Abstract
Glycosylation of proteins is an essential process in all eukaryotes and a great diversity in types of protein glycosylation exists in animals, plants and microorganisms. Mucin-type O-glycosylation, consisting of glycans attached via O-linked N-acetylgalactosamine (GalNAc) to serine and threonine residues, is one of the most abundant forms of protein glycosylation in animals. Although most protein glycosylation is controlled by one or two genes encoding the enzymes responsible for the initiation of glycosylation, i.e. the step where the first glycan is attached to the relevant amino acid residue in the protein, mucin-type O-glycosylation is controlled by a large family of up to 20 homologous genes encoding UDP-GalNAc:polypeptide GalNAc-transferases (GalNAc-Ts) (EC 2.4.1.41). Therefore, mucin-type O-glycosylation has the greatest potential for differential regulation in cells and tissues. The GalNAc-T family is the largest glycosyltransferase enzyme family covering a single known glycosidic linkage and it is highly conserved throughout animal evolution, although absent in bacteria, yeast and plants. Emerging studies have shown that the large number of genes (GALNTs) in the GalNAc-T family do not provide full functional redundancy and single GalNAc-T genes have been shown to be important in both animals and human. Here, we present an overview of the GalNAc-T gene family in animals and propose a classification of the genes into subfamilies, which appear to be conserved in evolution structurally as well as functionally.
Keywords: GalNAc-T, GalNAc-transferase, GALNT, monoclonal antibodies, O-glycoproteins, O-glycosylation
Mammalian protein O-glycosylation
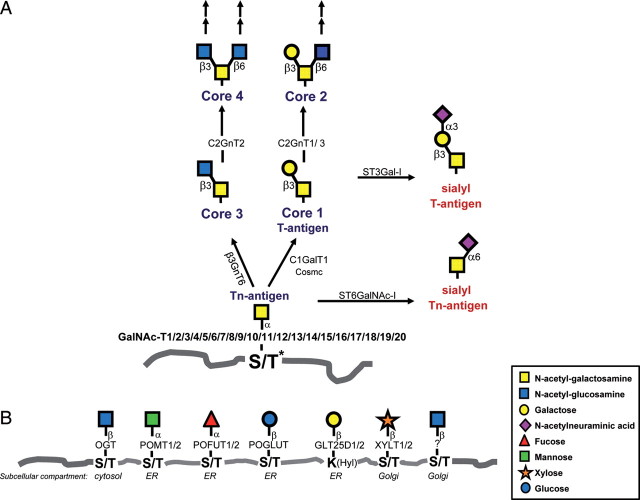
Mucin-type O-glycosylation is initiated by a large homologous polypeptide N-acetylgalactosamine (GalNAc)-transferase (GalNAc-T; GalNAc-Ts have also appeared in the literature as polypeptide ppGalNAcT, ppGalNAc-T, ppGalNAc T and GalNAcT) family that catalyzes the first step in the biosynthesis forming the GalNAcα1-O-serine (Ser)/threonine (Thr) linkage in O-glycoproteins. Thus, the large number of enzymes controlling the initiation step makes mucin-type O-glycosylation unique among other types of protein glycosylation. All other types of protein glycosylation are controlled by one or two isoenzymes or in the case of N-glycosylation a complex of proteins. The O-GalNAc residues are further processed by the addition of different monosaccharides catalyzed by 30 or more distinct glycosyltransferases (Figure 1). GalNAc O-glycosylation is initiated in the Golgi apparatus after most protein folding events have taken place (Figure 1B). N-Glycosylation and other types of O-glycosylation [including O-mannose, O-fucose, O-Glc (glucose) and O-Gal (galactose added to hydroxylysine, Hyl)] of proteins in the secretory pathway are initiated in the endoplasmic reticulum (ER). Only proteoglycan (O-Xyl) biosynthesis is initiated in the Golgi (Gotting et al. 2007). The abundant O-GlcNAc (N-acetylglucosamine) glycosylation occurs in the cytosol and nucleus and is in animals catalyzed by a single cytosolic enzyme without known homologs (Hu et al. 2010). This type of protein glycosylation is therefore not found on proteins processed in the secretory pathway, although a recent finding of the O-GlcNAc-type glycosylation on Notch is puzzling (Matsuura et al. 2008; Sakaidani et al. 2010).
Fig. 1.
Mammalian protein O-glycosylation pathways. (A) The common mucin-type O-glycosylation core 1–4 biosynthetic pathways. Mucin-type O-glycosylation is initiated by up to 20 GalNAc-Ts forming the Tn structure, which may be elongated by the core 1 synthase, C1Gal-T1, or the core 3 synthase, β3GnT6, and further branched by the core 2 synthases, C2GnT1-3. C1Gal-T1 function is dependent on the presence of the chaperone COSMC. The different core structures can be further elongated and branched by N-acetyllactosamine chains and/or terminated by blood group ABH-related structures, fucose and sialic acids. Sialylation may terminate chain elongation and branching as indicated by the action of ST3Gal-I on core 1, which produces the ST structure. Premature sialylation of the first GalNAc by ST6GalNAc-I leads to the cancer-associated structure STn. (Asterisk) Recent studies demonstrate that GalNAc may also be bound to Tyr (Halim et al. 2011; Steentoft et al. 2011). (B) Other known types of protein O-glycosylation in mammals and the initiating enzymes. These types include O-GlcNAc found on nuclear and cytoplasmic proteins, O-mannose found on α-dystroglycan, O-fucose and O-glucose found on EGF domains in membrane proteins, O-Gal linked to 5-Hyls found on collagens, O-xylose found on proteoglycans and recently identified O-GlcNAc found on extracellular proteins (Matsuura et al. 2008; Sakaidani et al. 2010). Glycosyltransferases involved in the formation of the structures depicted are indicated by their official name, and the subcellular compartments where these modifications are initiated are indicated.
These unique features of GalNAc O-glycosylation pose interesting possibilities with respect to regulation and functions of O-glycans as well as effects of genetic deficiencies in the large GalNAc-T gene family. Thus, the most pertinent and perhaps not mutually exclusive questions are whether the large number of GalNAc-T genes are used to dynamically regulate O-glycosylation to achieve differential modification of proteins and thereby serve specific functions or alternatively whether the large number of genes is needed to cover a wide spectrum of acceptor sequences and provide “back-up” in the face of deleterious mutations.
Deficiencies in genes controlling the initiation of other types of protein glycosylation generally cause severe phenotypes. Thus, deficiencies in the oligosaccharyltransferase complex are lethal in eukaryotes (Heesen et al. 1993; Kelleher and Gilmore 2006); deficiency in the O-mannosyltransferases (protein-O-mannosyltransferase T1 and T2 function in a heteromeric complex) leads to severe muscular dystrophies (Reeuwijk et al. 2006) and targeted disruption leads to embryonic lethality in mice (Lommel et al. 2010); deficiency in either of the two fucosyltransferases initiating O-fucosylation [protein O-fucosyltransferases 1 and 2 function with different substrates] is incompatible with life (Shi and Stanley 2003; Du et al., 2010); deficiency in the enzyme initiating O-glucosylation (POGLUT) is lethal (Stanley 2008; Acar et al. 2008; Fernandez-Valdivia et al. 2011); deficiency in protein O-xylosyltransferase 2 (XYLT2; one of the two xylosyltransferases XYLT1 and XYLT2) initiating proteoglycan chains leads to polycystic liver and kidney disease (Condac et al. 2007); and inactivation of lysyl hydroxylase 3 (one of three isoenzymes that precedes core Hyl galactosylation) causes early lethality in mice (Rautavuoma et al. 2004). In striking contrast, as will be discussed in the “GalNAcTs and disease” section, deficiencies in some of the vertebrate GalNAc-Ts produce only subtle phenotypes, suggesting a considerable degree of redundancy as well as unique functions of individual isoforms. Underscoring this is the observation that only one of 14 GalNAc-T isoforms in Drosophila, the pgant35A (dGalNAc-T1) gene, is essential for normal development beyond the larvae stage (Schwientek et al. 2002; Ten Hagen and Tran 2002).
There are a number of obstacles to deciphering unique functions of the GalNAc-T genes. Our knowledge of the spatio/temporal expression patterns of individual GalNAc-Ts has been hampered, in part, by a lack of appropriate immune reagents for determining specific distributions in tissues at the individual cell level as well as intracellular topology. Our understanding of the GalNAc O-glycoproteome has been largely limited to abundant glycoproteins, and, until recently, proteome-wide strategies to map sites of O-glycosylation have been missing. We hope that this situation will improve with the introduction of our recently described SimpleCell strategy, where O-glycosylation is simplified to GalNAc alone using zinc finger gene targeting in human cells (Steentoft et al. 2011). Another obstacle for the field has been a lack of general as well as isoform-specific substrate sequence motifs for predicting sites of O-glycosylation. Progress is also being made here with new strategies for in vitro studies of substrate specificities of individual GalNAc-Ts (Schwientek et al. 2007; Gerken et al. 2011). Thus, the field is advancing rapidly through the introduction of new technologies and hints of biological functions from genome-wide association studies (GWASs).
Here, we review the current state of the large GalNAc-T gene family, its evolution, the isoenzymes and their many functions.
The GalNAc-T enzymes
The GalNAc-Ts are classified as GT27 family members in the CAZy glycosyltransferase classification (http://www.cazy.org), which is based on sequence and structure similarities and today includes 94 distinct gene families. A total of 20 human GalNAc-T gene entries are available. Of these, 17 have already been reported in the literature, whereas the remaining three are reported in the accompanying manuscript by Raman et al. Most of these GalNAc-T genes have been found to encode active polypeptide GalNAc-Ts functioning in O-glycosylation (Table I). The GalNAc-T family is highly conserved throughout metazoan evolution and although the number of members varies, all completed genomes contain large families of highly homologous sequences indicating that the structure and function of GalNAc-Ts have been conserved.
Tabel I.
GalNAc-T genes in man, mouse, chimpanzee and fly (D. melanogaster)
| Alternative designation | Human | Intronsin ORF | Chromosomal locus |
Accession number |
Reference (human) | Identity (%;human vs mouse/human vs chimp) | Ortholog (D. melanogaster) | ||
|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Human | Mouse | ||||||
| GALNT1 | 10 | 18q12.1 | 18(B1) | X85018 | U73820 | White et al. (1995) | 91.1/100 | ||
| GALNT2 | 15 | 1q41-q42 | 8(E2) | X85019 | BC007172 | White et al. (1995) | 98.2/99 | cg3254(pgant2) | |
| GALNT3 | 9 | 2q24-q31 | 2(C3) | X92689 | U70538 | Bennett et al. (1996) | 95.7/99 | ||
| POC1B | GALNT4 | 0 | 12q21.33 | 10(C3) | Y08564 | U73819 | Bennett, Hassan et al. (1998) | 94.3/99 | cg30463 |
| GALNT5 | 9 | 2q24.1 | 2(C1.1) | AJ245539 | NM_172855 | Bennett, Hassan, Hollingsworth et al. (1999) | 85.6/99.2 | cg8182(pgant1) | |
| GALNT6 | 9 | 12q13 | 15(F3) | Y08565 | AJ133523 | Bennett, Hassan, Mandel et al. (1999) | 92.9/99.7 | ||
| GALNT7 | 11 | 4q34.1 | 8(B3.2) | AJ002744 | NM_144731 | Bennett, Hassan, Hollingsworth et al. (1999) | 93/99 | cg6394(pgant7) | |
| GALNT8 | 10 | 12p13.3 | —b | AJ271385 | —b | White et al. (2000) | —b/98 | ||
| GALNT9 | 10 | 12q24.33 | 5(5F) | AB040672 | AK032568 | Toba et al. (2000) | 95.2/99a | ||
| GALNT10f | 11 | 5q33.2 | 11(B1.3) | AJ505950 | BC016585 | Cheng et al. (2002) | 98.2/99.7 | cg2103 (pgant6) | |
| GALNT11 | 10 | 7q36.1 | 5(A3) | Y12434 | Y12435 | Schwientek et al. (2002) | 91.8/99.8 | cg7480(pgant35A) | |
| GALNT12 | 9 | 9q22.33 | 4(B1) | AJ132365 | AK042133 | Guo et al. (2002) | 91.1/100 | ||
| GALNT13 | 10 | 2q24.1 | 2(C1.1) | AJ505991 | AB082928 | Zhang et al. (2003) | 99.5/100 | cg31651(pgant5)c cg4445(pgant3) | |
| FLJ12691 | GALNT14 | 12 | 2p23.1 | 17(E2) | Y09324 | AK078292 | Wang et al. (2003) | 94.7/97.5 | |
| GALNTL2 | GALNT15 | 9 | 3p25.1 | 14(B1) | NM_054110 | AK005605 | Cheng et al. (2004) | 82.6/99.5 | cg10000 |
| GALNTL1 | GALNT16 | 13 | 14q24.1 | 12(C3) | AJ505951 | AB045325 | Peng et al. (2010) | 97.1/96 | |
| GALNTL6 | GALNT17g | 11 | 4q34.1 | 8(B3) | AJ626725 | AK015826 | Raman et al. (2011) | 97/100 | dcg7579/cg31776/cg31956(pgant4)/cg7297(pgant8)/cg7304 |
| GALNTL4 | GALNT18 | 10 | 11p15.3 | 7(F1) | AJ626724 | AAH24988 | Raman et al. (2011) | 97.4/99.8 | |
| GALNTL3 | GALNT19 | 10 | 7q11.23 | 5(G2) | AJ626726 | NM_145218 | Nakamura et al. (2005) | 99/100 | |
| GALNTL5 | GALNT20 | 7 | 7q36.1 | 5(A3) | NM_145292 | AK090330 | Raman et al. (2011) | 72.7/99e | |
aBased on partial chimp XM_001172345/XM_528857.2 overlapping transcripts.
bNo murine T8 identified.
cGroup with GALNT1/T13 Ia subfamily, see Figure 3.
dOrtholog's group in a clade separate from the GALNT7/T10/T17 IIb subfamily, see Figure 3.
eBased on overlapping chimp XR_023409.1/AC189698 sequences.
fOriginall published as mouse Galnt9 by Ten Hagen et al. (2001), but redesignated GALNT10 by Schwientek et al. (2002).
gGALNT17 has recently been published as GALNT20 by Peng et al. (2010), but we suggest this isoform to be assigned GALNT17 according to CAZy (http://afmb.cnrs-mrs.fr/fr/CAZY/) nomenclature.
