Abstract
Standard genetic approaches allow the production of protein composites by fusion of polypeptides in head-to-tail fashion. Some applications would benefit from constructions that are genetically impossible, such as the site-specific linkage of proteins via their N or C termini, when a remaining free terminus is required for biological activity. We developed a method for the production of N-to-N and C-to-C dimers, with full retention of the biological activity of both fusion partners and without inflicting chemical damage on the proteins to be joined. We use sortase A to install on the N or C terminus of proteins of interest the requisite modifications to execute a strain-promoted copper-free cycloaddition and show that the ensuing ligation proceeds efficiently. Applied here to protein–protein fusions, the method reported can be extended to connecting proteins with any entity of interest.
Keywords: antibodies, bioorthogonal, engineering, transacylation
The creation of protein–protein fusions, by genetic means, chemically, or enzymatically, has become an important tool to study cell biological and biochemical problems. Genetic fusions with fluorescent proteins are widely used to visualize their (sub)cellular localization in situ and in vivo (1). Coexpression of two orthogonally labeled chimeras allows the study of protein colocalization and dynamics of receptor dimerization. Moreover, protein fusions have been used to evaluate the biological relevance of otherwise transient protein complexes. Fusion or cross-linking of two or more of the interacting proteins can stabilize protein complexes and has been used to explore signaling and (hetero)dimerization of G-protein–coupled receptors (2, 3), chemokines, and cytokines (4–6).
Besides being useful biochemical tools, chimeric proteins are also promising as treatment options for cancer, autoimmune diseases, lysosomal storage diseases, and brain disorders (7–10). Toxins have been conjugated to antibodies, growth factors, and cytokines as a means of delivering these payloads to malignant cells that express the counterstructures recognized by such fusion proteins, to kill tumor cells while minimizing collateral damage (10–12). Bispecific antibodies, prepared by fusing two single-chain variable fragments (scFV) of immunoglobulins, may combine an antigen-binding domain specific for a tumor cell with a CD3 receptor-binding domain specific for T cells (13). These compounds then allow the T cells to exert cytotoxic activity or cytokine release locally and so elicit the desired antitumor response. Finally, protein fusion strategies have been used to prepare structurally defined biomaterials (14).
The production and purification of fusion proteins remains a biotechnological challenge. To obtain an active product, both domains of the chimera must adopt the native fold, without modification of residues and regions that are required for activity. The standard method to produce such proteins is by genetic fusion of the ORFs of the two proteins or protein fragments. Although recombinant expression often yields the correct product, partly folded and defective products cannot always be avoided.
If it were possible to conjugate by enzymatic means the natively folded, purified proteins by means of a minimal ligation tag, refolding steps could be avoided. Such methods exploit labeling at the C or N terminus of suitably modified protein substrates to produce the adducts of interest, exactly as if one were preparing the corresponding genetic fusions. Sortase-catalyzed transacylation reactions (15) allow such site-specific labeling of proteins, as well as the preparation of head-to-tail protein–protein fusions under native conditions, with excellent specificity and in near-quantitative yields (16, 17). Standard approaches fail to yield protein–protein fusions that are genetically impossible (N terminus to N terminus and C terminus to C terminus), although such unnatural liaisons would have great appeal for the construction of bispecific antibodies or their fragments. To accomplish constructions of this type, one has to resort to chemical ligation methods. Early chemical conjugation strategies relied on nonspecific cross-linking via amines or sulfhydryls (18). The lack of control over the site and stoichiometry of modification result in the formation of a heterogeneous product, limiting the usefulness of this approach. The rise of bioorthogonal chemistries combined with site-specific mutagenesis, native chemical ligation, intein-based ligation, and amber suppressor pyrrolysine tRNA technology has enabled the synthesis of nonnatural protein fusions, as applied to the production of bivalent and multivalent antibodies (19–21). Structural analogs of ubiquitin dimers were prepared by a combination of intein-based ligation, site-specific mutation, and copper-catalyzed click chemistry (22, 23). Site-specific incorporation of propargyloxyphenylalanine facilitated the synthesis of green fluorescent protein (GFP) dimers (19, 24).
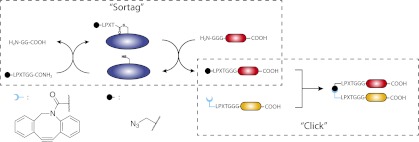
Nonetheless, the synthesis of bispecifics would benefit from a method that is orthogonal to the published methods and that allows easy access to modified native protein, as well as enables efficient nonnatural conjugation of protein termini. Moreover, the availability of orthogonal methods will make possible the synthesis of protein structures of even greater complexity (heterotrimers and higher-order complexes). We here report a versatile approach that allows the preparation of N-to-N and C-to-C fused proteins under native conditions. Click handles are introduced either N or C terminally, using a sortase-catalyzed reaction. The resulting modified proteins are then conjugated with a strain-promoted cycloaddition in an aqueous environment at neutral pH (Fig. 1).
