Abstract
β-propellers that bind polyphosphoinositides (PROPPINs), a eukaryotic WD-40 motif-containing protein family, bind via their predicted β-propeller fold the polyphosphoinositides PtdIns3P and PtdIns(3,5)P2 using a conserved FRRG motif. PROPPINs play a key role in macroautophagy in addition to other functions. We present the 3.0-Å crystal structure of Kluyveromyces lactis Hsv2, which shares significant sequence homologies with its three Saccharomyces cerevisiae homologs Atg18, Atg21, and Hsv2. It adopts a seven-bladed β-propeller fold with a rare nonvelcro propeller closure. Remarkably, in the crystal structure, the two arginines of the FRRG motif are part of two distinct basic pockets formed by a set of highly conserved residues. In comprehensive in vivo and in vitro studies of ScAtg18 and ScHsv2, we define within the two pockets a set of conserved residues essential for normal membrane association, phosphoinositide binding, and biological activities. Our experiments show that PROPPINs contain two individual phosphoinositide binding sites. Based on docking studies, we propose a model for phosphoinositide binding of PROPPINs.
Keywords: autophagy, protein-lipid interactions, X-ray crystallography, yeast
Phosphoinositides play important roles in membrane trafficking processes. At least 11 phosphoinositide binding domains, among them the FYVE, PH, and PX domains, have been identified (1). One class of phosphoinositide binding proteins that has not been structurally characterized so far is the β-propellers that bind polyphosphoinositides (PROPPINs) (2, 3). They are conserved from yeast to humans and interact with polyphosphoinositides PtdIns3P and PtdIns(3,5)P2. Lipid binding of PROPPINs depends on a conserved FRRG motif (2, 4, 5).
PROPPINs act as PtdIns3P binding partners during macroautophagy and its selective subtypes (4–12). Macroautophagy, a fundamental intracellular trafficking pathway related to numerous diseases, unselectively recycles cytosolic material (13–16). It starts at the preautophagosomal structure (PAS) with the formation of autophagosomes. These double-membraned vesicles finally fuse with lysosomes, where their content is degraded.
Saccharomyces cerevisiae contains three PROPPINs: Atg18, Atg21, and Hsv2 (homologous with swollen vacuole phenotype 2). They are highly homologous but have different autophagic subtype specificities. Atg18 binds PtdIns3P-dependent to the PAS. As a core autophagy protein, it is required for macroautophagy and its selective variants, including the cytoplasm-to-vacuole targeting (Cvt) pathway, which targets proaminopeptidase I to the vacuole, and piecemeal microautophagy of the nucleus (PMN), which removes part of the nucleus (5, 17, 18). Atg18 also binds PtdIns(3,5)P2-dependent to the vacuole, where it carries out nonautophagic functions, such as the regulation of the PtdIns3P 5-kinase Fab1, and vesicular transport from the vacuole to the Golgi (2, 19, 20). Atg21 typically functions in selective autophagy variants (4, 21, 22). The function of Hsv2 is unclear; only its requirement for efficient PMN has been described (21). Atg18, Atg21, and Hsv2 are further recruited to endosomal structures in a PtdIns3P-dependent manner (23).
Mammals contain four PROPPINs, named WIPI (WD-40 repeat-containing protein that interacts with PtdIns) (10). WIPI1 and WIPI2 have been assigned as Atg18 orthologs, and WIPI3 and WIPI4 form a clade with Hsv2, whereas Atg21 seems to be restricted to yeasts (12).
So far, it has remained elusive how PROPPINs bind PtdIns(3,5)P2 and PtdIns3P beyond the requirement of the FRRG motif. Our aim was to characterize the phosphoinositide binding mechanism of PROPPINs on a molecular level. Here, we report the 3.0-Å crystal structure of Kluyveromyces lactis Hsv2, which shares 37% sequence identity with its S. cerevisiae homolog. Using different in vivo and in vitro approaches, we define a set of conserved residues within two individual basic pockets essential for normal membrane association and biological activities of the PROPPINs. This shows that PROPPINs contain two distinct phosphoinositide binding sites. We finally propose a model for phosphoinositide binding of PROPPINs based on docking studies performed with PtdIns3P and PtdIns(3,5)P2 at both binding sites.
Results
Crystal Structure of K. lactis Hsv2.
We tried to crystallize different yeast PROPPINs and obtained crystals of the Hsv2 homolog from K. lactis. Native KlHsv2 crystals diffracted up to a resolution of 3.0 Å. The structure was determined by single-wavelength anomalous diffraction phasing with a 3.4-Å dataset collected from a selenomethionine substituted crystal (Table 1). The structure was refined to a Rwork/Rfree of 19.7%/24.3% and comprises residues 14–269 and 275–338.
Table 1.
X-ray crystallographic data collection and refinement statistics
| native KlHsv2 | SeMet KlHsv2 | KlHsv2 complete loop 6CD (malonate soak*) | |
| PDB entry† | 4AV9 | 4AV8 | |
| Space group | P4132 | P4132 | P4132 |
| Unit cell dimensions | a = b = c = 159.0 Å | a = b = c = 158.1 Å | a = b = c = 156.4 Å |
| α= β= γ= 90° | α= β= γ= 90° | α= β= γ= 90° | |
| Beamline | PXII (SLS) | PXII (SLS) | PXII (SLS) |
| Wavelength, Å | 1.0015 | 0.9791 | 0.9765 |
| Resolution, Å (high-resolution bin) | 50–3.0 (3.15–3.0) | 50–3.4 (3.55–3.4) | 40–3.35 (3.45–3.35) |
| No. of observed reflections/unique reflections | 120,154/14,269 | 123,154/17,554 | 94,262/ 9,859 |
| Completeness, %, total (high) | 99.6/98.3 | 100/100 | 99.5/100 |
| I, total (high) | 28.3/3.9 | 15.9/3.3 | 27.5/5.1 |
| Rsym‡, %, total (high) | 5.4/54.6 | 10.3/56.3 | 6.1/47.8 |
| Wilson B factor, Å2 | 84.5 | 95.3 | 102.4 |
| Refinement | |||
| Rwork/Rfree§, % | 19.7/24.3 | 20.0/24.9 | |
| Residues included in the model (total no. of protein atoms) | 14–269, 275–338 (2,536) | 14–338 (2,571) | |
| Sulfate ions | 3 | — | |
| Water molecules | 1 | — | |
| Overall B-factor, Å2 | 77.7 | 102.1 | |
| B-factor protein, Å2 | 77.7 | 102.1 | |
| B-factor sulfate, Å2 | 94.8 | — | |
| B-factor water, Å2 | 67.2 | — | |
| rmsd for bond length, Å | 0.005 | 0.004 | |
| rmsd for bond angles, ° | 1.114 | 1.15 |
SLS, Swiss Light Source.
