Abstract
Chemotaxis allows bacteria to follow gradients of nutrients, environmental stimuli, and signaling molecules, optimizing bacterial growth and survival. Escherichia coli has long served as a model of bacterial chemotaxis, and the signal processing by the core of its chemotaxis pathway is well understood. However, most of the research so far has focused on one branch of chemotactic signaling, in which ligands bind to periplasmic sensory domains of transmembrane chemoreceptors and induce a conformational change that is transduced across the membrane to regulate activity of the receptor-associated kinase CheA. Here we quantitatively characterize another, receptor-independent branch of chemotactic signaling that is linked to the sugar uptake through a large family of phosphotransferase systems (PTSs). Using in vivo characterization of intracellular signaling and protein interactions, we demonstrate that signals from cytoplasmic PTS components are transmitted directly to the sensory complexes formed by chemoreceptors, CheA and an adapter protein CheW. We further conclude that despite different modes of sensing, the PTS- and receptor-mediated signals have similar regulatory effects on the conformation of the sensory complexes. As a consequence, both types of signals become integrated and undergo common downstream processing including methylation-dependent adaptation. We propose that such mode of signaling is essential for efficient chemotaxis to PTS substrates and may be common to most bacteria.
Keywords: signal transduction, signal integration, fluorescence resonance energy transfer
In Escherichia coli and other bacteria, sensory complexes that consist of chemoreceptors, a kinase CheA, and an adapter protein CheW (Fig. 1A) are organized in clusters, which amplify and integrate signals through cooperative protein interactions (1, 2). Canonical chemoeffectors signal by binding to the periplasmic domains of specific receptors, either directly (3) or indirectly via periplasmic binding proteins (4). Attractant binding promotes the inactive conformation of the receptor–kinase complex and inhibits CheA autophosphorylation. As a consequence, cells that move up a gradient and experience an increase in attractant concentration have a lower level of the tumbling signal CheY-P, which extends their runs in the preferred direction. This initial inhibitory effect of attractant binding is opposed by a methyltransferase CheR, which together with the methylesterase CheB constitutes the adaptation system of the chemotaxis pathway. CheR recognizes the inactive conformation of the receptor dimer, gradually adding methyl groups to four specific glutamate residues at each receptor subunit until higher methylation increases receptor activity back to the prestimulus state. The comparatively slow process of adaptation provides swimming bacteria with a short-term memory for temporal comparisons of ligand concentration, which is key to bacterial chemotaxis strategy (2). Efficiency of adaptation is further enhanced by CheA-dependent phosphorylation of CheB, which increases its activity, although this negative feedback is not essential for adaptation (5).
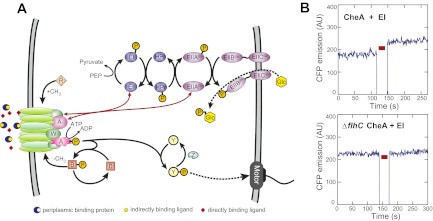
Fig. 1.
Interactions between the PTS and the chemotaxis pathway. (A) Overview of the PTS and the chemotaxis pathway (see text for details). Red arrows indicate interactions between components of both systems detected by an in vivo FRET screen (Fig. S1A). (B) Examples of a positive (Upper) and a negative (Lower) FRET measurement for CFP-CheA/EI-YFP pair in the wild-type and chemotaxis-negative (∆flhC) background, respectively. Red bar indicates a period of acceptor (YFP) photobleaching. AU, arbitrary units of fluorescence. Measurements for other positive FRET pairs and values of apparent FRET efficiency are shown in Fig. S2.
Less investigated are the details of receptor-independent signaling that is mediated by sugar uptake via phosphotransferase systems (PTSs) (6, 7). The PTS transporters (Fig. 1A) consist of the shared cytoplasmic components EI (gene ptsI) and HPr (gene ptsH), and of several sugar-specific EII components (8, 9). The major glucose-specific EII (EIIGlc) consists of the membrane component EIIBCGlc (gene ptsG) and the cytoplasmic protein EIIAGlc (gene crr). During their PTS-mediated transport across the membrane, sugar molecules become phosphorylated, which generates a constant flow of phosphate groups from phosphoenolpyruvate (PEP). The resulting reduction in phosphorylation of the cytoplasmic PTS proteins has a variety of regulatory functions, including catabolite repression and control of chemotaxis (8, 9). Signaling to the chemotaxis pathway further requires the core chemotaxis proteins CheA, CheW, and CheY (7, 10–12) but does not involve transphosphorylation between both systems (13). Instead it is likely to rely on allosteric inhibition of CheA by the unphosphorylated EI that was observed in vitro (10).