Fig. 1.
Schematic overview of the approach.
Results
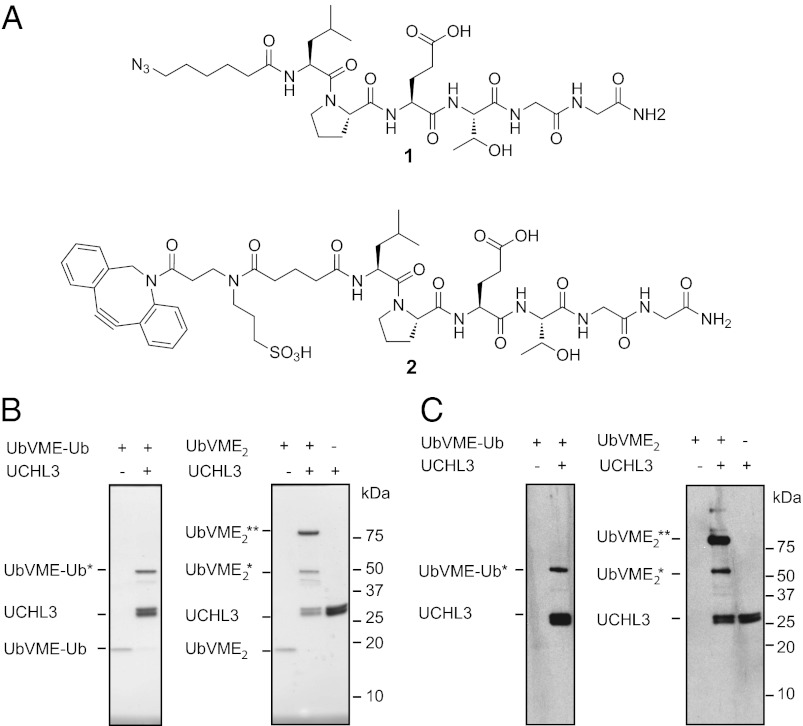
To construct N-to-N protein dimers, we synthesized LPETGG peptides 1 and 2, N-terminally equipped with an azidohexanoic acid or an aza-dibenzocyclooctyne (DIBAC) (25) (Fig. 1A; see SI Appendix for experimental details). Using sortase A from Staphylococcus aureus, we ligated these peptides to the N terminus of a substrate, G3-ubiquitin (G3Ub), with a suitably exposed short run of Gly residues to serve as the incoming nucleophile. Peptides 1 and 2 were transacylated efficiently onto G3-ubiquitin (SI Appendix and Fig. S1). With the modified proteins in hand, we established the requirements for dimerization. Azido-modified ubiquitin (80 μM) was mixed and incubated at 37 °C with a stoichiometric amount of ubiquitin equipped with a cyclooctyne. After 30 min, an ∼18-kDa polypeptide corresponding to the ubiquitin dimer was observed as revealed by Coomassie brilliant-blue staining and in an anti-ubiquitin immunoblot (Fig. S1). Extending the incubation time to 7 h resulted in ∼70% conversion to dimeric ubiquitin as quantified by SDS/PAGE using ImageJ. At lower concentrations (15 μM), the reaction still proceeded, albeit at a somewhat slower rate (∼70% conversion after 16 h).