*KlHsv2 crystals were soaked with inositol-1,3,5-triphosphate in a malonate-containing cryoprotectant to remove the sulfate from the crystallization buffer, which might otherwise interfere with ligand binding. However, we did not observe a bound PIP head group molecule in the electron density map.
†The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AV8 and 4AV9).
‡Rsym = ∑|I − 〈I〉|/∑I.
§Rwork = ∑|Fobs|− k|Fcalc|/∑|Fobs|. Calculation of Rfree involved 5% of randomly chosen reflections.
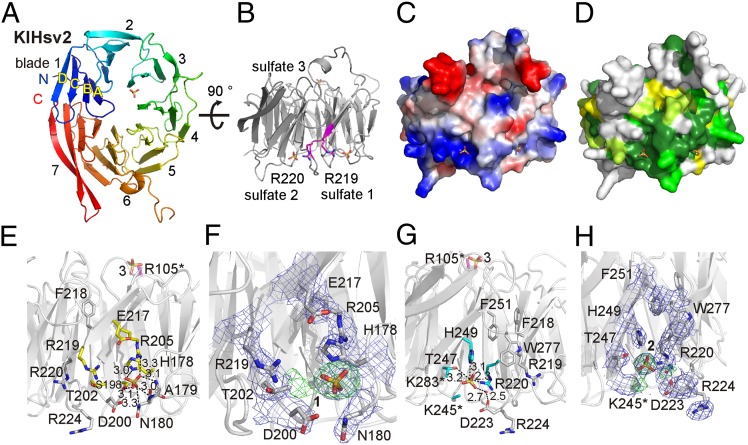
KlHsv2 forms a seven-bladed β-propeller. Each blade consists of four antiparallel β-strands, denoted as A to D from the inner to outer β-strand (Fig. 1A). KlHsv2 has a nonvelcro closure of the propeller, where the seventh blade is entirely formed by the C-terminal end of the protein. This topology is rare, but examples of propeller proteins displaying a nonvelcro closure have been observed before, for example, prolyl oligopeptidase and others (24–26). However, most of the known β-propeller structures have a velcro-like topology, where the N terminus contributes one or more strands to the last blade (27, 28).
Fig. 1.
Structure of KlHsv2. (A) KlHsv2 forms a seven-bladed β-propeller. (B) Two sulfate ions are bound right and left of the FRRG motif (magenta). (C) Electrostatic surface potential of the molecule in the same orientation as in B. (D) Conservation of KlHsv2 is shown, based on a sequence alignment of the Hsv2 homologs from K. lactis, S. cerevisiae, Ashbya gossypii, Candida glabrata, and Yarrowia lipolytica. The dark color corresponds to a high level of the conservation. Sequences were aligned with T-Coffee (50) and analyzed with the AMAS Server (51). (E) Sulfate binding site 1. Yellow residues were selected for mutagenesis studies. Polar contacts of the sulfate ion are shown, and the distances are given in angstroms. (F) Close-up view of sulfate site 1 with the overlaid 3.0-Å resolution 2mFo-DFc electron density map contoured at 1.0 σ (blue) and the difference (mFo-DFc) omit map at +3.0 σ (green). (G) Sulfate binding site 2. Cyan-colored residues were mutated to alanines. The side chains of K245, K283, and R105 were disordered (indicated by an asterisk), and these residues were modeled as alanines. R105 is localized opposite to both sulfate binding sites near the third sulfate ion and is colored magenta. (H) Close-up view of sulfate site 2 with the overlaid electron density map (blue) and the omit difference map contoured at +3.0 σ (green). This figure was prepared with PyMOL (52).
The β-strands within the blades of KlHsv2 are connected by short loop regions. An exception is blade 6, where a long loop comprising residues 254–278 connects strands C and D. Residues 270–274 within this loop are disordered in the 3.0-Å resolution structure. We also determined a KlHsv2 structure at a resolution of 3.35 Å, where these residues are visible in the electron density map, likely attributable to a tighter packing of the molecules in the crystal lattice, because this crystal had a smaller unit cell (Table 1).
The conserved FRRG motif, which is essential for phosphoinositide binding of PROPPINs is localized at the end of strand D in blade 5 and the loop connecting it with strand A of blade 6 (Fig. 1B). It is part of a positively charged and highly conserved region at the circumference of the β-propeller (Fig. 1 C and D).
In the 3.0-Å electron density map, we observed two spherical densities on either site of the FRRG motif. A third similarly shaped density was found near the center of the β-propeller (Fig. S1 A–C). Based on their shape and size, we modeled these densities as sulfate ions (Fig. 1 B, F, and H). The B-factors of sulfate ion 1 and sulfate ion 2 are 89.1 Å2 and 105.2 Å2, respectively, whereas the overall B-factor of the refined structure is 77.7 Å2. This indicates that the sulfate sites are not fully occupied; however, at a resolution of 3 Å, occupancy refinement cannot be done. To confirm that these sites are indeed occupied by sulfates, we also included water molecules at these positions, and in the case of site 2, the disordered side chain of K245 but positive density in the difference maps remained at these positions, confirming that sulfate ions are bound (Fig. S2 A–E).
These sulfate ions originated from the crystallization buffer, which contained 1.6 M magnesium sulfate. Sulfate ions often indicate the positions of the phosphates of phosphoinositide head groups (29–31). Sulfate site 3 is located in proximity to a negatively charged surface patch, which would be repulsive for membrane association, and thus is unlikely to represent a phosphoinositide binding site (Fig. S1C).
