Abstract
Monofunctional platinum(II) complexes of general formula cis-[Pt(NH3)2(N-heterocycle)Cl]Cl bind DNA at a single site, inducing little distortion in the double helix. Despite this behavior, these compounds display significant antitumor properties, with a different spectrum of activity than that of classic bifunctional cross-linking agents like cisplatin. To discover the most potent monofunctional platinum(II) compounds, the N-heterocycle was systematically varied to generate a small library of new compounds, with guidance from the X-ray structure of RNA polymerase II (Pol II) stalled at a monofunctional pyriplatin-DNA adduct. In pyriplatin, the N-heterocycle is pyridine. The most effective complex evaluated was phenanthriplatin, cis-[Pt(NH3)2(phenanthridine)Cl]NO3, which exhibits significantly greater activity than the Food and Drug Administration-approved drugs cisplatin and oxaliplatin. Studies of phenanthriplatin in the National Cancer Institute 60-cell tumor panel screen revealed a spectrum of activity distinct from that of these clinically validated anticancer agents. The cellular uptake of phenanthriplatin is substantially greater than that of cisplatin and pyriplatin because of the hydrophobicity of the phenanthridine ligand. Phenanthriplatin binds more effectively to 5′-deoxyguanosine monophosphate than to N-acetyl methionine, whereas pyriplatin reacts equally well with both reagents. This chemistry supports DNA as a viable cellular target for phenanthriplatin and suggests that it may avoid cytoplasmic platinum scavengers with sulfur-donor ligands that convey drug resistance. With the use of globally platinated Gaussia luciferase vectors, we determined that phenanthriplatin inhibits transcription in live mammalian cells as effectively as cisplatin, despite its inability to form DNA cross-links.
Keywords: cancer therapy, X-ray crystallography, DNA damage, transcription inhibition, metals in medicine
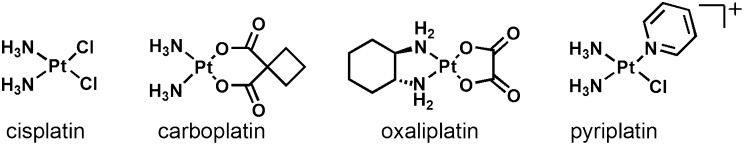
The Food and Drug Administration (FDA)-approved platinum-based antitumor drugs, cisplatin [cis-diamminedichloroplatinum(II)], carboplatin [cis-diammine(1,1′-cyclobutanedicarboxylato)-platinum(II)], and oxaliplatin [(R,R)-1,2-diaminocyclohexane)oxalatoplatinum(II)] (Fig. 1), are currently among the most effective chemotherapies in clinical use for the treatment of cancers (1). These Pt-based anticancer agents typically form bifunctional intra- and interstrand DNA cross-links through covalent bonds with purine nucleobases. These cross-links inhibit transcription and result in cell death (2, 3). Platinum-based drugs are limited by side effects and poor activity in certain types of cancer resulting from acquired or intrinsic resistance (1–3). These limitations evoke a need for new platinum-based chemotherapeutics with novel mechanisms of action.
Fig. 1.
Structures of cisplatin, carboplatin, oxaliplatin, and pyriplatin. The bifunctional platinum complexes (cisplatin, carboplatin, and oxaliplatin) are clinically approved anticancer drugs in the United States. Pyriplatin is an exploratory lead compound for a class of monofuntional, platinum-based anticancer drug candidates.
One approach that we have used to circumvent the shortcomings of classic bifunctional platinum-based drugs has been to revisit cationic, monofunctional platinum complexes previously demonstrated to display significant anticancer activity in animal tumor models (4). In contrast to monofunctional platinum(II) compounds, such as [Pt(dien)Cl]+ (dien = diethylenetriamine) (5) and [Pt(NH3)3Cl]+ (6), which early work proved to be inactive, the compound pyriplatin [cis-diamminepyridinechloroplatinum(II)] (Fig. 1) and several of its analogs have significant antineoplastic activity. Moreover, the profile of cellular response to these monofunctional complexes is different from those of the classic bifunctional, charge-neutral platinum-based drugs (7).
Pyriplatin binds to DNA in a monodentate fashion at the N7 position of guanine residues, with no significant distortion of the double helix (8). The resulting monofunctional DNA adducts inhibit transcription while significantly eluding repair (8). In addition, we identified pyriplatin as a substrate for organic cation transporters, which presents an opportunity for selective delivery to tumors, such as human colorectal cancer cells, which overexpress these transporters (8). More recently, X-ray crystallography was used to reveal the structure of RNA polymerase II (pol II) stalled at a pyriplatin adduct on a short DNA strand (9). This structure revealed that the pyriplatin-DNA adduct may prevent translocation from the active site and RNA chain elongation because of steric interactions and hydrogen bonding with the bridge helix, consistent with a mechanism of transcription inhibition by pyriplatin that differs from that of cisplatin (9). To further characterize pyriplatin, we recently reported transcription inhibition profiles for pyriplatin-DNA adducts in a variety of live mammalian cell lines (10).
The unique features of pyriplatin strongly encouraged further study of monofunctional Pt(II) compounds as anticancer drug candidates. Because pyriplatin is more than 10-fold less potent than cisplatin or oxaliplatin (7), it was of interest to explore analogs with improved activity. We therefore devised a strategy, based on our understanding of the structure of pyriplatin on DNA in pol II, to synthesize other monofunctional platinum compounds that might act more efficaciously as anticancer drugs and to investigate how cells process the specific lesions induced by these previously untried candidates.
