Abstract
The recent identification of metastasis suppressor genes, the products of which inhibit metastasis but not primary tumor growth, distinguishes oncogenic transformation and tumor suppression from a hallmark of malignancy, the ability of cancer cells to invade sites distant from the primary tumor. The metastasis suppressor CD82/KAI1 is a member of the tetraspanin superfamily of glycoproteins. CD82 suppresses metastasis by multiple mechanisms including inhibition of cell motility and invasion, promotion of cell polarity as well as induction of senescence and apoptosis in response to extracellular stimuli. A common feature of these diverse effects is CD82 regulation of membrane organization as well as protein trafficking and interactions, which affects cellular signaling and intercellular communication.
Keywords: endoplasmic reticulum-associated degradation (ERAD), gp78, epidermal growth factor receptor (EGFR), exosome, Duffy antigen receptor for chemokines (DARC)
Introduction
Metastasis remains by far the major cause of cancer-related mortality. This multi-step process has, as a first common initial feature, the movement of malignant cells from the primary tumor. This is followed by their entry into the circulation and their successful traversal of the circulation to a distant site where, after exiting the circulation, they establish themselves and proliferate to form overt metastases. An improved understanding of the details that allow for the completion of these multiple steps, and the molecular mechanisms by which they are naturally prevented, should lead to new approaches to cancer treatment. In the past decades, the discovery of genes that specifically suppress metastasis has offered new insights into the process of metastasis. The observation that metastasis suppressors exert little effect on primary tumor growth suggests that these gene products either directly or indirectly influence the interactions of the tumor cells with their microenvironments. Metastasis suppressor genes that interrupt different steps of the metastatic cascade have been described (see accompanying articles this issue). Herein, we discuss the functions of CD82/KAI1, focusing on its roles in regulating protein and membrane dynamics.
CD82 as a Metastasis Suppressor Gene
The metastasis suppressor gene KAI1 (Kangai1), located on chromosome 11p11.2, was originally identified in a screen for genes on chromosome 11 that suppressed metastasis of rat AT6.1 prostate cancer cells. Re-expression of KAI1 in AT6.1 reduced the formation of metastases without affecting primary tumor growth [1]. KAI1 was previously described as CD82, a lymphocyte cell surface protein involved in activation of T cell receptor [2]. CD82 is also known as antigens R2, C33, IA4 and 4F9 [3–6]. Subsequent studies confirmed the metastasis suppressor function of CD82 in hepatocarcinoma, melanoma, sarcoma, pancreatic and breast cancer cell lines [7–11]. The importance of CD82 in cancer progression is underscored by the observation that downregulation of CD82 mRNA and protein is associated with advanced stages of many malignancies (reviewed in [12]) including prostate, colon, lung, pancreatic, breast, ovarian and other cancers.
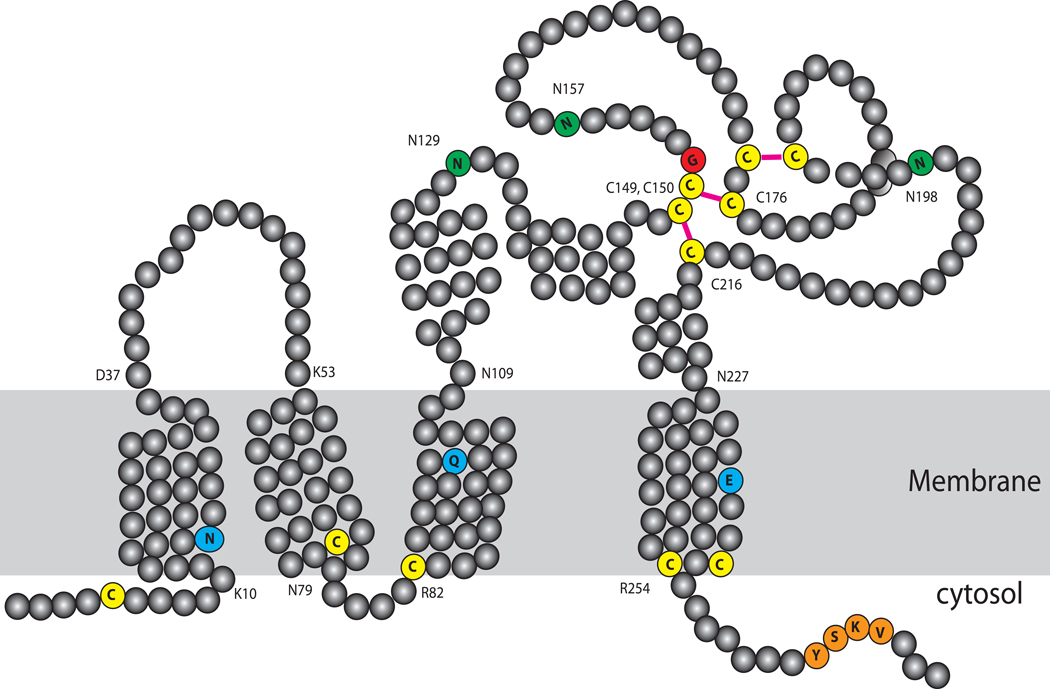
CD82 is a member of the tetraspanin superfamily of glycoproteins. Tetraspanins are characterized by four transmembrane domains with conserved polar residues, which may help to stabilize a tight tertiary structure [13–15] (Figure 1). They have short intracellular N- and C-termini, one short (ECL1) and one long (ECL2) extracellular hydrophilic loops and a very short intracellular loop. The intracellular juxtamembrane cysteines are usually modified by palmitoylation, which contributes to the formation of a tetraspanin web with other tetraspanins as well as non-tetraspanin proteins. Based on a modeled structure of CD81, a related tetraspanin, the N terminus likely forms a membrane-parallel amphipathic helix when palmitoylated whereas the C terminus likely adopts a random structure [13]. The short ECL1 appears to possess a β-strand that is enriched with hydrophobic residues and may interact with the hydrophobic groove of ECL2 [13]. ECL2 is the most variable region among the tetraspanins and provide antigenic specificity as most monoclonal antibodies map to this region. ECL2 also contains the signature Cys-Cys-Gly motif, which forms two disulfide bonds with two other conserved cysteines in ECL2. ECL2 can be divided into a constant region and a variable region: The constant region may contribute to oligomerization with other tetraspanins while the variable region may contribute to interactions with other non-tetraspanin proteins [13,16]. ECL2 of CD82 contains 3 consensus N-linked glycosylation sites. When overexpressed, CD82 sometimes show a wide range of molecular weights on SDS-PAGE (40–90 kDa) due to varying degrees of glycosylation. Glycosylation of tetraspanins is important not only for their transit through the endoplasmic reticulum (ER) during maturation but also for complex formation and cellular functions [15,17–18].
Figure 1. Schematic of CD82 structure.
