Abstract
Bacteroides fragilis can replicate in atmospheres containing ≤0.05% oxygen, but higher concentrations arrest growth by an unknown mechanism. Here we show that inactivation of a single gene, oxe (i.e., oxygen enabled) in B. fragilis allows for growth in concentrations as high as 2% oxygen while increasing the tolerance of this organism to room air. Known components of the oxidative stress response including the ahpC, kat, batA-E, and tpx genes were not individually important for microaerobic growth. However, a Δoxe strain scavenged H2O2 at a faster rate than WT, indicating that reactive oxygen species may play a critical role in limiting growth of this organism to low-oxygen environments. Clinical isolates of B. fragilis displayed a greater capacity for growth under microaerobic conditions than fecal isolates, with some encoding polymorphisms in oxe. Additionally, isolation of oxygen-enabled mutants of Bacteroides thetaiotaomicron suggests that Oxe may mediate growth arrest of other anaerobes in oxygenated environments.
Keywords: superoxide, peroxidase, anaerobiosis
When cyanobacteria developed the ability to strip electrons from water ∼2.5 billion years ago, they set in motion a series of environmental changes that would have profound effects on the evolution of life on this planet. The release of molecular oxygen (i.e., O2) as a byproduct of photosynthesis left many microbes vulnerable to oxidative damage, caused in part by the reactivity of key components of the metabolic networks used by these organisms. Free iron and flavin-containing proteins (1–5) can adventitiously donate electrons to oxygen to produce reactive oxygen species (ROS) like superoxide (i.e., O2−) or hydrogen peroxide. These endogenously generated ROSs can destroy important iron–sulfur clusters, inactivate key metabolic enzymes, and cause DNA lesions that may culminate in death of the organism (6–13). Primordial microbes facing extinction in this newly oxygenated environment were forced to retreat to protected niches in reducing sediments or to evolve mechanisms of protection from oxidative stress. These defenses included the elaboration of more robust ROS detoxifying enzymes like superoxide dismutase (14), rubrerythrins, catalases, and peroxidases (15), as well as metabolic enzymes fortified against ROS damage (16–18). With such protective measures in place, these bacteria could not only grow despite the presence of oxygen, but could further evolve to harness the enormous energy-generating potential of this powerful electron acceptor.
Obligately anaerobic bacteria cannot replicate in the presence of oxygen, and present-day species may have evolved from the sediment-dwelling ancient microbes. For this reason, obligate anaerobiosis was once thought to be rooted in the lack of ROS-detoxifying enzymes (19). Extensive evidence to the contrary has since negated this hypothesis, but the molecular mechanisms underlying anaerobiosis have remained elusive. Although the possibility remains that molecular oxygen itself can cause growth cessation in this class of organisms (20), a complete picture of O2 sensitivity has not yet taken shape.
Bacteroides fragilis provides a convenient model for testing theories of oxygen sensitivity. Previously classified as a “strict anaerobe,” this mammalian commensal has since been found to grow in and benefit from nanomolar concentrations of oxygen (21). In addition, even though it cannot replicate in room air, this organism is capable of mounting a strong response to aeration (22, 23), including the production of numerous ROS-scavenging enzymes (24–29) and other factors (30) that contribute to its impressive aerotolerance. The unique position of B. fragilis in the oxygen tolerance spectrum has allowed us to gain valuable insights into the nature of anaerobiosis. Herein we characterize derivatives of this bacterium that, as a result of mutation of a single gene (oxe), are capable of growth in as much as 2% oxygen, representing a 40-fold increase vs. WT B. fragilis. Additionally, an oxe mutant is even more tolerant of room air than its WT counterpart, thus providing an intriguing link between those bacteria that thrive in aerobic environments and those that do not.
Results
Isolation of Oxygen-Enabled Mutants.
Recent work has shown that B. fragilis can grow under atmospheres containing as much as 0.05% oxygen (500 nM dissolved O2) (21). Higher concentrations of O2, however, inhibit growth in a manner independent of increases in environmental redox potential (31). With these observations in mind, we sought to isolate mutants of B. fragilis capable of growth under normally restrictive oxygen concentrations. When WT B. fragilis was plated under microaerobic conditions (0.25–2% oxygen by volume), colonies were found to form at a frequency of ∼10−6 relative to an anaerobic control (Table 1). Purification of these colonies revealed that they were capable of robust growth under microaerobic conditions, suggesting that a spontaneous mutation had somehow given rise to “oxygen-enabled” variants of B. fragilis. To gain insights into the molecular nature of this microaerobic growth, we tested whether various mutations on the B. fragilis genome would alter the frequencies at which oxygen-enabled colonies arose.
Table 1.
Log10 efficiencies of plating for various strains of B. fragilis under microaerobic conditions
| Strain | Relevant characteristics | Log10 (efficiency of plating) |
| ADB77 | TM4000ΔthyA | −5.9 |
| ADB267 | ADB77Δoxe | −0.03 |
| ADB266 | ADB77ΔcydAB | −6.3 |
| BM37 | ADB77Δkat | −5.1 |
| BM28 | ADB77ΔahpC | −5.0 |
| IB263 | 638RoxyRC | −6.3 |
| MBD616 | TM4000 thyA2ΔsodA | −6.2 |
| YT135 | TM4000 batD::Tn4400′ | −5.9 |
| BM50 | ADB77ΔahpCΔkat | −5.7 |
| BM118 | ADB77ΔoxyR | −5.9 |
| BM95 | ADB77Δtpx | −4.9 |
| BM131 | ADB77 ΔahpCΔtpx | −5.2 |
| BM105 | ADB77ΔahpCΔkatΔtpx | ≤8.7 |
| BM134 | ADB77Δrubrerythrins | −5.4 |
| BM2 | ADB77Δrub | −4.9 |
Strains were grown anaerobically, serially diluted, and plated on BHIS. Test plates were transferred to microaerobic conditions, and control plates were transferred to anaerobic conditions. After several days, counts (in cfu) were enumerated. Log10 (efficiency of plating) is defined as the log10 (number of colonies arising in the presence of oxygen/number of colonies arising anaerobically).