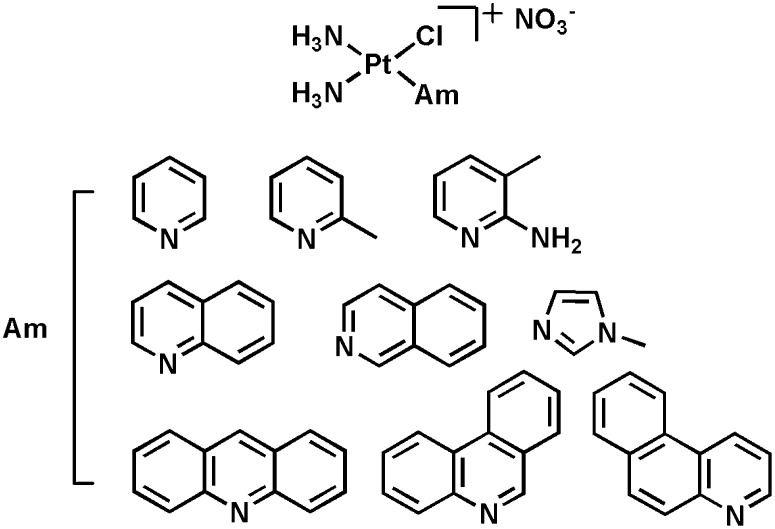
To improve the potency and cellular uptake of monofunctional Pt(II) complexes, various N-heterocyclic ligands (Am) were substituted for pyridine (Fig. 2) and the resulting compounds were fully characterized and evaluated for biological activity. Among them, phenanthriplatin, cis-[Pt(NH3)2(phenanthridine)Cl]NO3, was identified to exhibit greater efficacy than cisplatin and oxaliplatin in established human cancer cell lines. The cellular uptake, DNA binding, and transcription-inhibition properties of phenanthriplatin were investigated to provide insight into the high potency of this compound. The results are described in the present article.
Results and Discussion
Development of Unique Monofunctional Platinum(II) Compounds.
A series of monofunctional Pt(II) compounds was prepared in which the Am was systematically varied. The choice of ligands was guided by the X-ray structure of pol II stalled at a pyriplatin-DNA lesion, which suggested that increasing steric hindrance at platinum would more effectively block pol II progression along the template strand (9). We evaluated a variety of Am ligands, including 2-methylpyridine, 2-amino-3-methylpyridine, quinoline, isoquinoline, 1-methylimidazole, acridine, benzo[f]quinoline, and phenanthridine as substitutes for pyridine (Fig. 2). The monofunctional Pt(II) complexes were characterized using a variety of analytical methods, including 1H, 13C, and 195Pt NMR spectroscopy, electrospray ionization (ESI)-MS, and elemental analysis (see Materials and Methods and SI Appendix).
Fig. 2.
Structures of monofunctional platinum(II) complexes and N-heterocyclic ligands (Am) described in this study. These nine different N-heterocyclic ligands were selected to systematically modify the steric protection around the platinum center.
Crystallographic Characterization of Monofunctional Pt(II) Compounds.
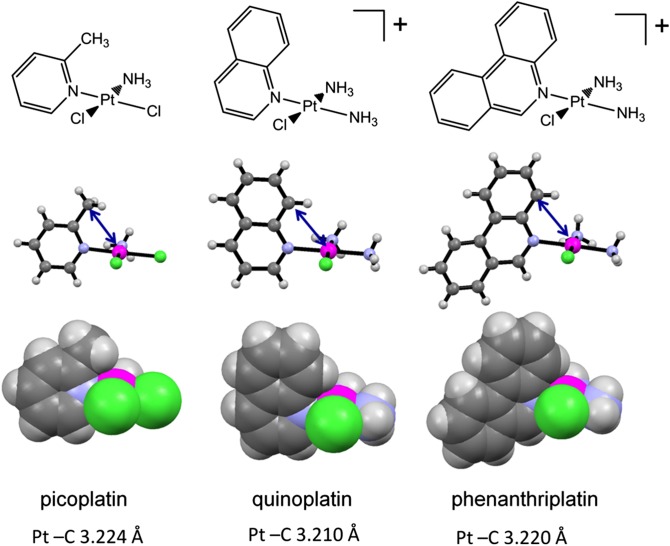
Two of the synthesized monofunctional platinum(II) complexes, cis-[Pt(NH3)2(quinoline)Cl]NO3 (quinoplatin) and cis-[Pt(NH3)2-(phenanthridine)Cl]NO3 (phenanthriplatin), were characterized structurally by X-ray crystallography. Depictions of these structures are included in the SI Appendix (SI Appendix, Figs. S1 and S2), together with tabulated refinement information and geometric parameters (SI Appendix, Tables S1 and S2). Both complexes adopt the expected square-planar coordination geometry with a cis arrangement of ammine ligands.