The two sulfate ions near the FRRG motif, which is required for phosphoinositide binding, however, indicate potential phosphoinositide binding site(s). Each of the FRRG arginines points to a sulfate ion. R220 interacts with sulfate 2, whereas the side chain of R219 is oriented toward sulfate site 1 (Fig. 1B). However, crystal packing contacts might affect the conformation of R219. W277, which is near R219, points toward sulfate 2 (Fig. 1 G and H). It is constrained in this conformation by stacking against the C-terminal end of a symmetry-related molecule in the crystal lattice (Fig. S1D). Without these packing contacts, the indol ring could swing out, leaving more space for R219, such that, in principle, R219 could point toward sulfate site 2. However, a 30-ns molecular dynamics (MD) simulation showed that R219 remains orientated toward sulfate binding site 1, despite the aromatic side chain of W277 moving away from sulfate site 2 (Fig. S1E).
Sulfate binding site 1 is freely accessible and not involved in crystal packing contacts. Sulfate 1 forms electrostatic interactions with R205 and H178. In addition, it forms hydrogen bonds with the side chains of S198 and N 180 and the backbone amides of A179 and N180 (Fig. 1 E and F).
Sulfate 2 is stabilized by electrostatic interactions with R220 of the FRRG motif and H249 (Fig. 1 G and H). Sulfate 2 also interacts with the side chain of T247 and a water molecule, which, in turn, forms a hydrogen bond with D223. Residues K245 and K283 are both located in proximity to sulfate 2. Their side chains are disordered in the crystal structure, and both residues were modeled as alanines as indicated by asterisks in Fig. 1G. The FRRG arginine R220 is locked in its conformation by a salt bridge with D223 and a network of hydrogen bonds between the guanidinium group and the carbonyl oxygens of G201 and D223. Larger residues than G221 (FRRG) would clash with the R220 side chain, which explains the requirement for a glycine at this position. Residues RRG of the FRRG motif are invariant, whereas the phenylalanine can be substituted by a leucine (2). The aromatic side chain of F218 (FRRG) participates in hydrophobic interactions between blades 5 and 6, and steric constraints are not as stringent at this position.
The two FRRG arginines point to the two sulfates irrespective of crystal packing contacts. Previously, two sulfate ions were detected in a single phosphoinositide binding site (31). However, the sulfates in our structure are 16.2 Å apart, whereas the distance between P1 and P3 in PtdIns3P is 6.5 Å, which excludes the possibility that they are part of the same binding site. Also, both sulfate sites are clearly separated by strand D of blade 5 and the loop connecting it to strand A of blade 6 (Fig. 1 B–D). Therefore, we speculated that either sulfate site 1 or 2 alone represents a phosphatidylinositol phosphate (PIP) binding site or, alternatively, the two sulfate sites correspond to two PIP binding sites.
We performed mutagenesis studies to probe the capabilities of sites 1 and 2 for phosphoinositide binding. We selected 10 basic and polar residues in proximity to the two sulfate binding sites, which are highly conserved among the Atg18 family members (Fig. S3), and mutated them to alanines in ScHsv2 (Fig. 1 E and G and Table 2). The KlHsv2 residue R105 is localized opposite to the two FRRG sulfate binding sites at the bottom of the β-propeller near sulfate 3 (Fig. 1 E and G). In the crystal structure, the side chain of R105 is disordered and the residue was modeled as an alanine, as marked by an asterisk in Fig. 1 E and G. We included this residue as a control in our mutagenesis studies. We first tested PIP binding of these mutants both in vitro and in vivo.
Table 2.
Summary of characterized PROPPIN mutants
| Residue KlHsv2 | Residue ScHsv2 | Residue ScAtg18 | Contacts with sulfates 1 and 2 in KlHsv2 structure | Phosphoinositide binding in docked KlHsv2 structure | Autophagic rate ScAtg18 | Requirement for membrane association microscopy ScAtg18 ScHsv2 | Requirement for membrane association liposome flotation assays ScHsv2 | Requirement for membrane association Rifs ScHsv2 |
| Site 1 | ||||||||
| H178 | H223 | H244 | + | + | 71% | − | − | |
| S198 | S243 | S264 | + | + | 65% | + | + | + |
| T202 | T247 | T268 | − | + | 61% | + | ± | |
| R205 | R250 | R271 | + | + | 64% | + | + | |
| E217 | E262 | Q283 | − | − | 88% | − | − | |
| R219 | R264 | R285 | − | + | 75% | + | + | + |
| Site 2 | ||||||||
| R220 | R265 | R286 | + | + | 44% | + | + | + |
| K245 | K290 | S311 | − | + | 61% | + | ± | |
| T247 | T292 | T313 | + | − | 85% | + | ± | |
| H249 | H294 | H315 | + | + | 49% | + | + | + |
| Control | ||||||||
| R105 | R123 | K102 | − | − | 101% | − | − | |
| R285 R286 | 26% | + | + | + | ||||
| S264 H315 | 34% | + | ||||||
| R285 H315 | 28% | + | ||||||
| S264 R286 | 34% | + | ||||||
| Penta | 28% |
Liposome Flotation Assays of ScHsv2 Mutants.
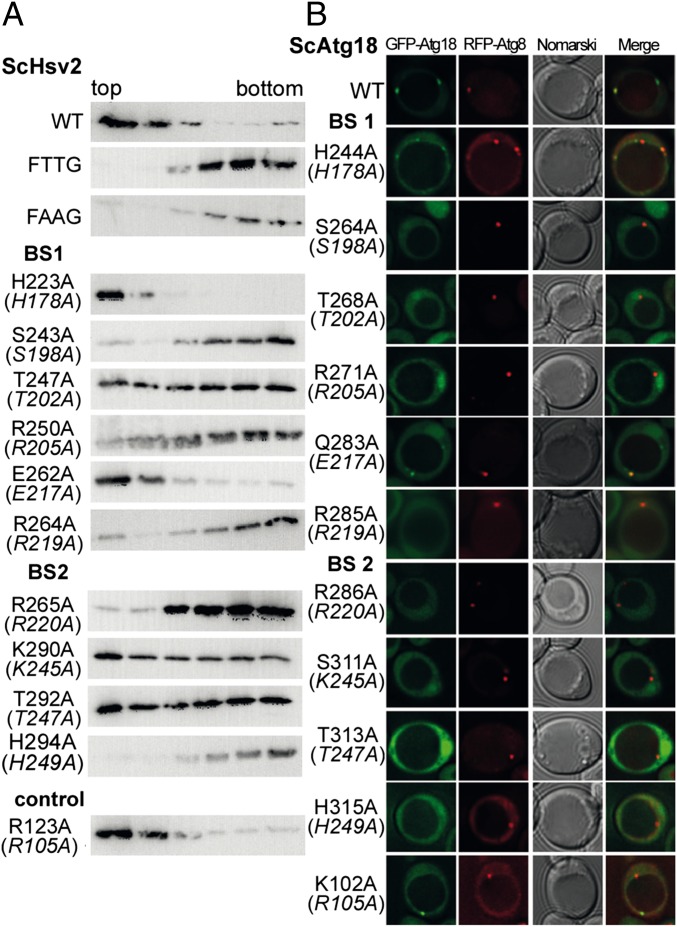
We analyzed binding of the recombinantly expressed ScHsv2 mutants to 2% (wt/wt) PtdIns3P containing liposomes with liposome flotation assays (Fig. 2A). Both WT ScHsv2 and the positive control ScHsv2R123A were only found in the light top fractions, whereas for the negative controls, the FTTG ScHsv2R264T R265T and FAAG double mutants, proteins were only present in the bottom fractions. In control experiments with PtdIns3P free liposomes, no binding was observed for WT and ScHsv2R123A.