These results demonstrate feasibility of the approach, but do the proteins joined in this click reaction retain their full biological activity as well? Therefore, we constructed a bivalent version (N-to-N fusion) of ubiquitin vinylmethylester (UbVME). UbVME is an active site-directed probe that covalently modifies a large number of ubiquitin-specific proteases (USPs) (26). The formation of these adducts is readily visualized by a shift in mobility upon analysis by SDS/PAGE. Modification of a USP with the bivalent version of UbVME should yield a complex that contains two UbVME units and two copies of the USP, with a corresponding increase in molecular weight of the adduct formed. The synthesis of the dimeric UbVME construct thus exploits the combined action of two bio-orthogonal reactions, an intein-based native ligation, to obtain the C-terminally modified version of ubiquitin bearing the vinylmethylester moiety (26), and the N-terminal sortagging reaction (27). Starting with G3-UbVME, prepared as described, we obtained the azido-modified and cyclooctyne-modified versions. By reacting equimolar amounts of azido- and cyclooctyne-modified UbVME and subsequent purification by reverse-phase HPLC to remove any unreacted UbVME monomers, we obtained the bivalent adduct. We evaluated the reactivity of this bivalent adduct using ubiquitin carboxyl-terminal hydrolase isozyme L3 (UCHL3), for which the crystal structure in complex with UbVME is known (28). As controls, we produced a dimeric construct in which one of the C termini is equipped with a reactive vinylmethyl ester and the other with a nonreactive carboxylic acid. The resulting UbVME-ubiquitin is therefore capable of binding a single UCHL3 molecule. Incubation of bivalent UbVME with an excess of N-terminally His-tagged UCHL3 (2 equivalents per vinylmethyl ester) (Fig. 2B) yielded the bivalent adduct bound to two UCHL3 molecules (∼67 kDa). When UCHL3 was incubated either with the control UbVME–ubiquitin constructs or with UbVME, the expected molecular weights shifts were observed, i.e., UCHL3 modified with an UbVME–ubiquitin dimer (∼47 kDa) and UCHL3 modified with an UbVME monomer (∼37 kDa) (Fig. S1), respectively. Immunoblotting for (His)6 (Fig. 2C) confirmed that the newly formed adduct indeed contains the (His)6 tag present in the UCHL3 input material. Both UbVME units in the bivalent adduct produced by the click reaction thus retain full activity, as evident from their ability to covalently modify the intended target.
Fig. 2.
Proof of concept for the synthesis of N-to-N fused proteins. (A) structures of the used N-terminal probes 1 and 2. (B and C) Labeling of his-tagged UCHL3 with dimeric UbVME. (B) Coomassie brilliant-blue–stained Tris-tricine gel. (C) Immunoblot using anti-His antibody. Ub-UbVME*, ubiquitin-UbVME bound to a single UCHL3; UbVME2*, dimeric UbVME bound to a single UCHL3 molecule; UbVME2**, dimeric UbVME bound to two UCHL3 molecules.
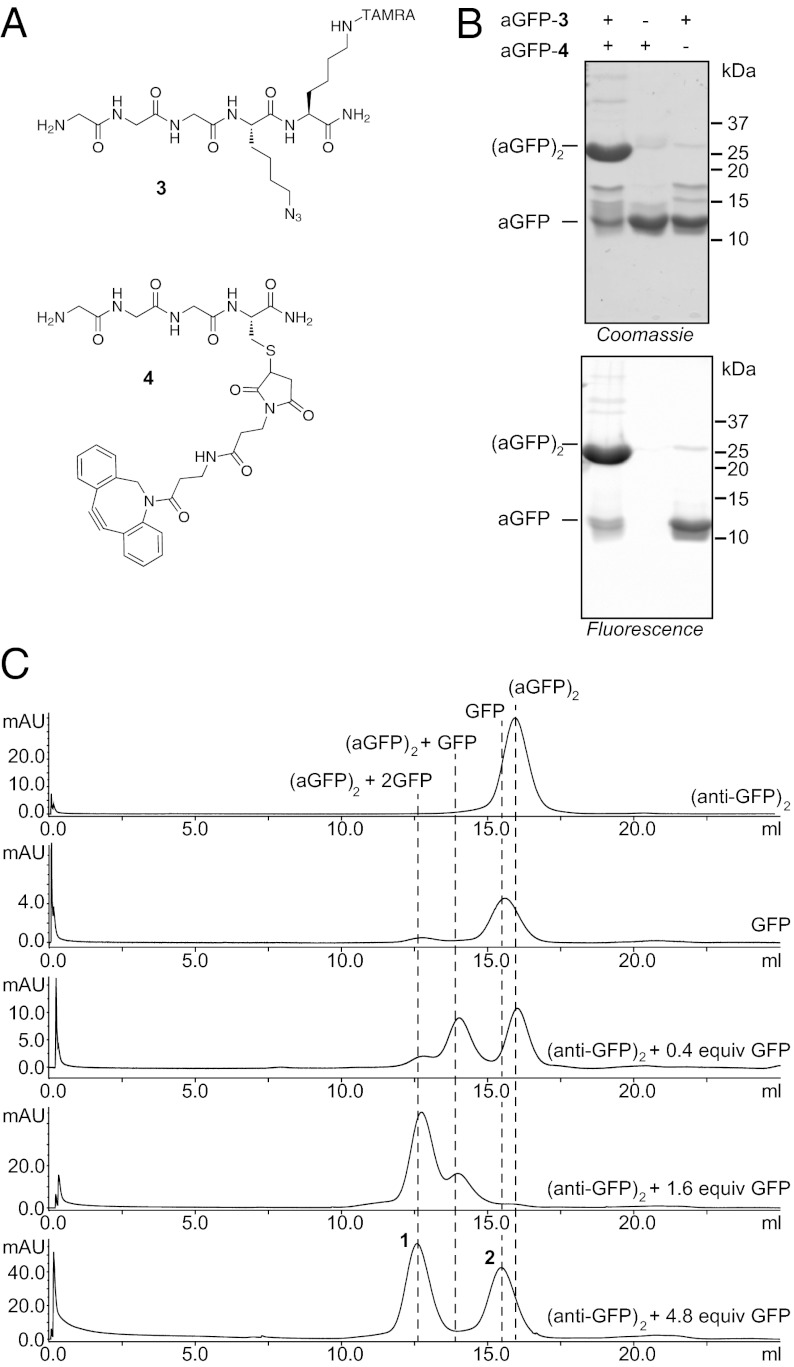