GalNAc-T activity was first identified and characterized in the bovine submaxillary gland and rat extracts (McGuire and Roseman 1967; Hagopian and Eylar 1968; Hagopian 1969), and the enzyme activity was subsequently partially purified and characterized from ovine submandibular glands (Hill et al. 1977). The enzyme activity had a broad pH optimum (6.5–8.6) and required divalent metal ions as cofactor (Mn2+ optimal) (Sugiura et al. 1982; Elhammer and Kornfeld 1986). Different strategies and sources were used for purification of the GalNAc-T enzyme activity, which resulted in isolation of two different enzymes, bovine GalNAc-T1 and human GalNAc-T2 with >80% sequence similarity (Hagen et al. 1993; Homa et al. 1993; White et al. 1995). Analysis of human gene sequences confirmed the existence of an ortholog of GalNAc-T1 with >95% sequence identity to bovine GalNAc-T1 and this was shown to encode an active enzyme, demonstrating that the human genome contained multiple GalNAc-T genes (Sørensen et al. 1995; Wandall et al. 1997). With the use of degenerate polymerase chain reaction (PCR)-based cloning strategies and emerging bioinformatics and EST (expressed sequence tagged) information, a number of additional distinct GalNAc-Ts were identified (Bennett et al. 1996; Clausen and Bennett 1996; Hagen et al. 1997; Bennett, Hassan, et al. 1998; Ten Hagen et al. 1998, 1999, 2001; Bennett, Hassan, Hollingsworth, et al. 1999; Bennett, Hassan, Mandel, et al. 1999), eventually leading to the appreciation of the existence of the large number of GalNAc-T genes present in mammals (Table I). Several recent reviews have provided updates on the continuously expanding GalNAc-T gene family and their biology (Clausen and Bennett, 1996; Hassan, Bennett et al. 2000; Ten Hagen et al. 2002; Tian and Ten Hagen 2008; Tabak 2010).
Considerable efforts have been devoted to analysis of the characteristics and functions of these enzymes and some general properties have emerged: (i) individual GalNAc-T isoforms have distinct acceptor peptide substrate specificities, although considerable overlap exists; (ii) acceptor peptide substrate specificities include both unmodified peptide sequences and partially glycosylated GalNAc-peptide substrates; (iii) a C-terminal GalNAc-T lectin domain modulates the specificity toward partially glycosylated GalNAc-peptide substrates and (iv) GalNAc-T isoforms are differentially expressed in cells and tissues during development and differentiation and marked changes in expression are found in diseases including cancer. These features suggest that GalNAc-Ts function in a coordinated fashion driven by their acceptor substrate specificities and that the cellular repertoire of GalNAc-Ts is regulated to accommodate the need for O-glycosylation of diverse protein substrates.
The domain structure of GalNAc-transferases: catalytic and lectin domains
GalNAc-Ts share the common type II membrane structure of Golgi glycosyltransferases with a short N-terminal cytoplasmic tail, a hydrophobic non-cleaved signal sequence serving as a membrane-spanning domain, a stem region of variable length and a luminal catalytic domain (Paulson and Colley 1989). However, GalNAc-Ts are unique among eukaryote glycosyltransferases in having a C-terminal ricin-like lectin domain (∼120 amino acids) in addition to a catalytic unit (Hazes 1996; Imberty et al. 1997). The subcellular topology of several GalNAc-Ts has been characterized by immunocytology; GalNAc-T1, -T2 and -T3 are present throughout the Golgi with isoform-specific differences in relative amounts in the different Golgi stacks (Rottger et al. 1998). The short cytoplasmic tails of all GalNAc-Ts contain basic residues that may be involved in interaction with peripheral Golgi membrane protein tethering complexes (Smith and Lupashin 2008). The stem regions are variable in length and range from around 90 amino acids (GalNAc-T1, -T13 and -T16) to 170 amino acids (GalNAc-T3, -T8 and -T15) with the exception of an extended 470-amino acid stem region found in GalNAc-T5. The stem regions are thought to displace the catalytic domains into the lumen of Golgi stacks, but more specific functions directed by the great variability in GalNAc-T stem regions remain to be determined.
The GalNAc-T catalytic domains (∼230 amino acids in length) contain a GT-A structural motif (Bourne and Henrissat 2001) characterized by two tightly interacting β–α–β Rossmann-like folds. The GT-A motif essentially corresponds to the previously described GT1 motif of the GalNAc-Ts (Hagen et al. 1999). The Rossmann-like folds contain residues which bind to the uracil moiety of the UDP-GalNAc donor substrate (Supplementary data, Figure S1; Fritz et al. 2006; Kubota et al. 2006). A DxH Mn2+ ion binding motif, which is conserved in all human isoforms and interacts with the diphosphate moiety of the donor substrate UDP leaving the group via the coordination of the Mn2+ ion, is located near the C-terminus of the second Rossmann fold. The functional role of the DxH motif has been demonstrated and, interestingly, the GalNAc-T DxH cannot be substituted for the more common DxD motif found in other glycosyltransferases using UDP sugars (Hagen et al. 1999). A Gal/GalNAc-T motif (Hagen et al. 1999) is shared among GalNAc-Ts and β4Gal-Ts, and this region interacts with the GalNAc moiety of the donor substrate (Fritz et al. 2004; Kubota et al. 2006; Figure 2 and Supplementary data, Figure S1). An additional GalNAc-binding pocket may also exist in the GalNAc-T10 catalytic domain, as initially proposed by Kubota et al. (2006) and further supported by subsequent investigations (Raman et al. 2008; Perrine et al. 2009). In the remaining part of the catalytic domain, the donor substrate-binding residues lie within or near a loop, which undergoes a large conformational change in response to UDP-GalNAc binding. Residues within this loop contribute to binding of acceptor peptide substrates; however, most of the acceptor substrate-binding residues lie along a pre-formed channel in the surface of the catalytic domain. A subset of these residues form a “proline pocket” (Fritz et al. 2006), which explains the preference of some GalNAc-T isoforms for acceptors containing a proline 3 residues to the C-terminus of the Thr or Ser to which GalNAc is added (O'Connell et al. 1991; Wilson et al. 1991; Gerken et al. 2011). Recent experiments using random peptide and glycopeptide libraries have begun to reveal additional acceptor substrate preferences for several GalNAc-T isoforms (Gerken et al. 2006, 2011; see the following section) and should lead to improved prediction of isoform-specific mucin-type O-glycosylation. An additional pocket accommodating the acceptor Thr methyl group was revealed by the crystal structure of a peptide bound to GalNAc-T2 (Fritz et al. 2006), which appears to explain why GalNAc-Ts often show preference for glycosylating Thr vs Ser residues. As with other retaining GT-A glycosyltransferases, definitive identification of GalNAc-T residues responsible for catalytic activity has not been made and the mechanism of retaining glycosyltransferases remains unclear (Lairson et al. 2008). A recent study (Soya et al. 2011) suggests that a double-displacement mechanism may be used by blood group A and B glycosyltransferases, but whether the findings of this study will apply to the GalNAc-Ts and other retaining glycosyltransferases will require additional investigation.
Fig. 2.
Phylogenetic and genomic analysis of the human GalNAc-T gene family. Left panel shows the unrooted tree derived from molecular phylogenetic anaylsis by the maximum likelihood method of Gblock (Talavera and Castresana 2007) curated ClustalW (http://www.ebi.ac.uk/FTP/) alignments. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011). In brief, the evolutionary history was inferred by the use of the maximum likelihood method based on the Dayhoff w/freq. model. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. There were a total of 206 positions in the final data set. The unrooted tree is based on amino acid analysis of the catalytic domain of all 20 human GalNAc-Ts. The catalytic domains were defined as previously described (Schwientek et al. 2002) using the accession numbers listed in Table I. The amino acid identities of the catalytic domains (%) are indicated between isoforms. The robustness of the predicted Ig phylogenetic branch was additionally confirmed by the analysis of separate trees generated for Ig (GalNAc-T15) with either subfamily I or II members (grouping Ig with, respectively, Ic or IIa with high bootstrapping confidence, data not shown). Right panel depicts genomic organization of the ORF for all 20 human GALNT genes. Intron positions are based on the gene alignments shown in Supplementary data, Figure S1. Exons are shown as boxes and isoforms grouped in the cladogram are shaded intermittently in grey and black. For GALNT5, the first exon encoding an extended stem region of ∼500 amino acids has been truncated as indicated by a break. Conserved intron/exon boundaries (boundaries shared among all genes except for GALNT4) are indicated by a solid line and labeled C1–C3 at the bottom. Intron boundaries shared between two or more genes are shown by a broken line. The intron phases (0, 1, 2) are indicated by numbers below introns (phase 0 introns are positioned between two codons, phase 1 introns after the first base of the codon and phase 2 introns are positioned after the second base of the codon). The position of the diagnostic introns unique for T5 (uT5a, b and c) and T15 (sT15) is indicated with arrows at their relative positions below. Based on the cladogram and similarities in genomic organizations we propose a classification of the GalNAc-Ts into seven subfamilies designated Ia–g and IIa and b. Members of subfamily I contain GalNAc-T isoforms that predominantly display peptide substrate specificity, and members of subfamily II contain GalNAc-T isoforms that predominantly display GalNAc-glycopeptide substrate specificity. The regions encoding the different domains of GalNAc-Ts (cytosolic/transmembrane/stem regions, catalytic and lectin domains) including positions of identified functional motifs are shown on the top.
The unique lectin domains of GalNAc-Ts belong to the Ricin-type lectin structural family (Dodd and Drickamer 2001). Hazes (1996) and Imberty et al. (1997) originally proposed the presence of lectin domains on GalNAc-Ts and these lectins have been classified as carbohydrate-binding module family 13 members in the CAZy database. This module is found in many kingdoms of life including prokaryotes (excluding archaea) and adopts a β-trefoil structure composed of three homologous repeats (α, β and γ) presumed to have evolved through a series of gene duplication events (Rutenber et al. 1987). The β-trefoil fold is found in superfamilies such as cytokines (interleukins and fibroblast growth factors; Ponting and Russell 2000), ricin B-like toxins (like agglutinins and hemolytic lectins; Hazes 1996), Kunitz protease inhibitors and actin-binding proteins (such as β-crystallin and hisactophilin-like actin-binding proteins; Mukhopadhyay 2000; Kureishy et al. 2002; Liu et al. 2002). General characteristics of β-trefoil folds are: (i) the presence of QxW and CLD motifs in each repeat of ∼40 amino acids; (ii) limited sequence similarity (Ornitz and Itoh 2001) and (iii) binding specificities as diverse as heparan sulfate, sulfated GalNAc, Xyl, Gal, Glc, lactose and GalNAc. The GalNAc-T lectin domain sequences are poorly conserved except for QxW and CLD motifs and six invariant cysteine residues that form disulfide bridges within each of the three α, β and γ repeats (Fritz et al. 2004). The carbohydrate-binding specificities of the GalNAc-T lectin domains studied have so far demonstrated rather high specificity for GalNAc and GalNAc glycopeptides (Wandall et al. 2007), but not for glycopeptides carrying elongated O-glycans such as Galβ1-3GalNAcα-O-Ser/Thr, NeuAcα2-6GalNAcα-O-Ser/Thr or GlcNAcβ1,3GalNAcα-O-Ser/Thr (Pedersen et al. 2011). Blocking experiments and inactivating mutations in the lectin domains affect GalNAc-glycopeptide substrate specificities of the enzymes. Mutational analyses have demonstrated that the α-repeat is important for GalNAc-T2, -T3 and -T4 lectin binding (Hassan et al. 2000; Wandall et al. 2007; Pedersen et al. 2011), whereas in GalNAc-T1 both the α- and β-repeats are important (Tenno et al. 2002) and in GalNAc-T10 it is the β-repeat that is important (Kubota et al. 2006). The lectin domains clearly function to modulate and improve the catalytic efficiency of GalNAc-Ts with partially GalNAc-glycosylated substrates containing a high density of acceptor sites such as those found in mucin tandem repeat sequences. How this translates to the glycosylation of large mucins in cells is unknown but it seems likely that the lectin domains serve to improve binding of GalNAc-Ts to mucin substrates in order to complete initiation of O-glycosylation before O-glycans are elongated (the processing step), as this would interfere with further addition of GalNAc residues. Since the initiation process occurs simultaneously with the processing step in the Golgi, the combined binding affinities of the catalytic and lectin domains may improve competition as long as there are acceptor sites available. This hypothesis is supported by one study demonstrating that the biosynthesis of the MUC5Ac mucin appears to go through an intermediate GalNAc-glycosylated glycoform before elongation starts (Sheehan et al. 2004). Further support may be found in a recent study demonstrating that the selective relocation of GalNAc-Ts to ER results in increased density of O-glycosylation (Gill et al. 2010). It thus seems possible that O-glycan density is in part secured by two subsets of GalNAc-Ts, one subset “initiating” on naked or low-density acceptors before another subset executes “follow-up” glycosylation by adding GalNAc to adjacent GalNAc occupied sites.
The catalytic and lectin domains are connected by a linker sequence 10–25 amino acids in length, which based on structural data display variability in flexibility. The GalNAc-T1 catalytic and lectin domains are closely associated and interact through a relatively large interfacial surface area (Fritz et al. 2004). The GalNAc-T1 lectin domain also contains an N-linked glycan at N552 (Wragg et al. 1997). Hydrogen bonding between the chitobiose core of the N-glycan and the amide nitrogen of linker residue Y428 and between residues R426 and E416 is likely to contribute to the stable association of the GalNAc-T1 lectin and catalytic domains. In contrast, the two domains of GalNAc-T2 show little to no interaction (Fritz et al. 2006). GalNAc-T2 lacks an N-linked glycan corresponding to that of the GalNAc-T1 lectin domain, but GalNAc-T10 has an N-linked glycan at N593. The N-linked glycans of GalNAc-T10 were removed by endoglycosidase H treatment prior to crystallization; thus, it remains unclear whether the N593 glycan on T10 affects linker flexibility of this isoform. Sequons for N-linked glycans corresponding to that in the GalNAc-T1 lectin domain are also present in human GalNAc-T13 and -T17. The relative positioning of the two domains has been attributed, in part, to differences in linker sequence properties. Peripheral GalNAc-T1 linker residues were shown to interact with both the catalytic and lectin domains, whereas a more stretched linker in GalNAc-T10 results in weaker interaction with the catalytic domain (Kubota et al. 2006). Therefore, linker flexibility may function to control the relative orientation of the lectin and catalytic domains and serve to drive lectin-mediated substrate specificities of GalNAc-Ts. This has been shown in experiments involving the replacement of the linker in GalNAc-T10 with the linker of GalNAc-T1, which resulted in altered substrate specificity of the chimeric enzyme from glycopeptide to broad peptide specificity (Kubota et al. 2006). However, in contrast to this, a similar study demonstrated that deleting the GalNAc-T10 lectin domain or exchanging it with the corresponding GalNAc-T2 lectin domain did not alter the specificity of the mutant chimeric isoform compared with normal wild-type GalNAc-T10 (Raman et al. 2008). This discrepancy in results may be due to different experimental conditions and acceptor substrates used or due to differences in the GalNAc-T10 linker region start positioning (Phe449 in the former and Pro455 in the later study). Hence, further studies are needed to clarify this.
Although studies of the binding specificities of GalNAc-T lectins analyzed so far demonstrate binding to the GalNAc monosaccharide residue, recent novel experimental approaches based on glycopeptide beads and microarrays have demonstrated differential binding specificities of GalNAc-T1, -T2, -T3 and -T4 lectins toward different subsets of small synthetic and recombinant GalNAc glycopeptides (Pedersen et al. 2011). This suggests that the lectins serve more complex roles in modulating functions of GalNAc-Ts.