B. fragilis encodes numerous oxidative stress defense enzymes that play a crucial role in protecting the organism from the damaging effects of ROS, and the expression of some of these proteins is activated at the transcriptional level by OxyR (23). A strain encoding a constitutively active allele of oxyR (32), as well as a ΔoxyR strain, were found to give rise to colonies at the same frequency as WT under microaerobic conditions (Table 1), indicating that the OxyR-mediated oxidative stress response (OSR) is neither sufficient nor essential for the oxygen-enabled phenotype. However, we reasoned that we might still be able to suppress the formation of oxygen-enabled colonies by deleting enzymes crucial to the reduction of oxygen or its reactive intermediates. As cytochrome oxidase (cydAB) is used during nanaerobic growth of B. fragilis and appears to be the major reducer of O2 under low-oxygen conditions (21), we hypothesized that it would also play a crucial role in microaerobic growth. However, a ΔcydAB strain plated with the same low efficiency as WT under microaerobic conditions, as did strains missing catalase (kat), superoxide dismutase (sod), alkylhydroperoxide reductase (ahpC), thioredoxin-dependent peroxidase (tpx), or the Bacteroides aerotolerance BAT operon (30) (Table 1), suggesting that a fully functional OSR was not necessary for the oxygen-enabled phenotype. Only when ahpC, kat, and tpx were deleted in combination were we unable to detect any oxygen-enabled colonies (Table 1).
One mutant, however, gave a surprising result. The BF638R_0963 gene (National Center for Biotechnology Information Gene ID 11704906) encodes a predicted flavoprotein with homology to the nitric oxide reductase NorV, as well as to rubredoxin:oxygen oxidoreductase (Roo), an enzyme found to protect Desulfovibrio vulgaris Hildenborough under 1% oxygen by reducing O2 to water (33). Based on this homology, we reasoned that deletion of BF638R_0963 would sensitize the organism to oxygen. Contrary to this prediction, we found that deletion of this gene resulted in a strain that plated with ∼100% efficiency under microaerobic conditions (Table 1). Our results therefore suggested a different function for this gene product in B. fragilis, prompting us to name the locus oxe (i.e., oxygen-enabled; Fig. 1).
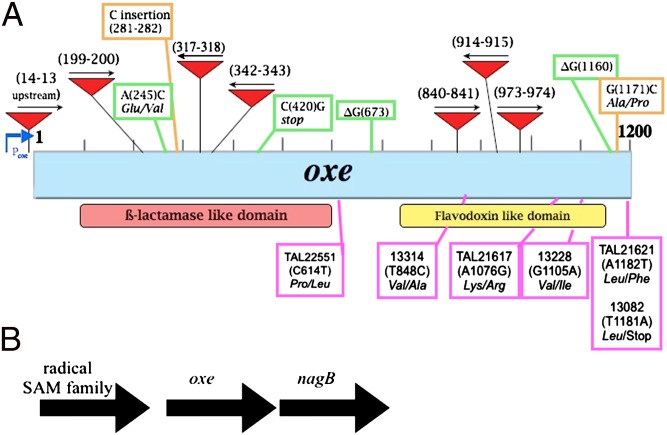
Fig. 1.
Map of the oxe region of B. fragilis. (A) Diagram of the oxe gene (blue box). Predicted protein domains are shown below. Red arrows depict IS4400′ insertions. Shown in the green boxes are the mutations found in spontaneous oxygen-enabled mutants of ADB77, with nucleotide positions in parentheses. Orange boxes denote mutations found in oxygen-enabled mutants from transposon-insertion pool. Purple boxes denote polymorphisms in Oxe sequence from clinical strains. (B) Genomic region surrounding the oxe gene. nagB, glucosamine 6-P deaminase.
To determine if mutations in other genes could give rise to O2-enabled strains, a pool of 8,000 Tn4400′ (30) transposon-insertion mutants was plated on brain heart infusion broth supplemented with 0.5% yeast extract and 15 μg/mL hematin (BHIS) under atmospheres containing as much as 2% oxygen. A PCR-based screen was used to determine if the enabled isolates had Tn4400′ insertions in oxe. Of 32 analyzed isolates, 23 showed insertions of the IS element from Tn4400′ within oxe. The other nine were found to fall into two groups that had insertions that lay outside of the oxe locus, but PCR amplification and sequencing of their oxe gene revealed that each had acquired spontaneous mutations in oxe in addition to the transposon insertion (Fig. 1). Importantly, introduction of a plasmid encoding WT oxe into these strains prevented growth under microaerobic conditions, indicating that the O2-enabled phenotype was a result of the mutations in oxe. Additionally, four isolates that arose from microaerobic plating of the WT B. fragilis strain were sequenced across the oxe locus. All were found to contain mutations in oxe (Fig. 1).
Δoxe Strain Plates at Unity in as Much as 2% Oxygen.