CD82 has four transmembrane domains with conserved polar residues (light blue), which may be important for structural integrity. The long extracellular loop (ECL2) contains six cysteine residues (yellow) predicted to form 3 disulfide bonds (magenta). The glycine residue in the characteristic Cys-Cys-Gly motif is highlighted in red. ECL2 also contains three asparagine (green) residues that are sites for N-linked glycosylation. Juxtamembrane cysteine residues (yellow) are usually palmitoylated. Palmitoylation of the N-terminal cysteine likely holds the N-terminus parallel to the membrane. The C-terminus contains a sorting motif (orange) important for localization to the endocytic pathway.
Tetraspanins form large multimeric complexes that consist of tetraspanins as well as other membrane and cytosolic proteins such as receptor tyrosine kinases, integrins, and adaptor proteins that are integral to signaling cascades. They are also enriched for specific lipids including gangliosides. These complexes are clustered in specialized membrane domains known as "tetraspanins-enriched microdomains" (TEM). CD82 does not have any evident enzymatic activity. As discussed in the following sections, CD82 seems to instead function by modulating the levels, trafficking or activity of its interacting partners in the TEM.
Mechanisms of CD82 Function
Unlike other metastasis suppressors, CD82 appears to inhibit multiple steps of the metastatic cascade including cell motility and invasion, proliferation, apoptosis and senescence. CD82 has an impact on the interaction of tumor cells with their microenvironments in ways that are non-permissive for survival and proliferation beyond the primary tumor. This broad range of effects can be achieved by the modulation of the activity and trafficking of proteins critical for metastasis through physical or functional interactions with CD82.
Cell-Cell Adhesion
One important function of CD82 is to promote homotypic cell-cell adhesion. Loss of cell adhesion plays an important role in epithelial-mesenchymal transition (EMT) and metastasis. In DU145 cells, transfection of CD82 induces homotypic cell-cell aggregation. This increase in cell-cell adhesion can be blocked with protein phosphatase 1, an inhibitor of Src kinase, or by overexpression of a kinase negative Src mutant [19], implicating activation of Src-mediated pathways in CD82-mediated effects. In non-small cell lung carcinoma, overexpression of CD82 promotes cell-cell adhesion [20], which depend on E-cadherin. CD82 does not alter the distribution of E-cadherin in these cells; instead, CD82 stabilizes E-cadherin/β-catenin complex formation, which can inhibit cancer cell dissemination from the primary tumor.
Cell Motility and Invasion
In vitro studies show that overexpression of CD82 inhibits cell motility and invasion [11,14–15,21–27]. The association of CD82 with α6 integrin results in decreased laminin adhesion and migration [28]. Integrins mediate cell adhesion to the extracellular matrix by binding matrix ligands such as laminin and fibronectin. CD82 downregulates α6 integrin by increasing its internalization resulting in reduced laminin adhesion. In H1299 lung carcinoma cells, CD82 reduces cell surface expression of b1 integrin by interfering with the b1 integrin glycosylation and maturation [24]. CD82 also associates with EWI-2/PGRL, a transmembrane immunoglobulin G superfamily protein [29]. Overexpression of EWI-2 in DU145 metastatic prostate cancer cells inhibits cell migration on both fibronectin- and laminin-coated substrata, indicating that EWI-2 directly regulates cell migration. The association of CD82 with EWI-2 enhances CD82 inhibition of cancer cell motility, suggesting that EWI-2 is required for CD82 function [29]. It is not clear how the CD82 interaction with EWI-2 affects the levels of integrins.
The signaling pathways responsible for CD82 inhibition of cell motility are not well understood. Overexpression of CD82 in DU145 upregulates the expression of focal adhesion kinase (FAK) and Lyn, a Src family tyrosine kinase [23]. The total amount of activated FAK and Lyn (as assessed by their phosphorylation), however, remains unchanged suggesting that their net activation is impaired by CD82. The levels of p130CAS, a downstream target of FAK-Lyn signaling, is also reduced by 2–4 fold. Consequently, the coupling of phosphorylated p130CAS and CrkII required for cell motility [30], is attenuated. The reduction in p130CAS is due to post-transcriptional effects as the amount of RNA is not affected [23]. Palmitoylation regulates CD82 localization and interaction with other tetraspanins, suggesting that CD82 interactions in the tetraspanin web are important for its motility-inhibitory effects. Interestingly, palmitoylation is required for CD82 inhibition on p130CAS-CrkII coupling. [31]. The precise mechanism by which CD82 lower the levels of p130CAS protein remains to be discovered.
An important aspect of the function of CD82 in inhibiting cell motility and invasion is by modulating the activity of receptor tyrosine kinases (RTKs). CD82 has been reported to attenuate epidermal growth factor receptor (EGFR) signaling by promoting internalization of the activated receptor [32]. This might, in part, be related to a CD82 endosomal-lysosomal sorting motif [33] and its localization to endocytic vesicles and exosomes [34] (see also below). The effect of CD82 on EGFR is inhibited by the ganglioside GD1a, which interferes with the ability of CD82 to recruit EGFR into TEMs [35]. In a study from a different group, the capacity of CD82 to inhibit EGFR appears to depend on PKC, caveolin-1 and the ganglioside GM3 [36]. In this model, EGFR internalization results from PKC phosphorylation of EGFR on Thr654. The effects of CD82 on EGFR therefore depend on components of the TEM.
Another example of RTK regulation is provided by CD82 suppression of c-Met [HGFR (hetatocyte growth factor receptor)] signaling in the PC3 prostate cancer cell line [37]. In these cells, CD82 reduces both integrin-dependent and HGF-induced activation of c-Met, although the levels of integrin are not altered. CD82 inhibits cell motility by inhibiting signaling to c-Met and Src, resulting in reduced activation of p130CAS [37]. In non-small cell lung carcinoma, CD82 specifically inhibits HGF-induced lamellipodial protrusion and cell migration but not HGF-induced proliferation [38], providing a potential explanation for the lack of effect of CD82 on primary tumor growth. CD82 does not interact directly with c-Met; instead, CD82 interferes with the recruitment of adaptor proteins Grb2 and p85 to selectively attenuate c-Met signaling via the Cdc42/Rac pathways. In contrast, c-Met mediated growth signaling via AKT and MAPK is unaffected [38]. How CD82 reduces Grb2 and p85 recruitment to c-Met is not known. As in the case for EGFR, gangliosides also affect CD82 suppression of c-Met signaling. In bladder cancer YTS1 cells, which do not express CD82, c-Met is constitutively activated. Transfection of CD82 inhibits c-Met signaling in the presence of GM2, indicating that the composition of gangliosides in the complex can regulate CD82 activity on RTKs and integrins [27].
A yeast two-hybrid screen identified Vangl1/Kitenin as a CD82-interacting protein [26] that binds to the C-terminus of CD82. Overexpression of Vangl1 in murine colon carcinoma cell line CT-26 enhances tumorigenecity and metastasis in vivo, as well as increases adhesion leading to invasion in vitro [26]. Interestingly, overexpression of Vangl1 restores invasiveness of CT-26 cells expressing CD82, suggesting that Vangl1 functions downstream of CD82. Vangl1 was identified in a separate screen for proteins phosphorylated in response to intestinal trefoil factor (ITF/TFF3), which protects intestinal epithelia promoting migration of epithelial cells during mucosal repair [39]. Consistent with its pro-migratory role, Vangl1 overexpression enhances wound closure of intestinal epithelial cells, whereas siRNA directed against Vangl1 inhibited the migratory response to ITF. ITF stimulates phosphorylation of Vangl1 and promotes its dissociation from E-cadherin at the plasma membrane. How CD82 regulates Vang1 is not clear. An inverse relationship is reported for levels of Vangl1 and CD82 in CT-26 cells. Notably, expression of a splice variant of CD82 lacking the C-terminus results in higher levels of Vangl1 [26]. One possible model for CD82 function is to regulate the trafficking and levels of Vangl1 by promoting its co-internalization.