Peptide and glycopeptide substrate specificities of GalNAc-transferases
The peptide and glycopeptide substrate specificities of the GalNAc-T family are still relatively poorly understood. Studies of the acceptor specificities of GalNAc-Ts have generally been limited to in vitro assays with a limited number of peptides. Nevertheless, substrate specificities identified with such peptides have been shown to correlate very well with functions on corresponding proteins in cells (Nehrke et al. 1998; DeFrees et al. 2006; Kato et al. 2006; Schjoldager et al. 2010). More recently, the application of random peptide and glycopeptide substrate libraries has allowed quantitative determination of neighboring residue preferences and emergence of GalNAc-T isoform-specific acceptor sequence motifs (Gerken et al. 2006, 2008, 2011; Perrine et al. 2009). This strategy is based on peptide substrates with the general motif: GAGA(X)nT(X)nGAGA (where X = subset of randomized AA residues including Ser-O-GalNAc and n = 3–5). These relatively short substrates are thought to reveal the catalytic domain peptide and glycopeptide specificity independent of the lectin domain. Presently, randomized peptide substrate preferences have been reported for GalNAc-T1, -T2, -T3, -T5, -T10 and -T12 and the fly orthologs of GalNAc-T1 and -T2 (pgant5 and pgant2, respectively; Gerken et al. 2008), whereas Ser/Thr-O-GalNAc preferences have been reported only for GalNAc-T1, -T2 and -T10 (Gerken et al. 2006, 2011; Perrine et al. 2009). Using these substrates, the mammalian and fly GalNAc-T1 and -T2 orthologs have been shown to have nearly identical peptide substrate specificities (Gerken et al. 2008). This approach suggests that the principal sequence motif determining the substrate specificity of GalNAc-Ts reside in the three neighboring N- and C-terminal residues flanking sites of glycosylation. Interestingly, all the transferases studied so far, except GalNAc-T10, display a roughly similar C-terminal sequence motif of Pro, Gly/Ala and Pro for the +1 to +3 positions relative to the site of glycosylation. This common proline motif correlates in these transferases with the presence of aromatic residues forming the previously mentioned “proline pocket” (Fritz et al. 2006). Several other uncharacterized GalNAc-Ts contain these residues, which are therefore expected to possess similar C-terminal proline motifs as well (Gerken et al. 2011). GalNAc-T10 lacks the key “proline pocket” residues and very strongly prefers Ser-O-GalNAc at the +1 position, clearly showing the catalytic domain of T10 directly binds glycopeptides (Perrine et al. 2009). This further suggests that the T10 lectin domain is dispensable for the “follow-up” glycosylation of certain glycopeptide substrates. Whether the remaining glycopeptide requiring GalNAc-Ts in subgroup IIb (see Figure 2 and the following section for a description of the subgroup definition), i.e. GalNAc-T7 and -T17, possess similar catalytic domain glycopeptide-binding properties has yet to be determined. In contrast to the C-terminal preferences, the N-terminal preferences show a wide range of isoform-specific enhancements with elevated preferences for Pro, Val and Tyr being the most common at the −3 to −1 positions (Gerken et al. 2011). Transferase specific enhancement values can be used to predict sites of glycosylation at the Isoform Specific O-glycosylation Prediction (ISOGlyP) web site, http://isoglyp.utep.edu.Thus, to date, it seems that for “proline pocket” containing transferases acceptor substrate specificity will be dominated by each isoform's different sensitivity to residues N-terminal of the site of glycosylation. It was recently shown that the overall preferences for charged residues vary among isoforms and this can be correlated with the calculated surface charge of the transferase (Gerken et al. 2011).
The human GALNT gene family
The 20 human GALNT genes localize to different chromosomal loci except for two sets of clustered genes (Table I). One cluster, GALNT7/T17, is separated by 100 kb and another, GALNT11/T20, is spaced 8.5 kb apart. In both cases, there are no predicted ORFs (open reading frames) between the clustered genes. GALNT5 and T13 are also located close to each other but the genes are interspaced by 1.5 Mb containing multiple ORFs encoding known proteins. The sizes of the individual gene loci for GALNTs vary from 5 kb for T4 to >1.2 Mb for T17. All genes contain multiple coding exons (8–16 exons) except for GALNT4. Analysis of GALNT intron/exon positioning reveals that three intron positions are conserved in all genes (conserved intron boundaries/positions C1–C3, Figure 2) and that others are conserved in subsets of genes (Bennett, Weghuis, et al. 1998). This suggests that all GALNT genes originate from one ancestral GALNT gene that contained at least these three introns and that additional introns were gained during the evolution of various paralogs. As described in the following section, the positioning of the C1–C3 introns has been conserved in lower species. The accumulation of introns observed during evolution thus suggests that a common ancestor of deuterostome GALNTs was intron poor and that introns were gained during the evolution of the mammalian GALNT gene family. For all GALNTs, the exons encoding the catalytic unit are flanked at the 5' end by the conserved C1 intron and at the 3' end by an intron that is semi-conserved in 16 of the GALNT genes. The majority of introns found within the lectin domains have been identically positioned in 16 of the GALNTs and in these cases the intron positions relative to the ORF (intron phase) are zero, i.e. the intron is positioned between two codons (Figure 2).
One gene, GALNT4, stands out among the genes by being encoded in a single exon (Figure 2). GALNT4 is unlikely to represent an ancestral gene since intronless GALNT genes are not found in lower organisms. We therefore previously proposed that GALNT4 arose through a retropositional event (Bennett, Hassan, et al. 1998). The most likely candidate gene giving rise to GALNT4 is GALNT12, which exhibits the highest sequence similarity and has nine introns (Figure 2). GALNT4 is located within the second intron of the WD40 repeat domain-containing cartwheel protein, Poc1, which is required for the structural maintenance of centrioles. Sequence alignments of the 5′- or 3′-untranslated regions (UTRs) of GALNT4 and T12 and with other GalNAc-T UTRs did not reveal any similarities. Analysis of repetitive elements flanking the GALNT4 and T12 loci showed that AluY elements (Batzer et al. 1996) are found flanking the single GALNT4 exon, whereas AluSx&j elements flank both GALNT4 and T12 transcription units. Since AluY elements have arisen late in evolution [25 million years ago (MYA); Batzer and Deininger 2002], GALNT4 must have arisen relatively late and not earlier than 25 MYA. These latter findings provide further support for the hypothesis that GALNT4 arose as a transposition event.
The GALNT genes have also given rise to a number of pseudogenes (psgenes). The first example described was a psgene derived from GALNT1 (Meurer et al. 1996) and we have identified additional four psgenes. One additional GALNT1 derived a psgene was identified within the HEATR6 locus on chromosome 17. Further psgenes were identified for GALNT11 in an intron of the LEPR gene on chromosome 1, GALNT13 on chromosome 3 and GALNT18 in an intron on chromosome 7. Retropositioning has been found to be the mechanism by which the vast majority (72%) of the more than 20,000 putative human psgenes have arisen (Torrents et al. 2003).
A classification of the GALNT gene family
The GALNT genes can be grouped into subfamilies. We originally reported the existence of a subfamily of two close GALNT paralogs, GALNT3 and T6, that exhibited very high sequence similarity throughout the coding region, identical genomic structure with nine intron/exon boundaries and encoded enzymes with similar substrate specificities (Bennett, Hassan, Mandel, et al. 1999). We have now surveyed all 20 GALNT genes and identified additional closely related subfamilies consisting of GALNT1/T13, GALNT2/T14/T16, GALNT7/T10/T17, GALNT4/T12, GALNT8/T9/T18/T19 and GALNT11/T20 (Figure 2). Currently, our understanding of the function of the 20 individual GalNAc-Ts is limited and characterization of the enzymes would benefit from grouping into subfamilies with related functions. Based on the example of the GALNT3 and T6 subfamilies (Bennett, Hassan, Mandel, et al. 1999), we therefore propose to classify all GALNT genes into two major groups, I and II, with predicted predominant peptide and GalNAc-peptide substrate specificities, respectively. Group I genes are further subdivided into seven distinct subfamilies (Ia, Ib, Ic, Id, Ie, If and Ig) and group II genes into two subfamilies (IIa and IIb). Paralogs within the classified subfamilies share identical intron number and positioning except for minor variations in introns positioned in the most 5′ region. These include subfamily Ib (GALNT2/T14/T16) with 15 introns in GALNT2 and 14 introns in T14 and T16 and subfamily Ie (GALNT8/T9/T18/T19), where the first intron in GALNT8 is shifted 111 bp relative to the first intron position in T9/T18/T19.
Two genes, GALNT5 and T15, do not show a significant relationship with these subfamilies. These two genes did not group into subfamilies in the phylogenetic analysis and contained unique intron positions. Thus, three unique introns in GALNT5 (uT5a/b/c) were identified at positions not found in any other GALNT gene. One specific intron boundary in GALNT15 (sT15) was also found only in GALNT7. This particular intron in GALNT7 is not conserved in position in the two other members belonging to the IIb subfamily (GALNT10/T17). On the other hand, all three members (GALNT7/T10/T17) share identical positioning of an intron (intron 8 in GALNT7) not found in GALNT15 (Figure 2).
GALNT20 lacks the exons encoding the lectin domain. GALNT20 colocalizes on chromosome 7q36 with GALNT11 and the two genes are spaced 8.5 kb apart. Experiments designed to demonstrate alternative splicing events suggest that alternative splicing between the two closely spaced transcriptional units does not occur (personal observation). GalNAc-T20 has an invariant glycine (G334R) residue in the Gal/GalNAc-T motif WG(G/R)EN (Supplementary data, Figure S1). The corresponding residues in the GalNAc-T1 crystal structure have been shown to be involved in essential UDP-GalNAc binding and are also found in the seven members of the β4Gal-T family. β4Gal-T7 also possess the invariant WG(G/R)EN mutation and, interestingly, this isoform has a different acceptor substrate specificity from the other members of the β4Gal-T family in that it transfers Gal to Xyl, which is essential for synthesis of the link region of proteoglycans (Almeida et al. 1999) instead of to GlcNAc. We have been unable to express the secreted GalNAc-T20 protein in insect cells and in COS7 cells, and C-terminally tagged full coding constructs failed to demonstrate Golgi localization (unpublished and accompanying manuscript by Raman et al.). Thus, GalNAc-T20 may represent: (i) a functional gene with yet unknown function and subcellular localization; (ii) a conserved expressed psgene or (iii) a gene with the potentiality for becoming a functional gene, a so-called potogene (Balakirev and Ayala 2003).
The currently available data on the enzymatic functions of GalNAc-Ts support the proposed subfamily classification, although the GALNT3/T6 subfamily is the best characterized (Bennett, Hassan, Mandel, et al. 1999). Within group I, seven subfamilies are found (Ia–Ig). Subfamily Ia (T1/T13) enzymes have similar peptide specificities, although one study has claimed that there are differences in the ability to glycosylate Syndecan-3 and MUC7-derived peptides (Zhang et al. 2003). However, we have not been able to confirm this (unpublished). In subfamily Ib (T2/T14/T16), GalNAc-T2 is the only well-characterized isoform in the literature, but our preliminary studies of these three enzymes show related functions with many substrates (unpublished and accompanying manuscript by Raman et al.). Subfamily Ie (T8/T9/T18/T19) enzymes are the least characterized. GalNAc-T9 was shown to have low activity with a small panel of peptides (Zhang et al. 2003), and in the accompanying paper GalNAc-T18 is also shown to have low activity with peptide substrates. In subfamily If (T11/T20) only GalNAc-T11 has been characterized and shown to have unique specificity compared with other GalNAc-Ts (Schwientek et al. 2002; Ten Hagen and Tran 2002). GALNT20 is different from all other GALNT genes in that the ORF lacks the C-terminal sequence encoding the lectin domain, and it remains to be verified that it does not represent a psgene. GalNAc-T5, constituting subfamily Id, and GalNAc-T15, constituting subfamily Ig, both show efficient activity with several peptide substrates (Ten Hagen et al. 1998; Cheng et al. 2004; Gerken et al. 2011).
Within group II, two enzyme subfamilies are found (IIa and IIb). Subfamily IIa GalNAc-T4 and T12 (T4/T12) share common GalNAc-glycopeptide substrates (unpublished). Interestingly, GalNAc-T4 is the only enzyme that uses GalNAc glycopeptides from the MUC1 tandem repeat, and hence the only known isoform that can complete the glycosylation of the MUC1 tandem repeat (Bennett, Hassan, et al. 1998; Hassan et al. 2000). GalNAc-T12 was originally reported to have very low activity toward an MUC5AC peptide substrate (Guo et al. 2002), but our preliminary studies clearly demonstrate that this enzyme primarily exhibits GalNAc-glycopeptide substrate specificity like T4 (unpublished). In subfamily IIb (T7/T10/T17), all three GalNAc-T isoforms are reported to have GalNAc-glycopeptide substrate specificities (Bennett, Hassan, Hollingsworth, et al. 1999; Ten Hagen et al. 2001; Peng et al. 2010). We predict that most of the GalNAc-Ts grouped in subfamilies will eventually turn out to have similar properties but with distinct subtle differences. As discussed later, the expression in cells and tissues of members of such subfamilies are different, so multiple members in subfamilies do not provide complete back-up.
Evolution of the GalNAc-T gene family
Studies on the evolution of GalNAc-Ts have calculated the substitution rates (between human and mouse) to be as low as those observed for histone and actin genes, and it was estimated that the GalNAc-T genes appeared among the first glycosyltransferases in early eukaryote evolution around 1200 MYA (Kaneko et al. 2000). Conservation of the intron phase (0) between the catalytic and lectin domains of the majority of GalNAc-transferases (Figure 2) suggests that the unique lectin domains on GalNAc-Ts arose via fusion with lectin modules early in evolution. A phylogram based on 102 putative GalNAc-T sequences from completed or preliminary assembled genomes depicts the size of the gene family in diverse species including Pan troglodytes (chimpanzee), Mus musculus (mouse), Gallus gallus (chicken), Xenopus tropicalis (frog), Caenorhabditis elegans (worm), Drosophila melanogaster (fly), Danio rerio (fish) and Toxoplasma gondii (microbe) (Figure 3, Table I). This analysis clearly demonstrates that all these organisms have a large number of GalNAc-transferases available and that there is a functional requirement for multiple isoforms with members from most of the classified subfamilies.
Fig. 3.