Here we used fluorescence resonance energy transfer (FRET) to follow in vivo the PTS-induced chemotactic response and to map interactions between PTS and chemotaxis proteins. Our results suggest that PTS signaling to the chemotaxis system is mediated through regulatory interactions of their core components at the chemosensory complexes. We further demonstrate that adaptation to PTS- and receptor-mediated signals relies on the same methylation-dependent mechanism, ensuring identical adaptation kinetics for all types of signaling in chemotaxis.
Results
Molecular Interactions Between PTS and Chemotaxis Proteins.
In our study, we focused on the PTS-mediated sensing of glucose, the preferred carbon source of E. coli. Glucose signals to the chemotaxis system both through the periplasmic glucose/galactose binding protein (GBP, gene mglB), which binds to the receptor Trg (4, 14), and via the PTS (6). To address the mechanism of signal transduction from the glucose-specific PTS to the chemotaxis pathway, we first tested in vivo all possible binary interactions or immediate proximities between components of the two systems using acceptor photobleaching FRET (15). In this screen, fusions of chemotaxis and PTS proteins to cyan and yellow fluorescent proteins (CFP and YFP, respectively) were coexpressed pairwise in wild-type E. coli cells, and FRET signals were detected by selectively photobleaching YFP (FRET acceptor) and following ensuing changes in the CFP (FRET donor) emission (Fig. 1B and Figs. S1A and S2). Our analysis yielded four positive pairs (Fig. 1A and Fig. S2A), with each of EI and EIIAGlc interacting with CheA and CheW. All observed interactions were comparatively weak, with apparent FRET efficiencies being between 0.5 and 1% (Fig. S2A), which is below the values observed in our assay for typical stable protein complexes (e.g., ∼2% for CheA/CheA) (15), but reproducibly above the negative control (Fig. 1B and Fig. S2C). The interactions between chemotaxis and PTS proteins were apparently stabilized by the presence of chemoreceptors, because neither of these interactions was observed in a strain deleted for all chemoreceptors or in a ∆flhC strain lacking the master activator of chemotaxis and flagellar gene expression (Fig. 1B and Fig. S2 B and C). EI and EIIAGlc may thus specifically bind to CheA and/or CheW assembled into the sensory complexes and not (or with lower affinity) to their unbound forms, likely explaining why the allosteric regulation of free CheA required an unphysiologically high excess of EI in vitro (10).
Consistent with the apparent weakness of their interactions, no localization of the PTS proteins to chemoreceptor clusters was observed under our experimental conditions (Fig. S3). Because fluorescent protein fusions to the cytoplasmic PTS components were functional for PTS chemotaxis (Fig. S4), we conclude that previously reported intracellular localization of PTS proteins (16) is not generally required for their function. Moreover, the observed interactions showed no apparent dependence on stimulation with glucose, suggesting that signal transmission from the PTS to the chemotaxis system relies on allosteric regulation within existing complexes rather than on phosphorylation-dependent changes in protein affinities.
PTS Complements Receptor-Mediated Response to Glucose.
To quantify the pathway response to glucose, we used a FRET-based reporter that relies on the phosphorylation-dependent interaction between CheY-YFP and its phosphatase CheZ-CFP to monitor the intracellular pathway activity in real time (Fig. S1B) (17, 18). Measurements of Trg- and PTS-mediated responses showed that these two modes of sensing operate at different glucose concentrations. In wild-type cells preequilibrated in buffer, where sensing is dominated by Trg, the glucose concentration at the half-maximal response (EC50) was ∼30 nM (Fig. 2A). The value for the PTS-mediated response in the absence of Trg was about 10-fold higher. Notably, saturating stimulation via the PTS also exerted weaker inhibitory effect on activity of the chemosensory complexes than receptor-mediated stimulation.
