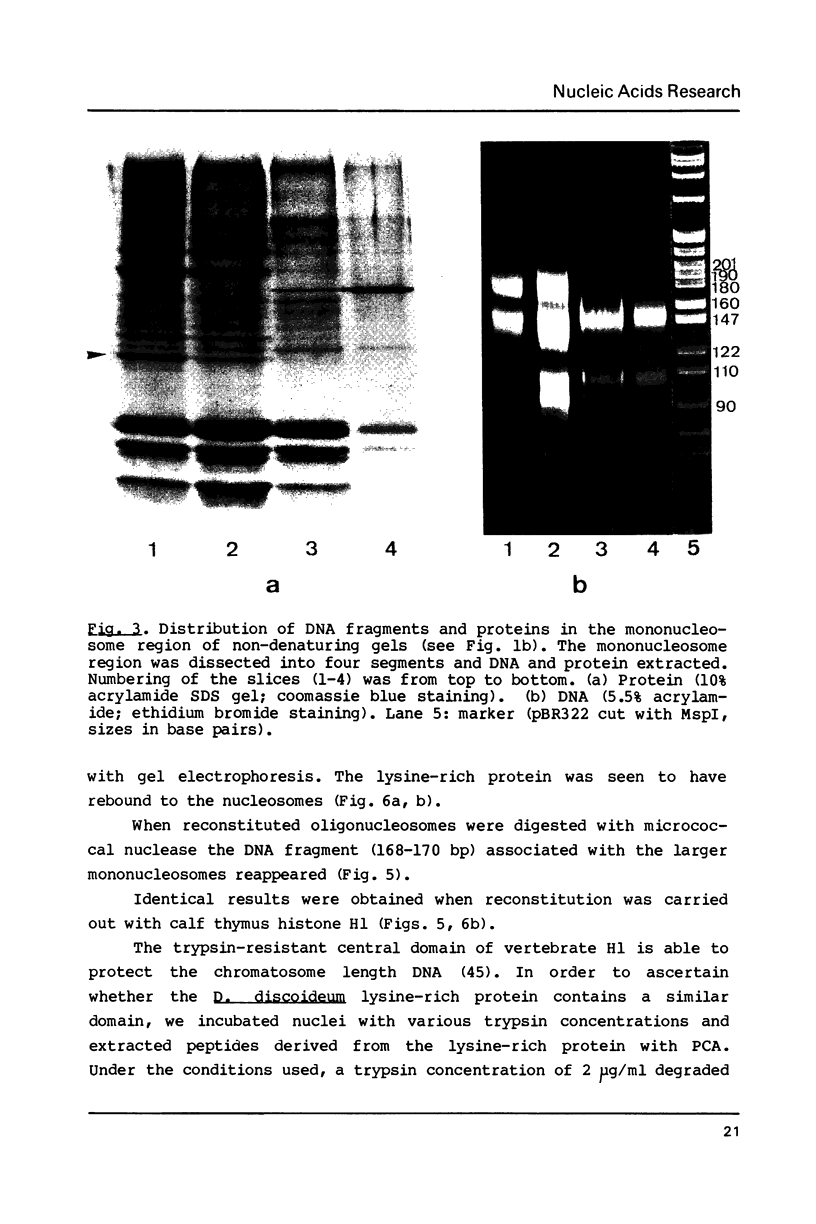
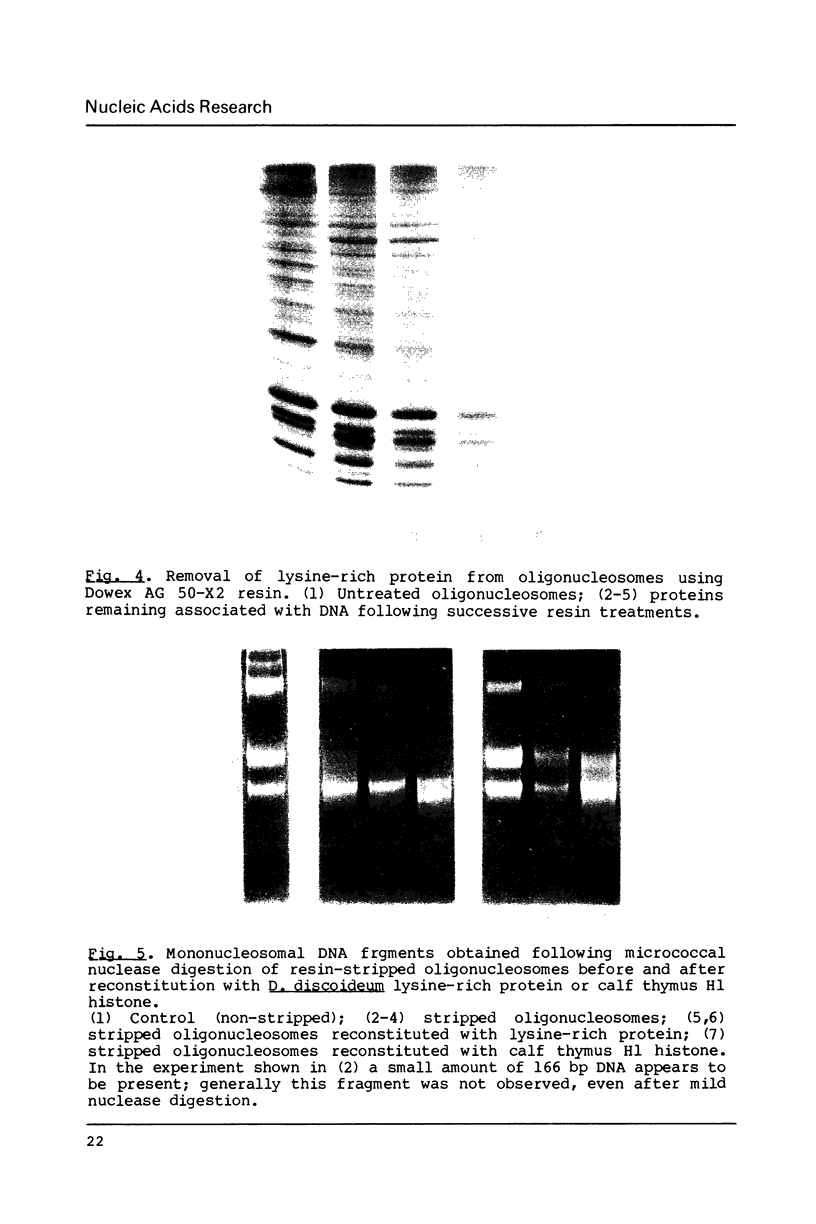
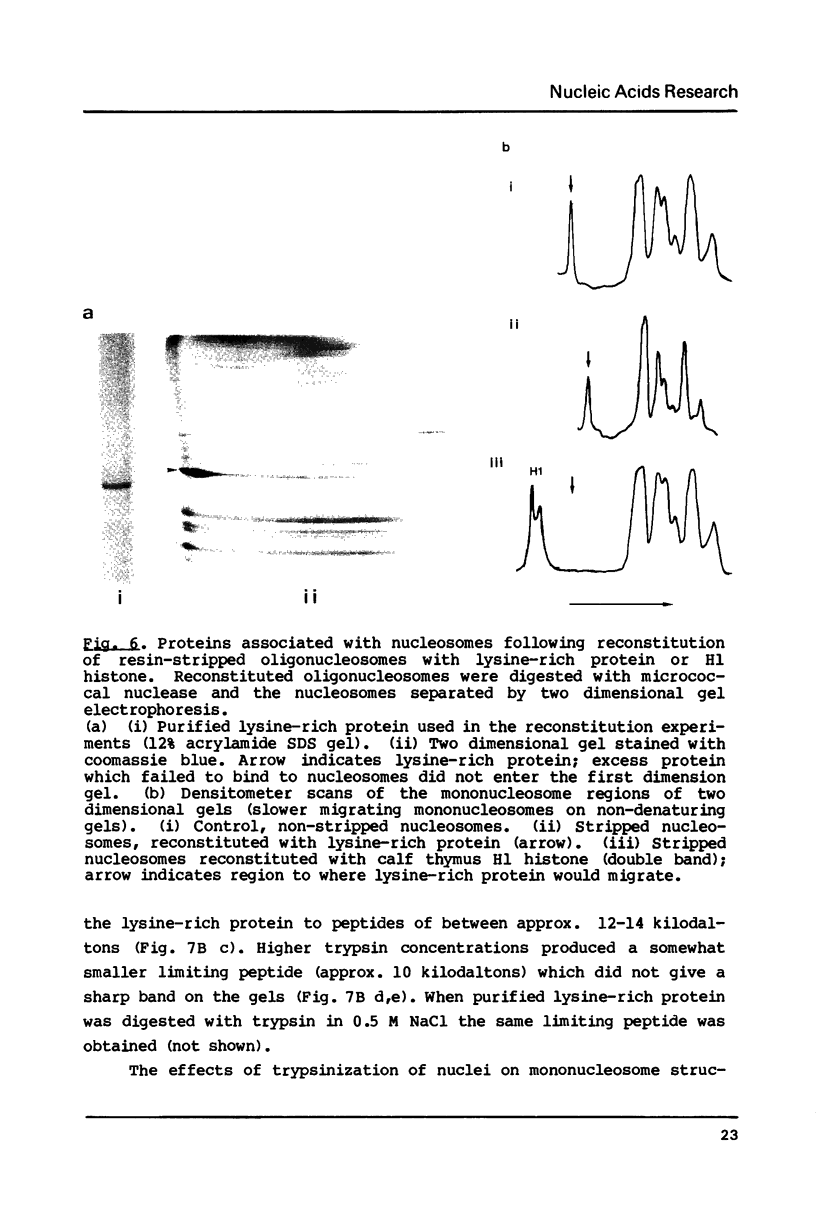
Abstract
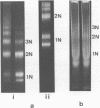
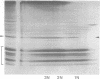
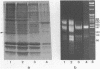
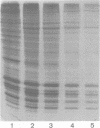
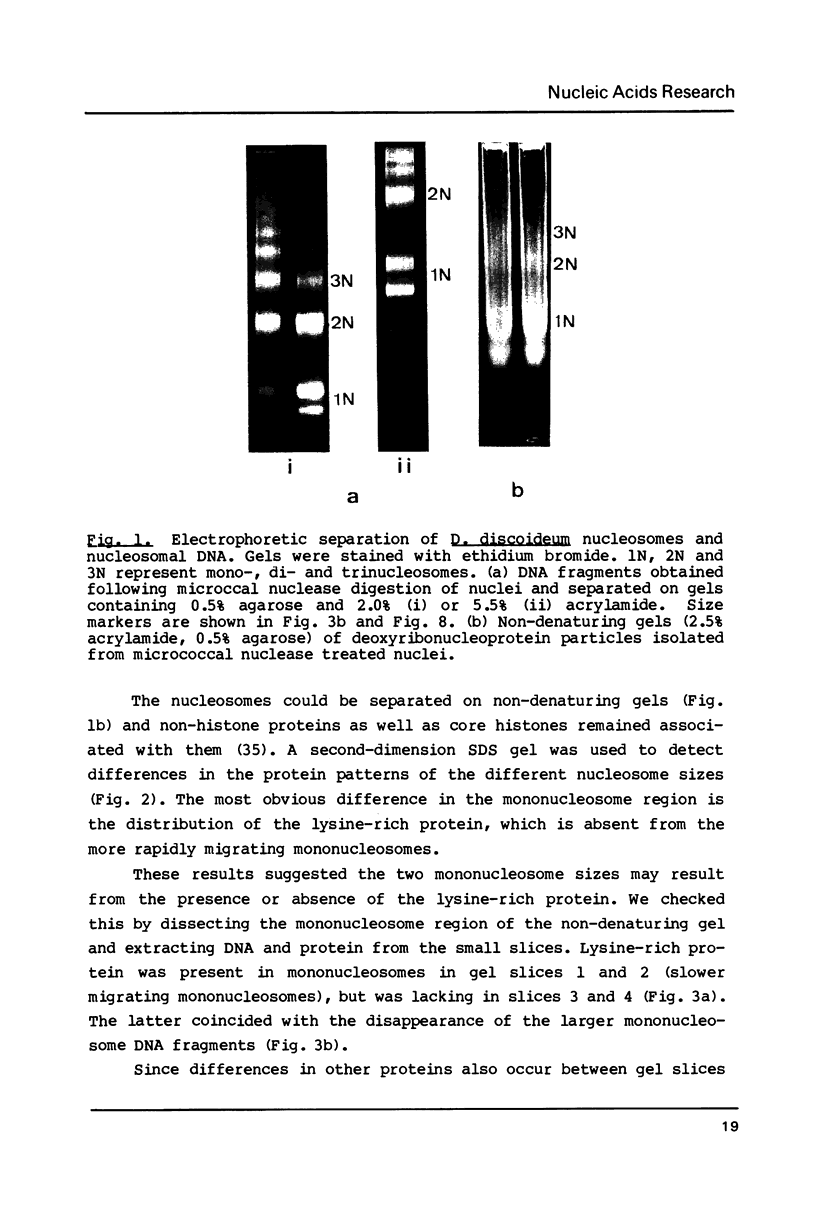
Mononucleosomes released from Dictyostelium discoideum chromatin by micrococcal nuclease contained two distinctive DNA sizes (166-180 and 146 bp). Two dimensional gel electrophoresis suggested a lysine-rich protein protected the larger mononucleosomes from nuclease digestion. This was confirmed by stripping the protein from chromatin with Dowex resin. Subsequently, only the 146 bp mononucleosome was produced by nuclease digestion. Reconstitution of the stripped chromatin with the purified lysine-rich protein resulted in the reappearance of the larger mononucleosomes. Two-dimensional gel electrophoresis showed the protein was associated with mononucleosomes. Hence, the protein functions as an H1 histone in bringing the two DNA strands together at their exit point from the nucleosome. Trypsin digestion of the lysine-rich protein in nuclei resulted in a limiting peptide of approx. 10 kilodaltons. Trypsin concentrations which degraded the protein to peptides of 12-14 kilodaltons and partially degraded the core histones did not change the DNA digestion patterns obtained with micrococcal nuclease. Thus, the trypsin-resistant domain of the lysine-rich protein is able to maintain chromatosome structure.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Bakke A. C., Wu J. R., Bonner J. Chromatin structure in the cellular slime mold Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1978 Feb;75(2):705–709. doi: 10.1073/pnas.75.