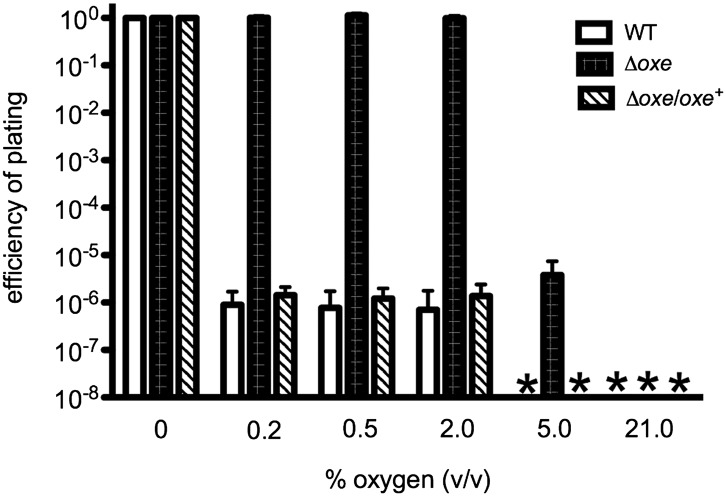
Although both WT and Δoxe strains plated with ∼100% efficiency in environments containing ≤0.05% oxygen compared with their respective anaerobic controls, they differed significantly in plating efficiencies at higher O2 concentrations (Fig. 2). WT B. fragilis gave rise to colonies at a frequency of ∼10−6 under 0.2% to 2% oxygen and failed to form detectable numbers of colonies in more oxygenated environments. The Δoxe strain, however, gave rise to similar numbers of colonies in all O2 levels as high as 2% (Fig. 2), and plated with a frequency of ∼10−6 under 5% oxygen. Importantly, when a plasmid-borne copy of oxe was introduced into the Δoxe strain, the increased plating efficiency with 2% O2 was lost, indicating that the phenotype could be complemented (Fig. 2).
Fig. 2.
Efficiency of plating under various percentages of oxygen. ADB77 (clear bars), ADB267 (black bars), and ADB267/pADB293 (hatched bars) were grown anaerobically to log phase and plated on BHIS under various oxygen concentrations. Procedure and calculations are as in Table 1. An asterisk indicates that values were below the limit of detection.
Δoxe Strain Is Auxotrophic for Amino Acids Under Microaerobic Conditions.
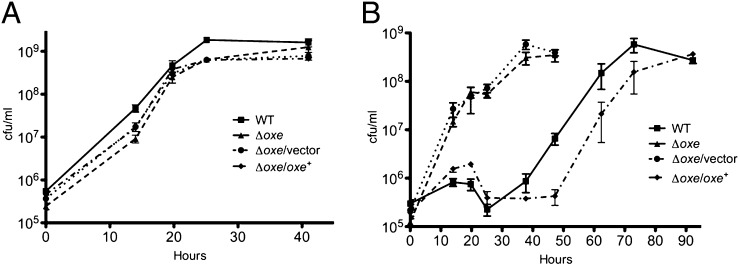
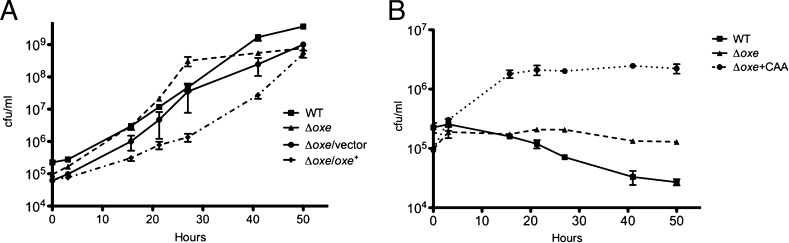
As shown in Fig. 3A and 4A, WT B. fragilis and the Δoxe strain grow equally well under anaerobic conditions in rich and minimal medium. However, when BHIS cultures were shaken under 1% oxygen, the Δoxe mutant began growing immediately whereas the WT exhibited a long lag phase lasting 20 to 30 h. Growth was similarly delayed for a Δoxe strain carrying a WT copy of oxe on a plasmid, but not for one carrying an empty vector (Fig. 3B). When the WT and complemented strains had initiated growth, they doubled at approximately the same rate as the Δoxe strain, and linear regression of the exponential growth rate suggested that this growth was a result of a variant population arising from a few bacteria present at the beginning of the experiment. Samples taken after 92 h of growth were clonally purified under anaerobic conditions, and isolates from this population were found to grow under microaerobic conditions, but failed to grow in room air. PCR amplification and sequence analysis of the oxe locus from six of these isolated colonies (three WT and three of the complemented strain) indicated that all had mutations within the oxe coding region. Thus, all the growth observed in BHIS with 1% O2 was due to the presence of oxe mutants. In minimal medium, neither WT nor the Δoxe strain grew when shaken under 1% oxygen, although supplementation with casamino acids restored some growth to the Δoxe mutant but not to WT (Fig. 4B).
Fig. 3.
Growth of B. fragilis Δoxe strains in rich medium. ADB77 (■), ADB267 (▲), ADB267/pJST61 (●), and ADB267/pADB293 (◆) were grown anaerobically to late log-phase in BHIS with or without erythromycin. These cultures were diluted 1:1,000 into fresh medium and shaken in flasks at 200 rpm in an anaerobic chamber (A) or in a chamber containing 1% oxygen (B). Samples were diluted in PBS solution. Aliquots (10 μL) were spotted to prereduced BHIS plates (with or without erythromycin) and incubated in the anaerobic chamber. Counts (in cfu) were enumerated after 24 to 36 h incubation by using a stage microscope. Results shown are the means of three experiments ± SEM.
Fig. 4.