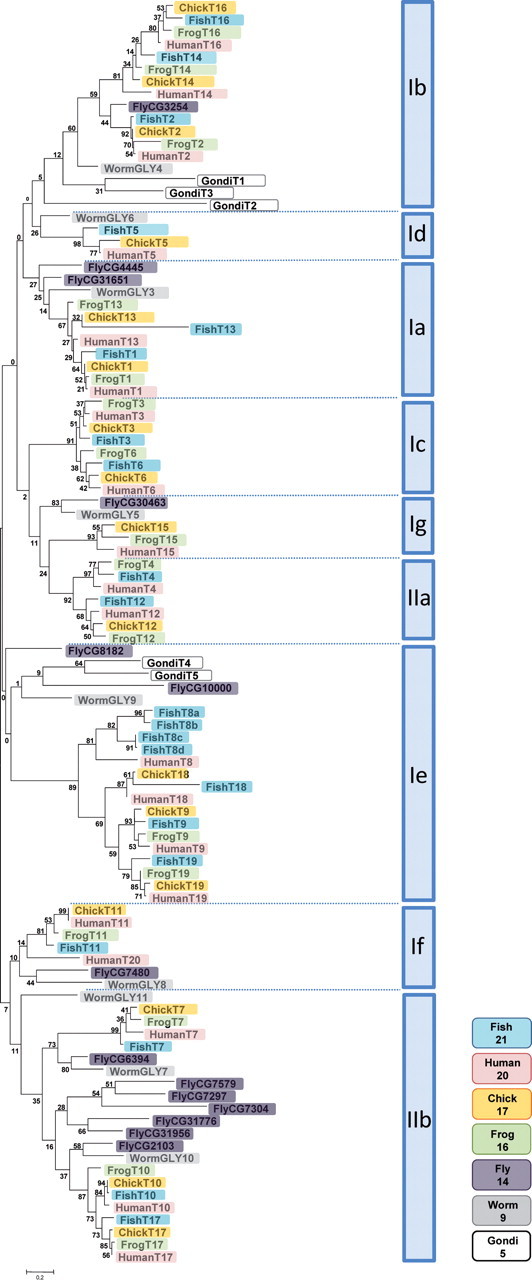
Phylogenetic tree of 102 GalNAc-Ts from human (Homo sapiens), frog (X. tropicalis), chicken (G. gallus), fish (D. rerio), fly (D. melanogaster), worm (C. elegans) and Gondi (T. gondii). The following Genbank accession numbers were used in addition to those of Table I. GALNT1: X. tropicalis NP_001025547.1, D. rerio XP_687472.2, D. melanogaster NP_001036338.1 (CG31651), G. gallus NP_001006381.1, C. elegans NP_498722.1 (gly-3). GALNT2: X. tropicalis XP_002931524.1, D. melanogaster NP_608773.2 (CG3254), G. gallus XP_419581.2, C. elegans NP_507850.2 (gly-4). GALNT3: X. tropicalis XP_002936794.1, D. rerio XM_003199248, D. melanogaster NP_725603.2 CG30463, G. gallus XP_422023.2. GALNT4: X. tropicalis NP_001072705.1, D. rerio NP_001038243.2, D. melanogaster NP_725603.2 (CG30463), C. elegans NP_001022851.1 (gly-5). GALNT5: D. rerio XP_001338929.2, G. gallus XP_422169.2. GALNT6: X. tropicalis XP_002933739.1, D. rerio NP_998361.1, G. gallus NP_001026749.1. GALNT7: X. tropicalis NP_001001200.1, D. rerio NP_001018477.1 and NP_573301.2 (CG6394), G. gallus XP_420521.2, C. elegans NP_503512.1 (gly-7). GALNT8: D. rerio XM_691878.2 (GALNT8a), XM_691987.2 (GALNT8b), XM_003198224.1 (GALNT8c), XM_003198223.1 (GALNT8d). GALNT9: X. tropicalis XP_002931923.1, D. rerio XP_001338018.1, G. gallus XP_415088.2. GALNT10: X. tropicalis NP_001072444.1, D. rerio NM_001076604.1, G. gallus XP_420520.2. GALNT11: X. tropicalis NP_001006904.1, D. rerio NP_001070030.1, D. melanogaster NP_652069.2, G. gallus XM_418541.2. GALNT12: X. tropicalis XM_002935135, D. rerio XP_688194.1, G. gallus XM_419065.2. GALNT13: X. tropicalis NP_001017277.1, D. rerio XM_002663311.2, D. melanogaster NM_136412 (CG4445), C. elegans NP_001022646.1 (gly-6), G. gallus XP_422165.2. GALNT14: X. tropicalis NP_001072369, D. rerio NP_001038460.1, G. gallus XM_419370.2. GALNT15: X. tropicalis XP_002932836.1, G. gallus XP_418741.2, D. melanogaster NP_648800 (CG7297), NP_996098 (CG7304), AAN10370.1 (CG31956), AAF51101.3 (CG31776), AF326979_1 (CG7579) and AAF56810.2 (CG10000). GALNT16: X. tropicalis NP_001039091.1, D. rerio XP_001339749.3, G. gallus XP_001231965.1. GALNT17: X. tropicalis XP_002933366.1, D. rerio NP_001139074.1, D. melanogaster NP_647749.2 (CG2103), C. elegans NP_001041037.1 (gly-10). GALNT18: D. rerio XP_689577.2, G. gallus XP_420966.2. GALNT19: X. tropicalis XP_002935847.1, D. rerio XP_696189.3, G. gallus XP_415728.2. GALNT20: C. elegans AAC13678.1 (gly-8), AAC13679 (GLY9) and NP_001022948 (gly-11), T. gondii XP_002365147.1 (T1), XP_002365091.1 (T2), XP_002369811.1 (T3), XP_002364915.1 (T4) and XP_002370555.1 (T5). Maximum likelihood phylogenetic analysis of Gblock curated ClustalW alignments was conducted as described in Figure 2. The subfamily classification is shown to the right. The different species are color coded and the number of genes identified in each species is indicated, according to the designations shown at the lower right.
The fish GalNAc-T gene family was found to be the largest, containing 21 members, followed by the human and chimp gene families with 20 members. Nineteen members were identified in rodents, 17 in chicken, 16 in frog, 14 in fly, 9 in worm and 5 in the microbial parasitic protozoan T. gondii. Notable differences in the size of individual subfamilies in different species include: (i) subfamily Id is missing (lacks a GALNT5 ortholog) in frog; (ii) Ie lacks GALNT8 orthologs in frog, chicken and mouse and GALNT18 is lacking in frog; (iii) subfamily Ig is missing (lacks a GALNT15 ortholog) in fish; (iv) subfamily If lacks GALNT20 orthologs in frog, chicken and fish and (v) subfamily IIa lacks a GALNT4 ortholog in chicken. Perhaps more significantly, subfamilies Ic and IIa are not found in worm and fly, and the low bootstrapping confidence at the in vertebrate Ie node in Figure 3 (1%) suggests that these genes evolved separate from the mammalian Ie subfamily. The microbial GALNT group in the Ib clade (Gondi-T1, -T2 and -T3) or Ie clade (Gondi-T4 and -T5). Due to the neglectable bootstrapping value (0) seen at the node branching the deuterostome and proteostome Ie lineages (Figure 3), it thus seems that the Ic, IIa and most likely the Ie lineages only arose in deuterostomes (including mammals). The mouse genome contains 19 GALNTs all located at syngenic human loci and these genes have identical genomic organization to the human genes. This variation in the number of paralogs and subfamilies in different species is likely to be a consequence of two evolutionary mechanisms: (i) lineage-specific paralog loss, which in a given species results in the specific loss of certain paralogs within a gene family and (ii) whole genome duplication (WGD) events that can result in species-specific genome duplication (Catchen et al. 2009). The fact that seven human subfamilies contain at least two members is in line with the proposed WGD model, which occurred twice during human evolution (Abi-Rached et al. 2002; McLysaght et al. 2002; Lundin et al. 2003; Dehal and Boore 2005). Time estimates have suggested that a primary duplication event occurred prior to the fish-tetrapod split followed by a second distinct genome duplication event early in vertebrate evolution (Jaillon et al. 2004). These proposed events are supported by studies establishing that the protostome (including nematodes/worm and arthropod/fly) and deuterostome lineages diverged 974 MYA and that mammal and actinopterygian fish (ray-finned fish) diverged 450 MYA (Hedges et al. 2004; Roger and Hug 2006). Thus, the human/fish and fly/worm GALNT families most likely evolved differently, which has resulted in a significant expansion of the Ie subfamily in fish with four GALNT8 paralogs, whereas this subfamily has been excluded in mouse and most likely in fly and worm too (Figure 3). The fish GALNT8 paralogs form two closely related pairs that reside on D. rerio chromosome 25 (GALNT8a/b) and chromosome 4 (GALNT8c/d) and are spaced 30 and 10 kb apart, respectively. The fish GALNT8c/d catalytic domain DNA sequences show 96% similarity. In addition, a fifth D. rerio GALNT8 ortholog may exist (BX004976.1), although this gene lacks the lectin domain encoded sequences and appears not to be transcribed (http://www.ensembl.org/). The D. melanogaster subfamily IIb is another example of species-specific subfamily expansion. The human IIb subfamily contains three genes (GALNT7/T10/T17), whereas 7 of the 14 GalNAc-Ts in D. melanogaster belong to subfamily IIb (Figure 3). The fly IIb subfamily contains the pgant8 cluster (CG7579, CG7304 and CG7297) colocalized to a 10-kb chromosome 3L locus and the pgant4 cluster (CG31956 and CG31776) c-localized to a 5-kb chromosome 2L locus. It should be noted that the CG7304 and CG7579 isoforms lack the essential DxH motif (found as DAQ and NGH, respectively) and they may represent non-functional gene elements. The enzymatic functions of two genes CG6394 (pgant7) and CG2103 (pgant6) within the fly subfamily IIb have been characterized and shown to exhibit the predicted glycopeptide substrate specificities (Ten Hagen et al. 2003).
Analysis of intron/exon positioning in human, fish, fly and worm support the model that GALNTs evolved from a common ancestral gene (Clausen and Bennett 1996). Of the three conserved human GALNT intron/exon boundaries, C1–C3 (Figure 2), both C1 and C2 are identically positioned in fish T1, T5, T11 and T14, as well as fly CG3254, CG31651, CG7480 and CG4445 genes (Supplementary data, Figure S2). C1 is identically positioned in worm Gly5 and 8, as well as fly CG7279, CG2103, CG31776, CG7579 and CG31956. C2 is identically positioned in fly CG30463 and worm Gly7 and Gly9 genes.
The prediction of true GalNAc-T orthologs among the different species analyzed in Figure 3 involves obvious uncertainties. Several studies have provided evidence that predicted orthologs from human, mouse and fly represent true functional orthologs (Schwientek et al. 2002; Ten Hagen and Tran 2002; Gerken et al. 2008). Interestingly, however, we found that the predicted orthologous genes in subfamily If, human GALNT11 and fly CG7480 (pgant35A), which were shown to have similar enzymatic functions (Schwientek et al. 2002), failed to complement each other in the lethal l(2)35Aa fly phenotype (Bennett et al. 2010).
Several parasitic genomes contain multiple GALNTs. Some of these have been expressed and shown to represent functional enzymes including ones from the protozoan apicomplexan parasite T. gondii (Stwora-Wojczyk et al. 2004). O-Linked GalNAc has been detected in the parasitic helminth Schistosoma mansoni (Nyame et al. 1987) and the protozoan parasite Trypanomsoma cruzi (Previato et al. 1998). Recent studies of T. cruzi genes have revealed the presence of two T. cruzi genes (TcOGNT1 and TcOGNT2) encoding enzymes distantly related to the mammalian GalNAc-T gene family. Both T. cruzi genes utilize UDP-GlcNAc and produce GlcNAcα1-O-Ser/Thr linkages (Heise et al. 2009).
Although GALNT orthologs were not identified in bacteria, plants or yeast, the presence of core mucin-type O-glycans in the fungus Cordyceps ophioglossoides has been reported (Kawaguchi et al. 1986).
Collectively, genetic, phylogenetic and functional information on GalNAc-Ts suggest that certain orthologs have been functionally conserved during evolution and that the evolution of the GalNAc-T family has resulted in both loss and expansion in the size and the number of subfamilies in different species.
Expression of GalNAc-transferases
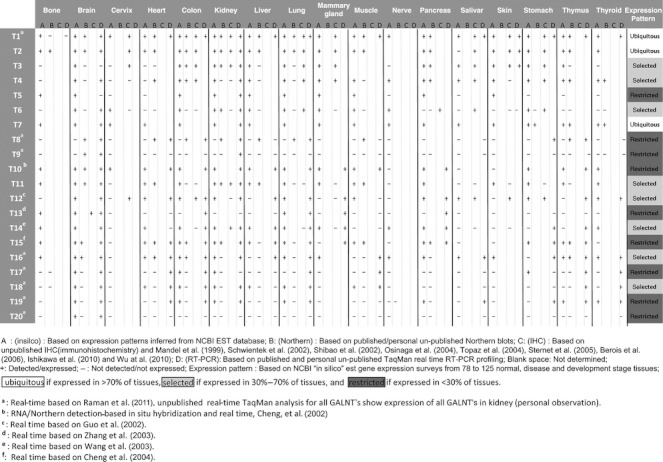
The GalNAc-Ts are differentially expressed in cells and tissues and marked changes in expression are found in cancer. Northern analyses of the first identified GalNAc-Ts showed that distinct isoforms have different expression patterns in mouse, rat (Hagen et al. 1997; Ten Hagen et al. 1998) and human organs (Bennett et al. 1996; Bennett, Hassan, et al. 1998). More comprehensive Northern studies indicate that some GalNAc-Ts are more ubiquitously expressed in organs, e.g. GALNT1 and T2 (Homa et al. 1993; White et al. 1995), whereas, for example, GALNT3, T4, T5, T7, T8 and T10 have more restricted expression patterns (Bennett et al. 1996; Bennett, Hassan, et al. 1998; Bennett, Hassan, Hollingsworth, et al. 1999; Ten Hagen et al. 1998, 2001; White et al. 2000; Table II). Northern analyses of human GALNT9 and rat GALNT19 indicate that expression of these genes is largely restricted to the brain (Toba et al. 2000; Nakamura et al. 2005), although human GALNT19 expression is also detected in a few other tissues (Table II; accompanying manuscript by Raman et al.). Northern and quantitative analyses of GALNT16 and T18 display broad expression profiles (unpublished and accompanying manuscript by Raman et al). Expression of GALNT20 by Northern analysis (unpublished) and quantitative reverse transcriptase (RT)–PCR analysis seems to be quite restricted to testis (accompanying manuscript by Raman et al.). Quantitative RT–PCR analysis of GALNT12 shows expression in several tissues (Guo et al. 2002), whereas GALNT13, T14 and T15 display broader expression patterns (Wang et al. 2003; Zhang et al. 2003; Cheng et al. 2004).
Table II.
Human GalNAc-T expression profiles based on in silico, Northern, IHC or RT–PCR
 |
Real-time RT–PCR expression profiling in mouse organs has corroborated a rather ubiquitous expression pattern for galnt1 and t2 and detected differential profiles for several of the other mouse genes such as galnt4 (detected in the colon, lung and sublingual gland), t12 (detected in the colon, spleen, thymus and sublingual gland) and t13 (not detected in any of the organs studied) (Young et al. 2003). In situ hybridization was used to profile the spatiotemporal expression pattern of seven murine galnts (galnt1/t2/t3/t4/t5/t7/t9) during mouse development, and in particular, galnt3, t5 and t7 were found to display unique patterns (Kingsley et al. 2000). RT–PCR and in situ hybridization have also been used to profile expression in Drosophila, and comprehensive semi-quantitative PCR analysis revealed expression of most of the eight isoforms tested during various developmental stages (Ten Hagen et al. 2003). Later studies of 12 of the 14 isoforms in the fly demonstrated unique expression patterns for CG3254 (pgant2), CG4445 (pgant3), CG7480 (pgant35A) and CG30463 during development. In particular, the CG3254 and CG30463 isoforms were found to display specific expression patterns related to eye development (Tian and Ten Hagen 2006).
Gene expression profiles can also be predicted “in silico” from the vast amount of information available from EST gene expression surveys. These surveys represent up to 125 normal, disease and development stage tissues and tissue-specific abundance information of individual human EST genes are provided (NCBI web site).
