Abstract
CpGH89 is a family 89 glycoside hydrolase with exo-α-d-N-acetylglucosaminidase activity that is produced by the human and animal pathogen Clostridium perfringens. This enzyme is active on the α-d-GlcpNAc-(1 → 4)-d-Galp motif that is displayed on the class III mucins within the gastric mucosa. Other members of this enzyme family, such as human NAGLU, are active on heparan. A truncated version of CpGH89 was rendered inactive through the mutation of two key catalytic residues, the protein crystallized and its structure determined in complex with α-d-GlcpNAc-(1 → 4)-d-Galp to reveal the molecular details of how this unique disaccharide is recognized by CpGH89. An analysis of this substrate complex not only provides insight into how this enzyme selects for its mucin-presented substrate but also advances our understanding of how its clinically relevant mammalian counterparts are specific for heparan.
Keywords: carbohydrate-active enzyme, glycoside hydrolase, heparin, mucin, NAGLU
Introduction
The extensive O-glycosylation of the glycoproteins that form the mucous layer in the gastrointestinal tracts of animals and humans results in a heavily hydrated and highly viscous protective barrier. This acts as a physical partition to protect the epithelia from harmful bacteria while also supporting innate and adaptive immunity (McGuckin et al. 2011). The carbohydrates present in mucosal layers are often highly complex and, consequently, bacteria that invade or normally inhabit the gut have evolved enzyme systems to degrade mucosal carbohydrates. Clostridium perfringens is one such human and veterinary pathogen that appears capable of degrading the complex carbohydrates of the protective mucosal barrier by using an extensive array of glycoside hydrolases (Kochetkov et al. 1976; Koutsioulis et al. 2008; Ficko-Blean et al. 2009; Fujita et al. 2011). These enzymes have a variety of distinct catalytic specificities and are notable for their complex modular structures (Ficko-Blean et al. 2009). A unique member of this bacterium's carbohydrate degrading toolbox is CpGH89 (locus tag CPF_0859 in the ATCC 13124 strain), which is a family 89 glycoside hydrolase with exo-α-d-N-acetylglucosaminidase activity (Ficko-Blean et al. 2008). This enzyme has recently been shown to have specificity for the terminal α-d-GlcpNAc-(1 → 4)-d-Galp motif that is unique to the class III mucins found in the deep gastric mucosal layers and, to date, is the only such enzyme known to have this activity (Fujita et al. 2011).
The enzyme activity more commonly associated with family 89 glycoside hydrolases is typified by the activity of human NAGLU (or HsGH89). This enzyme and its mammalian homologs are lysosomal enzymes involved in recycling of the glycosaminoglycan heparan; mutations in the human naglu gene can result in a lethal lysosomal storage disorder called mucopolysaccharidosis IIIB (MPSIIIB) or Sanfilippo syndrome (Yogalingam and Hopwood 2001). Its role is to cleave non-reducing terminal α-1,4-linked N-acetylglucosamine residues from heparan and thus shares the exo-α-d-N-acetylglucosaminidase activity with its bacterial counterparts. The monosaccharide that precedes GlcNAc in heparan is either β-d-glucuronic acid or α-l-iduronic acid highlighting the different substrate preferences of the mammalian enzymes compared with CpGH89.
The recently solved X-ray crystal structures of a truncated but catalytically active recombinant version of CpGH89 (referred to as GH89CM) in complex with the reaction product GlcNAc and with inhibitors are, at present, the only structures of a family 89 glycoside hydrolase (Ficko-Blean et al. 2008). These structures provided general insight into the molecular basis of MPSIIIB and into the nature of the non-reducing terminal GlcNAc-specific −1 subsite, which is highly conserved between CpGH89 and NAGLU; however, the molecular details of substrate recognition in the +1 subsite remain unknown. This particular subsite accommodates galactose in CpGH89 and β-d-glucuronic acid or α-l-iduronic acid in NAGLU and, presumably, must be a key in defining the different substrate specificities within the GH89 family. Here, we present the complex of a catalytically inactive mutant of GH89CM with the intact α-d-GlcpNAc-(1 → 4)-d-Galp disaccharide. A detailed structural view is provided of both the −1 and +1 subsites revealing how CpGH89 selects this particular carbohydrate component of mucin. This highlights the differences in the features of the +1 subsites of CpGH89 and NAGLU that allow NAGLU to recognize the terminal GlcNAc residues of heparan.