The plane of the aromatic heterocyclic ligand in both structures is approximately perpendicular to that of the platinum coordination plane. In this orientation, the quinoline and phenanthridine ligands provide steric protection against axial attack by an entering nucleophile perpendicular to the platinum coordination plane. Because the most efficacious ligand substitution reactions at platinum(II) centers occur by an associative mechanism at the axial positions, this steric protection should diminish the ligand substitution reaction rates of quinoplatin and phenanthriplatin compared with that of pyriplatin. This principle has been adopted for the related platinum anticancer complex cis-[Pt(NH3)(2-picoline)Cl2] (picoplatin), which is currently undergoing clinical trials (11). The reactions of picoplatin with water and other nucleophiles are much slower than those of cisplatin and related complexes lacking steric hindrance at the axial sites (12–14). The lesser reactivity of picoplatin, particularly with thiols, prevents undesired deactivation of the complex before it can reach DNA. The steric protection afforded by the quinoline and phenanthridine ligands of quinoplatin and phenanthriplatin, respectively, is comparable to that offered by picoplatin. Fig. 3 compares space-filling models of the structures of these three complexes, revealing protection of the platinum centers. The distances between the platinum atom and the overhanging carbon atoms (blue arrow in Fig. 3) of quinoplatin and phenanthriplatin (3.210 and 3.220 Å, respectively) are nearly identical to that of picoplatin (3.224 Å) (12). These results indicate that quinoplatin and phenanthriplatin should exhibit decreased reactivity with biological nucleophiles, a property that may be important in preventing their premature deactivation, as shown below. Finally, the large steric bulk of the phenanthridine and quinoline ligands revealed by these crystal structures will surely impede progression of pol II more effectively than the smaller pyridine ring of pyriplatin.
Fig. 3.
Comparison of the X-ray crystal structures of picoplatin, quinoplatin, and phenanthriplatin. The platinum-carbon distances shown reveal that these three complexes exhibit similar degrees of steric protection from incoming nucleophiles. Data for picoplatin are from reference 12.
Phenanthriplatin, the Most Potent Monofunctional Pt(II) Compound and Its Antiproliferative Effects in a Panel of Human Cancer Cell Lines.
To assess the biological activities of the newly synthesized monofunctional Pt(II) compounds, their in vitro cytotoxicities were determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay in various human cancer cell lines (SI Appendix, Table S3). The results indicate that the steric properties conveyed by the Am ligands in the monofunctional Pt(II) complexes play a significant role in modulating their cytotoxicities. More sterically demanding Am ligands give rise to monofunctional Pt(II) complexes with greater efficacy, as anticipated from the structural study of the pyriplatin–DNA–pol II ternary complex.
To directly compare phenanthriplatin with conventional Pt-based drugs in vitro, a panel of seven human cancer cell lines of different origin was treated with cisplatin, oxaliplatin, pyriplatin, or phenanthriplatin for 72 h, and cell viability was evaluated by the MTT assay. Table 1 reports IC50 (50% growth inhibition concentration) values for each of the seven cell lines and SDs obtained from at least three independent experiments, each performed in triplicate. The cytotoxicity of phenanthriplatin is substantially greater (4–40 times) than that of cisplatin or oxaliplatin.
Table 1.
IC50 values for cisplatin, oxaliplatin, pyriplatin, and phenanthriplatin in the seven-cell line panel
| IC50 (μM) |
|||||
| Cell line | Cancer type | Cisplatin | Oxaliplatin | Phenanthriplatin | Pyriplatin |
| A549 | Lung | 6.75 ± 0.38 | 6.79 ± 0.26 | 0.22 ± 0.01 | 52.1 ± 2.3 |
| HeLa | Cervix | 1.77 ± 0.72 | 11.8 ± 1.4 | 0.30 ± 0.02 | 31.3 ± 2.8 |
| MCF7 | Breast | 11.6 ± 0.6 | 17.9 ± 2.7 | 0.94 ± 0.09 | 109 ± 10 |
| U2OS | Bone | 7.15 ± 0.25 | 8.67 ± 0.59 | 0.59 ± 0.04 | 78.9 ± 6.7 |
| HT29 | Colorectal | 15.9 ± 1.5 | 1.81 ± 1.15 | 2.02 ± 0.04 | 144 ± 10 |
| NTera2 | Testis | 0.14 ± 0.03 | 1.12 ± 0.08 | 0.035 ± 0.002 | 5.16 ± 0.96 |
| PC3 | Prostate | 4.56 ± 0.52 | 13.2 ± 4.0 | 0.74 ± 0.04 | 47.9 ± 9.2 |
Data reflect the mean and SD of results from three separate experiments, each performed in triplicate.
The ultimate objective in cancer therapy is to find an anticancer drug that kills cancer cells selectively over healthy cells, thereby mitigating toxic side effects normally associated with chemotherapy. Normal lung fibroblasts (MRC5) and cancerous lung (A549) cells were used to evaluate the selectivity of phenanthriplatin for cancer vs. healthy cells. The A549 and MRC5 cell cultures were treated with cisplatin or phenanthriplatin for 72 h, after which cell viability was evaluated by the MTT assay (SI Appendix, Table S3). The ratio of IC50 values in healthy MRC5 cells to those in cancerous A549 cells was 0.9 for cisplatin compared with 3.9 for phenanthriplatin. The higher ratio obtained for phenanthriplatin reveals its selectivity for cancer cells, at least in the cellular monolayer assays used in this study.