Table II summarizes our current understanding of the expression patterns of GALNTs as evaluated by mRNA as well as more limited information on protein expression by immunohistochemistry. Overall, there is reasonable agreement between expression results obtained from conventional Northern and immunohistochemical analyses, but the latter provides further detailed information on specific cell-type expression as well as subcellular topology. Discrepancies are observed. For instance, according to immunohistochemical experiments, GalNAc-T4, -T6 and -T12 were not expressed in the kidney, whereas the corresponding RNAs appear to be expressed (Table II). Similarly, the GalNAc-T14 enzyme was not found in the lung (Stern et al. 2010), whereas RNA appears to be expressed. Several issues may give rise to the discrepancies observed, such as differences in detection sensitivity of the methods used, and biological issues related to RNA and protein transcriptional and translational control and stability, which underscore the need for further systematic studies in this area. Regardless, we have attempted to summarize GalNAc-T expression in Table II with a relative expression score for each of the GALNT genes.
Direct monitoring of enzyme protein in cells is clearly preferable in order to assess the actual enzyme expression level, topology and function. We and others have put efforts into developing antibodies suitable for immunohistochemistry. A general problem has been selecting antibodies that react with both the native and the denatured proteins, and only a few monoclonal antibodies (MAbs) have been identified with these characteristics. We have built a library of MAbs to human GalNAc-Ts (T1-T4, -T6, -T11, -T12 and -T14) and selected pairs of MAbs that react with either the native soluble enzymes or the denatured protein (Figure 4; Bennett, Hassan, et al. 1998; Bennett, Hassan, Mandel, et al. 1999; Mandel et al. 1999; Schwientek et al. 2002). Immunohistochemical studies with these antibodies on frozen sections and cell lines have extended our understanding of differential expression of GalNAc-Ts and demonstrated that even the more ubiquitously expressed isoforms such as GalNAc-T1 and -T2 indeed have cell-specific expression patterns. An illustrative example is the multilayered stratified squamous epithelium of oral mucosa with its well-defined differentiation pattern where GalNAc-Ts are differentially expressed. Thus, GalNAc-T2 is expressed in the lower immature proliferative cell layers, GalNAc-T1 in the differentiated superficial cell layers and GalNAc-T3 in all cell layers. The application of antibodies has, furthermore, clearly demonstrated that the repertoire of GalNAc-Ts in cells changes during malignant transformation (Mandel et al. 1999). In general, cells express multiple GalNAc-Ts. The transcriptome of the important Chinese hamster ovary cell line CHO-K1 was recently characterized and it was found that only four GalNAc-Ts (-T2, -T7, -T11 and -T19) are expressed (Xu et al. 2011). One exception to this may be sperm cells, where we have examined expression of six GalNAc-Ts (GalNAc-T1, -T2, -T3, -T4, -T6 and -T11) and found that only GalNAc-T3 is expressed in the acrosome of spermatozoa (Bennett, Hassan, Mandel, et al. 1999; Mandel et al. 1999). This may be one example where the existence of a subfamily of GalNAc-Ts (GalNAc-T3 and -T6) with similar properties cannot provide a complete functional back-up due to differential expression (Rajpert-De et al. 2007). Two mouse knockout models of galnt3 exhibit testicular calcifications and male infertility (Esapa et al. 2009; Ichikawa et al. 2009; Duncan et al. 2011), but it is unclear if this also applies to human. Deficiency in GALNT3 is found in the related diseases FTC (familial tumoral calcinosis) and HHS (hyperostosis hyperphosphotemia syndrome) and one patient studied has been reported to exhibit testicular microlithiasis and oligoazoospermia (Campagnoli et al. 2006). However, family studies appear to indicate that males with FTC have produced offspring (Ichikawa et al. 2005; Carmichael et al. 2009). Further studies are clearly needed to assess the function of GALNT3 in fertility.
Fig. 4.
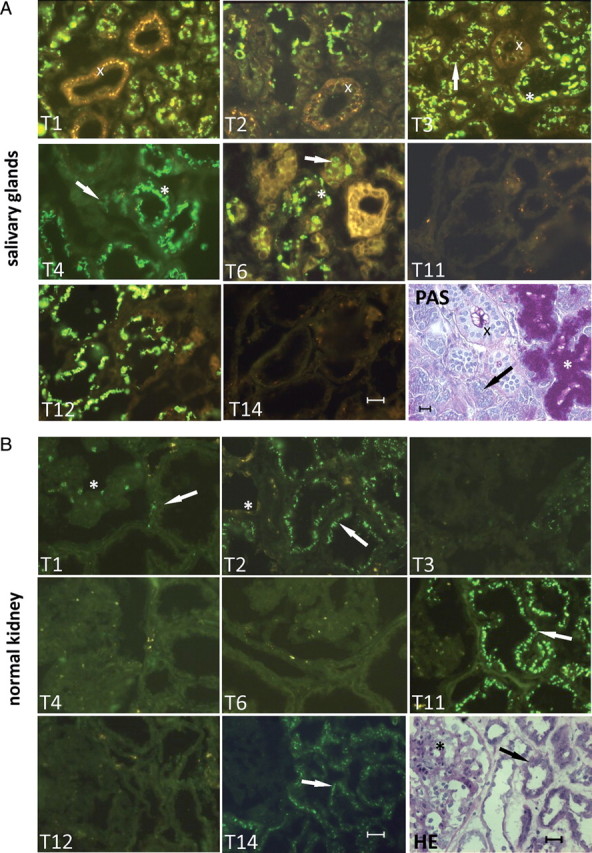
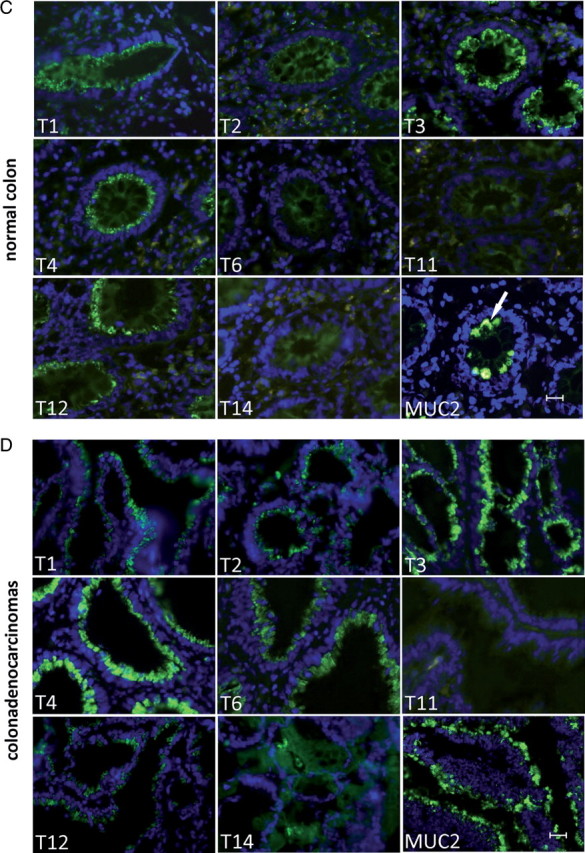
Illustration of expression patterns of eight GalNAc-T isoforms in normal (A) salivary glands, (B) kidney, (C) colon and in (D) colon adenocarcinomas evaluated by immunofluorescence histology using MAbs with well-characterized specificities. Each panel is labeled with the GalNAc-T isoform analyzed [GalNAc-T1 with MAb UH3 (4D8); GalNAc-T2 with MAb UH4 (4C4); GalNAc-T3 with MAb UH5 (2D10); GalNAc-T4 with MAb UH6 (4G2); GalNAc-T6 with MAb UH7 (2F3); GalNAc-T11 with MAb UH8 (1B2); GalNAc-T12 with MAb 1F9 (unpublished) and GalNAc-T14 with MAb 3D2 (unpublished)] (Bennett, Hassan, et al. 1998; Bennett, Hassan, Mandel, et al. 1999; Mandel et al. 1999; Schwientek et al. 2002), and the protocol used for staining fresh frozen sections was as described previously (Mandel et al. 1999). Positive FITC fluorescence is shown in green. (A) Neighboring sections were also stained with PAS, HE or MAb PMH1 to human GalNAc-glycosylated MUC2 as indicated (Reis et al. 1998). The PAS staining of salivary glands clearly marks mucous acini (indicated by asterisks), serous acini (indicated by arrows) and duct cells (indicated by crosss). (B) The HE staining of kidney marks glomeruli (indicated by asterisks) and tubules (indicated by arrows). (C and D) Staining for MUC2 in colon tissues marks goblet cells (indicated by an arrow). Colon tissues were counterstained with Dapi nuclear stain in blue. 20 μM scale bar is included in figures.
Examples of the exquisite cell-specific expression of GalNAc-Ts found in salivary glands, kidney and colon are illustrated in Figure 4. Although a large number of GalNAc-Ts are expressed in salivary glands (GalNAc-T11 and -T14 are not expressed), several isoforms show specific expression in different cell types. Thus, GalNAc-T3 is observed in the duct, serous and mucous cells, whereas GalNAc-T4 (Bennett, Hassan, et al. 1998) and -T6 are only weakly expressed in serous and strongly in mucous cells (Figure 4A). Similarly, in the kidney, GalNAc-T1 and -T2 are found in both tubules and glomeruli, whereas GalNAc-T11 (Schwientek et al. 2002) and -T14 are found only in tubules.
The repertoire of GalNAc-Ts also changes markedly in cancer. A number of studies have documented altered expression of GalNAc-Ts in cancer (Kohsaki et al. 2000; Shibao et al. 2002; Gu et al. 2004; Ishikawa et al. 2004; Landers et al. 2004; Miyahara et al. 2004; Yamamoto et al. 2004; Inoue et al. 2007). For example, GalNAc-T6 is not expressed in normal colon, but highly expressed in colon adenocarcinomas (Figure 4C and D). GalNAc-T6 has been associated with gastric carcinogenesis (Gomes et al. 2009), and T6 has also been associated with tumor stage ductal breast carcinomas (Berois et al. 2006) and is thus suggested as a new marker for breast cancer cell detection (Freire et al. 2006). In relation to this, a recent study has presented data proposing that elevated expression of GalNAc-T6 in breast cancer cells correlates with increased glycosylation and surface expression of the mucin MUC1 (Park et al. 2010). In a subsequent study, the same authors found that GalNAc-T6 was also important for the function of fibronectin-mediated adhesion (Park et al. 2011). Although these findings are intriguing, RNA knockdown strategies are known to have off-target effects and further studies are needed to support the conclusions. It is well-established that cancer cells produce aberrant glycosylation of proteins and lipids (Hakomori 1989, 1996; Dabelsteen 1996). However, the current information on aberrant glycosylation is generally limited to the structures of glycans and not to sites of attachment to proteins. Thus, our understanding of the effects of altered GalNAc-T expression in cancer is highly limited. It could be predicted that altered repertoire or topology of GalNAc-Ts in cells will result in altered density and patterns of O-glycans on proteins, but the means to address this have been limited. The only documented example of this is the apparent increased density of O-glycans on MUC1 in breast cancer cells (Muller et al. 1999; Muller and Hanisch 2002), but it is uncertain whether this relates to changes in the GalNAc-T repertoire or altered processing (Dalziel et al. 2001).
Evidence suggests that malignant transformation is associated with marked disorganization of the Golgi apparatus and that these changes may be responsible for the formation of cancer-associated glycosylation abnormalities (Kellokumpu et al. 2002). It has been put forward that a balanced signaling system monitors and controls Golgi vesicular trafficking rates. This signaling system involves protein chaperones, cytoskeletal proteins and a Golgi pool of Src kinases implicated in maintaining the dynamic equilibrium of the Golgi complex (Bard et al. 2003; Pulvirenti et al. 2008). It was recently shown that Src specifically regulates Golgi to ER retrograde traffic of GalNAc-Ts either directly (Gill et al. 2011) and/or through activation of the GTPase dynamin 2, a factor believed to be involved in Golgi vesiculation during secretion (Weller et al. 2010). Furthermore, a recent report suggests that certain GalNAc-T isoforms localize to the ER in cancer (Wang et al. 2011). Thus, the traditional view of an exclusive Golgi-localized subcellular topology of GalNAc-Ts is being challenged by these novel findings.
The regulatory events controlling GALNT expression still remain to be explored. So far, analysis of an upstream region of GALNT3 is the only report of the gene regulatory elements controlling GALNT gene expression (Nomoto et al. 1999). GALNT gene expression may also be modulated by microRNAs. One study proposed that the urothelium specific miR-129 targets pathways involved in cell death processes associated with poor bladder cancer outcome and one of the putative targets in this study was shown to be GALNT1 (Dyrskjot et al. 2009). Another study suggested that miR-378 involved in osteoblast differentiation targets GALNT7 (Kahai et al. 2009). More recently, miR-30d was shown to confer a pro-metastatic cancer effect through increased IL10 expression mediated by GALNT7 down regulation (Gaziel-Sovran et al. 2011). Interestingly, this study found that expression of multiple GALNTs was inversely correlated with miR-30d levels, suggesting that miR-30d regulates more isoforms.
Alternative splicing of GALNTs may affect functions. A large number of alternative splice variants are detected “in silico” (www.ensemble.com) for many GALNTs, but most of these are unlikely to encode functional proteins. In the accompanying paper, Raman et al. demonstrate that one GalNAc-T13V1 splice variant lacking part of its lectin domain had similar activity toward a panel of peptides and glycopeptides.
GalNAc-Ts and disease
A large number of congenital diseases of glycosylation (CDGs) have emerged in the last two decades. So far 45 CDGs have been described and most of these are found in the N-glycosylation pathway and in O-glycosylation pathways other than the mucin-type (Schachter and Freeze 2009; Jaeken 2010). Since the initiation steps of most types of protein glycosylation are catalyzed by only one or two glycosyltransferase genes, it is not surprising that defects in these genes have global effects on protein glycosylation and cause severe phenotypes. In contrast, the large number of GALNTs controlling the initiation step of mucin-type O-glycosylation may be expected to provide substantial biosynthetic back-up. Loss of a single GALNT gene may thus not produce discernable phenotypes or produce more discrete phenotypes in select organs. Early studies of GALNT-deficient mice confirmed the predicted redundancy, and several targeted GALNTs did not produce obvious phenotypes (Marth 1996; Lowe and Marth 2003). However, later investigations demonstrated that galnt1-deficient mice exhibit a bleeding disorder and have deficiency in B-cell maturation (Tenno et al. 2007). No overt phenotypes have been reported for mice deficient in galnt4 or t5 (Ten Hagen et al. 2002), deficient in both galnt4 and galnt5 (Mutant Mouse Regional Resource Centers stock number 029584-UCD) (Tabak 2010), deficient in galnt8 (Manzi et al. 2000), deficient in galnt10 (MMRC stock number 011647-UNC) or deficient in galnt14 (MMRC stock number 032320-UCD). Mice deficient in galnt13 exhibit decreased expression of Tn carbohydrate in brain tissues, but no apparent phenotype (Zhang et al. 2003). A more dramatic phenotype was observed by chance in Drosophila in an early study demonstrating that the recessive lethal mutations in l(2)35Aa (CG7480/pgant35A) could be rescued with genomic DNA encoding l(2)35Aa (Flores and Engels 1999). l(2)35Aa was shown to encode a functional enzyme (CG7480) orthologous to human GalNAc-T11, and the catalytic function of the enzyme was shown to be required for development (Schwientek et al. 2002; Ten Hagen and Tran 2002; Ten Hagen et al. 2009; Bennett et al. 2010). These studies demonstrated for the first time that individual GalNAc-T isoforms serve unique and essential functions and that the existence of a large GALNT gene family with 14 members in fly does not provide complete genetic and functional redundancy. Subsequent detailed analysis of the molecular mechanism causing the l(2)35Aa phenotype revealed that CG7480 is necessary for processing of tracheal cell glycoproteins that affect epithelial morphogenesis, which is needed to form an intact diffusion barrier in the Drosophila respiratory system (Tian and Ten Hagen 2007). More recently, deficiency in another Drosophila gene CG4445 (pgant3) was shown to cause a wing blistering phenotype (Zhang et al. 2010). Studies into the molecular mechanism behind this phenotype have determined that the extracellular matrix protein tiggrin is a specific substrate for the CG4445 enzyme and that the observed phenotype is caused by inappropriate secretion of extracellular matrix components, which alters cell adhesion events. Additionally, a recent study identified four Drosophila genes (CG31956, CG31651, CG6394 and CG30463) that are essential for viability, highlighting the essential developmental roles that are covered by O-glycosylation (Tran et al. 2011). A recent study of GalNAc-Ts in Xenopus has also proposed a specific function for the xgalntl1 isoform in neural and mesodermal tissue differentiation (GALNT16-predicted human ortholog. Herr et al. 2008). Although not clearly proven, this study suggests that xgalntl1 modulates the ActR-IIB receptor (TGF-β type II receptor family) at the molecular level, which implies that O-glycosylation may co-regulate TGF-β signaling in vertebrates. Xenopus left/right (LR) body patterning abnormalities were also observed in another Xenopus study demonstrating that xGalnt11 morpholino knockdown gave rise to abnormal heart and gut looping (Fakhro et al. 2011).