Fig. 2.
Liposome flotation assays with ScHsv2 mutants and fluorescence microscopy analysis of ScAtg18 mutants. (A) ScHsv2 mutants were added to liposomes consisting of 2% PtdIns3P, 73% phosphatidylcholine, 23% phosphatidylethanolamine, and 2% Texas Red–phosphatidylethanolamine and applied onto a Nycodenz (Progen) gradient. ScHsv2 protein was detected by immunoblotting with an anti-ScHsv2 antibody. For each ScHsv2 mutant, the corresponding KlHsv2 mutant is shown below in parentheses. (B) Fluorescence microscopic analysis of the membrane association of ScAtg18 mutants. The atg18Δ cells expressing the indicated GFP-ScAtg18 variants and RFP-Atg8 were grown to stationary phase in selective medium containing 0.3 mM methionine. Pictures were taken using a DeltaVision Spectris fluorescence microscope and deconvoluted using WoRx (Applied Precision) software. For each ScAtg18 mutant, the corresponding KlHsv2 mutant is shown below in parentheses.
Six residues from sulfate binding site 1 were analyzed (Fig. 2A and Table 2). Three mutants, ScHsv2R250A, ScHsv2S243A, and the FRRG mutant ScHsv2R264A, did not bind to the liposomes. A portion of ScHsv2T247A is found in both the top and bottom fractions, indicating that this residue is involved in effective PtdIns3P binding. In contrast, association of ScHsv2H223A and ScHsv2E262A with the liposomes was not diminished compared with WT ScHsv2, indicating that these two residues are not critical for PtdIns3P binding.
We tested four conserved residues of sulfate binding site 2. Liposome binding of ScHsv2H294A and the FRRG ScHsv2R265A mutant was completely abolished. ScHsv2K290A and ScHsv2T292A were found in both the top and bottom fractions, which implies that these residues contribute to PtdIns3P binding but are not essential.
Membrane Recruitment of GFP-ScHsv2 and GFP-ScAtg18 Mutants in S. cerevisiae Cells.
Next, we analyzed whether our in vitro results correlate with a loss of membrane association in vivo. For this purpose, we analyzed the membrane recruitment of both the GFP-ScHsv2 mutants and the corresponding GFP-ScAtg18 mutants by fluorescence microscopy. Both Atg18 and Hsv2 bind via PtdIns3P to endosomes. Atg18 is also recruited to the PAS in a PtdIns3P-dependent manner. These two PtdIns3P-positive structures can be distinguished using either the PAS marker RFP-Atg8 or the endosome marker Snf7-RFP. We coexpressed these markers in our studies to confirm the integrity of both PtdIns3P-dependent organelles.
The atg18Δ cells expressing WT GFP-ScAtg18 and the hsv2Δ cells expressing WT GFP-ScHsv2 showed the expected green punctate structures in fluorescence microscopy, indicating membrane association (Fig. 2B and Fig. S4 A and B). The positive controls GFP-ScHsv2R123A and GFP-ScAtg18K102A behaved like the WT proteins.
Four mutations of sulfate binding site 1 (ScAtg18: S264A, T268A, R271A, and R285A and ScHsv2: S243A, T247A, R250A, and R264A) resulted in a release of the protein into the cytosol (Fig. 2B, Fig. S4 A and B, and Table 2), suggesting an essential role of these amino acids for membrane association. In contrast, WT-like localization was observed for both GFP-Atg18H244A and GFP-Atg18Q283A (corresponding to GFP-Hsv2H223A and GFP-Hsv2E262A), which demonstrates that these residues are not important for membrane binding.
Similarly, we identified four amino acids near the sulfate binding site 2 that are essential for membrane association (ScAtg18: R286A, S311A, T313A, and H315A and ScHsv2: R265A, K290A, T292A, and H294A). These findings are in agreement with the liposome flotation assays. In fact, the loss of membrane association observed for three mutants, GFP-ScHsv2T247A (site 1) and GFP-ScHsv2K290A and GFP-ScHsv2T292A (both site 2), is more striking in vivo compared with the in vitro situation, where these mutants still remained partially associated with the liposomes (summarized in Table 2).
Taken together, we identified, in addition to the known double-arginine motif, a set of conserved residues within the two distinct basic pockets localized on both sites of the FRRG motif. These residues are essential for normal membrane association of PROPPINs in living cells. Our in vivo and in vitro results show that PROPPINs contain two PtdIns3P binding sites.
Requirement of Both Binding Sites for Effective Phosphoinositide Binding in Vitro.
We next investigated whether both binding sites might act in PtdIns3P binding. We prepared double mutants of ScHsv2 and compared their PIP binding properties with the corresponding single mutations.
As a first approach, we used protein lipid overlay assays (32). We selected the site 1 mutant ScHsv2S243A and the site 2 mutant ScHsv2H294A, which are both required for PIP binding (Table 2). Both single mutants, GST-ScHsv2S243A and GST-ScHsv2H294A, showed a reduced binding to PtdIns3P and PtdIns(3,5)P2 compared with the WT protein (Fig. S5). The corresponding double mutant, GST-Hsv2S243A H294A, almost completely lost binding to PtdIns3P and PtdIns(3,5)P2, as did our negative control, the FAAG mutant GST-Hsv2R264A R265A. These data suggest that both sites bind phosphoinositides and that they act in concert to increase the avidity of PROPPINs for membranes.