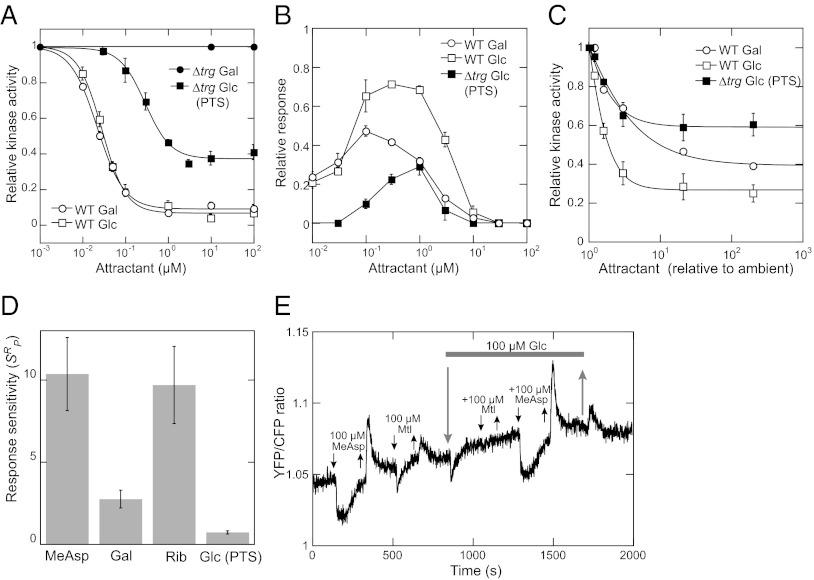
Fig. 2.
Characterization of the PTS-mediated response. Intracellular response of the chemotaxis pathway was measured using a CheY/CheZ FRET reporter of kinase activity in ∆(cheY cheZ) background [wild type (WT) for FRET] or in ∆trg ∆(cheY cheZ) background (∆trg). See Fig. S1 B and C for details of the assay. (A) Dose–response measurements. Buffer-adapted cells were stimulated by addition and subsequent removal of indicated concentration steps of glucose (Glc) or galactose (Gal). Kinase activity was plotted relative to the steady-state activity in the buffer. Zero activity was determined by a saturating stimulation with 100 μM α-methyl-DL-aspartate (MeAsp). Data were fitted using a Hill equation. Error bars here and throughout indicate SEs. (B) Dynamic range measurements. Cells were stimulated by stepwise addition of increasing amounts of attractant, allowing full adaptation before each subsequent stimulation. The response for each step was normalized to the response of buffer-adapted cells to 100 μM MeAsp. (C) Measurements of response sensitivity. Dose–response curves were measured as in A but for cells preadapted to an ambient concentration of 500 nM glucose or galactose, as indicated, and fitted using the Hill equation. Ligand concentration was normalized to the ambient concentration. (D) Response sensitivity at the peak of the dynamic range (SRP) for different attractants. SRP for the PTS-mediated response to glucose was calculated as the initial slope of the dependence in C. The values for ribose (Rib) and other attractants at the peaks of their respective dynamic range were measured previously and normalized to the fraction of ligand-specific receptors in the total receptor pool (19). (E) Effect of adaptation to glucose on the response to other ligands in ∆trg cells. The response was followed as a change in the ratio of YFP to CFP fluorescence due to FRET, with a higher ratio corresponding to higher FRET signal and therefore higher pathway activity. Addition and removal of mannitol (Mtl) and other attractants are indicated by down and up arrows, respectively. Gray bar indicates presence of glucose in the background, over which other stimuli were added. Gradual drift of the YFP/CFP ratio base line arises from a relatively faster loss of the CFP fluorescence over the time course of the measurement.