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Thomas J. O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981 Nov 25;9(22):5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyavsky A. V., Bavykin S. G., Goguadze E. G., Mirzabekov A. D. Primary organization of nucleosomes containing all five histones and DNA 175 and 165 base-pairs long. J Mol Biol. 1980 May 25;139(3):519–536. doi: 10.1016/0022-2836(80)90144-8. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Martinson H. G. The roles of H1, the histone core and DNA length in the unfolding of nucleosomes at low ionic strength. Nucleic Acids Res. 1980 Nov 11;8(21):4969–4987. doi: 10.1093/nar/8.21.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Regions of high and low cationic charge in a lysine-rich histone. J Biol Chem. 1970 Mar 25;245(6):1458–1466. [PubMed] [Google Scholar]
- Böhm L., Briand G., Sautière P., Crane-Robinson C. Proteolytic digestion studies of chromatin core-histone structure. Identification of limit peptides from histone H2B. Eur J Biochem. 1982 Apr 1;123(2):299–303. doi: 10.1111/j.1432-1033.1982.tb19767.x. [DOI] [PubMed] [Google Scholar]
- Böhm L., Briand G., Sautière P., Crane-Robinson C. Proteolytic digestion studies of chromatin core-histone structure. Identification of the limit peptides of histones H3 and H4. Eur J Biochem. 1981 Sep;119(1):67–74. doi: 10.1111/j.1432-1033.1981.tb05577.x. [DOI] [PubMed] [Google Scholar]
- Böhm L., Crane-Robinson C., Sautière P. Proteolytic digestion studies of chromatin core-histone structure. Identification of a limit peptide of histone H2A. Eur J Biochem. 1980 May;106(2):525–530. doi: 10.1111/j.1432-1033.1980.tb04599.x. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Charlesworth M. C., Parish R. W. Further studies on basic nucleoproteins from the cellular slime mold Dictyostelium discoideum. Eur J Biochem. 1977 May 2;75(1):241–250. doi: 10.1111/j.1432-1033.1977.tb11523.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth M. C., Parish R. W. The isolation of nuclei and basic nucleoproteins from the cellular slime mold Dictyostelium discoideum. Eur J Biochem. 1975 May;54(1):307–316. doi: 10.1111/j.1432-1033.1975.tb04141.x. [DOI] [PubMed] [Google Scholar]