Results and discussion
CpGH89, and by extension all members of glycoside hydrolase family 89 (GH89), catalyzes hydrolysis of the glycosidic bond through a retaining mechanism where the stereochemistry of the anomeric carbon involved in the glycosidic bond is retained in the product (Ficko-Blean et al. 2008). Structural analyses of this enzyme in complex with a reaction product and an inhibitor, interpreted in combination with site-directed mutagenesis of active site residues and kinetic analyses, strongly supports the identity of the general acid/base in this reaction as E483 with E601 acting as the nucleophile (Ficko-Blean et al. 2008). To generate a catalytically inactive version of GH89CM that maintained the ability to bind the substrate, we systematically mutated both residues to glutamine. Complete inactivity on both para-nitrophenyl α-N-acetyl-d-glucosaminide and α-d-GlcpNAc-(1 → 4)-d-Galp required the generation of an E483Q/E601Q double mutant (GH89CMmut). To provide additional insight into the specificity of CpGH89, the inactive double mutant, GH89CMmut, was screened for binding to glycan microarrays.
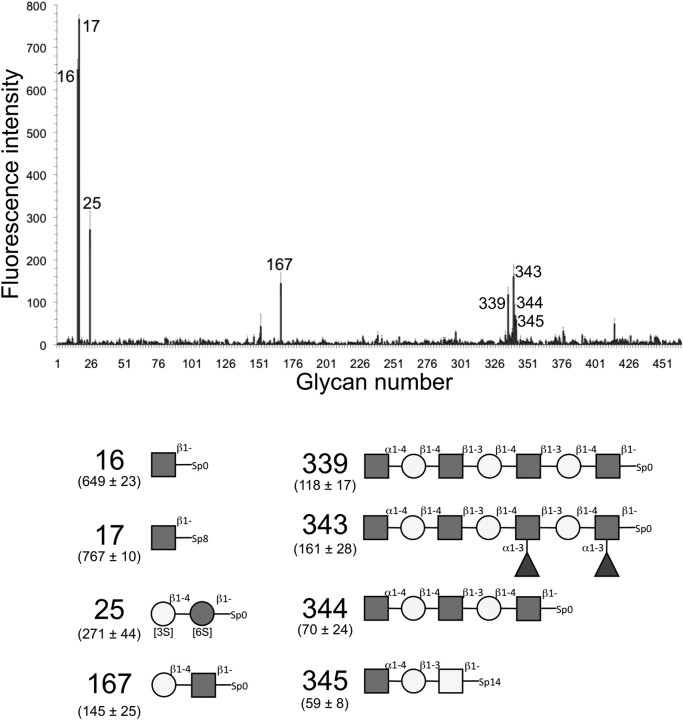
Consistent with the recent activity studies of CpGH89 (Fujita et al. 2011), GH89CMmut showed comparatively good binding to four glycans on the array that terminate in the α-d-GlcpNAc-(1 → 4)-d-Galp motif (glycans 339, 343, 344 and 345; Figure 1), but some binding to all seven of the glycans that terminate in this motif (glycans 339–345) was evident. Curiously, however, the compounds with the largest binding signal were both single GlcNAc residues that were β-linked, rather than α-linked, to their spacer arms (glycans 16 and 17). The protein also bound to β-d-Galp-(1 → 4)-d-GlcpNAc (N-acetyllactosamine or LacNAc; glycan 167) and sulfated lactose (glycan 25). These latter interactions are somewhat at odds with the results of Fujita et al. (2011) who showed CpGH89 to be specific for α-d-GlcpNAc-(1 → 4)-d-Galp (i.e. it was not active on β-linked substrates or other α-linked substrates). We suggest that the additional binding interactions with glycans 16, 17, 25 and 167 observed on the microarray are unlikely to be associated with recognition by the active site of the enzyme. The protein used for screening the array, GH89CMmut, included the N-terminal module, which has sequence identity with family 32 carbohydrate-binding modules (CBM32), and it is possible that this module is responsible for binding to the additional carbohydrate ligands. Indeed, though the overall results were considered inconclusive due to high background binding, independent