The B. fragilis Δoxe strain is auxotrophic for amino acids when grown microaerobically. ADB77, ADB267, ADB267/pJST61, and ADB267/pADB293 were grown anaerobically to late log phase in AMM with or without erythromycin. These cultures were diluted 1:2,000 into fresh medium and shaken in flasks at 200 rpm in an anaerobic chamber (A) or in a chamber containing 1% oxygen (B). Where appropriate, Casamino acids (CAA) were added to 200 μg/mL. Samples were diluted in PBS solution and plated on prereduced BHIS. Counts (in cfu) were enumerated after 24 to 36 h. Results shown are the means of three experiments ± SEM: (A) ADB77 (■), ADB267 (▲), ADB267/pJST61 (●), and ADB267pADB293 (◆); and (B) ADB77+CAA (■), ADB267 (▲), and ADB267+CAA (●).
Δoxe Retains Greater Viability Under Room Air than WT.
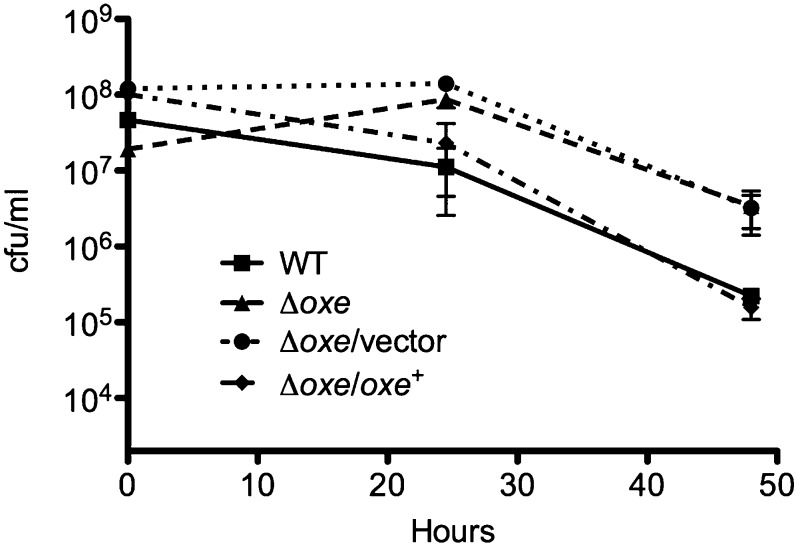
Although the Δoxe mutant clearly demonstrated a growth advantage vs. WT in a microaerobic environment, we wondered if the oxe deletion would increase aerotolerance when cells were shaken in room air. Fig. 5 shows that the Δoxe strain retains greater viability than WT over several days of exposure in PBS solution with glucose, culminating in a ∼10-fold advantage after 48 h. The increased aerotolerance of the Δoxe strain was lost when oxe was provided on a plasmid, but introduction of an empty vector had no effect.
Fig. 5.
B. fragilis Δoxe is more aerotolerant than WT. ADB77 (■), ADB267 (▲), ADB267/pJST61 (●), and ADB267/pADB293 (◆) cultures were grown anaerobically in BHIS to midlog phase. Cells were pelleted and washed with PBS solution, and then resuspended in 10 mL PBS solution plus thymine and 0.5% glucose in 125-mL foam-stoppered flasks. Flasks were shaken at 37 °C under room air. Samples were serially diluted in PBS solution. Aliquots (10 μL) were spotted to prereduced BHIS plates (with or without erythromycin) and incubated in the anaerobic chamber. Counts (in cfu) were enumerated after 2 d incubation by using a stage microscope. Shown are the means of three experiments ± SEM.
Oxe Is Not a Major Source of ROS.
oxe is predicted to encode a flavoprotein, a class of enzymes that uses flavins as cofactors in redox reactions. Electrons are funneled through this flavin moiety and onto a substrate. However, if the reduced flavin comes into contact with oxygen, the electrons can be transferred to O2 to generate ROS (1, 2). One model to explain the inability of WT B. fragilis to grow microaerobically, therefore, would posit that Oxe produces crippling amounts of ROS when cells are exposed to oxygen. To test this hypothesis, we deleted oxe from a strain with deletions in katB, ahpC, and tpx. We have previously shown (34) that this strain cannot effectively scavenge H2O2, so any peroxide made during exposure to oxygen can cross the cell membrane and be measured extracellularly. When the ΔkatΔahpCΔtpx strain was shaken under room air at an OD600 of 0.1, it accumulated H2O2 at a rate of 24 ± 1 nM/min. However, deleting oxe from this genetic background resulted in a strain that accumulated 29 ± 3 nM H2O2/min, a rate very similar to the parent strain, indicating that Oxe is not a major source of ROS under these conditions.
Δoxe Strain Scavenges H2O2 More Efficiently than WT.
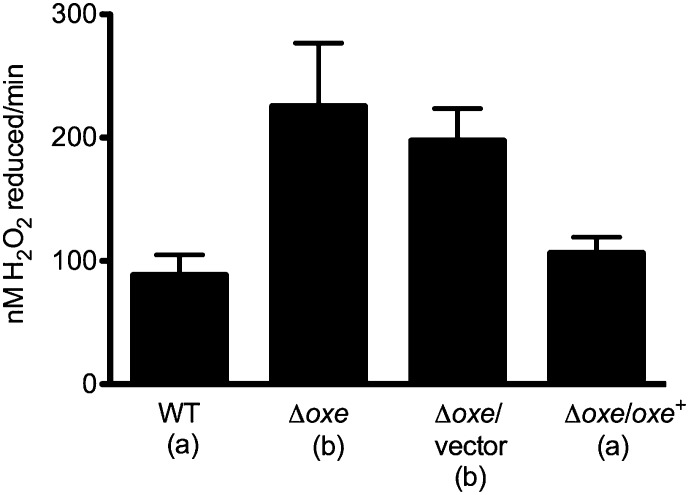
In addition to AhpC and Tpx, there are four other peroxidases and two rubrerythrins annotated in the B. fragilis genome, and it is possible that deleting oxe might activate them in some manner. We tested the H2O2 scavenging activity of WT and Δoxe strains by first shaking them aerobically and then adding exogenous peroxide to whole-cell suspensions. Fig. 6 shows that the Δoxe mutant did indeed scavenge H2O2 more effectively than WT, and that this scavenging rate is reduced when a WT copy of oxe is provided in trans.