That the Trg- and PTS-mediated modes of glucose sensing are complementary is even more apparent when comparing the dynamic range of concentrations over which cells can respond and adapt to a ligand (Fig. 2B). Measurements of the dynamic range were performed as before (19) by increasing sugar concentration in approximately threefold steps (Fig. S1C), which exposes the chemotaxis system to a nearly constant stimulus (20, 21). Because deletion of the PTS components affects expression of the chemotaxis genes via catabolite repression, the Trg-only response to glucose was approximated here by the wild-type response to galactose, which has the same signaling mode and a nearly identical binding affinity to GBP as glucose but is not a PTS substrate (4, 19) (Fig. 2A). We observed that the PTS-mediated response expands the dynamic range of glucose sensing by the chemotaxis system, peaking at concentrations where the Trg-mediated response decreases due to the saturation of the GBP (19). In the wild-type cells, the receptor- and PTS-mediated responses were approximately additive. The observed peak of the PTS-mediated response (∼1 μM) is consistent with the value deduced from studies of adaptation dynamics (11) and slightly lower than the apparent Km of the PTS-mediated glucose uptake (5 μM) (7). The dynamic range was similarly narrow for both the Trg-mediated and PTS-mediated response, suggesting that adaptation does not modulate the ligand binding affinity in either case (19).
Response Sensitivity and Integration of Receptor- and PTS-Mediated Stimuli.
We next characterized the response sensitivity (SR), defined as the ratio between the fractional change in kinase activity (∆A/A) and the fractional change in ligand concentration (∆L/L) (17, 19). The value of SR reflects the strength of the inhibition of sensory complexes by a given ligand (19). To determine this value, cells were preadapted to 500 nM glucose (or galactose), around the peak of the PTS-mediated response, and the initial slope of the normalized dose–response curves was determined (Fig. 2C). The calculated sensitivity for the wild-type response to glucose (1.00 ± 0.04) was approximately the sum of the sensitivities of the Trg-mediated response (0.31 ± 0.03) and the PTS-mediated response (0.74 ± 0.10), validating the additivity of these responses. Moreover, maximal sensitivity of the PTS-mediated response achieved at the peak of its dynamic range (SRP) was much lower than the sensitivity of the receptor-mediated response (19) (Fig. 2D), confirming relative weakness of the inhibition of the chemosensory complex activity via the PTS.
We further observed that adaptation to saturating concentrations of glucose in a ∆trg strain did not affect the response to the receptor-binding ligand MeAsp but eliminated the chemotactic response to other PTS ligands, such as mannitol (Fig. 2E). This result indicates that, whereas receptor- and PTS-mediated stimuli are independently sensed, individual PTS stimuli effectively compete with each other.
Receptor Methylation Controls PTS-Mediated Response.
To better understand the mechanism of adaptation in PTS-mediated signaling, we first compared adaptation kinetics for PTS- and receptor-mediated stimuli. When plotted against the response strength, the adaptation time for the PTS- and receptor-mediated response showed a similar dose dependence (Fig. 3A). This similarity was further demonstrated by a nearly identical adaptation time course for glucose and MeAsp stimuli of similar strength (Fig. 3A, Inset). These data strongly suggest that the same methylation-based adaptation operates for both types of stimuli. Nevertheless, the response and adaptation to the PTS-mediated stimuli was apparently not receptor specific, because either one of the major receptors, Tar or Tsr, was sufficient to mediate PTS signaling (Fig. 3B).
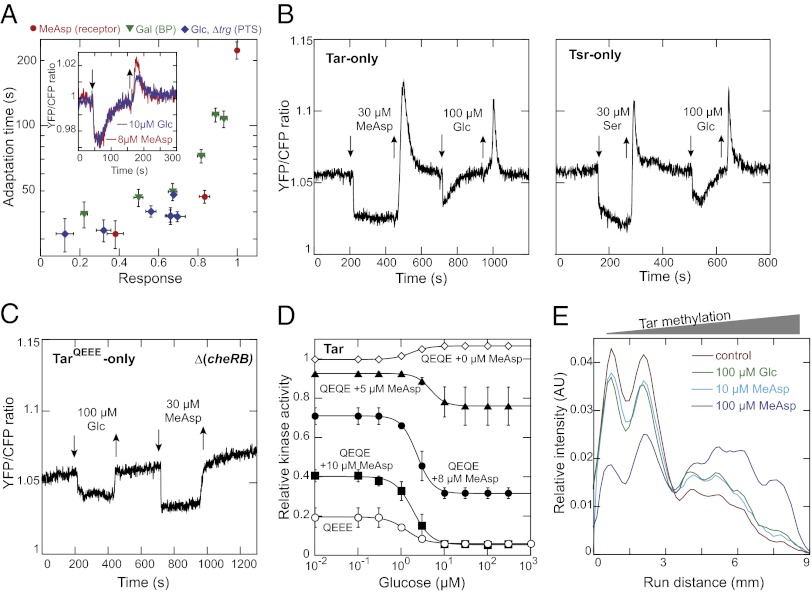
Fig. 3.