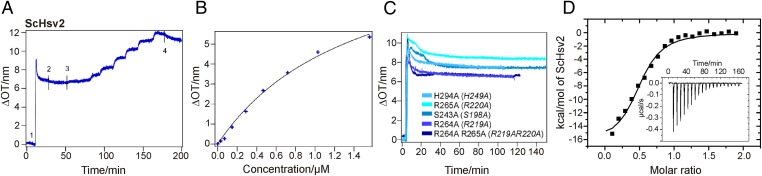
Next, we used reflectometric interference spectroscopy to measure PtdIns3P binding quantitatively (33, 34). In these experiments, silicon wafers with an oxide layer sufficiently thick for interference fringes were incubated with small unilamellar vesicles composed of 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC) and 3% PtdIns3P. This results in the formation of a lipid bilayer on the wafer. The coated wafers were then incubated with increasing concentrations of GST-ScHsv2 mutant proteins. Binding of the protein to the membrane bilayer was detected spectroscopically. The adsorption isotherm of WT ScHsv2 showed a Kd of 1.3 ± 0.2 μM in three independent experiments (Fig. 3 A and B). No significant binding was observed for membrane bilayers consisting of pure DOPC, confirming that membrane binding strictly depends on the presence of PtdIns3P. As expected, the ScHsv2R264A R265A mutant protein showed no significant membrane binding up to a concentration of 5 μM. We also selected four single-mutant proteins for measuring their PtdIns3P affinities by reflectometric interference spectroscopy. These four single-mutated proteins, ScHsv2R264A and ScHsv2S243A (both from site 1) and ScHsv2R265A and ScHsv2H294A (both from site 2), showed no detectable binding (Fig. 3C and Table 2). GST alone also did not yield any detectable binding. These measurements clearly show that both sites directly bind to PtdIns3P and that both sites are needed for effective membrane association.
Fig. 3.
Reflectometric interference spectroscopy and ITC measurements with ScHsv2. (A) Quantitative measurement of PtdIns3P binding of GST-ScHsv2 by reflectometric interference spectroscopy. Silicon wafers coated with a lipid bilayer consisting of DOPC and 3% PtdIns3P were incubated with increasing concentrations of GST-ScHsv2 WT protein. Binding of the protein was detected spectroscopically. The baseline was recorded in buffer. 1, formation of the DOPC bilayer [DOPC/PtdIns(3)P 97/3]; 2, rinsing with buffer; 3, stepwise addition of GST-ScHsv2 in increasing concentration; 4, rinsing with buffer. (B) Adsorption isotherm of binding of WT GST-ScHsv2 to PtdIns(3)P. A Kd value of 1.3 ± 0.2 μM was determined via a Langmuir-fit (black line). Measurements were done in triplicate. (C) Reflectometric interference spectroscopy measurements of GST-ScHsv2 mutants. After bilayer formation, each mutant protein was added in increasing concentrations (up to 5 μM) with pauses of 5 min in between. The GST-Hsv2 mutants showed no significant membrane binding. ΔOT, changes in optical thickness (nm). (D) For ITC measurements, ScHsv2 was titrated into 2% PtdIns3P containing liposomes. The integrated areas normalized to the amount of ScHsv2 injected (kcal/mol) vs. its molar ratio to PtdIns3P are shown. Inside is a figure showing the base line-corrected raw data with power plotted against time during the injections.
To determine the stoichiometry of PtdIns3P binding directly, we performed isothermal titration calorimetry (ITC) measurements. ScHsv2 was titrated into liposomes consisting of 2% PtdIns3P, 73% phosphatidylcholine, 23% phosphatidylethanolamine, and 2% Texas Red–phosphatidylethanolamine, which were prepared in the same way as the liposomes used for the liposome flotation assays. We observed an average molar ratio of 0.50 ± 0.06 from four measurements for protein binding to PtdIns3P, which corresponds to two PtdIns3P binding sites per PROPPIN molecule (Fig. 3D). The measured Kd is 0.67 ± 0.04 μM. The differences in Kd between the ITC and reflectometric interference spectroscopy measurements might be attributable to differences in membrane curvature (i.e., liposomes vs. a flat membrane) and differences in lipid composition of the membranes.
Relevance of the Two PIP Binding Sites for Macroautophagy and the Cvt Pathway.
After establishing that both PIP binding sites are needed for effective membrane recruitment in vitro, we wanted to investigate whether the two PIP binding sites are also of functional importance. We chose ScAtg18 for functional characterization because this core autophagy protein is essential for unselective macroautophagy and all selective autophagy subtypes (17, 18).
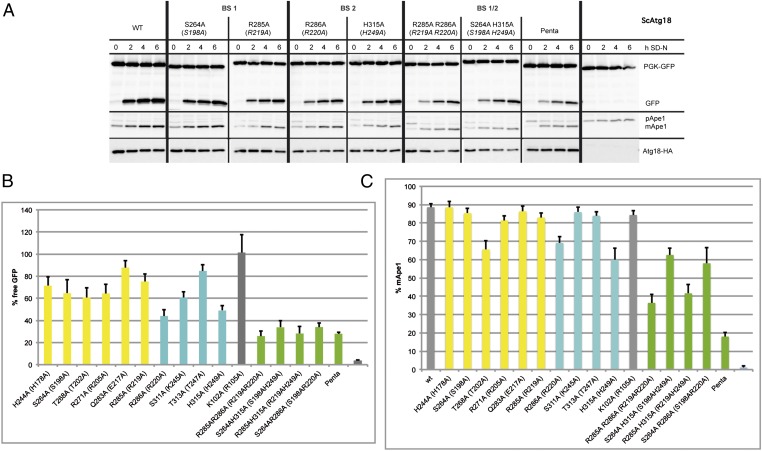
We measured the autophagic rate in atg18Δ cells expressing with the endogenous promoter the mutated Atg18 proteins tagged with an HA epitope. We used an established assay based on the degradation of a cytosolic GFP fusion of 3-phosphoglycerate kinase (Pgk1-GFP) (35). Here, autophagic breakdown of Pgk1-GFP yields proteolysis resistant GFP, which accumulates within the vacuole. The GFP level determined in Western blots thus reflects the autophagic rate. The autophagic rate of cells expressing WT Atg18-HA after 6 h of starvation was set to 100% (Fig. 4 A and B and Fig. S6). We measured the autophagic rate in five independent experiments and calculated the SEM for cells expressing the mutant proteins.