glycan microarray screening of the CBM32-like module showed significant binding to similar carbohydrates: sulfated galactose β-linked to the spacer, GlcNAc β-linked to the spacer, sulfated LacNAc and sulfated lactose (not shown). At present, it is unclear if the binding of the CBM-like modules is specific; however, its binding to these ligands supports the idea that the presence of this module may have resulted in the interaction of GH89CMmut with other glycans on the array. Overall, the consistent binding of CpGH89CMmut to α-d-GlcpNAc-(1 → 4)-d-Galp interpreted in the light of the results of Fujita et al. (2011) supports the contention that CpGH89 is specific for the terminal α-d-GlcpNAc-(1 → 4)-d-Galp disaccharide motif.
Fig. 1.
Glycan microarray analysis of carbohydrate binding by GH89CMmut. The carbohydrates giving the most significant signals are numbered and schematics of the structures of these glycans are given to the right of the array results. The glycan array number of the oligosaccharide is given to the left of the schematic with the raw fluorescence intensity from the array analysis given in parentheses beneath the glycan number. Dark gray boxes represent N-acetylglucosamine; light gray circles, galactose; dark gray circles, glucose; light gray boxes, N-acetylgalactosamine; triangles, fucose. The errors and error bars represent the standard error of the mean for measurements made in quadruplicate.
The X-ray crystal structure of GH89CMmut was solved in complex with α-d-GlcpNAc-(1 → 4)-d-Galp to 1.90 Å resolution. The electron density for the carbohydrate was well-defined revealing the disaccharide to be intact, i.e. not hydrolyzed (Figure 2A). The L-shaped carbohydrate is accommodated in the “sock-shaped” active site with the terminal GlcNAc residue in the −1 subsite and Gal in the +1 subsite (Figure 2B). The C1 hydroxyl group of the Gal residue is directed out of the active site, pointing into solution, suggesting a lack of additional subsites and an ability to accept the α-d-GlcpNAc-(1 → 4)-d-Galp disaccharide when presented at the terminus of a variety of glycans. Residue 483, which is normally a glutamate but in this case has been substituted for a glutamine, is suitably poised to donate a proton to the glycosidic bond of the disaccharide. The residue acting as the nucleophile, residue 601, which has likewise been altered from a glutamate to a glutamine, sits directly beneath C1 of the GlcNAc in the −1 subsite. This arrangement is consistent with the previous mechanistic assignments of a retaining catalytic mechanism (Ficko-Blean et al. 2008; Fujita et al. 2011).
Fig. 2.