To date, only one human GALNT gene, GALNT3, has been shown to underlie disease in humans (Topaz et al. 2004). The molecular mechanism underlying the FTC and HHS phenotypes caused by deficiency in GALNT3 was shown to be a lack of O-glycosylation of a single Thr glycosylation site in a proprotein convertase (PC) processing site of the phosphaturic factor FGF23, which leads to excessive processing and inactivation of FGF23 (Kato et al. 2006). Importantly, later studies demonstrated that mutations in FGF23 preventing its secretion also lead to FTC. Thus, defects in either GALNT3 or FGF23 lead to insufficient circulating active FGF23 and the same overall clinical manifestations. GalNAc-T3 has broader substrate specificity than glycosylation of FGF23, but it appears that the main essential function of this isoform in humans is to co-regulate processing and blood levels of FGF23. A more recent study showed that GalNAc-T3 expression appears to be regulated by Ca2+ and vitamin D levels (Chefetz et al. 2009), indicating a direct regulatory role of GalNAc-T3 mediated O-glycosylation in phosphate homeostasis. The murine orthologous gene, galnt3, appears to serve similar functions and a knockout mouse exhibited reduced intact FGF23 blood levels (Ichikawa et al. 2009). The deficient mice did not develop the classical FTC features, but maleGalnt3−/− null mice showed growth retardation, infertility and increased bone mineral density. However, conflicting results were obtained in another preliminary study demonstrating that a galnt3 homozygous mutant mouse model harboring a W589R lectin domain mutation phenotypically displays hyperphosphataemia and calcinosis (Esapa et al. 2009; Duncan et al. 2011). Thus, the phenotypes of these mutant mice resemble the human disease caused by inactivating GALNT3 mutations. The example of GALNT3 serves to shows that defects in this large gene family may result in subtle organ selective deficiencies in O-glycosylation and discrete phenotypes that may be difficult to identify. The example also shows that mucin-type O-glycosylation has highly specific regulatory functions in fundamental biological pathways such as PC processing, which exemplifies the cellular need for a large differentially regulated gene family regulating diverse-specific functions.
GWASs suggest that other GALNT genes may serve specific functions yet to be uncovered. Several studies have implicated GALNT2 as a candidate gene regulating plasma lipid levels and propose that its dysfunction links to cardiovascular disease (Kathiresan et al. 2008; Willer et al. 2008). We have recently identified a putative molecular mechanism for involvement of GalNAc-T2 in lipid metabolism, which may be similar to the way GALNT3 regulates proprotein processing of FGF23. Thus, GalNAc-T2 appears to control O-glycosylation just adjacent to a PC processing site (RAPR224↓TT) in angiopoietin-like protein 3 (ANGPTL3) that activates this inhibitor of lipoprotein lipases (Schjoldager et al. 2010). A direct role for GALNT2 in lipid metabolism was shown in recent knockdown and overexpression studies of murine GalNAc-T2, where targeting in the liver affected plasma lipids levels (Teslovich et al. 2010).
Several other studies have linked GALNTs to disease or susceptibility to diseases including GALNT3 with bone mineral density and fracture risk (Duncan et al. 2011), GALNT4 with acute coronary disease (O'Halloran et al. 2009) and GALNT14 to resistance to death-receptor-mediated apoptosis (Wagner et al. 2007). Inactivating somatic and germline mutations have been found in GALNT5, T12, T15, T16 and T17 in cancer patients that potentially impair function (Wood et al. 2007; Guda et al. 2009). A very recent report has linked rare GALNT11 exon deletions in congenital heart disease patients with heterotaxy caused by abnormalities in LR body patterning (Fakhro et al. 2011). Besides GALNT11, this study identified four additional genes not previously implicated in LR patterning and all five genes are believed to act in the same pathway.
Perspectives
Research efforts during the last two decades have identified and characterized the GALNT gene family as the largest glycosyltransferase gene family known catalyzing the formation of a single glycosidic linkage. Although progress has been made in understanding the functions of this large gene family, we believe that we have only begun to see the proverbial tip of the iceberg of the intricate biology of how site-specific protein O-glycosylation is regulated and the role of each of the GalNAc-T isoforms in biosynthesis of the O-glycoproteome. As discussed here, a number of new strategies are currently being applied to advance the field. We believe that these will eventually show that the GalNAc O-glycoproteome is vastly greater than currently understood and that individual GalNAc-Ts serve highly specific and dynamically regulated functions in producing the O-glycoproteome in health and disease. Site-specific O-glycosylation is likely to regulate many other biological processes similar to the abundant PC processing event. We therefore predict that the GALNT gene family underlies many diseases and susceptibilities to diseases, which seem to be supported by emerging GWAS and other linkage studies.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by funds from the University of Copenhagen (Excellence program), Danish Researchcouncils and the Carlsberg Foundation.
Note added in proof
After manuscript acceptance GalNAc-T18 was described by Li et al. (Glycobiology. 2011 Dec 14. Epub ahead of print). The nomenclature used by Li et al. for GalNAc-T18 follows the nomenclature suggested in this review.
Conflict on interest
None declared.
Abbreviations
CDG, congenital diseases of glycosylation; ER, endoplasmic reticulum; EST, expressed sequence tag; FTC, familial tumoral calcinosis; HHS, hyperostosis hyperphosphotemia syndrome; Gal, galactose; GalNAc, N-acetylgalactosamine; Glc, glucose; GlcNAc, N-acetylglucosamine; GalNAc-T, polypeptide GalNAc-transferase; GALNT, GalNAc-transferase gene; GWAS, genome-wide association study; Hyl, hydroxylysine; LR, left/right; MAb, monoclonal antibody; MYA, million years ago; ORF, open reading frame; PC, proprotein convertase; PCR, polymerase chain reaction; psgene, pseudogenes; RT, reverse transcriptase; Ser, serine; Thr, threonine; UTR, untranslated region; WGD, whole genome duplication; XYLT1/2, protein O-xylosyltransferase 1/2.
Supplementary Material
Acknowledgements
We would like to thank all present and prior members of our labs who have contributed to the work presented in this review. A special thank to Steven B. Levery and Maya Bonde Haaland for revision of the manuscript.
References
- Abi-Rached L, Gilles A, Shiina T, Pontarotti P, Inoko H. Evidence of en bloc duplication in vertebrate genomes. Nat Genet. 2002;31:100–105. doi: 10.1038/ng855. [DOI] [PubMed] [Google Scholar]
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ. Rumi Is a CAP10 Domain Glycosyltransferase that Modifies Notch and Is Required for Notch Signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R, Levery SB, Mandel U, Kresse H, Schwientek T, Bennett EP, Clausen H. Cloning and expression of a proteoglycan UDP-galactose:β-xylose β1, 4-galactosyltransferase I. A seventh member of the human β4-galactosyltransferase gene family. J Biol Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- Bard F, Mazelin L, Pechoux-Longin C, Malhotra V, Jurdic P. Src regulates Golgi structure and KDEL receptor-dependent retrograde transport to the endoplasmic reticulum. J Biol Chem. 2003;278:46601–46606. doi: 10.1074/jbc.M302221200. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin CM, Schmid CW, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Chen YW, Schwientek T, Mandel U, Schjoldager KB, Cohen SM, Clausen H. Rescue of Drosophila melanogaster l(2)35Aa lethality is only mediated by polypeptide GalNAc-transferase pgant35A, but not by the evolutionary conserved human ortholog GalNAc-transferase-T11. Glycoconj J. 2010;27:435–444. doi: 10.1007/s10719-010-9290-5. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Clausen H. cDNA cloning and expression of a novel human UDP-N-acetyl-α-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase, GalNAc-T3. J Biol Chem. 1996;271:17006–17012. doi: 10.1074/jbc.271.29.17006. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Hollingsworth MA, Clausen H. A novel human UDP-N-acetyl-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 1999;460:226–230. doi: 10.1016/s0014-5793(99)01268-5. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Hollingsworth MA, Akisawa N, Ikematsu Y, Merkx G, van Kessel AG, Olofsson S, Clausen H. Cloning and characterization of a close homologue of human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J Biol Chem. 1999;274:25362–25370. doi: 10.1074/jbc.274.36.25362. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, van Kessel AG, et al. Cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J Biol Chem. 1998;273:30472–30481. doi: 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Weghuis DO, Merkx G, van Kessel AG, Eiberg H, Clausen H. Genomic organization and chromosomal localization of three members of the UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase family. Glycobiology. 1998;8:547–555. doi: 10.1093/glycob/8.6.547. [DOI] [PubMed] [Google Scholar]
- Berois N, Mazal D, Ubillos L, Trajtenberg F, Nicolas A, Sastre-Garau X, Magdelenat H, Osinaga E. UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J Histochem Cytochem. 2006;54:317–328. doi: 10.1369/jhc.5A6783.2005. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Henrissat B. Glycoside hydrolases and glycosyltransferases: Families and functional modules. Curr Opin Struct Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- Campagnoli MF, Pucci A, Garelli E, Carando A, Defilippi C, Lala R, Ingrosso G, Dianzani I, Forni M, Ramenghi U. Familial tumoral calcinosis and testicular microlithiasis associated with a new mutation of GALNT3 in a white family. J Clin Pathol. 2006;59:440–442. doi: 10.1136/jcp.2005.026369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael KD, Bynum JA, Evans EB. Familial tumoral calcinosis: A forty-year follow-up on one family. J Bone Joint Surg Am. 2009;91:664–671. doi: 10.2106/JBJS.G.01512. [DOI] [PubMed] [Google Scholar]
- Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefetz I, Kohno K, Izumi H, Uitto J, Richard G, Sprecher E. GALNT3, a gene associated with hyperphosphatemic familial tumoral calcinosis, is transcriptionally regulated by extracellular phosphate and modulates matrix metalloproteinase activity. Biochim Biophys Acta. 2009;1792:61–67. doi: 10.1016/j.bbadis.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Tachibana K, Zhang Y, Guo J, Kahori Tachibana K, Kameyama A, Wang H, Hiruma T, Iwasaki H, Togayachi A, et al. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T10. FEBS Lett. 2002;531:115–121. doi: 10.1016/s0014-5793(02)03399-9. [DOI] [PubMed] [Google Scholar]
- Cheng L, Tachibana K, Iwasaki H, Kameyama A, Zhang Y, Kubota T, Hiruma T, Tachibana K, Kudo T, Guo JM, et al. Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T15. FEBS Lett. 2004;566:17–24. doi: 10.1016/j.febslet.2004.03.108. [DOI] [PubMed] [Google Scholar]
- Clausen H, Bennett EP. A family of UDP-GalNAc:polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology. 1996;6:635–646. doi: 10.1093/glycob/6.6.635. [DOI] [PubMed] [Google Scholar]
- Condac E, Silasi-Mansat R, Kosanke S, Schoeb T, Towner R, Lupu F, Cummings RD, Hinsdale ME. Polycystic disease caused by deficiency in xylosyltransferase 2, an initiating enzyme of glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 2007;104:9416–9421. doi: 10.1073/pnas.0700908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabelsteen E. Cell surface carbohydrates as prognostic markers in human carcinomas. J Pathol. 1996;179:358–369. doi: 10.1002/(SICI)1096-9896(199608)179:4<358::AID-PATH564>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Dalziel M, Whitehouse C, McFarlane I, Brockhausen I, Gschmeissner S, Schwientek T, Clausen H, Burchell JM, Taylor-Papadimitriou J. The relative activities of the C2GnT1 and ST3Gal-I glycosyltransferases determine O-glycan structure and expression of a tumor-associated epitope on MUC1. J Biol Chem. 2001;276:11007–11015. doi: 10.1074/jbc.M006523200. [DOI] [PubMed] [Google Scholar]
- DeFrees S, Wang ZG, Xing R, Scott AE, Wang J, Zopf D, Gouty DL, Sjoberg ER, Panneerselvam K, Brinkman-Van der Linden EC, et al. GlycoPEGylation of recombinant therapeutic proteins produced in Escherichia coli. Glycobiology. 2006;16:833–843. doi: 10.1093/glycob/cwl004. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd RB, Drickamer K. Lectin-like proteins in model organisms: Implications for evolution of carbohydrate-binding activity. Glycobiology. 2001;11:77R–79R. doi: 10.1093/glycob/11.5.71r. [DOI] [PubMed] [Google Scholar]
- Du J, Takeuchi H, Leonhard-Melief C, Shroyer KR, Dlugosz M, Haltiwanger RS, Holdener BC. O-Fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev. Biol. 2010;346:25–38. doi: 10.1016/j.ydbio.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, Nicholson GC, Eastell R, Prince RL, Eisman JA, Jones G, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrskjot L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- Elhammer A, Kornfeld S. Purification and characterization of UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase from bovine colostrum and murine lymphoma BW5147 cells. J Biol Chem. 1986;261:5249–5255. [PubMed] [Google Scholar]
- Esapa C, Head R, Chan E, Crane M, Cheeseman M, Hough T, McNally E, Carr A, Thomas G, Brwon M, et al. A mouse with a Trp589Arg mutation in N-acetylgalactosaminyltransferase 3 (Galnt3) provides a model for familial tumoral calcinosis. Endocr Abstr. 2009;19:19-OC31. [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci USA. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H. Regulation of the mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, Engels W. Microsatellite instability in Drosophila spellchecker1 (MutS homolog) mutants. Proc Natl Acad Sci USA. 1999;96:2964–2969. doi: 10.1073/pnas.96.6.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire T, Berois N, Sonora C, Varangot M, Barrios E, Osinaga E. UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a potential new marker for detection of bone marrow-disseminated breast cancer cells. Int J Cancer. 2006;119:1383–1388. doi: 10.1002/ijc.21959. [DOI] [PubMed] [Google Scholar]