The National Cancer Institute (NCI)-60 DTP (Developmental Therapeutics Program) Human Tumor Cell Line Screen has been used to evaluate the anticancer activities of many chemical compounds and natural product samples (15). This test uses 60 different human cancer cell lines representative of leukemia, nonsmall cell lung, colon, central nervous system, melanoma, ovarian, renal, prostate, and breast cancers (15). Anticancer compounds exhibit distinctive sensitivity and resistivity profiles in these cell lines that determine their spectrum of activity, which is often indicative of its cellular mechanism of action. The COMPARE program quantitatively matches the spectrum of activity of one compound to others in the NCI database (15). Given the potential utility of this screen in identifying novel drug candidates, phenanthriplatin was submitted to the NCI for evaluation in 2011. Phenanthriplatin showed significant growth inhibition of the 60 cell lines at a single dose of 10 μM. Based on its success in the single-dose screen, it was further tested against the 60-cell panel at five concentration levels. The detailed results and comparison with conventional bifunctional platinum-based antitumor drugs, such as cisplatin and oxaliplatin, are provided in SI Appendix, Fig. S3. More significantly, analysis by the online COMPARE algorithm revealed that phenanthriplatin could not be correlated with any other platinum anticancer agent. The highest correlation in the NCI database was for doxorubicin, with a correlation coefficient of 0.607. These results demonstrate that phenanthriplatin has a unique spectrum of activity compared with conventional platinum-based and most other anticancer drugs.
Cellular Uptake of Monofunctional Platinum(II) Compounds.
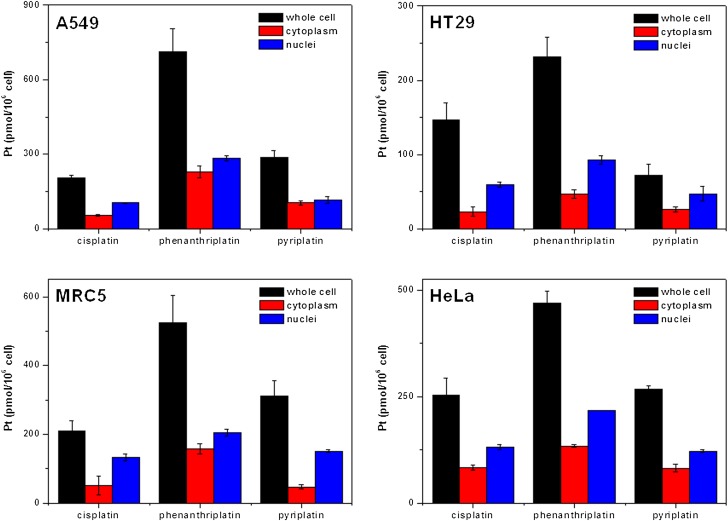
The activity of platinum drugs against cancer is mediated by a combination of processes, including cell entry, drug activation, DNA binding, and transcription inhibition (1–3). Cellular entry of Pt-based drugs is thought to occur by both passive diffusion and carrier-mediated active transport (1). To investigate the transport and distribution of monofunctional platinum(II) compounds in the cell, we measured the nuclear, cytosolic, and whole cell concentrations of platinum by atomic absorption spectroscopy after treatment of the cells with the compounds (Fig. 4). For this study, we used phenanthriplatin, as the most cytotoxic compound, pyriplatin for comparison, and cisplatin as a control. Four different cell lines, A549, HT29, MRC5, and HeLa, were treated with 5 μM platinum compound for 3 h.
Fig. 4.
Intracellular distribution of Pt in cancer cells treated with cisplatin, pyriplatin, or phenanthriplatin. Cells were treated with 5 μM platinum complex for 3 h. Platinum uptake is presented in units of pmol of Pt per 106 cells.
Even at this short exposure time, phenanthriplatin is taken up by cells more effectively than cisplatin or pyriplatin (Fig. 4). The results reflect the ability of the larger, hydrophobic heterocyclic phenanthridine ligand to facilitate uptake of the phenanthriplatin cation through the cytoplasmic membrane. Although the total uptake of platinum is quite different for each compound, the distribution of phenanthriplatin inside the cell is similar to that of pyriplatin and cisplatin. Most of the platinum is found in the nuclear rather than the cytoplasmic fraction. The remainder, ∼15–40%, is bound to the insoluble fraction, which consists primarily of cell membranes. The higher cellular uptake levels of phenanthriplatin probably contribute to its enhanced cytotoxicity compared with that of cisplatin and pyriplatin.
Reactivity with 5′-Deoxyguanosine Monophosphate and N-Acetyl Methionine.