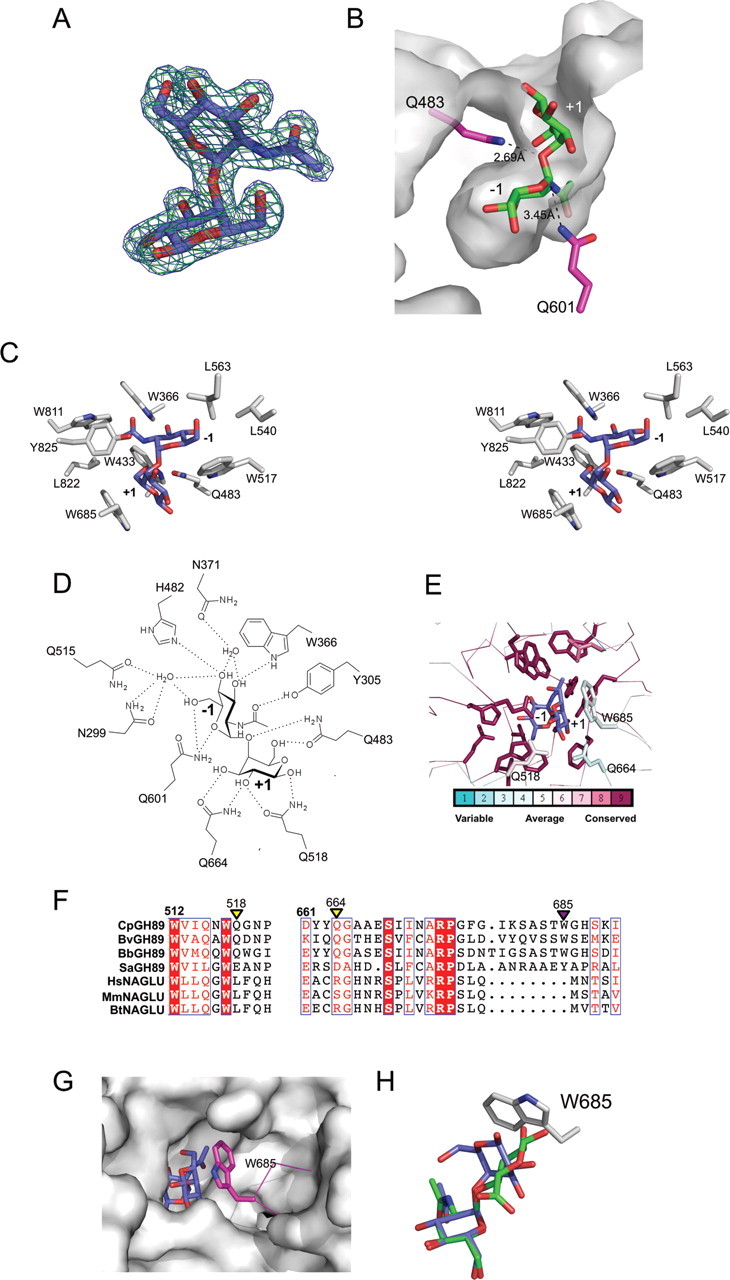
Structural analysis of GH89CMmut in complex with α-d-GlcpNAc-(1 → 4)-d-Galp. (A) Electron density for α-d-GlcpNAc-(1 → 4)-d-Galp in the active site of GH89CMmut. The electron density maps are maximum-likelihood (Murshudov et al. 1997)/σA (Read 1986) − weighted 2Fobs − Fcalc contoured at 1 σ (both at 0.29 e−/Å3) produced with the disaccharide omitted (green) and included (blue) in the refinement. (B) The cutaway view of the GH89CMmut active site surface showing accommodation of the α-d-GlcpNAc-(1 → 4)-d-Galp disaccharides. The mutated acid/base residue, Q483, and mutated nucleophile, Q601, are shown in stick representation with relevant distances shown as dashed lines and labeled. The subsites are labeled according to the convention established by Davies et al. (1997). (C) The divergent stereo view of the primary apolar interactions in the active site. (D) Schematic of the hydrogen-bonding pattern between the enzyme active site and the disaccharide substrate. (E) Conservation of functional residues determined using CONSURF (Ashkenazy et al. 2010) focusing on the active site of CpGH89. One hundred and forty-three unique amino acid sequences having greater than 28% amino acid sequence identity with the catalytic module of CpGH89 were used in the analysis. The color-coding scale for conservation is given immediately below the panel. (F) Sequence alignments of GH89, three randomly chosen bacterial examples of GH89 from Bacteroides vulgatus (BvGH89), Bifidobacterium bifidum (BbGH89) and Streptomyces avermitilis (SaGH89), and three mammalian GH89 enzymes from Homo sapiens (HsGH89; also referred to as NAGLU), Mus musculus (MmGH89) and Bos Taurus (BtGH89). These aligned segments show regions that correspond to the +1 subsite. Key residues involved in galactose recognition in the +1 subsite of CpGH89 are indicated with triangles and color coded to show hydrophobic interaction (purple) or hydrogen bonding (yellow). (G) The catalytic site of GH89CMmut superposed with a homology model of human NAGLU. The solvent accessible surface of human NAGLU is shown with the α-d-GlcpNAc-(1 → 4)-d-Galp disaccharide (blue sticks) from the GH89CMmut structure in the active site. W685 is shown in pink stick representation. (H) An overlay of α-d-GlcpNAc-(1 → 4)-d-Galp (blue) from the GH89CMmut structure with the GLYCAM (Kirschner et al. 2008)-modeled α-d-GlcpNAc-(1 → 4)-d-GlcAp (green) disaccharide present in heparan. W685, which is a key determinant of galactose recognition the +1 subsite, is shown in stick representation.