Metastasis requires not only cell motility but also activation of extracellular proteases that degrade the extracellular matrix and facilitate cell invasion. Expression of CD82 results in a 50-fold decrease in urokinase plasminogen activator surface receptor (uPAR) activity [40]. CD82 does not interact with uPAR directly. Instead, CD82 causes a redistribution of uPAR to focal adhesions, where it interacts with α5β1 integrin. This stable association with α5β1 prevents uPAR binding to its ligand uPA, resulting in reduced extracellular matrix proteolysis. CD82 can therefore regulate cell invasion by regulating the localization of extracellular proteases.
These studies suggest that CD82 suppresses cell migration and invasion by regulating the trafficking of signaling proteins. How CD82 achieves this through the tetraspanin web is not clear. Other tetraspanins have been shown to affect the biosynthesis and trafficking of their associated proteins [41]. CD82 can also inhibit receptor tyrosine kinases and integrin signaling by promoting their internalization. These dynamic interactions appear to impact cell motility and invasion by selectively targeting uPAR, Rho, PKC and Src pathways.
Cell Death and Survival
The most critical step(s) in the metastatic cascade regulated by CD82 is not known. While CD82 regulation of cell adhesion and migration are well established, it is clear that CD82 has additional functions in metastasis suppression. CD82 suppresses metastasis of tumor cells delivered by intravenous injection suggesting that CD82 functions are not limited to the primary tumor site. Furthermore, cells accumulating CD82 due to loss of its ubiquitin ligase, gp78, or transfected with CD82 do not show evidence of defects in their extravasation into the lungs when delivered by tail vein injection [8], suggesting that CD82 does not prevent movement out of the vasculature into distant sites (i.e. sites of potential metastases).
A major limiting factor in metastatic colonization is the survival and proliferation of tumor cells at the distant site. As tumor cells leave the primary tumor site, loss of adhesion can induce apoptosis (anoikis) in circulating tumor cells. Studies using the HT1080 human fibrosarcoma cell line have established that CD82 is regulated post-translationally by proteasomal degradation after being targeted by the ubiquitin ligase gp78 [8]. HT1080 cells in which CD82 levels are elevated by loss of gp78 show an increase in apoptosis in the lung shortly after being injected into the tail vein of mice [8]. This increased apoptosis results in higher rate of tumor cell attrition during early metastasis. In contrast, primary tumors of gp78-deficient cells do not show signs of increased apoptosis, consistent with metastasis suppression by CD82. These results suggest that CD82-dependent apoptosis depends on the tumor cell interaction with the micro-environment.
ER stress and oxidative stress signaling have been implicated in cancer progression. Both signaling pathways are coordinately regulated in cells. Cells activate a survival response called the unfolded protein response (UPR) during ER stress (reviewed in [42]). gp78 functions as a critical ubiquitin ligase in ER-associated degradation (ERAD), an important part of the UPR. Sustained ER stress activates cell death pathways possibly by accumulation of reactive oxygen species (ROS). The mitochondrion is a major source of ROS in cells. In addition, the ER also generates substantial ROS during oxidative folding of proteins involving disulfide bond formation. In the primary tumor, hypoxia upregulates survival pathways and suppresses mitochondrial function to reduce the accumulation of ROS, which may partially protect tumor cells from the apoptotic effect of CD82. Hypoxia also upregulates autophagy; increased autophagy appears to protect prostate cancer cells from apoptosis [43], providing another explanation for the lack of effect of CD82 on the primary tumor.
CD82 has also been shown to induce cell death in other studies [15,44–45]. The mechanisms of cell death may involve ER stress and autophagy [45] or oxidative stress associated with efflux of cellular glutathione [44]. However, it is not clear whether loss of intracellular glutathione is the cause or result of apoptosis [44]. The multidrug resistance protein MRP1, when properly localized to the plasma membrane, appears to mediate the efflux of cellular glutathione during apoptosis [46]. The effect of CD82 on the maturation or localization of MRP1 has not been studied. Overexpression of CD82 induces autophagic cell death in multiple myeloma cells [45] but only when it is GFP-tagged on the C-terminus [47]. In contrast, overexpression of N-terminal tagged CD82 inhibits cellular adhesion and migration but does not induce cell death. Upon closer examination, N-terminal tagged CD82 localizes to the plasma membrane of multiple myeloma cells whereas C-terminal tagged CD82 accumulates inside the cell reminiscent of intracellular inclusions [47]. These results suggest that cell death observed in [47] is likely caused by protein misfolding or improper trafficking of the overexpressed C-terminal tagged CD82 in multiple myeloma cells, which are sensitive to ER stress.
Tumor Cell Senescence
The blood group Duffy antigen receptor for chemokines (DARC) has been identified as a CD82-interacting protein [7]. DARC is expressed on the surface of erythrocytes and endothelial cells. Tumor cells expressing CD82 show enhanced adhesion to endothelial cells. The interaction of CD82 and DARC induces cellular senescence in the tumor cells in vitro, providing a possible explanation for the inhibitory effect of CD82 on metastasis, although tumor cell senescence in CD82-mediated metastasis suppression was not assessed in vivo [7]. Furthermore, the ability of CD82 to suppress metastasis of B16 melanoma cells is significantly compromised in DARC knockout mice, providing additional evidence that DARC may play a role in CD82-mediated metastasis suppression.
This provocative study provides insights into how CD82 specifically suppresses metastasis without inhibiting primary tumor growth. As tumor cells in the primary tumor do not contact DARC, which is highly expressed in cells of the vasculature, the presence of CD82 does not affect their proliferation. However, several issues remain unanswered. It is not known how CD82-DARC interaction induces tumor cell senescence. Moreover, sarcoma cells with elevated levels of CD82 do not show signs of senescence when cultured with endothelial cells [8], suggesting that the senescence signal is cell-type specific. Furthermore, the significance of CD82-DARC interaction induced senescence in metastasis suppression in vivo is not clear. Although tumor cells show increased metastasis in DARC−/− mice [7], the inability of CD82 to suppress metastasis in DARC-deficient mice could be related to increased angiogenesis in these animals [48–49]. DARC-deficient endothelial cells show enhanced angiogenesis in vitro as these cells escape senescence induced by DARC [49]. DARC has also been shown to remove angiogenic chemokines secreted by tumor cells [48]. Consequently, DARC-deficient mice show increased prostate tumor growth due to accumulation of angiogenic chemokines and increased angiogenesis [48]. Based on results from DARC-deficient mice, a role for DARC in prostate tumor progression has been postulated for African men as 70% of this population exhibit loss of DARC expression in erythrocytes [48]. However, a recent study finds little or no association between erythrocyte DARC expression and the risk or progression of prostate cancer in men of African descent [50]. Further study is clearly needed to address these questions.