Fig. 6.
B. fragilis Δoxe strain scavenges H2O2 at a faster rate than WT. Strains were grown to log phase in AMM (with or without erythromycin), and cells were pelleted by centrifugation. Pellets were resuspended in AMM lacking cysteine and shaken at 37 °C under room air for 1 h. Cells were again pelleted, washed with PBS solution, and resuspended to an OD600 of 0.3 in PBS solution containing thymine and 0.5% glucose. H2O2 (5 μM) was added to start the assay. Samples were removed over time and centrifuged briefly, and supernatants were assayed for [H2O2] by using Amplex Red. Rates are reported in nM H2O2 scavenged per minute. Shown are the means of at least three experiments ± SEM. ANOVA revealed a P value of 0.0066. Values for strains marked “a” differed significantly from those marked “b” with a P < 0.05 in a Newman–Keuls multiple comparison posttest.
Spontaneous Oxygen-Enabled Mutants Arise in Bacteroides thetaiotaomicron.
To test whether the oxygen-enabling phenomenon was specific to B. fragilis or could be recapitulated in other anaerobes, we attempted to isolate mutants of the closely related organism Bacteroides thetaiotaomicron that were capable of growth under microaerobic conditions. We were able to isolate colonies of B. thetaiotaomicron American Type Culture Collection 29741 capable of growth on BHIS under 0.5% O2 at a frequency of ∼10−7 relative to anaerobic controls. B. thetaiotaomicron encodes a protein with 97.2% homology to Oxe, and the gene is surrounded by loci encoding a putative radical SAM-family protein and NagB as in B. fragilis. Three colonies were purified on BHIS under 0.5% oxygen, and the region around the oxe locus was sequenced following PCR amplification. Two of the three predicted Oxe gene products showed amino acid changes (R193H and S244I) relative to WT. The third isolate encoded a WT Oxe, but showed a predicted amino acid change (T322I) in the product of the orf upstream of oxe, indicating that this putative radical SAM-family protein may play a role in an Oxe-dependent pathway.
Clinical Isolates of B. fragilis Display Greater Range of Microaerobic Plating Efficiencies than Fecal Isolates.
In the anaerobic environment of the mammalian intestine, B. fragilis is well protected from the toxic effects of oxygen. However, disruption of the intestinal wall can allow release of bacteria into more oxygenated tissue, posing a significant challenge to the survival of anaerobes. The peritoneum, for instance, contains 2% to 7% oxygen (35, 36). B. fragilis is capable of withstanding this microaerobic environment long enough to allow for the establishment of abscesses, which are eventually rendered anaerobic with the help of facultative anaerobes like Escherichia coli. However, we wondered if the transition between intestine and abscess might supply selective pressure for “oxygen-enabled” variants of B. fragilis, thus leading to an enrichment of these variants in the abscess. To test this idea, we determined the microaerobic plating efficiency of 26 independent clinical isolates of B. fragilis and compared them vs. eight fecal isolates from healthy volunteers. Strains were obtained from four different collections, and clinical specimens were isolated from various sites of infection. Ten clinical isolates plated with efficiencies of 10−5.5 or less under 0.5% to 1% oxygen, similar to ADB77. However, five isolates gave rise to colonies at frequencies of 10−5 to 10−2, and the remaining 11 plated with frequencies >10−2. In contrast, none of the fecal isolates plated with efficiencies >10−5.5.
The oxe gene from all strains was PCR amplified and sequenced. The predicted Oxe sequence of all fecal strains matched that of our reference strain, ADB77. Among those clinical strains that plated with low efficiency under microaerobic conditions, two encoded Oxe polymorphisms (V283A and V369I). Polymorphisms were also found in one clinical strain that plated with intermediate efficiency (L394stop), and three that grew with high efficiency (K359R, L394F, and P205L; Fig. 1A). Predicted Oxe sequences from the remaining strains matched ADB77.
Discussion
The discovery that B. fragilis can grow nanaerobically (21) suggested that its response to oxygen is more complex than previously thought. This complexity is further underscored by our finding that B. fragilis mutants capable of microaerobic growth can be readily isolated, and that all such oxygen-enabled mutants derived from B. fragilis TM4000 studied to date carry lesions in oxe. Clearly Oxe is not essential for anaerobic growth in vitro, yet microaerobic growth was partially dependent on the addition of Casamino acids. Earlier work with an E. coli sod mutant demonstrated a similar conditional sensitivity to oxygen (37), a phenotype later found to be the result of superoxide-mediated destruction of the iron-sulfur cluster of dihydroxyacid dehydratase, a key enzyme in the production of branched-chain amino acids (38). Therefore, the auxotrophies displayed by the Δoxe mutant under microaerobic conditions suggest that, although this strain is capable of growth in the presence of oxygen, it may be experiencing significant oxidative stress.