The α-d-GlcpNAc-(1 → 4)-d-Galp disaccharide is recognized by a complex network of polar and apolar interactions (Figure 2C and D). The −1 subsite binds the GlcNAc residue through a series of interactions that are identical to those described previously (Ficko-Blean et al. 2008). The residues making these interactions are invariant in GH89 (Figure 2E) consistent with a conserved specificity for terminal GlcNAc residues in this family. The +1 subsite accommodates galactose through a more limited set of interactions. The side chain of Q664 hydrogen bonds with the C3 and C2 hydroxyls of the galactose and the side chain of Q518 hydrogen bonds with the C2 hydroxyl group (Figure 2D). An additional hydrogen bond is made between Q483 and the C6 hydroxyl group of the galactose residue. The indole ring of W685 makes a key interaction with the galactose by lying parallel to the B face of the sugar ring in a classic carbohydrate–aromatic ring interaction (Figure 2C). This particular residue is the primary contributor to the unique angled contour of the active site. The significance of this is in recognizing the distinctive bent shape of the carbohydrate, which is afforded by the α-linkage to the axial O4 of the galactose residue. In contrast, any β-linkage or a α-linkage to an equatorial oxygen would result in a more planar relative orientation of the pyranose rings in the disaccharide that could not be accommodated in this active site. While GH89CMmut bound to a limited number of glycans on the microarray having β-linkages, it appears likely that this was attributable to the N-terminal CBM-like module and not due to interactions with the active site. Further supporting this structural interpretation, CpGH89 was unable to hydrolyze substrates where GlcNAc was β-linked to the para-nitrophenyl leaving group, α-linked either (1 → 2), (1 → 3) or (1 → 6) to galactose or α-(1 → 4)-linked to glucuronic acid (Fujita et al. 2011).