Involvement of chemoreceptor methylation in adaptation to PTS stimuli. (A) Dependence of adaptation time on the response amplitude for subsaturating stimulation with MeAsp and galactose in WT or glucose in ∆trg cells. Adaptation time was defined as the time required to regain 50% of the initial loss in FRET signal upon stimulation. Response amplitude was normalized to the maximal response elicited by 100 µM MeAsp. Inset shows an adaptation time course for MeAsp and glucose stimuli of similar strength, with down and up arrows indicating addition and removal of attractants, respectively. (B) PTS- and receptor-mediated responses in receptorless CheR+ CheB+ cells expressing Tar (Left) or Tsr (Right) from plasmids (Table S1). Note that incubation time with MeAsp or serine (Ser) was not long enough to allow full adaptation to saturating stimuli used here as a reference. (C) Response to glucose and MeAsp in receptorless ∆(cheR cheB) cells expressing single-modified TarQEEE. (D) Activity dependence of the PTS-mediated response. Receptorless ∆(cheR cheB) cells expressing half-modified TarQEQE were stimulated with glucose in the buffer or in presence of indicated concentrations of MeAsp (Fig. S5A). Response of cells expressing TarQEEE as in C is also shown. (E) Change of receptor methylation pattern in response to PTS- or receptor-mediated stimuli. Cells (∆trg ∆tsr) were stimulated by indicated amounts of MeAsp or glucose for 2 min and the distribution of Tar methylation levels was measured using immunoblotting (Fig. S6A). Intensity profiles reflect receptor mobility on the SDS/PAGE gel, with higher receptor mobility corresponding to higher methylation. For better comparison, each profile curve was normalized to the integral intensity of all bands within the respective lane after subtraction of the background.
We further observed that the PTS-mediated response to glucose strongly depends on the activity state of chemoreceptors. The response was clearly visible in a receptorless ∆(cheR cheB) strain expressing TarQEEE (Fig. 3C), which has one of four possible methylation sites replaced by a glutamine that is similar to methylated glutamate. This modification mimics the low levels of receptor methylation and activity in CheR+ CheB+ cells (22, 23). In contrast, no attractant-like response to glucose could be observed in the same background strain expressing higher-modified TarQEQE (Fig. 3D and Fig. S5A) that is more active (22); instead a weak but reproducible up-regulation of the CheY/CheZ interaction was observed. This latter effect was apparently independent of all other chemotaxis proteins (Fig. S5B) and might be explained by the glucose-stimulated up-regulation of the intracellular level of acetyl phosphate, a known phosphodonor for CheY (24). The inability of the PTS signal to inhibit the highly active TarQEQE receptors was likely due to the aforementioned weakness of the PTS signal, and indeed a clear response was observed when the activity of TarQEQE was lowered by prestimulation with subsaturating concentrations of MeAsp (Fig. 3D and Fig. S5A). Moreover, both amplitude and EC50 of the response to glucose changed with the level of receptor activity (Fig. 3D), confirming that receptors are directly involved in the processing of PTS stimuli.