The increased tolerance of the Δoxe strain to room air was similar to results seen with a D. vulgaris roo mutant (33) despite the increased sensitivity of this roo strain to a microaerobic environment. Clearly, the response to oxygen is varied among anaerobes, a point emphasized in work with Clostridium acetobutylicum that showed that activation of its OSR via the deletion of the gene encoding the PerR repressor gave rise to a more aerotolerant strain capable of some growth in room air (39). This result would seem to contrast with our finding that up-regulation of the B. fragilis OSR via a constitutively active OxyR did not allow for growth under higher O2 concentrations. Additionally, the authors note that this perR mutant exhibited a growth defect under anaerobic conditions, perhaps explaining why perR mutants do not arise spontaneously, unlike B. fragilis oxe mutants.
We have shown that Oxe is not a major source of ROS when B. fragilis is exposed to room air. If this enzyme is indeed a flavoprotein as BLAST analysis predicts, the flavin cofactor is most likely buried deep within the protein structure and thereby protected from autoxidation, unlike fumarate reductase (Frd), another flavin-containing enzyme that we have demonstrated accounts for ∼47% of the ROS generated by aerated B. fragilis (34). Interestingly, a frd mutant does not grow microaerobically unless oxe is also inactivated, indicating that a substantial reduction in ROS production is not enough to give rise to an oxygen-enabled strain.
Although deleting oxe did not lower the rate of production of endogenously generated ROS, it did increase H2O2 detoxification, thus allowing the Δoxe strain to more efficiently protect vulnerable metabolic enzymes under increased oxygen concentrations and thereby likely explaining the oxygen-enabled phenotype. The enhanced H2O2 detoxification is most likely not caused by the increased expression of peroxide scavenging enzymes, as quantitative PCR did not reveal any major differences in the expression of peroxide scavenging enzymes between WT and the Δoxe strain. We therefore believe that the Δoxe strain most likely accumulates excess reductant, and that peroxidases can tap into this reductant pool to more efficiently disproportionate H2O2. A similar model was proposed to explain the tolerance of a roo strain to room air (33), wherein the authors hypothesized that reduced rubredoxin normally consumed by the Roo-mediated reduction of oxygen might be funneled into ROS-scavenging reactions. Deletion of rubredoxin did not suppress the formation of oxygen-enabled mutants in our study (Table 1), so the identity of the key reductant is still unknown. However, the requirement of AhpC and Tpx for microaerobic growth suggests that NAD(P)H may play a critical role.
Our finding that clinical isolates of B. fragilis display a greater range of microaerobic plating efficiencies than fecal isolates suggests that passage from the intestine to oxygenated tissues might provide selective pressure for variants more capable of withstanding oxidative stress. Mutations in oxe might provide such an advantage, although the in vivo situation is obviously complex, as some enabled clinical strains did not have changes in Oxe sequence whereas others encoded polymorphisms in Oxe but did not plate with high efficiency under microaerobic conditions. The predicted changes in Oxe sequence in clinical strains are most likely not the result of laboratory manipulations made after isolation, as we have two clinical strains (TAL22551 and TAL22557) isolated from different sites in the same patient and both encode the same polymorphism (P205L), indicating that any potential mutations occurred in vivo. Additionally, clinical strains were passaged in the same manner as laboratory strains during the course of these experiments, yet ADB77 still plates at a frequency of ∼10−6 microaerobically.
The homology of Oxe to NorV (40) suggested that it might play a role in protection against nitric oxide. However, growth of WT and Δoxe was inhibited by similar concentrations of the nitric oxide donor DEANONOate, and extensive amperometric studies suggested that these strains could reduce nitric oxide at similar rates. We therefore do not know the function of Oxe in the anaerobic physiology of B. fragilis, although it appears to play an important role in vivo, as B. thetaiotaomicron strains carrying transposons in the oxe gene are outcompeted in the gnotobiotic mouse intestine (41). It is therefore possible that Bacteroides acquired Oxe during its evolution as a mammalian commensal, and that a progenitor was capable of growth in much higher concentrations of oxygen. The fact that B. fragilis mutants capable of growth under 5% oxygen can also be isolated further suggests that evolution in an aerobic world may not have always proceeded linearly with respect to degrees of aerotolerance.
Materials and Methods
Reagents.
H2O2 and horseradish peroxidase were purchased from Sigma, restriction enzymes were from New England Biolabs, and T4 DNA ligase and Amplex Red were from Invitrogen.
Growth Conditions.
Anaerobiosis was maintained by using a Coy anaerobic chamber (Coy Laboratory Products) containing 85% nitrogen, 10% hydrogen, and 5% (vol/vol) CO2. B. fragilis was grown in BHIS or anaerobic minimal medium (AMM) containing 0.5% glucose (42). In some cases, super-AMM plates containing 150 μg/mL hemin were used (43). For all thyA mutants, thymine was added to 50 μg/mL. Gentamicin (50 μg/mL), rifampicin (50 μg/mL), trimethoprim (80 μg/mL), erythromycin (8 μg/mL), and tetracycline (2.5 μg/mL) were added where appropriate. E. coli was grown aerobically in Luria broth, and chloramphenicol (25 μg/mL), ampicillin (100 μg/mL), and tetracycline (10 μg/mL) were added as appropriate.
Strains and Plasmids.
Bacterial strains and plasmids used in this study are described in Table S1. E. coli strain DH5α was used for cloning, and strain HB101/RK231 was used for mobilization of plasmids from DH5α to B. fragilis recipient strains. DH5α was made competent for transformation through use of the RbCl method previously described (44). After isolation, fecal and clinical strains of B. fragilis were grown in anaerobic chambers or jars containing an anaerobic atmosphere. Passaging and maintenance of these strains was performed in the same manner as for the laboratory strains.
DNA Manipulation.