We then explored a second example. Camelids produce unusual antibodies composed of heavy chains only (29). Their variable regions, when expressed recombinantly as single-domain constructs, also known as VHH, retain full antigen-binding capability (30). We synthesized bivalent single-domain VHH proteins by conjugating them via their C termini, using the combined sortagging-click strategy. We synthesized triglycine peptides containing an azide 3 or a cyclooctyne 4 (Fig. 3A) (see SI Methods for experimental details) and we produced recombinantly a synthetic version of a camelid VHH specific for GFP (31). This VHH was modified to contain a sortase substrate motif followed by a (His)6 tag to facilitate purification. Excellent conversion to anti-GFP VHH labeled with the click handles was achieved after incubating at 25 °C overnight as judged by SDS/PAGE and liquid chromatography (LC)/MS. Excess triglycine nucleophile was removed by size exclusion chromatography to avoid interference with the subsequent dimerization reaction (Fig. S2). Using these modified VHHs, we generated the corresponding C-to-C fused homodimer (Fig. 3B), which was purified to homogeneity by size exclusion chromatography (Fig. S2).
Fig. 3.
C-to-C homodimeric antibodies. (A) Structures of probes 3 and 4. (B) Dimerization of anti-GFP. (C) Size exclusion experiment demonstrating that both anti-GFPs bind GFP. From Top to Bottom: anti-GFP dimer; GFP; anti-GFP dimer + 0.4 equiv GFP; anti-GFP dimer + 1.6 equiv GFP; anti-GFP dimer + 4.8 equiv GFP (excess).
We incubated the VHH monomers and dimers with their target antigen, GFP, to assess complex formation. The modified VHH monomers, when incubated with GFP, show the expected increase in Stokes’ radius in a size exclusion chromatography experiment (Fig. S3). We then incubated the C-to-C VHH dimer with increasing concentrations of GFP and analyzed the complexes formed between the dimer and GFP by size exclusion chromatography (Fig. 3C). The results are unequivocal: The free VHH dimer is resolved from the dimer occupied by a single GFP at low concentrations of added GFP, which in turn is resolved from the dimer occupied by two GFP moieties at the higher GFP concentration. Peaks 1 and 2 (Fig. 3C) were concentrated and loaded on a native gel, showing a pattern of migration consistent with their molecular mass (Fig. S4).
This experiment shows that C-to-C fusion of an antibody fragment, in this case a single VHH domain, is readily achieved using sortase in combination with click chemistry. Not only is the conversion excellent (∼90%), but also the resulting products retain their full function. Because most of the nucleophiles used in the sortase reaction are water soluble, and all necessary functional groups that require harsh and/or nonselective reaction conditions are introduced during the synthesis of the nucleophile, this approach minimizes unwanted side reactions [such as acylation of available amino groups (18) and denaturation of proteins] that might affect biological activity.
We extended the above experiment to include an alpaca-derived VHH that is specific for mouse class II MHC products, VHH7, and linked it to the anti-GFP VHH via their C termini to create a heterobispecific product.