- Cruickshank H. M., Walker I. O. Cell-cycle-dependent dissociation of histone H1 from chromatin in nuclei of P. polycephalum. Nucleic Acids Res. 1981 Aug 11;9(15):3873–3885. doi: 10.1093/nar/9.15.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté S., Nadeau P., Neelin J. M., Pallotta D. Isolation and characterization of histones and other acid-soluble chromosomal proteins from Physarum polycephalum. Can J Biochem. 1982 Mar;60(3):263–271. doi: 10.1139/o82-031. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Laemmli U. K. Cell cycle changes in Physarum polycephalum histone H1 phosphate: relationship to deoxyribonucleic acid binding and chromosome condensation. Biochemistry. 1980 May 13;19(10):2240–2246. doi: 10.1021/bi00551a038. [DOI] [PubMed] [Google Scholar]
- Goff C. G. Histones of Neurospora crassa. J Biol Chem. 1976 Jul 10;251(13):4131–4138. [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H., Johns E. W. Microheterogeneity in a non-histone chromosomal protein. FEBS Lett. 1976 May 1;64(2):412–414. doi: 10.1016/0014-5793(76)80339-0. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Ohba Y., Yasuda H., Yamada M. The interaction of H1 histone with nucleosome core. J Biochem. 1981 Jun;89(6):1881–1888. doi: 10.1093/oxfordjournals.jbchem.a133390. [DOI] [PubMed] [Google Scholar]
- Ishimi Y., Yasuda H., Ohba Y., Yamada M. The reassociation with chromatin of H1 fragments bisected with thrombin. J Biol Chem. 1981 Aug 25;256(16):8249–8251. [PubMed] [Google Scholar]
- Klingholz R., Straätling W. H. Digestion of chromatin to H1-depleted 166 basepair particles by Ca2+/Mg2+-dependent endonuclease. FEBS Lett. 1982 Mar 8;139(1):105–108. doi: 10.1016/0014-5793(82)80497-3. [DOI] [PubMed] [Google Scholar]
- Klingholz R., Strätling W. H. Reassociation of histone H1 to H1-depleted polynucleosomes. J Biol Chem. 1982 Nov 10;257(21):13101–13107. [PubMed] [Google Scholar]