Exosomes
Exosomes are small vesicles that are derived from the multivesicular body and released into the extracellular space upon fusion with the plasma membrane (reviewed in [51]). Exosomes can serve to transfer both genetic materials and proteins to distant cells. Interestingly these structures are enriched in tetraspanins including CD82 [34].
Highly provocative findings on a potential role for CD82 acting as a metastasis suppressor through exosome formation comes from a study on the Wnt signaling pathway. The Wnt signaling pathway, which is mediated in part through β-catenin and downstream effectors, is implicated in cell growth and metastasis. CD82 as well as CD9, another tetraspanin, suppress β-catenin-mediated Wnt signaling activity and induce a significant decrease in β-catenin levels [52]. This decrease occurs by induction of β-catenin export via exosomes, which can be blocked by a sphingomyelinase inhibitor. CD82 fails to induce exosome release of β-catenin in cells that express low levels of E-cadherin. These results suggest that CD82 and CD9 down-regulate the Wnt signaling pathway through the exosomal clearance of β-catenin. This is interesting in light of the finding that a β-catenin/Reptin complex represses CD82 transcription [53]. These results suggest the existence of a feed forward mechanism whereby an increase in β-catenin inhibits CD82 expression to further enhance Wnt signaling. The significance of this pathway to CD82 metastasis suppression remains to be determined. Conversely, exosomal removal of β-catenin could relieve repression of CD82 expression by β-catenin, possibly leading to further increase in exosome release and attenuation of Wnt signaling.
Regulation of CD82 Expression
A challenge to activating CD82 for therapy requires an understanding of how CD82 is regulated in normal and cancerous tissues. There is little evidence for gene mutation or loss of heterozygosity, promoter mutation or promoter hypermethylation to account for loss of CD82 expression [54–58]. Loss of CD82 during cancer progression may result from changes in transcriptional regulation in tumor cells. An alternatively spliced variant of CD82, which produces a dominant negative protein, has also been described. More recently, post-translational modification has been shown to play a role in regulating the levels of CD82 in tumor cells.
Transcriptional Regulation
Three regulatory regions are defined in the human CD82 promoter: an enhancer region, a negative regulatory region and a minimal promoter region [59–60]. Expression of CD82 can be induced with cytokines, growth factors, phorbol esters and drugs [61–65]. A p53 regulatory element has been described in the enhancer region [62]. However, several studies correlating p53 loss and CD82 expression do not support a simple model for CD82 regulation [66–67]. One way to reconcile these differences is provided by the finding that CD82 expression is regulated by p53, JunB and AP2 as described in [60,68]. Furthermore, JunB induction of CD82 expression can be enhanced by ATF3 [69]. The requirement for more than one transcription factor at the promoter may explain the contradictory data concerning the role of p53 in regulating CD82 expression [62,66,70–71].
CD82 expression by NF-κB is induced by pro-inflammatory cytokines such as IL-1β, IL-4, IL-6, IFN-γ and TNF-α [72]. The CD82 promoter has a binding site for NF-κB p50 subunit. In non-metastatic cells, binding of p50 and a NCOR1-TAB2-HDAC3 co-repressor complex inactivates transcription. IL-1β stimulation recruits a Tip60/Fe65/Pontin co-activator complex that displaces the NCOR1 complex [53]. In metastatic cells, Tip60 is downregulated and a β-catenin/Reptin complex replaces the Tip60/Pontin complex. Unlike Tip60/Pontin complex, the β-catenin/Reptin complex represses NF-κB activation of CD82 expression [53]. The repressor function of Reptin requires sumoylation on Lys456; a Reptin mutant that lacks functional sumoylation relieves transcriptional repression [73]. Similarly, overexpression of a de-sumoylating enzyme SENP1 reverses the repressive function of Reptin [73]. These data provide a direct link for Reptin post-translational modification by sumoylation to cancer metastasis.
Recent studies suggest that hypoxia induces CD82 expression [74]. A hypoxia-response element is identified within the 500bp upstream region of mouse CD82 gene. Hypoxia-dependent induction of CD82 is mediated by hypoxia-inducible factor (HIF)-1α, which binds to the CD82 promoter. Consistent with this hypothesis, hypoxia-driven upregulation of CD82 is abrogated in HIF-1α null mouse embryonic fibroblasts. Ischemic tissues show increased CD82 protein synthesis suggesting that CD82 is a hypoxia target gene in vivo. Hypoxia also can induce CD82 expression indirectly; hypoxia induces the expression of another metastasis suppressor, NDRG1 [75]. NDRG1 inhibits metastasis partly by downregulating the expression of the transcription factor ATF3 [76], which interacts with NF-κB p50 to repress CD82 expression [69].
CD82 transcription can also be regulated epigenetically [77]. This is exemplified by the Special AT-rich sequence-binding protein 1 (SATB1), which recruits chromatin remodeling enzymes to regulate chromatin structure and gene expression. SATB1 downregulates the transcription of CD82, as well as other metastasis suppressor genes, through chromatin modification. RNAi -mediated knockdown of SATB1 in the highly aggressive breast cancer cell line MDA-MB-231 strikingly restores breast-like acinar polarity and inhibits tumor growth and metastasis in vivo. On the other hand, ectopic overexpression of SATB1 increases the metastatic potential of the relatively non-aggressive breast cancer cell line SKBR3 [77].
Post-translational Regulation
Besides transcription, the levels of CD82 can also be regulated by post-translational modifications. CD82 undergo modification of its N-linked glycans in the secretory pathways and is subject to endoplasmic reticulum quality control mechanisms [78]. The ubiquitin ligase gp78 plays an important role in ERAD [79]. RNAi-mediated knockdown of gp78 inhibits sarcoma metastasis but not primary tumor growth, an effect that mirrors overexpression of metastasis suppressors [8]. Loss of gp78 accumulates CD82 due to reduced degradation of the metastasis suppressor. Consistent with this idea, the metastatic activity of gp78 requires its ubiquitin ligase activity [8] but not its putative function as a G-protein coupled receptor for the human autocrine motility factor (AMF; phosphoglucose isomerase), which was proposed [80–81] prior to its characterization as a ubiquitin ligase involved in ERAD [82]. gp78 promotes the degradation of CD82, improving cell viability during metastasis. Furthermore, levels of CD82 correlate inversely with gp78 in a sample of sarcomas [8]. The study of metastasis suppressors has relied on their overexpression in cells that normally lack expression of the genes. Although a powerful approach, overexpression studies sometimes raise questions about the function of the endogenous protein. This study in sarcoma cells [8] provides direct evidence that raising the levels of endogenous CD82, by preventing it ubiquitination and proteasomal degradation, suppresses metastasis but does not affect primary tumor growth, confirming the metastasis suppressor function of CD82.