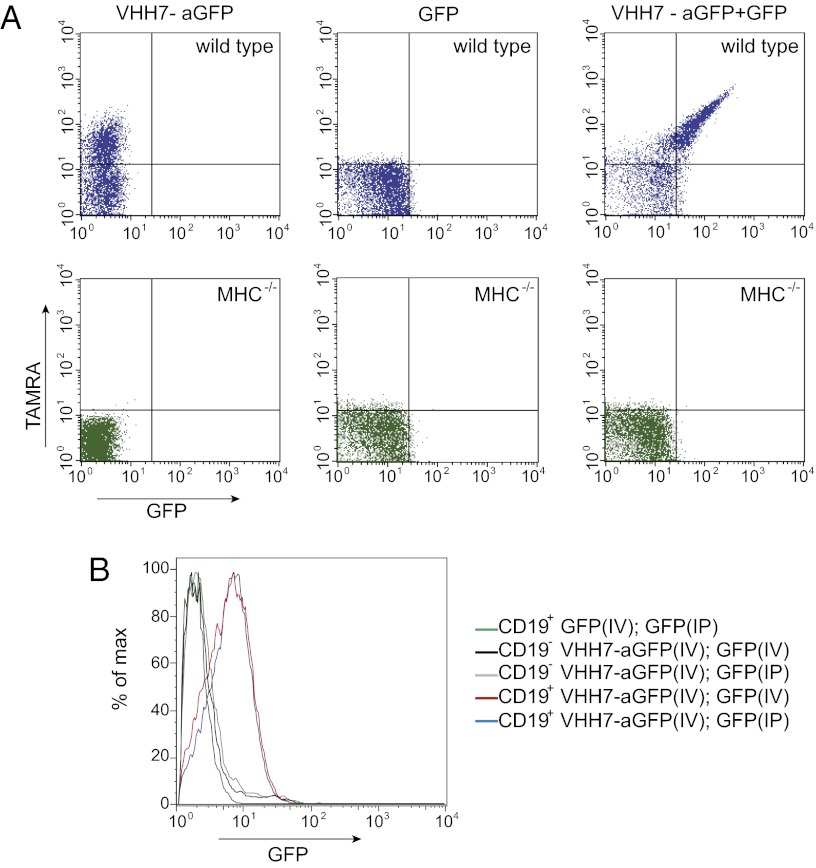
Two adducts were prepared as described above, one containing a tetramethylrhodamine (TAMRA) fluorophore at the junction (using peptide 3) and the other a nonfluorescent conjugate (using peptide 5) (see Fig. S5 for structure). The two adducts were purified to obtain the fluorescent and nonfluorescent bispecific VHH preparations (Fig. S5) and added to mouse lymph node cells, the B cells among which are uniformly positive for class II MHC products. When cells were exposed to the bispecific fluorescent VHH, we observed specific staining of B cells in the TAMRA channel (Fig. 4A). No staining was detected for the nonfluorescent bispecific antibody (Fig. S6). We then added GFP to cells exposed to bispecific VHHs, resulting in staining in the GFP channel for both bispecifics. We conclude that in this case, too, each of the fusion partners retains its activity and specificity. Lymph node cells of a MHC class II knockout mouse failed to stain with the bispecific VHHs, demonstrating specificity.
Fig. 4.
(A) FACs staining of mouse lymph node cells with anti-MHC II–anti-GFP antibodies. (Upper) Staining observed in wild-type cells. (Lower) Staining of MHC class II-deficient cells. (B) In vivo delivery of GFP. Mice were injected with 50 μg bispecific antibodies and either received directly intraperitoneally or after 1 h i.v. 50 μg GFP. Stained cells were analyzed by flow cytometry.
As a proof-of-principle experiment to demonstrate the use of such bispecific antibody derivatives for the creation of a deep tissue reservoir (32), we injected the anti–GFP-VHH7 bispecific construct i.v. with the goal to first target a relevant cell population (B cells) with this reagent. We directly administered a single bolus of recombinant GFP (50 μg) either intraperitoneally or 1 h later i.v. and killed the animals 5.5 h later. Splenocytes were harvested and analyzed by flow cytometry (Fig. 4B). Most CD19+ cells (B cells) were GFP+, indicating successful capture of GFP in vivo. Administration of GFP into control animals that had not received the bispecific construct showed no GFP staining on CD19+ or CD19− cells. This experiment thus shows that a bispecific construct can be used to first target a cell population of interest, which can then be addressed with a ligand for the remaining free second binding site. Construction of bispecific reagents of this type would allow the targeted delivery of biologicals in a manner that might avoid acute toxicity, as is observed for systemic interleukin-2 (IL2) administration.
With these tools in hand, it becomes possible, in principle, to connect any entity proved to be a substrate in a sortase reaction and in all possible topologies. We successfully C-terminally conjugated human IL2 and IFN-α to anti-GFP and VHH-7 using this approach (Fig. S7), thus showing the general applicability of these tools.