The residues comprising the +1 subsite of CpGH89 are not as well conserved within the family as the residues in the −1 subsite, which is coherent with the hypothesis that the +1 subsites of GH89 enzymes are likely responsible for imparting specificity to members of this family (Figure 2E). Supporting this, a comparison of randomly selected bacterial GH89 enzymes with three heparan recycling GH89 enzymes from mammals reveals the key residues in the +1 subsite to be quite well conserved among the bacterial enzymes but they are not conserved with the mammalian ones (Figure 2F). Significantly, the loop containing W685 in CpGH89, the key residue for the recognition of α-d-GlcpNAc-(1 → 4)-d-Galp, is absent in the mammalian enzymes (Figure 2F). Analysis of a homology model of human NAGLU that was based on the structure of CpGH89 (Ficko-Blean et al. 2008) suggests that this shortened loop creates a depression in the surface of NAGLU that would otherwise be filled by the loop containing W685 (Figure 2G). This may create a +1 subsite in NAGLU having a different architecture in order to accommodate a carbohydrate with a different trajectory of entry into the enzyme active site. Indeed, a GLYCAM (Kirschner et al. 2008)-generated model of α-d-GlcpNAc-(1 → 4)-d-GlcAp, the substrate for NAGLU, suggests that by virtue of the glycosidic oxygen being equatorial to the glucuronic acid it adopts a more linear conformation than α-d-GlcpNAc-(1 → 4)-d-Galp, where the glycosidic oxygen is axial to the galactose (Figure 2H). The α-d-GlcpNAc-(1 → 4)-l-IdoAp motif in heparan also adopts a linear conformation similar to that of α-d-GlcpNAc-(1 → 4)-d-GlcAp (not shown). Under these conditions, the β-d-glucuronic acid or α-l-iduronic acid would clash with W685 of CpGH89 preventing the sugar from binding to this enzyme. The putative +1 subsite of NAGLU, therefore, appears to be contoured to accommodate the more linear conformation of its substrate and the resulting changed trajectory of entry into the active site.
GH89 is a comparatively small family yet it contains members from both prokaryotes and eukaryotes. CpGH89, which is the only prokaryotic GH89 whose catalytic activity has been characterized, is active on the α-d-GlcpNAc-(1 → 4)-d-Galp motif found on the carbohydrates of type III mucins, while the characterized eukaryotic examples, such as mammalian NAGLU, are known to process the non-reducing terminal GlcNAc residue from heparan. Though the structure of a eukaryotic GH89 is yet to be determined, the structure of CpGH89 in complex with α-d-GlcpNAc-(1 → 4)-d-Galp suggests that significant modifications between the architectures of the +1 subsites of bacterial and mammalian enzymes are responsible for their different substrate specificities. This analysis, while giving new insight into the mode of carbohydrate recognition by the mucin-active bacterial enzymes, also advances our understanding of heparan recognition by mammalian GH89 enzymes.
Materials and methods
Cloning
The procedure for cloning the gene fragment encoding a catalytically active fragment (nucleotides 76–2748 encoding amino acids 26–916; here called GH89CM) of the family 89 glycoside hydrolase from C. perfringens ATCC 13124 was described previously (Ficko-Blean et al. 2008). Site-directed mutants were made using a “mega-primer” PCR method (Barik 1996). The E483Q mutation was first introduced using primers 1–4 (Table I). This mutated gene fragment was then used as a template to introduce the E601Q substitution using primers 1, 2, 5 and 6 (Table I). Using the standard cloning procedures, the amplified fragment of DNA carrying the mutations was cloned into pET28a(+) (Novagen) via NheI and XhoI restriction sites present in the 5′ and 3′ oligonucleotide primers (primers 1 and 2; Table I). The resulting construct comprised a gene encoding GH89CM with the E483Q and E601Q mutations, referred to here as GH89CMmut and with a hexa-histadine tag fused to the N terminus through a thrombin protease cleavage site. The sequence fidelity of the construct was verified by bi-directional DNA sequencing. The protein was produced and purified as described previously for GH89CM (Ficko-Blean et al. 2008). Protein concentration was determined at 280 nm using a calculated extinction coefficient of 207,060 M−1 cm−1 (Gasteiger et al. 2003).
Table I.