Fig. 4.
Analysis of the functional relevance of a set of conserved residues for autophagy. Cells deleted for ATG18 and cotransformed with pRS316-PGK1-GFP and pRS313-Atg18-HA, the relevant Atg18-HA mutants, or an empty plasmid as a negative control were grown to stationary phase in selection medium and shifted for 6 h to SD-(N) medium to induce macroautophagy. Samples are taken every 2 h. Only Western blots of representative strains are shown; residual blots are shown in Fig. S6. (A) (Top) Pgk1-GFP and GFP are detected using a GFP antibody; the amount of free GFP represents the autophagic rate. (Middle) Samples were reprobed to follow the Cvt pathway using an Ape1 antibody, which detects pApe1 and mApe1. (Bottom) Samples were further reprobed with an HA antibody to demonstrate the stability of the Atg18-HA proteins. (B) Autophagic rate is determined by quantifying the amount of free GFP using AIDA software (Raytest). The amount of free GFP of the Atg18-HA WT at 6 h is set to 100%. (C) To monitor the Cvt pathway, the percentage of mApe1 has been calculated from the total amount of pApe1 and mApe1 before starvation using AIDA software. Mutants in binding site 1 are labeled yellow, mutants of binding site 2 are shown in cyan, and mutants carrying mutations in both binding sites are shown in green. The WT, the negative control with empty plasmid, and the control mutant (K102A) with a mutation opposite to the binding sites are labeled in gray.
As expected, the positive control Atg18K102A-HA showed WT-like autophagic activity (101%) (Fig. 4B and Table 2). In agreement with their previously established clear cytosolic localization, the four mutations affecting binding site 1 (S264A, 65%; T268A, 61%; R271A, 64%; and R285A, 75%) and four additional mutations in the second binding site (R286A, 44%; S311A, 61%; T313A, 85%; and H315A, 49%) of Atg18 led to a reduced autophagic rate (Fig. 4B). We assume that the only partial reduction of macroautophagy is attributable to protein-protein interactions within the living cell, which lead to a small residual amount of membrane-associated Atg18 proteins. In fluorescence microscopy, the large amount of protein released into the cytosol prevents detection of such faint structures, whereas accessory proteins were absent in our in vitro assays.
In general, the second binding site seems to have a stronger effect on the autophagic activity of Atg18. To test whether both binding sites act in concert, we also measured double mutants containing one mutation in either of the two PIP binding sites. Indeed, Atg18R285A R286A-HA (26%), Atg18S264A H315A-HA (34%), Atg18R285A H315A-HA (28%), and Atg18S264A R286A-HA (34%) showed a more severe reduction of the macroautophagic rate. Even Atg18penta-HA (28%), containing five mutations divided between both binding sites (S264A, R285A, and H244A of site 1 and R286A and H315A of site 2), showed no complete macroautophagy block. This is in line with previous findings with the Atg18FTTG mutant, which is strongly affected in phosphoinositide binding (4, 5) but might still be partially recruited to the PAS by its interaction partner Atg2 or other components (8, 9, 36). Two mutations at binding site 1 (Atg18H244A and Atg18Q283A), which are not essential for membrane association of the proteins, showed a somewhat reduced autophagic rate (71% and 88%, respectively). The atg18Δ cells showed the expected complete autophagy block.
To dissect the roles of both PIP binding sites for selective autophagy, we further analyzed the Cvt pathway. The Cvt pathway is a constitutive transport pathway active under nutrient-rich conditions, which selectively targets proaminopeptidase I (pApeI) to the vacuole, where it is proteolytically matured. We determined the extent of pApeI maturation under both logarithmic (OD600 = 1) and stationary growth conditions (OD600 = 6). As shown in Fig. 4C, the effects of single mutations in both binding sites were less severe than their effects on macroautophagy. Similar to the measurements of the macroautophagic rate, combining mutations in both binding sites resulted in a more severe phenotype. To exclude the possibility that the observed effects on macroautophagy and the Cvt pathway resulted from instability of the Atg18 mutant proteins, the samples were reprobed with an HA antibody. Compared with the WT protein, the levels of the mutant proteins were not significantly altered. Our data show that both PIP binding sites are of functional importance for macroautophagy and the selective Cvt pathway.
Requirement of the Two Binding Sites for Vacuole Homeostasis, a Nonautophagic Function of ScAtg18.
Because of the PtdIns(3,5)P2-dependent nonautophagic role of Atg18 in vacuole homeostasis, atg18Δ cells show a characteristic single enlarged vacuole (2, 19, 20). Expression of either Atg18S264A-HA or Atg18R285A-HA, mutated within the first binding site, or Atg18H315A-HA or Atg18R286A-HA, mutated within the second binding site, resulted in a mixture of cells with WT-like fragmented multilobed vacuoles and those with a single large vacuole, indicating at least partial functionality of the mutated proteins (Fig. S7). The double-mutated proteins Atg18S264A H315A-HA and Atg18R264A R265A-HA did not restore normal vacuole morphology. These findings are in good agreement with the effects measured on macroautophagic activity and demonstrate that both binding sites are also relevant for a cellular function of Atg18 dependent on PtdIns(3,5)P2 binding and that they act in concert.
In Silico Analyses of the Phosphoinositide Binding Sites and Membrane Association.