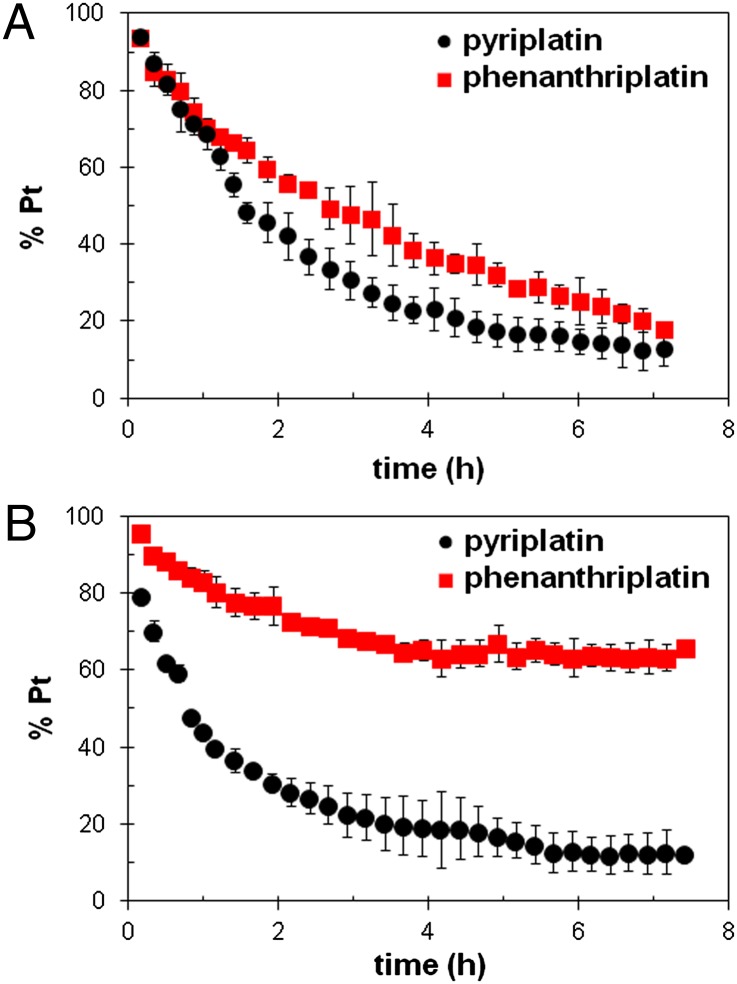
Platinum-based drugs are activated by the leaving of labile chloride ligands and aquation. The activated, cationic platinum-aqua complexes bind readily to DNA and other nucleophiles (1–3). The aquation rates of pyriplatin and phenanthriplatin in D2O at 37 °C were investigated by 1H NMR spectroscopy (SI Appendix, Fig. S4). Under these conditions, pyriplatin and phenanthriplatin aquate at similar rates; after 1 h the reaction is complete and both of these complexes are in equilibrium with their aqua analogs (SI Appendix, Fig. S5). The equilibrium constant for aquation is 0.1 mM for both species. To simulate the interaction of nucleobases on DNA with cationic, monofunctional Pt(II) compounds, pyriplatin and phenanthriplatin were treated with 16 equivalents of 5′-deoxyguanosine monophosphate (5′-dGMP) and monitored by 1D 1H NMR spectroscopy (SI Appendix, Fig. S6) at 37 °C in PBS, pH 7.4. Under these pseudo first-order conditions, the reactivity of pyriplatin and phenanthriplatin with 5′-dGMP is similar (Fig. 5A). Following a pseudo first-order treatment, the rate constants were computed to be 0.22 h−1 and 0.29 h−1 for phenanthriplatin and pyriplatin, respectively. The corresponding half-lives of 3.2 h and 2.4 h for phenanthriplatin and pyriplatin (SI Appendix, Fig. S7) suggest that the increased steric bulk supplied by the phenanthridine ligand does not retard binding of phenanthriplatin to N7-guanosine compared with pyriplatin.
Fig. 5.
Progress of reactions of (A) pyriplatin and phenanthriplatin with 16 equiv of 5′-dGMP at 37 °C or (B) 1 equiv of N-acetyl methionine (N-AcMet) at 37 °C, both in 10 mM PBS buffer (pH* = 7.4) monitored by 1H NMR spectroscopy. pH* refers to a pH measurement uncorrected for the effect of deuterium on the electrode.
Sulfur-containing molecules, which are widely distributed in cellular systems, play an important role in the cellular chemistry of platinum drugs, including their uptake, distribution, and efflux (16). Because of their high binding affinity for platinum, many intracellular sulfur-containing molecules, such as metallothionein and glutathione (GSH), bind to platinum before it reaches the nucleus (16). In this manner they can decrease DNA platination levels and lower the efficacy of Pt compounds. To gain information about the interactions of sulfur-containing compounds with monofunctional Pt(II) compounds, we tested the reactivity of phenanthriplatin and pyriplatin with an equimolar concentration of N-acetyl methionine (N-AcMet) at 37 °C (SI Appendix, Fig. S8). (Although GSH is arguably a more biologically relevant nucleophile than N-AcMet, initial 1H NMR data obtained for the reaction of phenanthriplatin or pyriplatin with GSH were too complicated for a straightforward kinetic analysis. Kinetic analyses were hampered by overlapping peaks and possibly the formation of multiple products. The use of N-AcMet produced data that were amenable for direct comparison of the reactivities of phenanthriplatin versus pyriplatin.) In this experiment, phenanthriplatin reacted much more slowly with N-AcMet than pyriplatin (Fig. 5B). When the kinetic data for phenanthriplatin were fit to an expression for a second-order rate law, the derived rate constant was 0.034 mM-1 h−1, corresponding to a half-life of 15.0 h (SI Appendix, Fig. S9). In the case of pyriplatin, the calculated rate constant was 0.56 mM-1 h−1 with a half-life of 1.04 h, which suggests that the reaction proceeds >10-fold more rapidly than that for phenanthriplatin. ESI-MS data of these reaction mixtures after 48 h revealed the presence of molecular ion peaks corresponding to [Pt(ND3)(Am)(N-AcMet)Cl]+, where Am represents pyridine or phenanthridine for pyriplatin and phenanthriplatin, respectively (SI Appendix, Fig. S10). These species originate from replacement of an ammine ligand trans to N-AcMet, a consequence of the strong trans effect of the sulfur-donor ligand, and therefore represent inactive metabolites of the parent complexes. Although the reaction products of both complexes are similar, the kinetic data reveal that phenanthriplatin is relatively inert to N-AcMet, but exhibits reactivity toward 5′-dGMP similar to that of pyriplatin. The bulky phenanthridine ligand thus inhibits reaction with N-AcMet more effectively than with 5′-dGMP. Although the reasons for this kinetic preference of phenanthriplatin to bind 5′-dGMP over N-AcMet are not entirely clear, the trend suggests that phenthriplatin will bind guanosine nucleosides on DNA efficiently, as required for pol II inhibition, but react less readily with cytoplasmic sulfur-containing nucleophiles, which might promote cellular resistance to the compound.