In a MMTV-gp78 transgenic mouse model, overexpression of gp78 in mammary tissue results in selective loss of CD82 and mammary gland hyperplasia [83], reinforcing the importance of post-translational regulation of CD82 levels in vivo. These studies in which gp78 levels are manipulated with resultant effects on CD82 levels establish an important role for post-translational regulation of metastasis suppressors and provide an example of how metastasis suppressor activity can be lost without loss of gene expression [8, 80]. The possibility of significant post-translational regulation of metastasis suppressor genes should also caution against drawing conclusions based on studies that focus simply on changes in gene transcription during metastasis.
Post-translational regulation of CD82 also plays a role in regulating trophoblast invasion. Decidual cells show strong expression of CD82 [63] and silencing of CD82 increases trophoblast invasion [84]. In decidualizing endometrial stromal cells, CD82 is induced by cAMP. Interestingly, induction of CD82 by cAMP analog results not from increased gene transcription but rather increased CD82 protein stability. In contrast, the cAMP analog 8-bromo-cAMP induces accelerated degradation of CD82 in breast cancer cells [63]. Understanding the changes in post-translational regulation of CD82 in cancer cells may lead to new targets for activating CD82 in advanced stages of the disease.
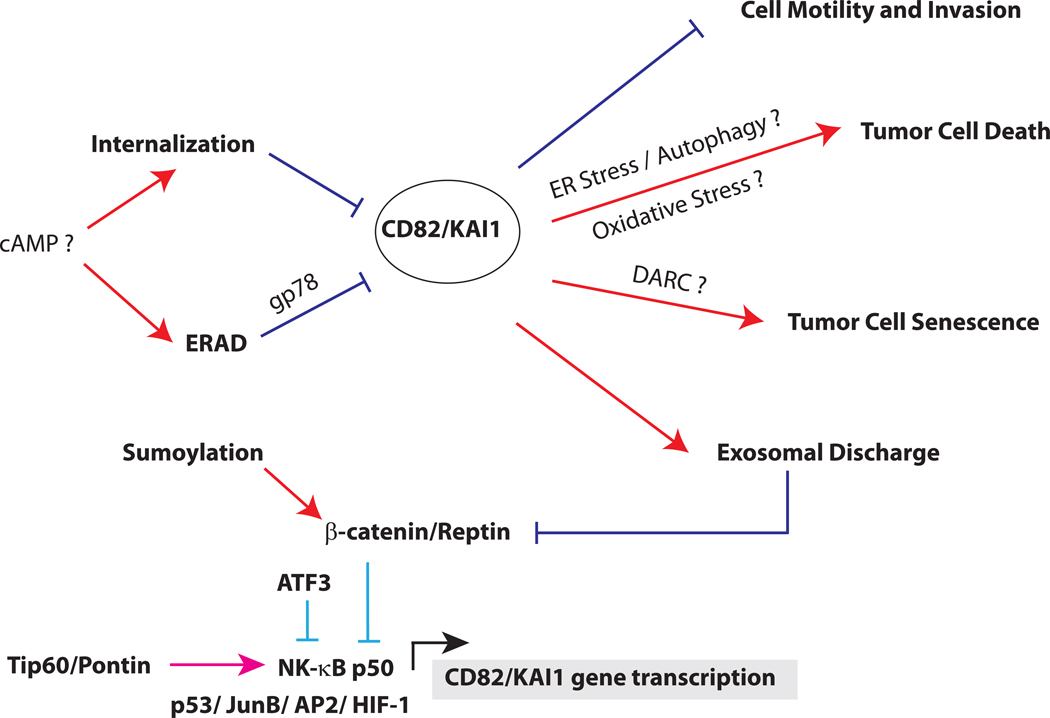
Besides ubiquitination, CD82 is also regulated by palmitoylation and glycosylation, both of which are required for CD82 function [15,31]. It is tempting to speculate that loss of these post-translational modifications may directly, or indirectly promote CD82 clearance by the cellular quality control systems and thereby inactivate the metastasis suppressor functions of CD82. A schematic of CD82 regulation and functions in metastasis suppression is summarized in Figure 2.
Figure 2. Schematic of CD82 regulation and functions.
CD82 can be transcriptionally activated by p53, JunB, AP2, NF-κB p50 and HIF-1 (below). CD82 expression by p50 is regulated by the alternative recruitment of Tip60/Pontin or β-catenin/Reptin complex. In normal cells, CD82 transcription is repressed by a NCOR/TAB2/HDAC3 complex (not shown), which is displaced by a Tip60/Pontin co-activator complex for p50 in the presence of cytokines. In cancer cells, Tip60 is downregulated and replaced with a β-catenin/Reptin complex, which represses CD82 transcription. Sumoylation of Reptin is required for its repressor function, which can be inactivated by de-sumoylation. ATF3 can also repress p50 induction of CD82. After translation (above), CD82 protein is regulated by quality control mechanisms in the endoplasmic reticulum. This process, known as ER-associated degradation (ERAD), depends on the ubiquitin ligase gp78. In some cancer cells, cAMP promotes CD82 degradation either by enhancing its internalization or accelerating its degradation from the ER. CD82 inhibits metastasis by multiple mechanisms, including inhibiting cell motility and invasion, promoting apoptosis, inducing tumor cell senescence and enhancing exosomal discharge of β-catenin.
Conclusion
CD82 suppresses metastasis at multiple steps of the metastatic cascade. CD82 does not appear to have intrinsic enzymatic activity; however, it has become clear in recent years that CD82 has an impact on multiple signaling pathways by regulating the trafficking of partner proteins and by influencing cell membrane dynamics. Although the role of CD82 on cell surface dynamics has been studied extensively, recent studies suggest that CD82 may play additional metastasis suppressor functions in the secretory pathway. Despite these advances, much remains to be understood about how CD82 functions at a molecular level and how it suppresses metastasis. Most studies to date have focused on the effect of overexpressing CD82 on specific cell lines, measuring changes in properties such as cell motility, adhesion or senescence in vitro. Whether these effects are important in vivo remain to be determined. Furthermore, it is not clear that results obtained in specific cell lines can be generalized to other malignancies. Indeed it is possible that CD82 plays distinct roles in different cell types, possibly explaining some of the contradictory findings in the field. As we gain a better understanding of CD82 function in normal and cancer cells, some of these issues would hopefully be resolved. For efficient targeting of CD82 in therapy, it is important to learn about the mechanisms by which CD82 changes during cancer progression, which could lead to novel methods of activating the metastasis suppressor. Besides transcriptional changes, it is now clear that CD82 is regulated by post-translational mechanisms. Changes in post-transcriptional regulation of CD82 in metastatic cells may well play a critical role in disease progression. It is also critical to investigate the effects of activating CD82 during different stages of cancer progression. This can be studied by regulating the expression of CD82 in animal models. Such experiments should also further clarify the precise steps in metastasis regulated by CD82. Finally, the relationship of CD82 to other metastasis suppressor has not been studied. If signaling from CD82 overlaps with those of other metastasis suppressors, novel targets can be identified that provide a common node for treating metastasis.