Oligonucleotide primers used for amplification and cloning
| Primer number | Name | Sequence |
|---|---|---|
| 1 | GH89CMF | CAT ATG GCT AGC GGT GTT GAA ATT ACG GAA G |
| 2 | GH89CMR | GAA TTC CTC GAG TTA TGA TTC ATT TTC ACC TAA |
| 3 | GH89CME483QF | GAG TAG ATC CAT TCC ATC AGG GTG GAA ATA CTG GTG |
| 4 | GH89CME483QR | CAC CAG TAT TTC CAC CCT GAT GGA ATG GAT CTA CTC |
| 5 | GH89CME601QF | GCT AAT TCA GAG CAC ATG GTT GGT ATA GGT ATT ACA CCA CAG GCT ATA AAT ACA AAT CC |
| 6 | GH89CME601QR | GGA TTT GTA TTT ATA GCC TGT GGT GTA ATA CCT ATA CCA ACC ATG TGC TCT GAA TTA GC |
Glycan microarray screening
The primary amines of GH89CMmut were labeled covalently using the FluoReporter FITC Protein Labeling Kit (Molecular Probes) according to the manufacturer's recommendations. The protein was separated from free fluorophore and exchanged into phosphate-buffered saline using a PD-10 column (GE Biosciences). Glycan microarray screening was performed by Core H of the Consortium for Functional Glycomics (www.functionalglycomics.org/). Printed glycan microarrays (version 4.1) were probed with fluorescein isothiocyanate-labeled GH89CMmut at 200 μg/mL in binding buffer (20 mM Tris–HCl, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.5% Tween-20, 1% bovine serum albumin) using the standard procedures.
Crystallography
GH89CMMut (20 mg/mL) was crystallized by the hanging drop vapor diffusion method at 18°C in 2.0 M (NH4)2SO4 with 3% (v/v) glycerol. The complex was obtained by adding a small amount of solid α-d-GlcpNAc-(1 → 4)-d-Galp directly to the crystallization drop and incubating for 10 min. These crystals were cryoprotected in crystallization solution supplemented with 20% glycerol. Diffraction data were collected using a Rigaku R-AXIS IV++ area detector coupled to an MM-002 X-ray generator with Osmic “blue” optics and an Oxford Cryostream 700. The diffraction data were processed using D*trek (Pflugrath 1999; data collection statistics are given in Table II). The structure of GH89CMmut in complex with α-d-GlcpNAc-(1 → 4)-d-Galp was determined using the existing structure of the non-mutated version of this construct [Protein Data Bank (PDB) id 2vcc; Ficko-Blean et al. 2008] as a starting point. The model was manually corrected and completed with COOT (Emsley and Cowtan 2004) and refined with REFMAC (Murshudov et al. 1997). Water molecules were added using COOT:FINDWATERS and manually inspected after refinement. Five percent of the observations were flagged as “free” and used to monitor refinement procedures (Brunger 1992). Model validation was performed with MOLPROBITY (Chen et al. 2010). Structure and refinement statistics are shown in Table II. Coordinates and structure factors have been deposited in the PDB with code 4a4a.
Table II.
X-ray data collection, refinement and model statistics
| Data collection statistics | GH89CMmut in complex with α-d-GlcpNAc-(1 → 4)-d-Galp |
|---|---|
| Space group | P61 |
| Resolution | 30.00–1.90 (1.95–1.90)a |
| Cell dimension a, b, c (Å) | 91.00, 91.00, 252.86 |
| Rsymb | 0.076 (0.349) |
| Completeness (%) | 99.4 (97.3) |
| 〈I/σI〉 | 9.7 (2.5) |
| Redundancy | 4.40 (2.60) |
| Total reflections | 405,475 |
| Unique reflections | 92,199 |
| Wilson B-factor (Å2) | 25.7 |
| Refinement statistics | |
| Rcryst (%)c | 16.9 |
| Rfree (%)c | 21 |
| RMSD | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 1.483 |
| Average B-factors (Å2) | |
| Protein | 24.07 |
| Water molecules | 32.55 |
| Carbohydrate ligand | 17.73 |
| Other ligands/ions | 66.85 |
| Number of atoms | |
| Protein atoms chain A | 7188 |
| Water molecules | 811 |
| Carbohydrate Ligand | 26 |
| Other ligands/ions | 74 |
| Ramachandran statistics | |
| Most favored (%) | 97.4 |
| Additional allowed (%) | 2.5 |
| Disallowed (%) | 0.1 |
aValues in parentheses are for the highest resolution bin.