Ethidium Bromide DNA-Binding Competition Studies.
Ethidium bromide, a phenanthridine-based dye, is a well-known DNA intercalator. Because the phenanthridine ligand of phenanthriplatin is the same as that in ethidium bromide, we considered the possibility of an intercalative DNA binding mode of the platinum-bound molecule. To investigate the primary DNA-binding mode of phenanthriplatin, the affinity of ethidium bromide for calf thymus DNA in the presence of different platinum compounds was investigated, and the data were subjected to a Scatchard analysis (17). Using this approach, it is possible to determine whether the inhibition of ethidium binding is competitive (type A), noncompetitive (type D), or both (type B) (17). No inhibition of ethidium binding is labeled as type C (17). Scatchard plots obtained after a 1-min incubation period for cisplatin, pyriplatin, or phenanthriplatin revealed type C behavior (SI Appendix, Fig. S11A), indicating that, at this short incubation time, none of the platinum complexes inhibit ethidium intercalation, presumably because of the slow kinetics that characterizes the formation of covalent adducts. After a 12-h incubation period of DNA with the platinum compounds, type C behavior was still observed for cisplatin and pyriplatin, whereas type D behavior occurred for phenanthriplatin (SI Appendix, Fig. S11B). This result clearly indicates that phenanthriplatin inhibits ethidium binding noncompetitively (17) and, therefore, that the binding mode of phenanthriplatin to DNA is not intercalative. Covalent adducts of phenanthriplatin are most likely responsible for the noncompetitive inhibition of ethidium binding.
Transcription Inhibition of Monofunctional Pt-DNA Adducts in Live Mammalian Cells.
Investigations of the cellular processing of Pt-DNA lesions are important for understanding the mechanism of action of platinum drugs. One of the major consequences of Pt-DNA damage is transcription inhibition, the extent of which dictates the efficacy of Pt drugs (18). We therefore investigated the transcription inhibitory properties of phenanthriplatin and compared the results to those for cisplatin and pyriplatin using live mamalian cells and our recently described protocol (19). Globally platinated Gaussia luciferase (GLuc) expression vectors were generated by treating pGLuc with varying concentrations of cisplatin, pyriplatin, or phenanthriplatin in Hepes buffer. A549 or HT29 cells were transfected using the platinated transcription probes, and transcription levels were investigated by determining GLuc expression, as measured by fluorescence, following addition of coelenterazine as a substrate for the exported enzyme. The emission intensities were normalized against unplatinated plasmids as a control. Transcription profiles were obtained by plotting normalized levels of GLuc expression against platination levels (Pt/plasmid ratio) at five different time points (SI Appendix, Fig. S12).
After 60 h, the transcription levels were substantially restored in both cell lines, indicating repair of monofunctional Pt(II)-DNA and cisplatin-DNA adducts. For example, at a Pt/DNA ratio of 24.3 for phenanthriplatin, the transcription level recovered from 19.5% at 12 h to 51.1% at 60 h in A549 cells, whereas the recovery was 25.8% to 60.2% for cisplatin and 55.2 to 66.8% for pyriplatin. In HT29 cells, the transcription recovered from 28.1% at 12 h to 37.6% at 60 h at a Pt/DNA ratio of 24.3 for phenanthriplatin, whereas the recovery was 15.6% to 42.9% for cisplatin and 55.3% to 75.2% for pyriplatin (SI Appendix, Fig. S12). D0 values, defined as the number of Pt lesions per plasmid required to reduce transcription levels to 37% of control (20), were computed to quantitate transcription inhibition in the two cell lines (Table 2). An increase in D0 value at different time points represents restoration of transcription. Phenanthriplatin inhibits transcription in A549 and HT29 cells as efficiently as cisplatin. The transcription inhibition by pyriplatin is less efficient than that of either cisplatin or phenanthriplatin by a factor of two. The more effective transcription inhibition of phenanthriplatin-DNA adducts compared with those of pyriplatin, as shown here, is most likely a significant factor contributing to its increased cytotoxicity.
Table 2.
D0 values of globally platinated probes with cisplatin, phenanthriplatin, or pyriplatin assayed at different time intervals after transfection for A549 and HT29 cells
| A549 (Pt/plasmid) |
HT29 (Pt/plasmid) |
|||||
| Time after transfection (h) | Cisplatin | Phenanthriplatin | Pyriplatin | Cisplatin | Phenanthriplatin | Pyriplatin |
| 12 | 16.8 | 14.2 | 39.2 | 9.9 | 18.9 | 35.6 |
| 24 | 20.0 | 17.3 | 58.5 | 14.5 | 21.1 | 40.5 |
| 36 | 29.5 | 22.2 | 68.8 | 19.2 | 21.9 | 42.9 |
| 48 | 36.4 | 31.6 | 85.2 | 22.1 | 23.5 | 44.5 |
| 60 | 42.6 | 39.8 | 89.1 | 30.0 | 25.0 | 49.3 |
Conclusions
Among the series of recent monofunctional, cationic platinum(II) compounds, phenanthriplatin destroys cancer cells with greater efficacy than either cisplatin or oxaliplatin. Our data indicate that the higher cytotoxicity of phenanthriplatin originates from its efficient cellular uptake and strong transcription inhibitory properties. Studies of its reactivity toward cellular components reveal efficient binding to nucleobases, typically required of an active platinum drug, but retarded reactivity toward a sulfur-containing nucleophile, such as those associated with cellular resistance mechanisms. An important feature of phenanthriplatin is that its spectrum of activity against panels of cells does not match those of the FDA-approved platinum-based drugs. Phenanthriplatin may therefore be effective against cancer types that are typically resistant to platinum therapy.