Most importantly, the PTS-mediated response in ∆(cheR cheB) strains was nonadaptive (Fig. 3C and Fig. S5A), demonstrating that receptor methylation is in fact required for adaptation to PTS stimuli. Consistent with the involvement of the methylation system, adaptation of CheR+ CheB+ cells to a saturating PTS-mediated glucose stimulation influenced the level of receptor methylation, as observed by the increased mobility of higher-modified receptors in an SDS/PAGE gel (Fig. 3E and Fig. S6). In a strain expressing Tar as the only receptor, the change in methylation was similar to that elicited by a MeAsp stimulus of comparable magnitude (Fig. 3E and Fig. S6A). In the wild-type cells, methylation of both Tar and Tsr was affected (Fig. S6 B and C), although the exact magnitude of changes for individual receptors was difficult to assess because of a partial overlap between Tsr and Tar bands on the gel. Although these results appear to contradict a previous study reporting methylation-independent adaptation to PTS substrates (25), we believe that the high sugar concentration used in those experiments (10 mM) might have led to PTS-independent effects on the chemotaxis system, such as those mediated by energy taxis (26).
CheB Phosphorylation Is Not Essential for Adaptation to PTS Stimuli.
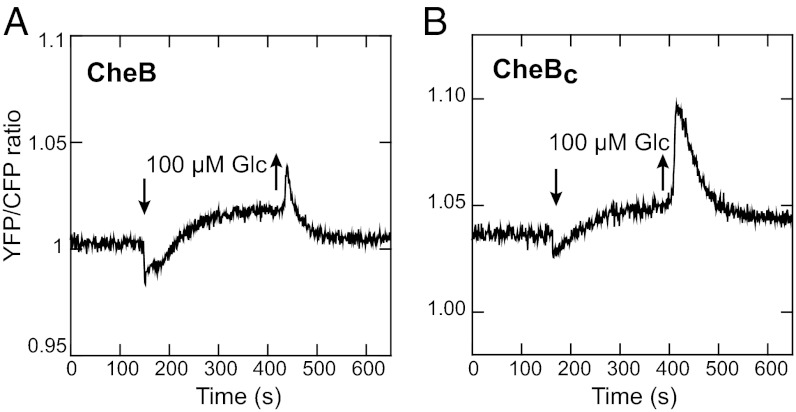
In principle, PTS stimuli can promote increased receptor methylation either directly, by inactivating receptors to make them better substrates for CheR or indirectly, by reducing CheB phosphorylation and thereby downregulating the rate of demethylation. We ruled out the second mechanism by showing that CheB phosphorylation is not required for adaptation to the PTS-mediated glucose response. Adaptation was clearly visible in a ∆trg ∆cheB strain that expresses either the wild-type CheB (Fig. 4A) or CheBC (Fig. 4B), the catalytic domain of CheB that cannot be phosphorylated and is constitutively active (27). This result unambiguously demonstrates that the regulatory interactions between the PTS and the chemotaxis pathway must directly affect the activity state of chemoreceptors, leading to subsequent methylation-dependent adaptation.
Fig. 4.
CheB phosphorylation and adaptation to PTS stimuli. Comparison of FRET after addition of glucose in a Δtrg ΔcheB strain expressing wild-type CheB (A) or CheBC, a constitutively active form of CheB lacking the regulatory domain (B), from plasmids (Table S1). The higher level of FRET in the glucose-adapted cells most likely results from CheA-independent phosphorylation of CheY (Fig. 3D and Fig. S4B).
Discussion
Although the involvement of PTS-mediated sugar transport in chemotaxis has been long established (6, 7), the interplay between the PTS- and receptor-mediated chemotactic signaling remained largely unclear. Consistent with the previously observed allosteric inhibition of CheA activity by unphosphorylated EI (10), our in vivo data suggest that signaling from the PTS is mediated by interactions of EI, and possibly EIIAGlc, with core components of the chemosensory complexes, CheA and CheW (Fig. 1A and Fig. S7). These interactions apparently depend on the formation of ternary receptor-CheW-CheA complexes, which may explain why a large excess of EI was needed to regulate the activity of free CheA in vitro (10). Most importantly, we provide evidence that dephosphorylation of the PTS proteins bound at sensory complexes lowers the activity of the entire complex, rather than of CheA alone as assumed in previous models. Because sensory complexes are believed to switch between active and inactive states as single cooperative units (2), inhibitory effects on CheA and CheW can be transmitted to receptors favoring their inactive conformation (Fig. S7B), much the same way as ligand binding to receptors favors the inactive conformation of CheA (Fig. S7A). As a consequence, adaptation to PTS stimuli can be mediated by the same regulatory feedback from receptor activity to the methylation system. Such common processing of receptor- and PTS-mediated signals is of fundamental importance for chemotaxis, because it inherently ensures the same (optimal) relation between the strength of the initial response and the duration of subsequent adaptation for both types of stimuli, as well as the high precision of adaptation for PTS stimuli—two important features of the overall chemotaxis strategy (2, 28).