Primers used in this study were synthesized by Integrated DNA Technologies and are listed in Table S2. Genomic sequence for B. fragilis NCTC9343 (GenBank accession no. CR626927.1) was provided by the Sanger Centre (www.sanger.ac.uk/Projects/B_fragilis) or by Pedant3 (http://pedant.gsf.de). Genomic DNA was amplified by using HotStarTaq Master Mix (Invitrogen). Plasmid and PCR product purifications were performed with QIAprep spin columns (Qiagen). Primers oxe1 and oxe4 were used in combination with primer 646J to amplify oxe::IS4400 junction fragments by using the PCR method. DNA sequencing was performed by the Tufts University Core Facility.
Strain Construction.
All deletion mutants were constructed using a double-crossover technique as previously described (45). DNA fragments for cloning were obtained by PCR amplification of chromosomal DNA from strain ADB77. To build a deletion construct for oxe, primers oxe1 and oxe2 were used to amplify a fragment consisting of 724 bp of oxe upstream sequence and 86 bp of oxe amino-terminal coding sequence. The downstream fragment was created by PCR amplifying a region consisting of 44 bp of oxe carboxyl-terminal coding sequence and 836 bp of downstream sequence with primers oxe3 and oxe4. Purified PCR products were digested with HindIII/NcoI and NcoI/BamHI, purified, and ligated to pADB242 that had been digested with BamHI and HindIII to create pADB267. A similar scheme was used to build deletion constructs for oxyR, rubredoxin, and rubrerythrins with the appropriate primers.
The suicide plasmid pADB267 was delivered to the recipient B. fragilis strains as previously described (46). TetR colonies were screened for the appropriate cointegration event by using primers 61RAB and oxe5. Following recombination and resolution of the integrated plasmid, TetS colonies were screened via PCR with primers oxe5 and oxe4 to identify Δoxe clones.
The oxe complementing plasmid was created by PCR amplifying a region from 270 bp upstream of the oxe translational start site to 77 bp downstream of the oxe coding sequence by using primers oxe6 and oxe7. The purified PCR product was digested with BamHI and ligated to BglII-digested pJST61 to create pADB293.
Growth Curves.
Dense cultures of WT and Δoxe strains were subcultured as indicated into fresh medium. When appropriate, casamino acids were added to 200 μg/mL. Cultures were grown at 37 °C with shaking (200 rpm) under anaerobic or microaerobic conditions (0.25–1% oxygen). Samples were serially diluted in reduced PBS solution or growth medium, and 10-μL aliquots were spotted on BHIS plates that had been reduced overnight in the anaerobic chamber. Counts (in cfu) were enumerated under a stage microscope after 1 to 2 d. Oxygen concentration was measured by using the pyrogallol method (21) or with a Coy oxygen analyzer ESD (model 630) and adjusted as necessary by addition of anaerobic gas mix.
Efficiency of Plating.
B. fragilis strains were grown anaerobically to midlog phase in BHIS, serially diluted, and spread on BHIS plates that had been reduced in an anaerobic chamber overnight. Plates were transferred anaerobically to AnaeroPack boxes (Mitsubishi Gas Chemical) fitted with side ports, and sealed boxes were removed from anaerobic chamber. O2 was injected through the side port to achieve the appropriate oxygen concentrations. O2 concentrations described for experiments involving AnaeroPack boxes refer to that which was initially present. An oxe+ strain was included in all experiments as a negative control to ensure that sufficient oxygen was retained in these boxes throughout the experiments. Alternatively, plates were placed in an anaerobic chamber from which the O2-scrubbing catalyst had been removed and the appropriate amount of oxygen had been added. O2 concentrations were monitored throughout the course of the experiments and adjusted as necessary. Counts (in cfu) were enumerated after 3 to 5 d at 37 °C.
Room Air Challenge.
Strains were grown in BHIS broth under anaerobic conditions until midlog phase. Cultures were centrifuged, and pellets were washed once with PBS solution. After a second centrifugation, pellets were resuspended in 10 mL oxygenated PBS solution, and 0.5% glucose and 50 μg/mL thymine were added. Samples were taken immediately and serially diluted, and a 10-μL aliquot was spotted onto prereduced BHIS plates (with or without erythromycin), which were incubated in the anaerobic chamber. Cell suspensions were transferred to 125-mL foam-stoppered flasks and shaken at 250 rpm and 37 °C under room air. Samples were processed as above. Counts (in cfu) were enumerated after 2 d of incubation.
H2O2 Detection.
Cultures were grown anaerobically to midlog phase in AMM and treated as previously described (34). After shaking in room air, cells were washed twice and resuspended to an OD600 of 0.3 in 10 mL PBS solution containing 50 μg/mL thymine and 0.5% glucose. H2O2 (5 μM) was added to cell suspensions to start the assay, and the H2O2 concentration was followed over time by using Amplex red (34).
Supplementary Material
Acknowledgments
We thank Dr. Carol Kumamoto for assistance in the fluorometric studies and Tufts Medical Center, the Tufts Anaerobe Laboratory, Dr. Laurie Comstock, Dr. David Hecht, and Dr. Jeffrey Smith for providing strains. This research was supported by National Institute of Allergy and Infectious Disease/National Institutes of Health Grant AI-19497.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203796109/-/DCSupplemental.