Materials and Methods
Details regarding the preparation of cis-[Pt(NH3)2(Am)Cl]NO3, cell culture, the MTT assay, and fluorescence Scatchard plots are presented in the SI Appendix.
Synthesis of Phenanthriplatin, cis-[Pt(NH3)2(phenanthridine)Cl]NO3.
A detailed procedure for preparing phenanthriplatin is described in the SI Appendix. X-ray quality crystals were obtained from methanol/diethyl ether. White solid. Yield: 59%. ESI-MS m/z calculated (M+): 443.06, found: 443.1. 1H NMR (400 MHz, DMSO-d6): δ 4.43 (3H, broad), 4.60 (3H, broad), 7.93 (2H, q), 8.02 (1H, t), 8.14 (1H, t), 8.46 (1H, d), 8.93 (2H, q), 9.78 (1H, d), 9.95 (1H, s). 13C NMR (100 MHz, DMSO-d6): δ 122.5, 123.3, 125.2, 126.2, 129.0, 129.4, 130.2, 131.7, 134.2, 142.4, 160.1. 195Pt NMR (86 MHz, DMSO-d6): δ -2299. Analysis calculated for C13H15ClN4O3Pt: C, 30.87; H, 2.99; N, 11.08; found: C, 31.08; H, 3.02; N, 11.03.
X-Ray Crystallographic Studies.
Crystallographic data for phenanthriplatin and quinoplatin have been deposited at the Cambridge Structural Database (CSD) under CSD reference nos. CCDC 875229 and CCDC 875230. Single crystals were mounted in Paratone oil on cryoloops and frozen under a 110 K or 100 K KRYO-FLEX nitrogen cold stream. Data were collected on a Bruker APEX CCD X-ray diffractometer with graphite-monochromated Mo-Kα radiation (λ = 0.71073 Å) controlled by the APEX2 software package (21). Absorption corrections were applied using SADABS (21, 22). The structures were solved using direct methods and refined on F2 with the SHELXTL-97 software package (23, 24). Structures were checked for higher symmetry using PLATON (25). All nonhydrogen atoms were located and refined anisotropically. Unless otherwise stated, hydrogen atoms were placed in idealized locations and given isotropic thermal parameters equivalent to either 1.5 (terminal CH3 or NH3 hydrogen atoms) or 1.2 times the thermal parameter of the atom to which they were attached. Structure refinement was carried out using established strategies (26). Further details regarding the refinement of quinoplatin and phenanthriplatin can be found in SI Appendix along with a table of data collection and refinement parameters (SI Appendix, Table S1).
Cellular Uptake of Platinum.
The cellular accumulation of platinum was determined as previously described (27, 28), with some modifications. The detailed procedure is described in the SI Appendix.
Kinetic Studies.
NMR spectra were collected on a Varian 500 spectrometer equipped with a triple-resonance broadband inverse probe and a variable temperature unit. The 1D 1H NMR kinetic studies were performed in duplicate as a standard time-arrayed experiment using a variable delayed list. Incremented 1D spectra were processed in exactly the same way and signals of aromatic amine ligands from platinum compounds were integrated. The relative concentrations of the platinum compound at each time point were calculated from peak integrals. The aquation of pyriplatin and phenanthriplatin was investigated at 37 °C by NMR spectroscopy in D2O solutions containing 2 mM of the Pt compound with dioxane as an internal standard. Reactions of platinum compounds with N-AcMet were performed in NMR tubes containing 2 mM of the Pt complex and 2 mM (1 equiv) of N-AcMet in 10 mM PBS buffer, D2O, pH* 7.4 at 37 °C. Reactions of platinum compounds with 5′-dGMP were performed in NMR tubes containing 2 mM of the platinum compounds and 32 mM (16 equiv) of 5′-dGMP in 10 mM PBS buffer, D2O, pH* 7.4 at 37 °C. Deuterated 3-(trimethylsilyl)propionic acid sodium salt (TMS-PFASS) was used as an internal standard. The pH* values are the measured pH values without correction for the effect of deuterium on the electrode.
GLuc Luminometry Assay.
pGLuc plasmid was obtained using commercially available pCMV-GLuc vector and globally platinated plasmids were prepared as previously reported (19). Transfection of transcription probes into A549 and HT29 cells with the pGLuc plasmid was carried out using liposomal transfecting agents. Determination of expression levels was tested by Luciferase assays monitored by a luminometer (19). Detailed procedures for GLuc Luminometry assay are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the Developmental Therapeutics Program of the National Cancer Institute (NCI) for performing the NCI-60 cell line screening and the COMPARE analysis for phenanthriplatin. This work was supported by National Cancer Institute Grant CA034992 and a Misrock Postdoctoral Fellowship (to G.Y.P.). Spectroscopic instrumentation at the Massachusetts Institute of Technology Department of Chemistry Instrumentation Facility is maintained with funding from National Institutes of Health Grant 1S10RR13886-01.