We further conclude that receptor- and PTS-mediated signals are additive and that adaptation to a receptor-mediated stimulus does not affect response sensitivity to a PTS stimulus and vice versa, as already reported for different receptor-mediated stimuli (19). However, because chemotactic adaptation to PTS stimuli occurs “downstream” of the PTS, preexposure to saturating levels of one PTS substrate inhibits the response to other PTS ligands (Fig. S7B), similar to the mutual inhibition observed for competitive binding of two ligands to the same binding site on a receptor (Fig. S7A). The PTS signal to the chemotaxis pathway thus provides an already “integrated” readout of the overall PTS-mediated sugar uptake. This mode of signaling may allow cells to establish preferences for individual sugars within the PTS network itself and accumulate in regions of maximal PTS-mediated sugar uptake.
Because of the observed dependence of the PTS–chemotaxis interactions on ternary complex formation, it remains unclear whether EI and EIIAGlc directly interact with both CheA and CheW or whether interaction with one of these proteins (e.g., CheA) serves as a scaffold to bring the respective PTS component into the immediate vicinity of the other protein (e.g., CheW) to yield a FRET signal. Furthermore, it cannot be excluded that EI and EIIAGlc directly interact with the conserved cytoplasmic tip of the receptor dimer, because large molecular distance may preclude energy transfer between a fusion protein bound at the cytoplasmic tip and the C-terminal fluorophore tag of the receptor (15). In any case, the observed interactions between chemosensory complexes and PTS proteins appear to be weak, consistent with the apparent lack of localization of the PTS components to chemosensory clusters and with the weakness of chemosensory complex inhibition by PTS signals. Because no apparent dependence on glucose stimulation could be observed for these interactions, regulatory signals are likely to be transmitted from the PTS to the chemotaxis system via phosphorylation-induced conformational changes within protein complexes rather than by strong changes in protein binding affinities (Fig. S7B).
From the perspective of cell physiology, the involvement of EI and EIIAGlc in signaling to the chemotaxis pathway is consistent with their central roles in the PTS network and propensity to regulate cellular functions (8, 9). EI provides a convergence point for all sugar-specific PTS branches, and the level of EI phosphorylation thus reflects the overall rate of sugar transport through the PTS network. And although EIIAGlc is a more specific component of the PTS branches responsible for uptake of glucose, maltose, and trehalose, the uptake of other sugars also affects EIIAGlc phosphorylation because phosphotransfer reactions within the PTS are reversible (9).
Methods
Strains and Plasmids.
All plasmids and strains used in this study are listed in Table S1. Deletions of trg, flhC, and pts genes were performed by P1 transduction using RP1131 or the listed strains from the Keio collection (29) as donors. For KG28, the kanamycin resistance cassette was flipped out using the temperature-sensitive plasmid pCP20 that encodes FLP recombinase (30).
Soft Agar Plates.
Functionality of fluorescent protein fusions to the PTS components was tested on Tanaka minimal medium (31) supplemented with 0.3% (wt/vol) agar, 1 mM glucose, 100 µg/mL ampicillin, and 20 μM IPTG to induce expression of plasmid-encoded fusion proteins.
Preparation of Cells.
Bacteria were grown as before (19) at 34 °C and 275 rpm in 10 mL tryptone broth (TB) supplemented with appropriate antibiotics (100 μg/mL ampicillin, 34 μg/mL chloramphenicol, 50 μg/mL kanamycin) and specific inducers (Table S1) to OD600 of 0.45. Cells were then harvested by centrifugation (10 min at 5,000 × g) and resuspended in 10 mL tethering buffer (10 mM KPO4, 0.1 mM EDTA, 1 μM methionine, 10 mM lactic acid, 67 mM NaCl, pH 7).