Discussion
The ability to fuse proteins via their N or C termini creates immediate opportunities for the production of molecules not accessible by standard genetic means. We show that proteins connected in this manner retain functionality for N-to-N and for C-to-C fusions. As an instructive example, we demonstrate the ability to create C-terminally fused bispecific camelid-derived VHH constructs with full retention of the binding capacity of both fusion partners. Possible applications extend to other fusions as well. For those situations where the desired combination demands that both C or both N termini remain available for proper function, standard genetic approaches fall short. Click chemistry has developed to the point where off-the-shelf reagents suitable for solid-phase peptide synthesis allow ready access to the peptides that enable these types of fusion. Although demonstrated here for protein–protein fusions, further modifications of the click handles used to connect the two proteins allow installation of yet other functionalities, such as fluorophores, or pharmacologically active small molecules. Ease of modification of proteins of interest, ready access to recombinant sortases of different origin, and the flexibility afforded in nucleophile design through use of standard peptide synthetic methodology add to the versatility of sortase-mediated transacylation.
Protein fusions not easily accessed by other means are within easy reach using the technology described here. Of note, Hudak et al. described the use of an aldehyde tag in combination with strain-promoted click chemistry to achieve similar goals and produced hIgG fused to human growth hormone and maltose-binding protein (33). This approach is orthogonal to our sortagging strategy and immediately suggests the possibility of combining methods such as the aldehyde tag-click chemistry method developed by Hudak et al. with the chemo-enzymatic method developed here to access even more challenging protein–protein fusions.
Methods
N-Terminal Sortagging.
Sortase A of S. aureus (150 μM final concentration, 4.5× stock in 50 mM Tris, pH 7.4, 150 mM NaCl) and probe 1 or 2 (0.5 mM final concentration, 10× stock) were added to UbVME (58 μM final concentration) in sortase buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 10 mM CaCl2). The resulting mixture was incubated at 37 °C for 3 h. Next, the solution was acidified with 1% TFA in H2O and purified by reverse-phase HPLC [30→45% (vol/vol) B in 20 min, 3 mL/min]. The resulting purified protein was neutralized with saturated aqueous (sat. aq.) NaHCO3 concentrated in vacuo, redissolved in H2O, and quantified by gel electrophoresis. The protein was analyzed by LC/MS: N3-UbVME, Rt = 7.70 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI (electrospray ionization)/MS, m/z = 9,714 (M+H)+; DIBAC-UbVME, Rt = 7.54 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 9,360 (M+H)+.
C-Terminal Sortagging.
Sortase A of S. aureus (150 μM final concentration, 4.5× stock in 50 mM Tris, pH 7.4, 150 mM NaCl) and probe (0.5 mM final concentration, 10× stock) were added to the VHH (15 μM final concentration) in sortase buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 10 mM CaCl2). The resulting mixture was incubated at 25 °C overnight. The protein was purified by size exclusion on a Superdex 75. The resulting purified protein was concentrated in centrifugal filter units and analyzed by gel electrophoresis and LC/MS. Anti–GFP-3: Rt = 6.02 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 14,330 (M+H)+. Anti GFP-4: Rt = 7.90 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 14,170 (M+H)+. VHH7-3: Rt = 7.20 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 15,549 (M+H)+. VHH7-5: Rt = 7.00 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 15,139 (M+H)+.
Synthesis of Dimeric UbVME Constructs.
A mixture of azido-modified UbVME (42.5 μL, 80 μM) and cyclooctyne-modified UbVME (42.5 μL, 70 μM) was incubated overnight and subsequently purified by reverse-phase HPLC [30→45% (vol/vol) B in 20 min, 3 mL/min]. After purification, the solution was neutralized with sat. aq. NaHCO3 and concentrated in vacuo. Dimeric ubiquitin constructs containing only one reactive vinylmethyl ester were obtained by incubating either azido-modified ubiquitin (42.5 μL, 80 μM) with cyclooctyne-modified UbVME (42.5 μL, 70 μM) or azido-modified UbVMe (42.5 μL, 80 μM) with cyclooctyne-modified ubiquitin (42.5 μL, 70 μM). After dimerization, the proteins were purified and handled as described above.
Labeling of UCHL3 with Dimeric UbVME Constructs.
Purified dimeric constructs (0.5 μg, 24.5 pmol) were diluted in 20 μL Tris buffer [20 mM Tris, pH 8, 100 mM NaCl, 0.1 mM TCEP (tris(2-carboxyethyl)phosphine)] in the presence or absence of UCHL3 (94 pmol). The resulting mixture was incubated for 2 h, denatured with Laemmli sample buffer (4×) ,and loaded on a Tris-tricine gel. The proteins were either directly analyzed by Coomassie brilliant-blue staining or transferred to a PVDF membrane. The membrane was blocked with 4% (wt/vol) BSA in PBS/Tween (0.1% vol/vol). Penta-His HRP (1:12,500) was added and the membrane was agitated for 30 min at room temperature. The membrane was washed four times with 0.1% vol/vol Tween in PBS before the proteins were visualized using ECL plus.