Footnotes
Conflict of interest statement: S.J.L. has financial interest in Blend Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Cambridge Structural Database (http://www.ccdc.cam.ac.uk/products/csd/), Cambridge Crystallographic Data Centre, Cambridge CB2 1EZ, United Kingdom (CSD reference nos. CCDC 875229 and CCDC 875230).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207670109/-/DCSupplemental.
References
- 1.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 3.Jung YW, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 4.Hollis LS, Amundsen AR, Stern EW. Chemical and biological properties of a new series of cis-diammineplatinum(II) antitumor agents containing three nitrogen donors: cis-[Pt(NH3)2(N-donor)Cl]+ J Med Chem. 1989;32:128–136. doi: 10.1021/jm00121a024. [DOI] [PubMed] [Google Scholar]
- 5.Pinto AL, Lippard SJ. Sequence-dependent termination of in vitro DNA-synthesis by cis-diamminedichloroplatinum(II) and trans-diamminedichloroplatinum(II) Proc Natl Acad Sci USA. 1985;82:4616–4619. doi: 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bursova V, Kasparkova J, Hofr C, Brabec V. Effects of monofunctional adducts of platinum(II) complexes on thermodynamic stability and energetics of DNA duplexes. Biophys J. 2005;88:1207–1214. doi: 10.1529/biophysj.104.051771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovejoy KS, et al. Spectrum of cellular responses to pyriplatin, a monofunctional cationic antineoplastic platinum(II) compound, in human cancer cells. Mol Cancer Ther. 2011;10:1709–1719. doi: 10.1158/1535-7163.MCT-11-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lovejoy KS, et al. cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects. Proc Natl Acad Sci USA. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Zhu G, Huang XH, Lippard SJ. X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct. Proc Natl Acad Sci USA. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu G, Myint M, Ang WH, Song L, Lippard SJ. Monofunctional platinum-DNA adducts are strong inhibitors of transcription and substrates for nucleotide excision repair in live mammalian cells. Cancer Res. 2012;72:790–800. doi: 10.1158/0008-5472.CAN-11-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheate NJ, Walker S, Craig GE, Oun R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010;39:8113–8127. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Guo Z, Parsons S, Sadler PJ. Stereospecific and kinetic control over the hydrolysis of a sterically hindered platinum picoline anticancer complex. Chem Eur J. 1998;4:672–676. [Google Scholar]
- 13.Holford J, et al. Chemical, biochemical and pharmacological activity of the novel sterically hindered platinum co-ordination complex, cis-[amminedichloro(2-methylpyridine)] platinum(II) (AMD473) Anticancer Drug Des. 1998;13:1–18. [PubMed] [Google Scholar]
- 14.Chen Y, Guo Z, Parkinson JA, Sadler PJ. Kinetic control of reactions of a sterically hindered platinum picoline anticancer complex with guanosine 5′-monophosphate and glutathione. J Chem Soc Dalton Trans. 1998:3577–3585. [Google Scholar]
- 15.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 16.Boulikas T, Vougiouka M. Cisplatin and platinum drugs at the molecular level. (Review) Oncol Rep. 2003;10:1663–1682. [PubMed] [Google Scholar]
- 17.Howe-Grant M, Wu KC, Bauer WR, Lippard SJ. Binding of platinum and palladium metallointercalation reagents and antitumor drugs to closed and open DNAs. Biochemistry. 1976;15:4339–4346. doi: 10.1021/bi00664a031. [DOI] [PubMed] [Google Scholar]
- 18.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ang WH, Myint M, Lippard SJ. Transcription inhibition by platinum-DNA cross-links in live mammalian cells. J Am Chem Soc. 2010;132:7429–7435. doi: 10.1021/ja101495v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mello JA, Lippard SJ, Essigmann JM. DNA adducts of cis-diamminedichloroplatinum(II) and its trans isomer inhibit RNA polymerase II differentially in vivo. Biochemistry. 1995;34:14783–14791. doi: 10.1021/bi00045a020. [DOI] [PubMed] [Google Scholar]
- 21.2008. APEX2 2008-4.0. (B.A., Inc. Madison, WI)
- 22.Sheldrick GM. 2008 SADABS Area-Detector Absorption Correction (University of Göttingen, Göttingen, Germany) [Google Scholar]
- 23.Sheldrick GM. 2000 SHELXTL-97, 6.14 (University of Göttingen, Göttingen, Germany) [Google Scholar]
- 24.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 25.Spek AL. 2008 PLATON, a multipurpose crystallographic tool (Utrecht University, Utrecht, The Netherlands) [Google Scholar]
- 26.Müller P. Practical suggestions for better crystal structures. Crystallogr Rev. 2009;15:57–83. [Google Scholar]
- 27.Huxley M, et al. An androgenic steroid delivery vector that imparts activity to a non-conventional platinum(II) metallo-drug. Dalton Trans. 2010;39:11353–11364. doi: 10.1039/c0dt00838a. [DOI] [PubMed] [Google Scholar]
- 28.Dhar S, Lippard SJ. Mitaplatin, a potent fusion of cisplatin and the orphan drug dichloroacetate. Proc Natl Acad Sci USA. 2009;106:22199–22204. doi: 10.1073/pnas.0912276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.