ACKNOWLEDGEMENTS
Research in the authors’ laboratory is supported by the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barrett JC. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268:884–886. doi: 10.1126/science.7754374. [DOI] [PubMed] [Google Scholar]
- 2.Lebel-Binay S, Lagaudriere C, Fradelizi D, Conjeaud H. CD82, member of the tetra-span-transmembrane protein family, is a costimulatory protein for T cell activation. J Immunol. 1995;155:101–110. [PubMed] [Google Scholar]
- 3.Nojima Y, et al. The 4F9 antigen is a member of the tetra spans transmembrane protein family and functions as an accessory molecule in T cell activation and adhesion. Cell Immunol. 1993;152:249–260. doi: 10.1006/cimm.1993.1285. [DOI] [PubMed] [Google Scholar]
- 4.Fukudome K, Furuse M, Imai T, Nishimura M, Takagi S, Hinuma Y, Yoshie O. Identification of membrane antigen C33 recognized by monoclonal antibodies inhibitory to human T-cell leukemia virus type 1 (HTLV-1)-induced syncytium formation: altered glycosylation of C33 antigen in HTLV-1-positive T cells. J Virol. 1992;66:1394–1401. doi: 10.1128/jvi.66.3.1394-1401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebel-Binay S, et al. Further characterization of CD82/IA4 antigen (type III surface protein): an activation/differentiation marker of mononuclear cells. Cell Immunol. 1994;154:468–483. doi: 10.1006/cimm.1994.1092. [DOI] [PubMed] [Google Scholar]
- 6.Imai T, Yoshie O. C33 antigen and M38 antigen recognized by monoclonal antibodies inhibitory to syncytium formation by human T cell leukemia virus type 1 are both members of the transmembrane 4 superfamily and associate with each other and with CD4 or CD8 in T cells. J Immunol. 1993;151:6470–6481. [PubMed] [Google Scholar]
- 7.Bandyopadhyay S, et al. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat Med. 2006;12:933–938. doi: 10.1038/nm1444. [DOI] [PubMed] [Google Scholar]
- 8.Tsai YC, et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 9.Xu JH, Guo XZ, Ren LN, Shao LC, Liu MP. KAI1 is a potential target for anti-metastasis in pancreatic cancer cells. World J Gastroenterol. 2008;14:1126–1132. doi: 10.3748/wjg.14.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang JM, Peng ZH, Si SH, Liu WW, Luo YH, Ye ZY. KAI1 gene suppresses invasion and metastasis of hepatocellular carcinoma MHCC97-H cells in vitro and in animal models. Liver Int. 2008;28:132–139. doi: 10.1111/j.1478-3231.2007.01620.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Wei LL, Tang C, Slack R, Mueller S, Lippman ME. Overexpression of KAI1 suppresses in vitro invasiveness and in vivo metastasis in breast cancer cells. Cancer Res. 2001;61:5284–5288. [PubMed] [Google Scholar]
- 12.Tonoli H, Barrett JC. CD82 metastasis suppressor gene: a potential target for new therapeutics? Trends Mol Med. 2005;11:563–570. doi: 10.1016/j.molmed.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys J. 2006;90:212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bari R, Zhang YH, Zhang F, Wang NX, Stipp CS, Zheng JJ, Zhang XA. Transmembrane interactions are needed for KAI1/CD82-mediated suppression of cancer invasion and metastasis. Am J Pathol. 2009;174:647–660. doi: 10.2353/ajpath.2009.080685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono M, Handa K, Withers DA, Hakomori S. Motility inhibition and apoptosis are induced by metastasis-suppressing gene product CD82 and its analogue CD9, with concurrent glycosylation. Cancer Res. 1999;59:2335–2339. [PubMed] [Google Scholar]
- 16.Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono M, Handa K, Withers DA, Hakomori S. Glycosylation effect on membrane domain (GEM) involved in cell adhesion and motility: a preliminary note on functional alpha3, alpha5-CD82 glycosylation complex in ldlD 14 cells. Biochem Biophys Res Commun. 2000;279:744–750. doi: 10.1006/bbrc.2000.4030. [DOI] [PubMed] [Google Scholar]
- 18.Scholz CJ, Sauer G, Deissler H. Glycosylation of tetraspanin Tspan-1 at four distinct sites promotes its transition through the endoplasmic reticulum. Protein Pept Lett. 2009;16:1244–1248. doi: 10.2174/092986609789071234. [DOI] [PubMed] [Google Scholar]
- 19.Jee B, Jin K, Hahn JH, Song HG, Lee H. Metastasis-suppressor KAI1/CD82 induces homotypic aggregation of human prostate cancer cells through Src-dependent pathway. Exp Mol Med. 2003;35:30–37. doi: 10.1038/emm.2003.5. [DOI] [PubMed] [Google Scholar]
- 20.Abe M, Sugiura T, Takahashi M, Ishii K, Shimoda M, Shirasuna K. A novel function of CD82/KAI-1 on E-cadherin-mediated homophilic cellular adhesion of cancer cells. Cancer Lett. 2008;266:163–170. doi: 10.1016/j.canlet.2008.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Takaoka A, Hinoda Y, Sato S, Itoh F, Adachi M, Hareyama M, Imai K. Reduced invasive and metastatic potentials of KAI1-transfected melanoma cells. Jpn J Cancer Res. 1998;89:397–404. doi: 10.1111/j.1349-7006.1998.tb00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takaoka A, Hinoda Y, Satoh S, Adachi Y, Itoh F, Adachi M, Imai K. Suppression of invasive properties of colon cancer cells by a metastasis suppressor KAI1 gene. Oncogene. 1998;16:1443–1453. doi: 10.1038/sj.onc.1201648. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XA, He B, Zhou B, Liu L. Requirement of the p130CAS-Crk coupling for metastasis suppressor KAI1/CD82-mediated inhibition of cell migration. J Biol Chem. 2003;278:27319–27328. doi: 10.1074/jbc.M303039200. [DOI] [PubMed] [Google Scholar]
- 24.Jee BK, Lee JY, Lim Y, Lee KH, Jo YH. Effect of KAI1/CD82 on the beta1 integrin maturation in highly migratory carcinoma cells. Biochem Biophys Res Commun. 2007;359:703–708. doi: 10.1016/j.bbrc.2007.05.159. [DOI] [PubMed] [Google Scholar]
- 25.Jee BK, Park KM, Surendran S, Lee WK, Han CW, Kim YS, Lim Y. KAI1/CD82 suppresses tumor invasion by MMP9 inactivation via TIMP1 up-regulation in the H1299 human lung carcinoma cell line. Biochem Biophys Res Commun. 2006;342:655–661. doi: 10.1016/j.bbrc.2006.01.153. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Park SR, Chay KO, Seo YW, Kook H, Ahn KY, Kim YJ, Kim KK. KAI1 COOH-terminal interacting tetraspanin (KITENIN), a member of the tetraspanin family, interacts with KAI1, a tumor metastasis suppressor, and enhances metastasis of cancer. Cancer Res. 2004;64:4235–4243. doi: 10.1158/0008-5472.CAN-04-0275. [DOI] [PubMed] [Google Scholar]