Dimerization of Nanobodies.
Homodimeric anti-GFP nanobody was prepared by incubating anti–GFP-3 (100 μL, 80 μM) and anti–GFP-4 (100 μL, 85 μM) overnight at room temperature. Heterodimeric VHH7-3–anti-GFP-4 and VHH7-5–anti-GFP-4 were obtained by reacting either VHH7-3 (200 μL of a 20-μM solution) or VHH7-5 (200 μL of a 60-μM solution) with anti–GFP-4 (100 μL of a 120-μM solution) overnight at 25 °C. The dimeric nanobodies were purified by size exclusion on a Superdex 75. Fractions were collected and concentrated in centrifugal filter units. The purified dimers were analyzed on 15% SDS/PAGE.
(Anti-GFP)2: Rt = 10.07 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 28,526 (M+H2O+H)+. VHH7-3–anti-GFP-4: Rt = 10.91 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 29,755 (M+ H2O+H)+. VHH7-5–anti-GFP-4: Rt = 10.87 min, linear gradient 5→45% (vol/vol) B in 20 min; ESI/MS, m/z = 29,329 (M+H2O+H)+.
Functionality Assay of Homodimeric Nanobodies.
Homodimeric anti-GFP nanobody (20 μL, 25 μM) was incubated with GFP (2.5 μL, 10 μL ,and 30 μL of an 80-μM solution). The formed nanobody–GFP complex was subjected to size exclusion on a Superdex 200.
Functionality Assay of Heterodimeric Nanobodies.
Lymph node cells were harvested from C57BL/6 (Jackson Laboratories) or MHCII-deficient mice (Jackson Laboratories), washed, and incubated for 10 min with VHH7–anti-GFP, GFP, and VHH7–anti-GFP+GFP at 4 °C. The cells were collected by centrifugation, washed with PBS, and analyzed by flow cytometry.
In Vivo Delivery Assay.
For the delivery assays, BALB/c mice (Jackson Laboratories) were injected in the tail vein with either the bispecific antibody or GFP (50 μg per mouse). The mice receiving the bispecific antibody either directly received GFP (50 μg) intraperitoneally or received GFP (50 μg) i.v. after 1 h. After 5.5 h, blood was harvested, the mice were killed, and cells were isolated from lymph nodes, thymus, and spleen. Cells were washed with PBS and incubated with anti–CD19-APC (BD Pharmingen) and 7-AAD (Viaprobe; BD) for 10 min at 4 °C. The cells were washed with PBS and analyzed by flow cytometry. Mice were housed at the Whitehead Institute for Biomedical Research, and all studies were approved by the Massachusetts Institute of Technology Committee on Animal Care.
Supplementary Material
Acknowledgments
We acknowledge funding from The Netherlands Organisation for Scientific Research (to M.D.W.) and the Human Frontiers Science Program (to J.J.C.). This work was supported by National Institutes of Health R01 Awards AI033456 and AI087879. We are grateful to Dr. Charles B. Shoemaker (R01 Award U54 AI57159) for his help with developing a VHH that is specific for mouse class II MHC (VHH7).
Footnotes
Conflict of interest statement: The authors declare that a patent application on the production of N-to-N and C-to-C fusions has been filed.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205427109/-/DCSupplemental.