- Labhart P., Banz E., Ness P. J., Parish R. W., Koller T. A structural concept for nucleoli of Dictyostelium discoideum deduced from dissociation studies. Chromosoma. 1984;89(2):111–120. doi: 10.1007/BF00292894. [DOI] [PubMed] [Google Scholar]
- Labhart P., Ness P., Banz E., Parish R., Koller T. Model for the structure of the active nucleolar chromatin. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):557–564. doi: 10.1101/sqb.1983.047.01.065. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. The nucleotide and amino acid coding sequence of a gene for H1 histone that interacts with euchromatin. The early embryonic H1 gene of the sea urchin Strongylocentrotus purpuratus. J Biol Chem. 1982 Aug 25;257(16):9438–9443. [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Macleod A. R., Wong N. C., Dixon G. H. The amino-acid sequence of trout-testis histone H1. Eur J Biochem. 1977 Aug 15;78(1):281–291. doi: 10.1111/j.1432-1033.1977.tb11739.x. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Orientation of the nucleosome within the higher order structure of chromatin. Cell. 1980 Nov;22(1 Pt 1):87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- Mirzabekov A. D. Nucleosomes structure and its dynamic transitions. Q Rev Biophys. 1980 May;13(2):255–295. doi: 10.1017/s0033583500001670. [DOI] [PubMed] [Google Scholar]
- Mohberg J., Rusch H. P. Isolation of the nuclear histones from the Myxomycete, Physarum polycephalum. Arch Biochem Biophys. 1969 Nov;134(2):577–589. doi: 10.1016/0003-9861(69)90320-8. [DOI] [PubMed] [Google Scholar]
- Ness P. J., Labhart P., Banz E., Koller T., Parish R. W. Chromatin structure along the ribosomal DNA of Dictyostelium. Regional differences and changes accompanying cell differentiation. J Mol Biol. 1983 May 25;166(3):361–381. doi: 10.1016/s0022-2836(83)80090-4. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Osipova T. N., Pospelov V. A., Svetlikova S. B., Vorob'ev V. I. The role of histone H1 in compaction of nucleosomes. Sedimentation behaviour of oligonucleosomes in solution. Eur J Biochem. 1980 Dec;113(1):183–188. doi: 10.1111/j.1432-1033.1980.tb06153.x. [DOI] [PubMed] [Google Scholar]