bRsym = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi|〈Ii(hkl)〉|, where Ii(hkl) is the intensity measured for the ith reflection and áI(hkl)ñ is the average intensity of all reflections with indices hkl.
cRcryst = Σhkl||Fobs(hkl)| − |Fcalc(hkl)||/Σhkl|Fobs(hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
Funding
This work was supported by a Canadian Institutes for Health Research grant (FRN 68913). We are grateful to Core H of the Consortium for Functional Glycomics for performing the glycan array experiments. The resources and collaborative efforts provided by The Consortium for Functional Glycomics were funded by NIGMS (GM62116). A.B.B. is a Canada Research Chair in Molecular Interactions and a Michael Smith Foundation for Health Research Scholar.
Conflict of interest
None declared.
Abbreviations
CBM32, family 32 carbohydrate-binding modules; GH89, Glycoside hydrolase family 89; LacNAc, N-acetyllactosamine; MPSIIIB, mucopolysaccharidosis IIIB
References
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. doi:10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. Site-directed mutagenesis in vitro by megaprimer PCR. Methods Mol Biol. 1996;57:203–215. doi: 10.1385/0-89603-332-5:203. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. doi:10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. doi:10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321(Pt 2):557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. doi:10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ficko-Blean E, Gregg KJ, Adams JJ, Hehemann JH, Czjzek M, Smith SP, Boraston AB. Portrait of an enzyme, a complete structural analysis of a multimodular β-N-acetylglucosaminidase from Clostridium perfringens. J Biol Chem. 2009;284:9876–9884. doi: 10.1074/jbc.M808954200. doi:10.1074/jbc.M808954200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficko-Blean E, Stubbs KA, Nemirovsky O, Vocadlo DJ, Boraston AB. Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc Natl Acad Sci USA. 2008;105:6560–6565. doi: 10.1073/pnas.0711491105. doi:10.1073/pnas.0711491105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Tsuchida A, Hirata A, Kobayashi N, Goto K, Osumi K, Hirose Y, Nakayama J, Yamanoi T, Ashida H, et al. Glycoside hydrolase family 89 α-N-acetylglucosaminidase from Clostridium perfringens specifically acts on GlcNAcα1,4Galβ1R at the non-reducing terminus of O-glycans in gastric mucin. J Biol Chem. 2011;286:6479–6489. doi: 10.1074/jbc.M110.206722. doi:10.1074/jbc.M110.206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. doi:10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. doi:10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetkov NK, Derevitskaya VA, Arbatsky NP. The structure of pentasaccharides and hexasaccharides from blood group substance H. Eur J Biochem. 1976;67:129–136. doi: 10.1111/j.1432-1033.1976.tb10641.x. doi:10.1111/j.1432-1033.1976.tb10641.x. [DOI] [PubMed] [Google Scholar]
- Koutsioulis D, Landry D, Guthrie EP. Novel endo-α-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology. 2008;18:799–805. doi: 10.1093/glycob/cwn069. doi:10.1093/glycob/cwn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. doi:10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. doi:10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 10):1718–1725. doi: 10.1107/s090744499900935x. doi:10.1107/S090744499900935X. [DOI] [PubMed] [Google Scholar]
- Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr A. 1986;42:140–149. doi:10.1107/S0108767386099622. [Google Scholar]
- Yogalingam G, Hopwood JJ. Molecular genetics of mucopolysaccharidosis type IIIA and IIIB: Diagnostic, clinical, and biological implications. Hum Mutat. 2001;18:264–281. doi: 10.1002/humu.1189. doi:10.1002/humu.1189. [DOI] [PubMed] [Google Scholar]