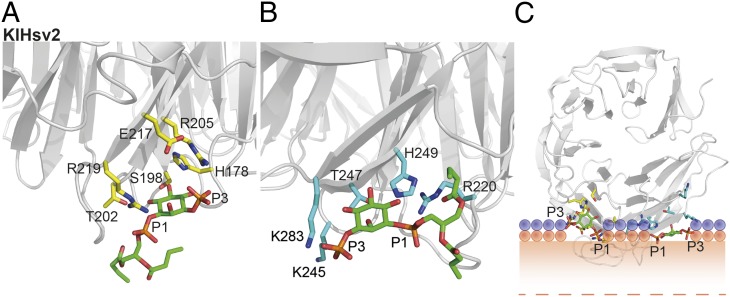
To gain further insights into PtdIns3P and PtdIns(3,5)P2 binding, we performed in silico docking. A 30-ns MD simulation of solvated KlHsv2 using the coordinates of a 3.35-Å crystal structure, which includes the complete loop connecting strands C and D in blade 6, showed that this loop is the most flexible region of the molecule and adopted different conformations. Overlaying the crystal structure with the coordinates of the simulation end point showed release of W277 from its crystal packing contacts, freeing binding site 2 (Fig. S1B). We further docked PtdIns3P and PtdIns(3,5)P2 into both binding pockets with AutoDock 4.2 (37). KlHsv2 residues H178, S198, D200, T202, R205, and R219 are involved in PtdIns3P binding in site 1 (Fig. 5A and Fig. S8C). Salt bridges formed between phosphates P1 and R219 and between P3 and R205. In a similar manner R220, K245, H249, and K283 were important for PtdIns3P binding in site 2 (Fig. 5B and Fig. S8C). Phosphate P1 formed a salt bridge with H249 and R220, whereas phosphate P3 interacted with K245 and K283. However, docking for site 2 might not be as precise as for site 1, because the side chains of K245 and K283 are disordered in the crystal structure. For PtdIns(3,5)P2, H178 and H249 needed to be protonated for successful docking. Although protonation of these residues did not change docking of PtdIns3P, PtdIns(3,5)P2 was recognized by the same residues as PtdIns3P with additional salt bridges between phosphates P5 and H178 in site 1 and K283 in site 2 (Fig. S8 A and B).
Fig. 5.
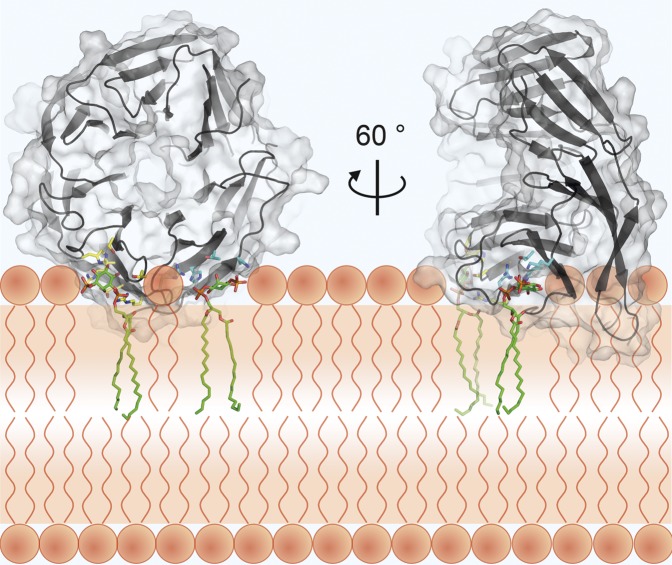
Docking of PtdIns3P into KlHsv2 and model for membrane binding of PROPPINs. Docking of PtdIns3P into binding site 1 (A) and docking into site 2 (B). Residues important for PIP binding in binding site 1 are shown in yellow and in cyan for site 2. (C) Model for membrane recognition of PROPPINs. The head groups of the two PtdIns3P molecules are shown as green sticks. The hydrophobic tails were omitted for clarity. Residues involved in PIP binding of site 1 are represented with yellow sticks, and those involved in PIP binding of site 2 are shown in cyan. The positions of the phosphates of the membrane phospholipids are shown through orange circles, whereas the violet circles represent the polar groups of these phospholipids. The shaded orange bar depicts the length of the phospholipid fatty acid chains from one leaflet of the bilayer, and the dashed orange line marks the center of the bilayer. The model was drawn approximately to scale. Loops between β-strands 6C and 6D and 7C and 7D are likely to insert into the membrane.
Based on PtdIns3P docked in both sites, we propose a model for PROPPIN membrane recognition (Fig. 5C). When defining the membrane as the horizontal plane cutting through the two phosphates P1 of the PtdIns3P, the two loops connecting strands 6C and 6D and 7C and 7D, respectively, would insert into the bilayer. The propeller then binds perpendicular to the membrane.
Discussion
Previous studies uncovered the crucial role of an FRRG motif for phosphoinositide binding of PROPPINs (2, 5, 9). In our KlHsv2 crystal structure, these arginines are localized on the circumference of the β-propeller (Fig. 1B) within two distinct conserved basic pockets. A similar arrangement of two consecutive arginines was observed in the crystal structure of the PH domain of Slm1 (synthetic lethal with MSS4 protein 1). Here, the two arginines, which are both essential for lipid binding, point toward two predicted lipid binding sites: the canonical phosphoinositide binding site and a second putative dihydrosphingosine-1-phosphate binding site (38).
Our ITC measurements show that PROPPINs have two phosphoinositide binding sites. We mutated conserved residues of PROPPINs within both lipid binding sites and identified seven residues (for ScAtg18: S264, T268, R285 and R271 in binding site 1 and R286, S311 and H315 in binding site 2; Table 2) critical for membrane association both in vivo and in vitro. In contrast, biological activities, such as macroautophagy and the Cvt pathway, were only partially affected, most likely attributable to residual protein-protein interactions. The additive effects of double mutations within the two sites demonstrated that both are needed for normal membrane association and function.
Comparison of the in silico docked structures of KlHsv2 with known phosphoinositide binding domains showed that the head group is oriented similar to that of the PtdIns3P head group in the EEA1-FYVE domain with the phosphate P3 pointing outside of the binding pocket (39). Remarkably, most FYVE domains must dimerize for efficient binding to endosomes to allow coordinated binding of two PtdIns3P molecules within the same membrane (40–43). This suggests that for normal membrane association of these proteins, the affinity of a single phosphoinositide binding site might not be sufficient. In agreement, mutation of a single critical residue in either site 1 or 2 of PROPPINs abolished membrane binding. This explains why PROPPINs have two PIP binding sites.
Based on our docking studies, we propose a model for membrane recognition of PROPPINs, where the β-propeller adopts a perpendicular orientation toward the membrane (Fig. 5C). This model further predicts the insertion of two loops into the membrane, which would synergistically enhance the membrane association of PROPPINs. Participation of membrane insertion loops and nonspecific electrostatic interactions in membrane binding has already been demonstrated for several phosphoinositide binding proteins (1, 43), for example, the PtdIns3P binding PX domain of Vam7p (44) and FYVE domains (45).