- Parish R. W., Schmidlin S., Fuhrer S., Widmer R. Electrophoretic isolation of nucleosomes from Dictyostelium nuclei and nucleoli: proteins associated with monomers and dimers. FEBS Lett. 1980 Feb 11;110(2):236–240. doi: 10.1016/0014-5793(80)80081-0. [DOI] [PubMed] [Google Scholar]
- Parish R. W., Stalder J., Schmidlin S. Biochemical evidence for a DNA repeat length in the chromatin of Dictyostelium discoideum. FEBS Lett. 1977 Dec 1;84(1):63–66. doi: 10.1016/0014-5793(77)81057-0. [DOI] [PubMed] [Google Scholar]
- Pruitt S. C., Grainger R. M. A repeating unit of higher order chromatin structure in chick red blood cell nuclei. Chromosoma. 1980;78(3):257–274. doi: 10.1007/BF00327387. [DOI] [PubMed] [Google Scholar]
- Reeck G. R., Isackson P. J., Teller D. C. Domain structure in high molecular weight high mobility group nonhistone chromatin proteins. Nature. 1982 Nov 4;300(5887):76–78. doi: 10.1038/300076a0. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L., Eder G., Lurz R. In vitro core particle and nucleosome assembly at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3284–3288. doi: 10.1073/pnas.76.7.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Smith B. J., Harris M. R., Sigournay C. M., Mayes E. L., Bustin M. A survey of H1o-and H5-like protein structure and distribution in higher and lower eukaryotes. Eur J Biochem. 1984 Jan 16;138(2):309–317. doi: 10.1111/j.1432-1033.1984.tb07916.x. [DOI] [PubMed] [Google Scholar]
- Sommer Yeast chromatin: search for histone H1. Mol Gen Genet. 1978 May 31;161(3):323–331. doi: 10.1007/BF00331008. [DOI] [PubMed] [Google Scholar]
- Spiker S., Mardian J. K., Isenberg I. Chomosomal HMG proteins occur in three eukaryotic kingdoms. Biochem Biophys Res Commun. 1978 May 15;82(1):129–135. doi: 10.1016/0006-291x(78)90586-7. [DOI] [PubMed] [Google Scholar]
