CC, Romano LJ, Ellenberger T. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc Natl Acad Sci USA. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem Res Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 7.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J Environ Sci Health, Part C-Environ Carcinogen Ecotoxicol Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 8.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L, Rajabzadeh G, Traficante DD, Cho BP. Conformational heterogeneity of arylamine-modified DNA: 19F NMR evidence. J Am Chem Soc. 1997;119:5384–5389. [Google Scholar]
- 10.Cho BP, Beland FA, Marques MM. NMR structural studies of a 15-mer duplex from a ras protooncogene modified with the carcinogen 2-aminofluorene: conformational heterogeneity. Biochemistry. 1994;33:1373–1384. doi: 10.1021/bi00172a013. [DOI] [PubMed] [Google Scholar]
- 11.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluorene-DNA adducts. Chem Res Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 12.Meneni SR, D’Mello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meneni S, Shell SM, Zou Y, Cho BP. Conformation-specific recognition of carcinogen-DNA adduct in E. coli nucleotide excision repair. Chem Res Toxicol. 2007;20:6–10. doi: 10.1021/tx600273h. [DOI] [PubMed] [Google Scholar]
- 14.Meneni SR, Shell SM, Gao L, Jurecka P, Lee W, Sponer J, Zou Y, Chiarelli MP, Cho BP. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. doi: 10.1021/bi700858s. in press. [DOI] [PubMed] [Google Scholar]
- 15.Meneni SR, Liang F, Cho BP. Examination of the long-range effects of aminofluorene-induced conformational heterogeneity and its relevance to the mechanisms of translesion DNA synthesis. J Mol Biol. 2007;366:1387–1400. doi: 10.1016/j.jmb.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 17.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc Natl Acad Sci U S A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmquist CE, Wang F, Stover JS, Stone MP, Rizzo CJ. Conformational differences of the C8-Deoxyguanosine adduct of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) within the NarI recognition sequence. Chem Res Toxicol. 2007:445–454. doi: 10.1021/tx060229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions. J Am Chem Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme. Biochemistry. 2007;46:8498–8516. doi: 10.1021/bi700361u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann GR, Fuchs RPP. Mechanisms of frameshift mutations: Insight from aromatic amines. Chem Res Toxicol. 1997;4:347–359. doi: 10.1021/tx960128n. [DOI] [PubMed] [Google Scholar]
- 22.Shibutani S. Requirements for frame-shift deletion during translesion synthesis. Environ Mutagen Res. 2004;26:135–141. [Google Scholar]
- 23.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J Biol Chem. 2004;279:45360–45368. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 24.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–15935. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs RPP, Fujii S. Translesion synthesis in Escherichia coli: Lessons from the NarI mutation hot spot. DNA Repair (Amst) 2007;6:1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 26.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;27:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 27.Burnouf D, Koehl P, Fuchs RPP. Single adduct mutagenesis: Strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc Natl Acad Sci U S A. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 29.Shibutani S, Suzuki N, Grollman AP. Mutagenic specificity of (Acetylamino)fluorine-derived DNA adducts in mammalian cells. Biochemistry. 1998;37:12034–12041. doi: 10.1021/bi981059+. [DOI] [PubMed] [Google Scholar]
- 30.Koffel-Schwartz N, Fuchs RPP. Sequence determinants for -2 frameshift mutagenesis at NarI-derived hot spots. J Mol Biol. 1995;252:507–513. doi: 10.1006/jmbi.1995.0515. [DOI] [PubMed] [Google Scholar]
- 31.Broschard TH, Koffel-Schwartz N, Fuchs RPP. Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots. J Mol Biol. 1999;288:191–199. doi: 10.1006/jmbi.1999.2667. [DOI] [PubMed] [Google Scholar]
- 32.Gill JP, Romano LJ. Mechanism for N-acetyl-2-aminofluorene-induced frameshift mutagenesis by Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2005;44:15387–15395. doi: 10.1021/bi051437s. [DOI] [PubMed] [Google Scholar]
- 33.Stover JS, Chowdhury G, Zang H, Guengerich FP, Rizzo CJ. Translesion synthesis past the C8- and N2-deoxyguanosine adducts of the dietary mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the NarI recognition sequence by prokaryotic DNA polymerases. Chem Res Toxicol. 2006;19:1506–1517. doi: 10.1021/tx0601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-external conformer of the anti-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 35.Mao B, Gorin A, Gu Z, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a -2 deletion site in the NarI hot spot sequence context. Biochemistry. 1997;36:14479–14490. doi: 10.1021/bi972205z. [DOI] [PubMed] [Google Scholar]
- 36.Friebolin H. In Basic one- and two-dimensional NMR spectroscopy. 3. Wiley-VCH; 1998. pp. 301–329. [Google Scholar]
- 37.Koehl P, Valladier P, Lefèvre JF, Fuchs RPP. Strong structural effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Nucleic Acids Res. 1989;17:9531–9541. doi: 10.1093/nar/17.23.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution conformation of [AF]dG opposite a -2 deletion site in a DNA duplex: intercalation of the covalently attached aminofluorene ring into the helix with base displacement of the C8-modified syn guanine and adjacent unpaired 3′-adenine into the major groove. Biochemistry. 1995;34:16641–16653. doi: 10.1021/bi00051a012. [DOI] [PubMed] [Google Scholar]
- 39.Roy D, Hingerty BE, Shapiro R, Broyde S. A slipped replication intermediate model is stabilized by the syn orientation of N-2-aminofluorene- and N-2-(Acetyl)aminofluorene-modified guanine at a mutational hotspot. Chem Res Toxicol. 1998;11:1301–1311. doi: 10.1021/tx980106w. [DOI] [PubMed] [Google Scholar]
- 40.Hansen PE, Dettman HD, Sykes BD. Solvent-induced deuterium isotope effects on fluorine-19 chemical shifts of some substituted fluorobenzenes. Formation of inclusion complexes. J Magn Reson. 1985;62:487–496. [Google Scholar]
- 41.Xu R, Mao B, Amin S, Geacintov NE. Bending and circularization of site-specific and stereoisomeric carcinogen-DNA adducts. Biochemistry. 1998;37:769–778. doi: 10.1021/bi971785x. [DOI] [PubMed] [Google Scholar]
- 42.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, Van Houten B, Geacintov NE. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006– 7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 43.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 45.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 46.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2′-Deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.