References
- 1.Massey V, et al. The production of superoxide anion radicals in the reaction of reduced flavins and flavoproteins with molecular oxygen. Biochem Biophys Res Commun. 1969;36:891–897. doi: 10.1016/0006-291x(69)90287-3. [DOI] [PubMed] [Google Scholar]
- 2.Massey V. Activation of molecular oxygen by flavins and flavoproteins. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 3.Korshunov S, Imlay JA. Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol Microbiol. 2010;75:1389–1401. doi: 10.1111/j.1365-2958.2010.07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- 5.Woodmansee AN, Imlay JA. Reduced flavins promote oxidative DNA damage in non-respiring Escherichia coli by delivering electrons to intracellular free iron. J Biol Chem. 2002;277:34055–34066. doi: 10.1074/jbc.M203977200. [DOI] [PubMed] [Google Scholar]
- 6.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 7.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 8.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babior BM, Curnutte JT, Kipnes RS. Biological defense mechanisms. Evidence for the participation of superoxide in bacterial killing by xanthine oxidase. J Lab Clin Med. 1975;85:235–244. [PubMed] [Google Scholar]
- 11.Farr SB, D’Ari R, Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridovich I. Oxygen toxicity: A radical explanation. J Exp Biol. 1998;201:1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- 13.Henle ES, Linn S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 14.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 15.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vita N, Hatchikian EC, Nouailler M, Dolla A, Pieulle L. Disulfide bond-dependent mechanism of protection against oxidative stress in pyruvate-ferredoxin oxidoreductase of anaerobic Desulfovibrio bacteria. Biochemistry. 2008;47:957–964. doi: 10.1021/bi7014713. [DOI] [PubMed] [Google Scholar]
- 17.Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liochev SI, Fridovich I. Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch Biochem Biophys. 1993;301:379–384. doi: 10.1006/abbi.1993.1159. [DOI] [PubMed] [Google Scholar]
- 19.McCord JM, Keele BB, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan N, Imlay JA. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol Microbiol. 2001;39:1562–1571. doi: 10.1046/j.1365-2958.2001.02343.x. [DOI] [PubMed] [Google Scholar]
- 21.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 22.Rocha ER, Herren CD, Smalley DJ, Smith CJ. The complex oxidative stress response of Bacteroides fragilis: The role of OxyR in control of gene expression. Anaerobe. 2003;9:165–173. doi: 10.1016/S1075-9964(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 23.Sund CJ, et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: The role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 24.Herren CD, Rocha ER, Smith CJ. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene. 2003;316:167–175. doi: 10.1016/s0378-1119(03)00759-5. [DOI] [PubMed] [Google Scholar]
- 25.Rocha ER, Selby T, Coleman JP, Smith CJ. Oxidative stress response in an anaerobe, Bacteroides fragilis: A role for catalase in protection against hydrogen peroxide. J Bacteriol. 1996;178:6895–6903. doi: 10.1128/jb.178.23.6895-6903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha ER, Smith CJ. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate Anaerobe bacteroides fragilis. J Bacteriol. 1999;181:5701–5710. doi: 10.1128/jb.181.18.5701-5710.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha ER, Tzianabos AO, Smith CJ. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J Bacteriol. 2007;189:8015–8023. doi: 10.1128/JB.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sund CJ, Greg Wells W, Jeffrey Smith C. The Bacteroides fragilis P20 scavengase homolog is important in the oxidative stress response but is not controlled by OxyR. FEMS Microbiol Lett. 2006;261:211–217. doi: 10.1111/j.1574-6968.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- 29.Privalle CT, Gregory EM. Superoxide dismutase and O2 lethality in Bacteroides fragilis. J Bacteriol. 1979;138:139–145. doi: 10.1128/jb.138.1.139-145.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang YP, Dallas MM, Malamy MH. Characterization of the Batl (Bacteroides aerotolerance) operon in Bacteroides fragilis: Isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol Microbiol. 1999;32:139–149. doi: 10.1046/j.1365-2958.1999.01337.x. [DOI] [PubMed] [Google Scholar]
- 31.Onderdonk AB, Johnston J, Mayhew JW, Gorbach SL. Effect of dissolved oxygen and Eh and Bacteroides fragilis during continuous culture. Appl Environ Microbiol. 1976;31:168–172. doi: 10.1128/aem.31.2.168-172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha ER, Owens G, Jr, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/jb.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wildschut JD, Lang RM, Voordouw JK, Voordouw G. Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris hildenborough under microaerophilic conditions. J Bacteriol. 2006;188:6253–6260. doi: 10.1128/JB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan BM, Malamy MH. Fumarate reductase is a major contributor to the generation of reactive oxygen species in the anaerobe Bacteroides fragilis. Microbiology. 2012;158:539–546. doi: 10.1099/mic.0.054403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitt MD. Oxygen tension in the gut. N Engl J Med. 1970;282:1039–1040. doi: 10.1056/NEJM197004302821814. [DOI] [PubMed] [Google Scholar]
- 36.Sawyer RG, Spengler MD, Adams RB, Pruett TL. The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann Surg. 1991;213:253–260. doi: 10.1097/00000658-199103000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo CF, Mashino T, Fridovich I. alpha, beta-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J Biol Chem. 1987;262:4724–4727. [PubMed] [Google Scholar]
- 39.Hillmann F, Fischer RJ, Saint-Prix F, Girbal L, Bahl H. PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol Microbiol. 2008;68:848–860. doi: 10.1111/j.1365-2958.2008.06192.x. [DOI] [PubMed] [Google Scholar]
- 40.Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 41.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baughn AD, Malamy MH. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: Implications for the evolution of the mitochondrial Krebs cycle. Proc Natl Acad Sci USA. 2002;99:4662–4667. doi: 10.1073/pnas.052710199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varel VH, Bryant MP. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl Microbiol. 1974;28:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 45.Guiney DG, Hasegawa P, Davis CE. Plasmid transfer from Escherichia coli to Bacteroides fragilis: Differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci USA. 1984;81:7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson JS, Malamy MH. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.