- Suchiliené S. P., Gineitis A. A. Histones from Saccharomyces cerevisiae. Exp Cell Res. 1978 Jul;114(2):454–458. doi: 10.1016/0014-4827(78)90508-6. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Unravelled nucleosomes, nucleosome beads and higher order structures of chromatin: influence of non-histone components and histone H1. J Mol Biol. 1981 Jul 15;149(4):709–733. doi: 10.1016/0022-2836(81)90354-5. [DOI] [PubMed] [Google Scholar]
- Thoma F., Losa R., Koller T. Involvement of the domains of histones H1 and H5 in the structural organization of soluble chromatin. J Mol Biol. 1983 Jul 5;167(3):619–640. doi: 10.1016/s0022-2836(83)80102-8. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J. W., Rykowski M., Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983 Dec;35(3 Pt 2):711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]
- Weber S., Isenberg I. High mobility group proteins of Saccharomyces cerevisiae. Biochemistry. 1980 May 13;19(10):2236–2240. doi: 10.1021/bi00551a037. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Allen J. R., Riedel G., Van Holde K. E. The effects of salt concentration and H-1 depletion on the digestion of calf thymus chromatin by micrococcal nuclease. Nucleic Acids Res. 1979;6(5):1843–1862. doi: 10.1093/nar/6.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Simpson R. T. Localization of the sites along nucleosome DNA which interact with NH2-terminal histone regions. J Biol Chem. 1977 Sep 25;252(18):6516–6520. [PubMed] [Google Scholar]
- Whitlock J. P., Jr, Stein A. Folding of DNA by histones which lack their NH2-terminal regions. J Biol Chem. 1978 Jun 10;253(11):3857–3861. [PubMed] [Google Scholar]
- Widmer R., Fuhrer S., Parish R. W. Biochemical evidence for a distinctive chromatin structure in nucleoli of Dictyostelium. FEBS Lett. 1979 Oct 15;106(2):363–369. doi: 10.1016/0014-5793(79)80533-5. [DOI] [PubMed] [Google Scholar]