- Fritz TA, Hurley JH, Trinh LB, Shiloach J, Tabak LA. The beginnings of mucin biosynthesis: The crystal structure of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-T1. Proc Natl Acad Sci USA. 2004;101:15307–15312. doi: 10.1073/pnas.0405657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- Gaziel-Sovran A, Segura MF, Di MR, Collins MK, Hanniford D, Vega-Saenz de ME, Rakus JF, Dankert JF, Shang S, Kerbel RS, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Hagen KGT, Jamison O. Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase orthologue pairs. Glycobiology. 2008;18:861–870. doi: 10.1093/glycob/cwn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Jamison O, Perrine CL, Collette JC, Moinova H, Ravi L, Markowitz SD, Shen W, Patel H, Tabak LA. Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J Biol Chem. 2011;286:14493–14507. doi: 10.1074/jbc.M111.218701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken TA, Raman J, Fritz TA, Jamison O. Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem. 2006;281:32403–32416. doi: 10.1074/jbc.M605149200. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189:843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DJ, Clausen H, Bard F. Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011;21(3):149–158. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Gomes J, Marcos NT, Berois N, Osinaga E, Magalhaes A, Pinto-de-Sousa J, Almeida R, Gartner F, Reis CA. Expression of UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal metaplasia, and gastric carcinoma. J Histochem Cytochem. 2009;57:79–86. doi: 10.1369/jhc.2008.952283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotting C, Kuhn J, Kleesiek K. Human xylosyltransferases in health and disease. Cell Mol Life Sci. 2007;64:1498–1517. doi: 10.1007/s00018-007-7069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Oyama T, Osaki T, Li J, Takenoyama M, Izumi H, Sugio K, Kohno K, Yasumoto K. Low expression of polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in lung adenocarcinoma: Impact on poor prognosis and early recurrence. Br J Cancer. 2004;90:436–442. doi: 10.1038/sj.bjc.6601531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci USA. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JM, Zhang Y, Cheng L, Iwasaki H, Wang H, Kubota T, Tachibana K, Narimatsu H. Molecular cloning and characterization of a novel member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, pp-GalNAc-T12. FEBS Lett. 2002;524:211–218. doi: 10.1016/s0014-5793(02)03007-7. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Hagen KG, Beres TM, Balys MM, VanWuyckhuyse BC, Tabak LA. cDNA cloning and expression of a novel UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1997;272:13843–13848. doi: 10.1074/jbc.272.21.13843. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Hazes B, Raffo R, deSa D, Tabak LA. Structure-function analysis of the UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. Essential residues lie in a predicted active site cleft resembling a lactose repressor fold. J Biol Chem. 1999;274:6797–6803. doi: 10.1074/jbc.274.10.6797. [DOI] [PubMed] [Google Scholar]
- Hagen FK, Van Wuyckhuyse B, Tabak LA. Purification, cloning, and expression of a bovine UDP-GalNAc:polypeptide N-acetyl-galactosaminyltransferase. J Biol Chem. 1993;268:18960–18965. [PubMed] [Google Scholar]
- Hagopian A, Eylar E . Glycoprotein biosynthesis: The purification and characterization of a polypeptide. N-acetylgalactosaminyl transferase from bovine submaxillary glands. Arch Biochem Biophys. 1969;129:515–524. doi: 10.1016/0003-9861(69)90209-4. [DOI] [PubMed] [Google Scholar]
- Hagopian A, Eylar EH. Glycoprotein biosynthesis: Studies on the receptor specificity of the polypeptidyl:N-acetylgalactosaminyl transferase from bovine submaxillary glands. Arch Biochem Biophys. 1968;128:422–433. doi: 10.1016/0003-9861(68)90048-9. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Aberrant glycosylation in tumors and tumor-associated carbohydrate antigens. Adv Cancer Res. 1989;52:257–331. doi: 10.1016/s0065-230x(08)60215-8. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996;56:5309–5318. [PubMed] [Google Scholar]
- Halim A, Brinkmalm G, Ruetschi U, Westman-Brinkmalm A, Portelius E, Zetterberg H, Blennow K, Larson G, Nilsson J. Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid β-peptides in human cerebrospinal fluid. Proc Natl Acad Sci USA. 2011;108:11848–11853. doi: 10.1073/pnas.1102664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H, Bennett EP, Mandel U, Hollingsworth MA, Clausen H. Control of Mucin-Type O-Glycosylation: O-Glycan Occupancy is Directed by Substrate Specificities of Polypeptide GalNAc-Transferases. 2000:p273–292. Wiley-VCH Publishers. [Google Scholar]
- Hassan H, Reis CA, Bennett EP, Mirgorodskaya E, Roepstorff P, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, Clausen H. The lectin domain of UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J Biol Chem. 2000;275:38197–38205. doi: 10.1074/jbc.M005783200. [DOI] [PubMed] [Google Scholar]
- Hazes B. The (QxW)(3) domain: A flexible lectin scaffold. Protein Sci. 1996;5:1490–1501. doi: 10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesen Ste, Knauer R, Lehle L, Aebi M. Yeast Wbp1p and Swp1p form a protein complex essential for oligosaccharyl transferase activity. EMBO J. 1993;12:279–284. doi: 10.1002/j.1460-2075.1993.tb05654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise N, Singh D, van der Wel H, Sassi SO, Johnson JM, Feasley CL, Koeller CM, Previato JO, Mendonca-Previato L, West CM. Molecular analysis of a UDP-GlcNAc:polypeptide α-N-acetylglucosaminyltransferase implicated in the initiation of mucin-type O-glycosylation in Trypanosoma cruzi. Glycobiology. 2009;19:918–933. doi: 10.1093/glycob/cwp068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr P, Korniychuk G, Yamamoto Y, Grubisic K, Oelgeschlager M. Regulation of TGF-β signalling by N-acetylgalactosaminyltransferase-like 1. Development. 2008;135:1813–1822. doi: 10.1242/dev.019323. [DOI] [PubMed] [Google Scholar]
- Hill HD, Jr, Reynolds JA, Hill RL. Purification, composition, molecular weight, and subunit structure of ovine submaxillary mucin. J Biol Chem. 1977;252:3791–3798. [PubMed] [Google Scholar]
- Homa FL, Hollander T, Lehman DJ, Thomsen DR, Elhammer AP. Isolation and expression of a cDNA clone encoding a bovine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1993;268:12609–12616. [PubMed] [Google Scholar]
- Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010;584:2526–2538. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- Ichikawa S, Lyles KW, Econs MJ. A novel GALNT3 mutation in a pseudoautosomal dominant form of tumoral calcinosis: Evidence that the disorder is autosomal recessive. J Clin Endocrinol Metab. 2005;90:2420–2423. doi: 10.1210/jc.2004-2302. [DOI] [PubMed] [Google Scholar]
- Ichikawa S, Sorenson AH, Austin AM, Mackenzie DS, Fritz TA, Moh A, Hui SL, Econs MJ. Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology. 2009;150:2543–2550. doi: 10.1210/en.2008-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A, Piller V, Piller F, Breton C. Fold recognition and molecular modeling of a lectin-like domain in UDP-GalNac:polypeptide N-acetylgalactosaminyltransferases. Protein Eng. 1997;10:1353–1356. doi: 10.1093/protein/10.12.1353. [DOI] [PubMed] [Google Scholar]
- Inoue T, Eguchi T, Oda Y, Nishiyama K, Fujii K, Izumi H, Kohno K, Yamaguchi K, Tanaka M, Tsuneyoshi M. Expression of GalNAc-T3 and its relationships with clinicopathological factors in 61 extrahepatic bile duct carcinomas analyzed using stepwise sections—special reference to its association with lymph node metastases. Mod Pathol. 2007;20:267–276. doi: 10.1038/modpathol.3800700. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Kitayama J, Nariko H, Kohno K, Nagawa H. The expression pattern of UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase-3 in early gastric carcinoma. J Surg Oncol. 2004;86:28–33. doi: 10.1002/jso.20042. [DOI] [PubMed] [Google Scholar]
- Jaeken J. Congenital disorders of glycosylation. Ann N Y Acad Sci. 2010;1214:190–198. doi: 10.1111/j.1749-6632.2010.05840.x. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS One. 2009;4:1–14. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Nishihara S, Narimatsu H, Saitou N. The evolutionary history of glycosyltransferase genes. Trends Glycosci Glycotechnol. 2000;13:147–155. [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Jeanneau C, Tarp MA, Benet-Pages A, Lorenz-Depiereux B, Bennett EP, Mandel U, Strom TM, Clausen H. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J Biol Chem. 2006;281:18370–18377. doi: 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi N, Ohmori T, Takeshita Y, Kawanishi G, Katayama S, Yamada H. Occurrence of Gal β(1, 3) GalNAc-Ser/Thr in the linkage region of polygalactosamine containing fungal glycoprotein from Cordyceps ophioglossoides. Biochem Biophys Res Commun. 1986;140:350–356. doi: 10.1016/0006-291x(86)91097-1. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47–62. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Kellokumpu S, Sormunen R, Kellokumpu I. Abnormal glycosylation and altered Golgi structure in colorectal cancer: Dependence on intra-Golgi pH. FEBS Lett. 2002;516:217–224. doi: 10.1016/s0014-5793(02)02535-8. [DOI] [PubMed] [Google Scholar]
- Kingsley PD, Hagen KGT, Maltby KM, Zara J, Tabak LA. Diverse spatial expression patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyl-transferase family member mRNAs during mouse development. Glycobiology. 2000;10:1317–1323. doi: 10.1093/glycob/10.12.1317. [DOI] [PubMed] [Google Scholar]
- Kohsaki T, Nishimori I, Nakayama H, Miyazaki E, Enzan H, Nomoto M, Hollingsworth MA, Onishi S. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isozymes T1 and T2 in human colorectal cancer. J Gastroenterol. 2000;35:840–848. doi: 10.1007/s005350070021. [DOI] [PubMed] [Google Scholar]
- Kubota T, Shiba T, Sugioka S, Furukawa S, Sawaki H, Kato R, Wakatsuki S, Narimatsu H. Structural basis of carbohydrate transfer activity by human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J Mol Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC. Fascins, and their roles in cell structure and function. Bioessays. 2002;24:350–361. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- Landers KA, Burger MJ, Tebay MA, Purdie DM, Scells B, Samaratunga H, Lavin MF, Gardiner RA. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int J Cancer. 2004;114:950–956. doi: 10.1002/ijc.20760. [DOI] [PubMed] [Google Scholar]
- Liu C, Gaspar JA, Wong HJ, Meiering EM. Conserved and nonconserved features of the folding pathway of hisactophilin, a beta-trefoil protein. Protein Sci. 2002;11:669–679. doi: 10.1110/ps.31702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M, Willer T, Cruces J, Strahl S. POMT1 is essential for protein O-mannosylation in mammals. Methods Enzymol. 2010;479:323–342. doi: 10.1016/S0076-6879(10)79018-2. [DOI] [PubMed] [Google Scholar]
- Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics. 2003;3:53–63. [PubMed] [Google Scholar]
- Mandel U, Hassan H, Therkildsen MH, Rygaard J, Jakobsen MH, Juhl BR, Dabelsteen E, Clausen H. Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: Immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology. 1999;9:43–52. doi: 10.1093/glycob/9.1.43. [DOI] [PubMed] [Google Scholar]
- Manzi AE, Norgard-Sumnicht K, Argade S, Marth JD, van Halbeek H, Varki A. Exploring the glycan repertoire of genetically modified mice by isolation and profiling of the major glycan classes and nano-NMR analysis of glycan mixtures. Glycobiology. 2000;10:669–689. doi: 10.1093/glycob/10.7.669. [DOI] [PubMed] [Google Scholar]
- Marth JD. Complexity in O-linked oligosaccharide biosynthesis engendered by multiple polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 1996;6:701–705. doi: 10.1093/glycob/6.7.701. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J Biol Chem. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- McGuire EJ, Roseman S. Enzymatic synthesis of the protein-hexosamine linkage in sheep submaxillary mucin. J Biol Chem. 1967;242:3745–3747. [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Meurer JA, Drong RF, Homa FL, Slightom JL, Elhammer AP. Organization of a human UDP-GalNAc:polypeptide, N-acetylgalactosaminyltransferase gene and a related processed pseudogene. Glycobiology. 1996;6:231–241. doi: 10.1093/glycob/6.2.231. [DOI] [PubMed] [Google Scholar]
- Miyahara N, Shoda J, Kawamoto T, Furukawa M, Ueda T, Todoroki T, Tanaka N, Matsuo K, Yamada Y, Kohno K, et al. Expression of UDP-N-acetyl-α-D-galactosamine-polypeptide N-acetylgalactosaminyltransferase isozyme 3 in the subserosal layer correlates with postsurgical survival of pathological tumor stage 2 carcinoma of the gallbladder. Clin Cancer Res. 2004;10:2090–2099. doi: 10.1158/1078-0432.ccr-1024-03. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D. The molecular evolutionary history of a winged bean alpha-chymotrypsin inhibitor and modeling of its mutations through structural analyses. J Mol Evol. 2000;50:214–223. doi: 10.1007/s002399910024. [DOI] [PubMed] [Google Scholar]
- Muller S, Alving K, Peter-Katalinic J, Zachara N, Gooley AA, Hanisch FG. High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J Biol Chem. 1999;274:18165–18172. doi: 10.1074/jbc.274.26.18165. [DOI] [PubMed] [Google Scholar]
- Muller S, Hanisch FG. Recombinant MUC1 probe authentically reflects cell-specific O-glycosylation profiles of endogenous breast cancer mucin. High density and prevalent core 2-based glycosylation. J Biol Chem. 2002;277:26103–26112. doi: 10.1074/jbc.M202921200. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Toba S, Hirai M, Morishita S, Mikami T, Konishi M, Itoh N, Kurosaka A. Cloning and expression of a brain-specific putative UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase gene. Biol Pharm Bull. 2005;28:429–433. doi: 10.1248/bpb.28.429. [DOI] [PubMed] [Google Scholar]
- Nehrke K, Hagen FK, Tabak LA. Isoform-specific O-glycosylation by murine UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-T3, in vivo. Glycobiology. 1998;8:367–371. doi: 10.1093/glycob/8.4.367. [DOI] [PubMed] [Google Scholar]
- Nomoto M, Izumi H, Ise T, Kato K, Takano H, Nagatani G, Shibao K, Ohta R, Imamura T, Kuwano M, et al. Structural basis for the regulation of UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase-3 gene expression in adenocarcinoma cells. Cancer Res. 1999;59:6214–6222. [PubMed] [Google Scholar]
- Nyame K, Cummings RD, Damian RT. Schistosoma mansoni synthesizes glycoproteins containing terminal O-linked N-acetylglucosamine residues. J Biol Chem. 1987;262:7990–7995. [PubMed] [Google Scholar]
- O'Connell B, Tabak LA, Ramasubbu N. The influence of flanking sequences on O-glycosylation. Biochem Biophys Res Commun. 1991;180:1024–1030. doi: 10.1016/s0006-291x(05)81168-4. [DOI] [PubMed] [Google Scholar]
- O'Halloran AM, Patterson CC, Horan P, Maree A, Curtin R, Stanton A, McKeown PP, Shields DC. Genetic polymorphisms in platelet-related proteins and coronary artery disease: Investigation of candidate genes, including N-acetylgalactosaminyltransferase 4 (GALNT4) and sulphotransferase 1A1/2 (SULT1A1/2) J Thromb Thrombolysis. 2009;27:175–184. doi: 10.1007/s11239-008-0196-z. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:1–12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Katagiri T, Chung S, Kijima K, Nakamura Y. Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia. 2011;13:320–326. doi: 10.1593/neo.101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- Paulson JC, Colley KJ. Glycosyltransferases. Structure, localization, and control of cell type-specific glycosylation. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- Pedersen JW, Bennett EP, Schjoldager K, Sjoberg A, Levery SB, Meldal M, Clausen H, Wandall HH. Lectin domains of polypeptide GalNAc-Ts exhibit glycopeptide binding specificity. J Biol Chem. 2011;286:32684–32696. doi: 10.1074/jbc.M111.273722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Togayachi A, Kwon YD, Xie C, Wu G, Zou X, Sato T, Ito H, Tachibana K, Kubota T, et al. Identification of a novel human UDP-GalNAc transferase with unique catalytic activity and expression profile. Biochem Biophys Res Commun. 2010;402:680–686. doi: 10.1016/j.bbrc.2010.10.084. [DOI] [PubMed] [Google Scholar]