References
- 1.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 2.Seifert R, Wenzel-Seifert K, Kobilka BK. GPCR-Galpha fusion proteins: Molecular analysis of receptor-G-protein coupling. Trends Pharmacol Sci. 1999;20:383–389. doi: 10.1016/s0165-6147(99)01368-1. [DOI] [PubMed] [Google Scholar]
- 3.Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688–695. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leong SR, et al. IL-8 single-chain homodimers and heterodimers: Interactions with chemokine receptors CXCR1, CXCR2, and DARC. Protein Sci. 1997;6:609–617. doi: 10.1002/pro.5560060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasser MW, et al. Differential activation and regulation of CXCR1 and CXCR2 by CXCL8 monomer and dimer. J Immunol. 2009;183:3425–3432. doi: 10.4049/jimmunol.0900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drury LJ, et al. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci USA. 2011;108:17655–17660. doi: 10.1073/pnas.1101133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boado RJ, et al. Genetic engineering, expression, and activity of a chimeric monoclonal antibody-avidin fusion protein for receptor-mediated delivery of biotinylated drugs in humans. Bioconjug Chem. 2008;19:731–739. doi: 10.1021/bc7004076. [DOI] [PubMed] [Google Scholar]
- 8.Lu JZ, Hui EK-W, Boado RJ, Pardridge WM. Genetic engineering of a bifunctional IgG fusion protein with iduronate-2-sulfatase. Bioconjug Chem. 2010;21:151–156. doi: 10.1021/bc900382q. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Q-H, Boado RJ, Lu JZ, Hui EK-W, Pardridge WM. Re-engineering erythropoietin as an IgG fusion protein that penetrates the blood-brain barrier in the mouse. Mol Pharm. 2010;7:2148–2155. doi: 10.1021/mp1001763. [DOI] [PubMed] [Google Scholar]
- 10.Pastan I, Hassan R, Fitzgerald DJ, Kreitman RJ. Immunotoxin therapy of cancer. Nat Rev Cancer. 2006;6:559–565. doi: 10.1038/nrc1891. [DOI] [PubMed] [Google Scholar]
- 11.Osusky M, Teschke L, Wang X, Wong K, Buckley JT. A chimera of interleukin 2 and a binding variant of aerolysin is selectively toxic to cells displaying the interleukin 2 receptor. J Biol Chem. 2008;283:1572–1579. doi: 10.1074/jbc.M706424200. [DOI] [PubMed] [Google Scholar]
- 12.Rafei M, et al. A MCP1 fusokine with CCR2-specific tumoricidal activity. Mol Cancer. 2011;10:121. doi: 10.1186/1476-4598-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res. 2009;69:4941–4944. doi: 10.1158/0008-5472.CAN-09-0547. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair JC, Davies KM, Vénien-Bryan C, Noble MEM. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat Nanotechnol. 2011;6:558–562. doi: 10.1038/nnano.2011.122. [DOI] [PubMed] [Google Scholar]
- 15.Popp MW, Ploegh HL. Making and breaking peptide bonds: Protein engineering using sortase. Angew Chem Int Ed Engl. 2011;50:5024–5032. doi: 10.1002/anie.201008267. [DOI] [PubMed] [Google Scholar]
- 16.Guimaraes CP, et al. Identification of host cell factors required for intoxication through use of modified cholera toxin. J Cell Biol. 2011;195:751–764. doi: 10.1083/jcb.201108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: A versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Raines RT. Dibromobimane as a fluorescent crosslinking reagent. Anal Biochem. 1995;225:174–176. doi: 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]
- 19.Schellinger JG, et al. A general chemical synthesis platform for crosslinking multivalent single chain variable fragments. Org Biomol Chem. 2012;10:1521–1526. doi: 10.1039/c0ob01259a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natarajan A, et al. Construction of di-scFv through a trivalent alkyne-azide 1,3-dipolar cycloaddition. Chem Commun (Camb) 2007;(7):695–697. doi: 10.1039/b611636a. [DOI] [PubMed] [Google Scholar]
- 21.Xiao J, Hamilton BS, Tolbert TJ. Synthesis of N-terminally linked protein and peptide dimers by native chemical ligation. Bioconjug Chem. 2010;21:1943–1947. doi: 10.1021/bc100370j. [DOI] [PubMed] [Google Scholar]
- 22.Weikart ND, Sommer S, Mootz HD. Click synthesis of ubiquitin dimer analogs to interrogate linkage-specific UBA domain binding. Chem Commun (Camb) 2012;48:296–298. doi: 10.1039/c1cc15834a. [DOI] [PubMed] [Google Scholar]
- 23.Weikart ND, Mootz HD. Generation of site-specific and enzymatically stable conjugates of recombinant proteins with ubiquitin-like modifiers by the Cu(I)-catalyzed azide-alkyne cycloaddition. ChemBioChem. 2010;11:774–777. doi: 10.1002/cbic.200900738. [DOI] [PubMed] [Google Scholar]
- 24.Bundy BC, Swartz JR. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug Chem. 2010;21:255–263. doi: 10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- 25.Debets MF, et al. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem Commun (Camb) 2010;46:97–99. doi: 10.1039/b917797c. [DOI] [PubMed] [Google Scholar]
- 26.Borodovsky A, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 27.Antos JM, et al. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J Am Chem Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misaghi S, et al. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 29.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 30.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 31.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 32.Schellens JHM. In: Cancer Clinical Pharmacology. Schellens JHM, McLeod HL, Newell DR, editors. New York: Oxford Univ Press; 2005. pp. 30–39. [Google Scholar]
- 33.Hudak JE, et al. Synthesis of heterobifunctional protein fusions using copper-free click chemistry and the aldehyde tag. Angew Chem Int Ed Engl. 2012;51:4161–4165. doi: 10.1002/anie.201108130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.