- 27.Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2-tetraspanin CD82 complex inhibits met and its cross-talk with integrins, providing a basis for control of cell motility through glycosynapse. J Biol Chem. 2007;282:8123–8133. doi: 10.1074/jbc.M611407200. [DOI] [PubMed] [Google Scholar]
- 28.He B, Liu L, Cook GA, Grgurevich S, Jennings LK, Zhang XA. Tetraspanin CD82 attenuates cellular morphogenesis through down-regulating integrin alpha6-mediated cell adhesion. J Biol Chem. 2005;280:3346–3354. doi: 10.1074/jbc.M406680200. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003;63:2665–2674. [PubMed] [Google Scholar]
- 30.Klemke RL, Leng J, Molander R, Brooks PC, Vuori K, Cheresh DA. CAS/Crk coupling serves as a "molecular switch" for induction of cell migration. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 32.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 2000;10:1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- 33.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 34.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 35.Odintsova E, Voortman J, Gilbert E, Berditchevski F. Tetraspanin CD82 regulates compartmentalisation and ligand-induced dimerization of EGFR. J Cell Sci. 2003;116:4557–4566. doi: 10.1242/jcs.00793. [DOI] [PubMed] [Google Scholar]
- 36.Wang XQ, Yan Q, Sun P, Liu JW, Go L, McDaniel SM, Paller AS. Suppression of epidermal growth factor receptor signaling by protein kinase C-alpha activation requires CD82, caveolin-1, and ganglioside. Cancer Res. 2007;67:9986–9995. doi: 10.1158/0008-5472.CAN-07-1300. [DOI] [PubMed] [Google Scholar]
- 37.Sridhar SC, Miranti CK. Tetraspanin KAI1/CD82 suppresses invasion by inhibiting integrin-dependent crosstalk with c-Met receptor and Src kinases. Oncogene. 2006;25:2367–2378. doi: 10.1038/sj.onc.1209269. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi M, Sugiura T, Abe M, Ishii K, Shirasuna K. Regulation of c-Met signaling by the tetraspanin KAI-1/CD82 affects cancer cell migration. Int J Cancer. 2007;121:1919–1929. doi: 10.1002/ijc.22887. [DOI] [PubMed] [Google Scholar]
- 39.Kalabis J, Rosenberg I, Podolsky DK. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J Biol Chem. 2006;281:6434–6441. doi: 10.1074/jbc.M512905200. [DOI] [PubMed] [Google Scholar]
- 40.Bass R, Werner F, Odintsova E, Sugiura T, Berditchevski F, Ellis V. Regulation of urokinase receptor proteolytic function by the tetraspanin CD82. J Biol Chem. 2005;280:14811–14818. doi: 10.1074/jbc.M414189200. [DOI] [PubMed] [Google Scholar]
- 41.Hu CC, Liang FX, Zhou G, Tu L, Tang CH, Zhou J, Kreibich G, Sun TT. Assembly of urothelial plaques: tetraspanin function in membrane protein trafficking. Mol Biol Cell. 2005;16:3937–3950. doi: 10.1091/mbc.E05-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai YC, Weissman AM. The Unfolded Protein Response, Degradation from Endoplasmic Reticulum and Cancer. Genes Cancer. 2010;1:764–778. doi: 10.1177/1947601910383011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu CY, et al. Overexpression of KAI1 induces autophagy and increases MiaPaCa-2 cell survival through the phosphorylation of extracellular signal-regulated kinases. Biochem Biophys Res Commun. 404:802–808. doi: 10.1016/j.bbrc.2010.12.063. [DOI] [PubMed] [Google Scholar]
- 44.Schoenfeld N, Bauer MK, Grimm S. The metastasis suppressor gene C33/CD82/KAI1 induces apoptosis through reactive oxygen intermediates. FASEB J. 2004;18:158–160. doi: 10.1096/fj.03-0420fje. [DOI] [PubMed] [Google Scholar]
- 45.Zismanov V, Lishner M, Tartakover-Matalon S, Radnay J, Shapiro H, Drucker L. Tetraspanin-induced death of myeloma cell lines is autophagic and involves increased UPR signalling. Br J Cancer. 2009;101:1402–1409. doi: 10.1038/sj.bjc.6605291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammond CL, Marchan R, Krance SM, Ballatori N. Glutathione export during apoptosis requires functional multidrug resistance-associated proteins. J Biol Chem. 2007;282:14337–14347. doi: 10.1074/jbc.M611019200. [DOI] [PubMed] [Google Scholar]
- 47.Tohami T, Drucker L, Shapiro H, Radnay J, Lishner M. Overexpression of tetraspanins affects multiple myeloma cell survival and invasive potential. FASEB J. 2007;21:691–699. doi: 10.1096/fj.06-6610com. [DOI] [PubMed] [Google Scholar]
- 48.Shen H, Schuster R, Stringer KF, Waltz SE, Lentsch AB. The Duffy antigen/receptor for chemokines (DARC) regulates prostate tumor growth. FASEB J. 2006;20:59–64. doi: 10.1096/fj.05-4764com. [DOI] [PubMed] [Google Scholar]
- 49.Xu L, Ashkenazi A, Chaudhuri A. Duffy antigen/receptor for chemokines (DARC) attenuates angiogenesis by causing senescence in endothelial cells. Angiogenesis. 2007;10:307–318. doi: 10.1007/s10456-007-9084-y. [DOI] [PubMed] [Google Scholar]
- 50.Elson JK, Beebe-Dimmer JL, Morgenstern H, Chilkuri M, Blanchard J, Lentsch AB. The Duffy Antigen/Receptor for Chemokines (DARC) and prostate-cancer risk among Jamaican men. J Immigr Minor Health. 2011;13:36–41. doi: 10.1007/s10903-010-9330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 52.Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JH, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 54.Tagawa K, Arihiro K, Takeshima Y, Hiyama E, Yamasaki M, Inai K. Down-regulation of KAI1 messenger RNA expression is not associated with loss of heterozygosity of the KAI1 gene region in lung adenocarcinoma. Jpn J Cancer Res. 1999;90:970–976. doi: 10.1111/j.1349-7006.1999.tb00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong JT, Suzuki H, Pin SS, Bova GS, Schalken JA, Isaacs WB, Barrett JC, Isaacs JT. Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res. 1996;56:4387–4390. [PubMed] [Google Scholar]
- 56.Liu FS, Dong JT, Chen JT, Hsieh YT, Ho ES, Hung MJ. Frequent down-regulation and lack of mutation of the KAI1 metastasis suppressor gene in epithelial ovarian carcinoma. Gynecol Oncol. 2000;78:10–15. doi: 10.1006/gyno.2000.5801. [DOI] [PubMed] [Google Scholar]