- Perrine CL, Ganguli A, Wu P, Bertozzi CR, Fritz TA, Raman J, Tabak LA, Gerken TA. Glycopeptide-preferring polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol Chem. 2009;284:20387–20397. doi: 10.1074/jbc.M109.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Russell RB. Identification of distant homologues of fibroblast growth factors suggests a common ancestor for all beta-trefoil proteins. J Mol Biol. 2000;302:1041–1047. doi: 10.1006/jmbi.2000.4087. [DOI] [PubMed] [Google Scholar]
- Previato JO, Sola-Penna M, Agrellos OA, Jones C, Oeltmann T, Travassos LR, Mendonca-Previato L. Biosynthesis of O-N-acetylglucosamine-linked glycans in Trypanosoma cruzi. Characterization of the novel uridine diphospho-N-acetylglucosamine:polypeptide N-acetylglucosaminyltransferase-catalyzing formation of N-acetylglucosamine alpha O-threonine. J Biol Chem. 1998;273:14982–14988. doi: 10.1074/jbc.273.24.14982. [DOI] [PubMed] [Google Scholar]
- Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, San PE, Beznoussenko GV, Mironov AA, Turacchio G, et al. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol. 2008;10:912–922. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]
- Rajpert-De ME, Poll SN, Goukasian I, Jeanneau C, Herlihy AS, Bennett EP, Skakkebaek NE, Clausen H, Giwercman A, Mandel U. Changes in the profile of simple mucin-type O-glycans and polypeptide GalNAc-transferases in human testis and testicular neoplasms are associated with germ cell maturation and tumour differentiation. Virchows Arch. 2007;451:805–814. doi: 10.1007/s00428-007-0478-4. [DOI] [PubMed] [Google Scholar]
- Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Lu M, Tabak LA. The catalytic and lectin domains of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem. 2008;283:22942–22951. doi: 10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J, Guan Y, Perrine CL, Gerken TA, Tabak LA. UDP-N-Acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyltransferases: Completion of the Family Tree. Glycobiology. 2011 Dec 20 doi: 10.1093/glycob/cwr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautavuoma K, Takaluoma K, Sormunen R, Myllyharju J, Kivirikko KI, Soininen R. Premature aggregation of type IV collagen and early lethality in lysyl hydroxylase 3 null mice. Proc Natl Acad Sci USA. 2004;101:14120–14125. doi: 10.1073/pnas.0404966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeuwijk Jv, Maugenre S, van den Elzen C, Verrips A, Bertini E, Muntoni F, Merlini L, Scheffer H, Brunner HG, Guicheney P, et al. The expanding phenotype of POMT1 mutations: From Walker-Warburg syndrome to congenital muscular dystrophy, microcephaly, and mental retardation. Hum Mutat. 2006;27:453–459. doi: 10.1002/humu.20313. [DOI] [PubMed] [Google Scholar]
- Reis CA, Sorensen T, Mandel U, David L, Mirgorodskaya E, Roepstorff P, Kihlberg J, Hansen JE, Clausen H. Development and characterization of an antibody directed to an alpha-N-acetyl-D-galactosamine glycosylated MUC2 peptide. Glycoconj J. 1998;15:51–62. doi: 10.1023/a:1006939432665. [DOI] [PubMed] [Google Scholar]
- Roger AJ, Hug LA. The origin and diversification of eukaryotes: Problems with molecular phylogenetics and molecular clock estimation. Philos Trans R Soc Lond B Biol Sci. 2006;361:1039–1054. doi: 10.1098/rstb.2006.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111:45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- Rutenber E, Ready M, Robertus JD. Structure and evolution of ricin B chain. Nature. 1987;326:624–626. doi: 10.1038/326624a0. [DOI] [PubMed] [Google Scholar]
- Sakaidani Y, Furukawa K, Okajima T. O-GlcNAc modification of the extracellular domain of Notch receptors. Methods Enzymol. 2010;480:355–373. doi: 10.1016/S0076-6879(10)80016-3. [DOI] [PubMed] [Google Scholar]
- Schachter H, Freeze HH. Glycosylation diseases: Quo vadis? Biochim Biophys Acta. 2009;1792:925–930. doi: 10.1016/j.bbadis.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Vester-Christensen MB, Bennett EP, Levery SB, Schwientek T, Yin W, Blixt O, Clausen H. O-glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3: Possible role of polypeptide GalNAc-transferase-2 in regulation of concentrations of plasma lipids. J Biol Chem. 2010;285:36293–36303. doi: 10.1074/jbc.M110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schafer MA, et al. Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in drosophila. J Biol Chem. 2002;277:22623–22638. doi: 10.1074/jbc.M202684200. [DOI] [PubMed] [Google Scholar]
- Schwientek T, Mandel U, Roth U, Muller S, Hanisch FG. A serial lectin approach to the mucin-type O-glycoproteome of Drosophila melanogaster S2 cells. Proteomics. 2007;7:3264–3277. doi: 10.1002/pmic.200600793. [DOI] [PubMed] [Google Scholar]
- Sheehan JK, Kirkham S, Howard M, Woodman P, Kutay S, Brazeau C, Buckley J, Thornton DJ. Identification of molecular intermediates in the assembly pathway of the MUC5AC mucin. J Biol Chem. 2004;279:15698–15705. doi: 10.1074/jbc.M313241200. [DOI] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibao K, Izumi HNY, Ohta R, Nagata N, Nomoto M, Matsuo K, Yamada Y, Kitazato K, Itoh H, et al. Expression of UDP-N-acetyl-α-D-galactosamine-polypeptide galNAc N-acetylgalactosaminyl transferase-3 in relation to differentiation and prognosis in patients with colorectal carcinoma. Cancer. 2002;94:1939–1946. doi: 10.1002/cncr.10423. [DOI] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen T, White T, Wandall HH, Kristensen AK, Roepstorff P, Clausen H. UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1995;270:24166–24173. doi: 10.1074/jbc.270.41.24166. [DOI] [PubMed] [Google Scholar]
- Soya N, Fang Y, Palcic MM, Klassen JS. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology. 2011;21:547–552. doi: 10.1093/glycob/cwq190. [DOI] [PubMed] [Google Scholar]
- Stanley P. Glucose: A novel regulator of notch signaling. ACS Chem Biol. 2008;3:210–213. doi: 10.1021/cb800073x. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Stern HM, Padilla M, Wagner K, Amler L, Ashkenazi A. Development of immunohistochemistry assays to assess GALNT14 and FUT3/6 in clinical trials of dulanermin and drozitumab. Clin Cancer Res. 2010;16:1587–1596. doi: 10.1158/1078-0432.CCR-09-3108. [DOI] [PubMed] [Google Scholar]
- Stwora-Wojczyk MM, Kissinger JC, Spitalnik SL, Wojczyk BS. O-glycosylation in Toxoplasma gondii: Identification and analysis of a family of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Int J Parasitol. 2004;34:309–322. doi: 10.1016/j.ijpara.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Sugiura M, Kawasaki T, Yamashina I. Purification and characterization of UDP-GalNAc:polypeptide N-acetylgalactosamine transferase from an ascites hepatoma, AH 66. J Biol Chem. 1982;257:9501–9507. [PubMed] [Google Scholar]
- Tabak LA. The role of mucin-type O-glycans in eukaryotic development. Semin Cell Dev Biol. 2010;21:616–621. doi: 10.1016/j.semcdb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen KG, Bedi GS, Tetaert D, Kingsley PD, Hagen FK, Balys MM, Beres TM, Degand P, Tabak LA. Cloning and characterization of a ninth member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, ppGaNTase-T9. J Biol Chem. 2001;276:17395–17404. doi: 10.1074/jbc.M009638200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2002;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Hagen FK, Balys MM, Beres TM, Van Wuyckhuyse B, Tabak LA. Cloning and expression of a novel, tissue specifically expressed member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family. J Biol Chem. 1998;273:27749–27754. doi: 10.1074/jbc.273.42.27749. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tetaert D, Hagen FK, Richet C, Beres TM, Gagnon J, Balys MM, VanWuyckhuyse B, Bedi GS, Degand P, et al. Characterization of a UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase that displays glycopeptide N-acetylgalactosaminyltransferase activity. J Biol Chem. 1999;274:27867–27874. doi: 10.1074/jbc.274.39.27867. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tran DT. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J Biol Chem. 2002;277:22616–22622. doi: 10.1074/jbc.M201807200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Tran DT, Gerken TA, Stein DS, Zhang Z. Functional characterization and expression analysis of members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family from Drosophila melanogaster. J Biol Chem. 2003;278:35039–35048. doi: 10.1074/jbc.M303836200. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Zhang L, Tian E, Zhang Y. Glycobiology on the fly: Developmental and mechanistic insights from Drosophila. Glycobiology. 2009;19:102–111. doi: 10.1093/glycob/cwn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenno M, Ohtsubo K, Hagen FK, Ditto D, Zarbock A, Schaerli P, von Andrian UH, Ley K, Le D, Tabak LA, et al. Initiation of protein O glycosylation by the polypeptide GalNAcT-1 in vascular biology and humoral immunity. Mol Cell Biol. 2007;27:8783–8796. doi: 10.1128/MCB.01204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenno M, Saeki A, Kezdy FJ, Elhammer AP, Kurosaka A. The lectin domain of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1 is involved in O-glycosylation of a polypeptide with multiple acceptor sites. J Biol Chem. 2002;277:47088–47096. doi: 10.1074/jbc.M207369200. [DOI] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. Expression of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology. 2006;16:83–95. doi: 10.1093/glycob/cwj051. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J Biol Chem. 2007;282:606–614. doi: 10.1074/jbc.M606268200. [DOI] [PubMed] [Google Scholar]
- Tian E, Ten Hagen KG. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J. 2008;26:325–334. doi: 10.1007/s10719-008-9162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S, Tenno M, Konishi M, Mikami T, Itoh N, Kurosaka A. Brain-specific expression of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase (GalNAc-T9) Biochim Biophys Acta. 2000;1493:264–268. doi: 10.1016/s0167-4781(00)00180-9. [DOI] [PubMed] [Google Scholar]
- Topaz O, Shurman DL, Bergman R, Indelman M, Ratajczak P, Mizrachi M, Khamaysi Z, Behar D, Petronius D, Friedman V, et al. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat Genet. 2004;36:579–581. doi: 10.1038/ng1358. [DOI] [PubMed] [Google Scholar]
- Torrents D, Suyama M, Zdobnov E, Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DT, Zhang L, Zhang Y. Earl L, Ten Hagen KG TE. Additional members of the polypeptide GalNAc transferase family are essential for viability in Drosophila. J Biol Chem. 2011 doi: 10.1074/jbc.M111.306159. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von GM, Yee SF, Totpal K, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Hassan H, Mirgorodskaya E, Kristensen AK, Roepstorff P, Bennett EP, Nielsen PA, Hollingsworth MA, Burchell J, Taylor-Papadimitriou J, et al. Substrate specificities of three members of the human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J Biol Chem. 1997;272:23503–23514. doi: 10.1074/jbc.272.38.23503. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Irazoqui F, Tarp MA, Bennett EP, Mandel U, Takeuchi H, Kato K, Irimura T, Suryanarayanan G, Hollingsworth MA, et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate binding specificity for GalNAc: Lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology. 2007;17:374–387. doi: 10.1093/glycob/cwl082. [DOI] [PubMed] [Google Scholar]
- Wang J, Li X, Kubota T, Narimatsu H, Zhang Y. A vertebrate-specific Y subfamily of UDP-N-acetyl-α-D-galactosamine:polypeptide N-acetyl-galactosaminyltransferases. Glycoconj J. 2011;28:317. [Google Scholar]
- Wang H, Tachibana K, Zhang Y, Iwasaki H, Kameyama A, Cheng L, Guo Jm, Hiruma T, Togayachi A, Kudo T. Cloning and characterization of a novel UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, pp-GalNAc-T14*1. Biochem Biophys Res Commun. 2003;300:738–744. doi: 10.1016/s0006-291x(02)02908-x. [DOI] [PubMed] [Google Scholar]
- Weller SG, Capitani M, Cao H, Micaroni M, Luini A, Sallese M, McNiven MA. Src kinase regulates the integrity and function of the Golgi apparatus via activation of dynamin 2. Proc Natl Acad Sci USA. 2010;107:5863–5868. doi: 10.1073/pnas.0915123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bennett EP, Takio K, Sørensen T, onding N, lausen H. Purification and cDNA cloning of a human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. J Biol Chem. 1995;270:24156–24165. doi: 10.1074/jbc.270.41.24156. [DOI] [PubMed] [Google Scholar]
- White KE, Lorenz B, Evans WE, Meitinger T, Strom TM, Econs MJ. Molecular cloning of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T8, and analysis as a candidate autosomal dominant hypophosphatemic rickets (ADHR) gene. Gene. 2000;246:347–356. doi: 10.1016/s0378-1119(00)00050-0. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IB, Gavel Y, von HG. Amino acid distributions around O-linked glycosylation sites. Biochem J. 1991;275:529–534. doi: 10.1042/bj2750529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Wragg S, Hagen FK, Tabak LA. Identification of essential histidine residues in UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T1. Biochem J. 1997;328:193–197. doi: 10.1042/bj3280193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Nakamori S, Tsujie M, Takahashi Y, Nagano H, Dono K, Umeshita K, Sakon M, Tomita Y, Hoshida Y, et al. Expression of uridine diphosphate N-acetyl-α-D-galactosamine:polypeptide N-acetylgalactosaminyl transferase 3 in adenocarcinoma of the pancreas. Pathobiology. 2004;71:12–18. doi: 10.1159/000072957. [DOI] [PubMed] [Google Scholar]
- Young WW, Jr, Holcomb DR, Ten Hagen KG, Tabak LA. Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR: A new view of a large family. Glycobiology. 2003;13:549–557. doi: 10.1093/glycob/cwg062. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iwasaki H, Wang H, Kudo T, Kalka TB, Hennet T, Kubota T, Cheng L, Inaba N, Gotoh M, et al. Cloning and characterization of a new human UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes GalNAc alpha-aerine/threonine antigen. J Biol Chem. 2003;278:573–584. doi: 10.1074/jbc.M203094200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tran DT, Ten Hagen KG. An O-glycosyltransferase promotes cell adhesion during development by influencing secretion of an extracellular matrix integrin ligand. J Biol Chem. 2010;285:19491–19501. doi: 10.1074/jbc.M109.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.