Spatial distribution of phosphoinositides is one criterion to define an intracellular compartment (1). Selective recognition of phosphoinositides is thus a key determinant for protein localization. Studies on PROPPINs, including those from S. cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, and humans, indicate that some of them are able to bind both PtdIns(3,5)P2 and PtdIns3P (4–7, 12, 46). Indeed, our docking studies suggested that PtdIns3P and PtdIns(3,5)P2 fit into both binding pockets. Docking of PtdIns(3,5)P2 was only successful when H178 and H249 of KlHsv2 were protonated. This is reminiscent of the histidine switch observed for PtdIns3P binding of the EEA1-FYVE domain (47) and PtdIns(3,4,5)P3 binding of the GRP1 PH domain (48). We thus speculate that PtdIns(3,5)P2 binding might also depend on pH. Indeed, PtdIns3P is typically restricted to endosomes and the PAS, whereas PtdIns(3,5)P2 is found at the vacuolar membrane. So far, a PtdIns(3,5)P2-dependent biological function has been documented only for ScAtg18a (2, 19, 20).
PROPPINs are members of the family of WD-40 propeller proteins, which typically act as scaffolds for protein-protein interactions (28, 49). We expect that binding of ScAtg18 to PtdIns(3,5)P2 and PtdIns3P at different loci is mediated by additional protein-protein interactions. At the PAS, a protein complex of Atg18 with the integral membrane protein Atg9 and the peripheral membrane protein Atg2 was detected (36). Apparently, a stable Atg18–Atg2 complex is formed in the cytosol and then recruited by the transmembrane protein Atg9 to the PAS (9), although an Atg18-containing protein complex regulating the PtdIns3P 5 kinase Fab1 was reported at the vacuolar membrane (19, 20). Taken together, both protein-lipid and protein-protein interactions may determine the correct in vivo localization of PROPPINs and, compared with our in vitro measurements, may yield even higher affinities for membrane binding.
Materials and Methods
A detailed description of cloning, mutagenesis, and protein purification procedures is provided in SI Materials and Methods. The crystallization and structure determination of KlHsv2; liposome flotation assays; protein lipid overlay assays; reflectometric interference, ITC measurements and the MD simulation; and docking studies are also described there. Detailed procedures of the fluorescence microscopy analysis; measurements of the macroautophagic and Cvt pathway rates are also given.
Supplementary Material
Acknowledgments
Diffraction data were collected at beamline X10SA (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland). We thank the beamline staff for their help during data collection. We thank Esra Demircioglu and Danilo Meyer for their help with the ITC measurements. We further thank Michaela Hellwig, Petra Schlotterhose, and Ursel Ries for excellent technical assistance. We also thank Reinhard Jahn for his support. This work was funded by the Deutsche Forschungsgemeinschaft through a grant of the SFB860 (to M.T. and K.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AV8 and 4AV9).
See Author Summary on page 11906 (volume 109, number 30).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205128109/-/DCSupplemental.
References
- 1.Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dove SK, et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 4.Strømhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krick R, Tolstrup J, Appelles A, Henke S, Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–4638. doi: 10.1016/j.febslet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 6.Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell. 2004;15:2652–2663. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Q, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285:11476–11488. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proikas-Cezanne T, et al. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 11.Mauthe M, et al. Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy. 2011;7:1448–1461. doi: 10.4161/auto.7.12.17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polson HE, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:6. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 13.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farré JC, Krick R, Subramani S, Thumm M. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 17.Barth H, Meiling-Wesse K, Epple UD, Thumm M. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 2001;508:23–28. doi: 10.1016/s0014-5793(01)03016-2. [DOI] [PubMed] [Google Scholar]
- 18.Guan J, et al. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell. 2007;18:4232–4244. doi: 10.1091/mbc.E07-04-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krick R, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiling-Wesse K, et al. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J Biol Chem. 2004;279:37741–37750. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- 23.Krick R, Henke S, Tolstrup J, Thumm M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy. 2008;4:896–910. doi: 10.4161/auto.6801. [DOI] [PubMed] [Google Scholar]
- 24.Bosch J, Tamura T, Tamura N, Baumeister W, Essen LO. The beta-propeller domain of the trilobed protease from Pyrococcus furiosus reveals an open Velcro topology. Acta Crystallogr D Biol Crystallogr. 2007;63:179–187. doi: 10.1107/S0907444906045471. [DOI] [PubMed] [Google Scholar]
- 25.Brandstetter H, Kim JS, Groll M, Huber R. Crystal structure of the tricorn protease reveals a protein disassembly line. Nature. 2001;414:466–470. doi: 10.1038/35106609. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc Natl Acad Sci USA. 2011;108:16571–16576. doi: 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neer EJ, Smith TF. G protein heterodimers: New structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- 28.Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karathanassis D, et al. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoebel S, Blankenfeldt W, Goody RS, Itzen A. High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep. 2010;11:598–604. doi: 10.1038/embor.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayan K, Lemmon MA. Determining selectivity of phosphoinositide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gauglitz G, Brecht A, Kraus G, Nahm W. Chemical and biochemical sensors based on interferometry at thin (multi-)layers. Sens Actuators B Chem. 1993;11:21–27. [Google Scholar]
- 34.Proll G, Markovic G, Steinle L, Gauglitz G. Reflectometric interference spectroscopy. Methods Mol Biol. 2009;503:167–178. doi: 10.1007/978-1-60327-567-5_8. [DOI] [PubMed] [Google Scholar]
- 35.Welter E, Thumm M, Krick R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010;6:794–797. doi: 10.4161/auto.6.6.12348. [DOI] [PubMed] [Google Scholar]
- 36.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 37.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallego O, et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumas JJ, et al. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- 40.Gaullier JM, et al. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 41.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayakawa A, et al. Structural basis for endosomal targeting by FYVE domains. J Biol Chem. 2004;279:5958–5966. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- 43.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 44.Lee SA, et al. Molecular mechanism of membrane docking by the Vam7p PX domain. J Biol Chem. 2006;281:37091–37101. doi: 10.1074/jbc.M608610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutateladze TG, et al. Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 46.Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: Metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Lee SA, et al. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc Natl Acad Sci USA. 2005;102:13052–13057. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J, et al. Molecular mechanism of membrane targeting by the GRP1 PH domain. J Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35:565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 51.Livingstone CD, Barton GJ. Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 52.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System (Schrodinger, Portland, OR), Version 1.3r1.