- 57.Miyazaki T, et al. Mutation and expression of the metastasis suppressor gene KAI1 in esophageal squamous cell carcinoma. Cancer. 2000;89:955–962. doi: 10.1002/1097-0142(20000901)89:5<955::aid-cncr3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 58.Jackson P, Millar D, Kingsley E, Yardley G, Ow K, Clark S, Russell PJ. Methylation of a CpG island within the promoter region of the KAI1 metastasis suppressor gene is not responsible for down-regulation of KAI1 expression in invasive cancers or cancer cell lines. Cancer Lett. 2000;157:169–176. doi: 10.1016/s0304-3835(00)00483-3. [DOI] [PubMed] [Google Scholar]
- 59.Gao AC, Lou W, Dong JT, Barrett JC, Danielpour D, Isaacs JT. Defining regulatory elements in the human KAI1 (CD 82) metastasis suppressor gene. Prostate. 2003;57:256–260. doi: 10.1002/pros.10309. [DOI] [PubMed] [Google Scholar]
- 60.Marreiros A, Dudgeon K, Dao V, Grimm MO, Czolij R, Crossley M, Jackson P. KAI1 promoter activity is dependent on p53, junB and AP2: evidence for a possible mechanism underlying loss of KAI1 expression in cancer cells. Oncogene. 2005;24:637–649. doi: 10.1038/sj.onc.1208216. [DOI] [PubMed] [Google Scholar]
- 61.Mashimo T, Bandyopadhyay S, Goodarzi G, Watabe M, Pai SK, Gross SC, Watabe K. Activation of the tumor metastasis suppressor gene, KAI1, by etoposide is mediated by p53 and c-Jun genes. Biochem Biophys Res Commun. 2000;274:370–376. doi: 10.1006/bbrc.2000.3139. [DOI] [PubMed] [Google Scholar]
- 62.Mashimo T, Watabe M, Hirota S, Hosobe S, Miura K, Tegtmeyer PJ, Rinker-Shaeffer CW, Watabe K. The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci U S A. 1998;95:11307–11311. doi: 10.1073/pnas.95.19.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gellersen B, et al. Expression of the metastasis suppressor KAI1 in decidual cells at the human maternal-fetal interface: Regulation and functional implications. Am J Pathol. 2007;170:126–139. doi: 10.2353/ajpath.2007.060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sigala S, Faraoni I, Botticini D, Paez-Pereda M, Missale C, Bonmassar E, Spano P. Suppression of telomerase, reexpression of KAI1, and abrogation of tumorigenicity by nerve growth factor in prostate cancer cell lines. Clin Cancer Res. 1999;5:1211–1218. [PubMed] [Google Scholar]
- 65.Akita H, Iizuka A, Hashimoto Y, Kohri K, Ikeda K, Nakanishi M. Induction of KAI-1 expression in metastatic cancer cells by phorbol esters. Cancer Lett. 2000;153:79–83. doi: 10.1016/s0304-3835(00)00352-9. [DOI] [PubMed] [Google Scholar]
- 66.Jackson P, Grimm MO, Kingsley EA, Brosius U, Antalis T, Yardley G, Russell PJ. Relationship between expression of KAI1 metastasis suppressor gene, mRNA levels and p53 in human bladder and prostate cancer cell lines. Urol Oncol. 2002;7:99–104. doi: 10.1016/s1078-1439(01)00175-2. [DOI] [PubMed] [Google Scholar]
- 67.Jackson P, Ow K, Yardley G, Delprado W, Quinn DI, Yang JL, Russell PJ. Downregulation of KAI1 mRNA in localised prostate cancer and its bony metastases does not correlate with p53 overexpression. Prostate Cancer Prostatic Dis. 2003;6:174–181. doi: 10.1038/sj.pcan.4500634. [DOI] [PubMed] [Google Scholar]
- 68.Marreiros A, Czolij R, Yardley G, Crossley M, Jackson P. Identification of regulatory regions within the KAI1 promoter: a role for binding of AP1, AP2 and p53. Gene. 2003;302:155–164. doi: 10.1016/s0378-1119(02)01101-0. [DOI] [PubMed] [Google Scholar]
- 69.Liu W, et al. KAI1 Gene Is Engaged in NDRG1 Gene-mediated Metastasis Suppression through the ATF3-NF{kappa}B Complex in Human Prostate Cancer. J Biol Chem. 286:18949–18959. doi: 10.1074/jbc.M111.232637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duriez C, Falette N, Cortes U, Moyret-Lalle C, Puisieux A. Absence of p53-dependent induction of the metastatic suppressor KAI1 gene after DNA damage. Oncogene. 2000;19:2461–2464. doi: 10.1038/sj.onc.1203580. [DOI] [PubMed] [Google Scholar]
- 71.Wu Q, et al. Role of tumor metastasis suppressor gene KAI1 in digestive tract carcinomas and cancer cells. Cell Tissue Res. 2003;341:237–249. doi: 10.1007/s00441-003-0781-6. [DOI] [PubMed] [Google Scholar]
- 72.Lebel-Binay S, Lagaudriere C, Fradelizi D, Conjeaud H. CD82, tetra-span-transmembrane protein, is a regulated transducing molecule on U937 monocytic cell line. J Leukoc Biol. 1995;57:956–963. doi: 10.1002/jlb.57.6.956. [DOI] [PubMed] [Google Scholar]
- 73.Kim JH, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 74.Kim B, et al. Identification of the KAI1 metastasis suppressor gene as a hypoxia target gene. Biochem Biophys Res Commun. 393:179–184. doi: 10.1016/j.bbrc.2010.01.118. [DOI] [PubMed] [Google Scholar]
- 75.Salnikow K, Davidson T, Zhang Q, Chen LC, Su W, Costa M. The involvement of hypoxia-inducible transcription factor-1-dependent pathway in nickel carcinogenesis. Cancer Res. 2003;63:3524–3530. [PubMed] [Google Scholar]
- 76.Bandyopadhyay S, et al. The tumor metastasis suppressor gene Drg-1 down-regulates the expression of activating transcription factor 3 in prostate cancer. Cancer Res. 2006;66:11983–11990. doi: 10.1158/0008-5472.CAN-06-0943. [DOI] [PubMed] [Google Scholar]
- 77.Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 78.Cannon KS, Cresswell P. Quality control of transmembrane domain assembly in the tetraspanin CD82. EMBO J. 2001;20:2443–2453. doi: 10.1093/emboj/20.10.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shimizu K, et al. The autocrine motility factor receptor gene encodes a novel type of seven transmembrane protein. FEBS Lett. 1999;456:295–300. doi: 10.1016/s0014-5793(99)00966-7. [DOI] [PubMed] [Google Scholar]
- 81.Watanabe H, Carmi P, Hogan V, Raz T, Silletti S, Nabi IR, Raz A. Purification of human tumor cell autocrine motility factor and molecular cloning of its receptor. J Biol Chem. 1991;266:13442–13448. [PubMed] [Google Scholar]
- 82.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Joshi B, Li L, Nabi IR. A role for KAI1 in promotion of cell proliferation and mammary gland hyperplasia by the gp78 ubiquitin ligase. J Biol Chem. 285:8830–8839. doi: 10.1074/jbc.M109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li MQ, Hou XF, Lv SJ, Meng YH, Wang XQ, Tang CL, Li DJ. CD82 gene suppression in endometrial stromal cells leads to increase of the cell invasiveness in the endometriotic milieu. J Mol Endocrinol. 2011 doi: 10.1530/JME-10-0165. [DOI] [PubMed] [Google Scholar]