Fluorescence Imaging and FRET.
Fluorescence imaging and FRET experiments were performed as described previously (15, 17–19). For acceptor photobleaching FRET, bacteria were applied to a thin agarose pad (1% agarose in tethering buffer) and YFP photobleaching was achieved by a brief 20-s illumination using a 532-nm laser. Integral CFP fluorescence of a field of several hundred cells was recorded before and after bleaching with 1-s integration time using photon counters. For stimulus-dependent FRET measurements cells were attached to a polylysine-coated coverslip and placed into a flow chamber of 50 μL volume, which was kept under constant flow of tethering buffer (500 μL/min) by a syringe pump that was stopped briefly to add and remove attractants. Changes in FRET upon stimulation were calculated from changes in the YFP/CFP ratio of a field of cells (18, 23).
Immunoblotting.
Immunoblotting and data analysis were performed as before (19, 23). Samples were separated on an 8% (wt/vol) SDS polyacrylamide gel for 17 h, blotted on a nitrocellulose membrane, and analyzed using primary polyclonal anti-Tar antibody and goat antirabbit IRDye 800 conjugated secondary antibody (Rockland), both diluted 1:10,000. Membranes were scanned with an Odyssey Imager (LI-COR) and intensity profiles of the protein distribution within a lane were evaluated using ImageJ software (http://rsbweb.nih.gov/ij).
Supplementary Material
Acknowledgments
We thank G. Schwarz and D. Kentner for help with cloning and Ned S. Wingreen and Abiola Pollard for helpful comments on the manuscript. This work was supported by Grant SO 421/7-2 from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205307109/-/DCSupplemental.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sourjik V, Wingreen NS. Responding to chemical gradients: Bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesibov R, Adler J. Chemotaxis toward amino acids in Escherichia coli. J Bacteriol. 1972;112:315–326. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler J, Hazelbauer GL, Dahl MM. Chemotaxis toward sugars in Escherichia coli. J Bacteriol. 1973;115:824–847. doi: 10.1128/jb.115.3.824-847.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 6.Adler J, Epstein W. Phosphotransferase-system enzymes as chemoreceptors for certain sugars in Escherichia coli chemotaxis. Proc Natl Acad Sci USA. 1974;71:2895–2899. doi: 10.1073/pnas.71.7.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengeler J, Auburger AM, Mayer R, Pecher A. The phosphoenolpyruvate-dependent carbohydrate: Phosphotransferase system enzymes II as chemoreceptors in chemotaxis of Escherichia coli K 12. Mol Gen Genet. 1981;183:163–170. doi: 10.1007/BF00270156. [DOI] [PubMed] [Google Scholar]
- 8.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: Phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol. 2009;16:65–87. doi: 10.1159/000219373. [DOI] [PubMed] [Google Scholar]
- 10.Lux R, Jahreis K, Bettenbrock K, Parkinson JS, Lengeler JW. Coupling the phosphotransferase system and the methyl-accepting chemotaxis protein-dependent chemotaxis signaling pathways of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11583–11587. doi: 10.1073/pnas.92.25.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lux R, et al. Elucidation of a PTS-carbohydrate chemotactic signal pathway in Escherichia coli using a time-resolved behavioral assay. Mol Biol Cell. 1999;10:1133–1146. doi: 10.1091/mbc.10.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowsell EH, Smith JM, Wolfe A, Taylor BL. CheA, CheW, and CheY are required for chemotaxis to oxygen and sugars of the phosphotransferase system in Escherichia coli. J Bacteriol. 1995;177:6011–6014. doi: 10.1128/jb.177.20.6011-6014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson MS, Rowsell EH, Taylor BL. Investigation of transphosphorylation between chemotaxis proteins and the phosphoenolpyruvate:sugar phosphotransferase system. FEBS Lett. 1995;374:161–164. doi: 10.1016/0014-5793(95)01097-x. [DOI] [PubMed] [Google Scholar]
- 14.Kondoh H, Ball CB, Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci USA. 1979;76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kentner D, Sourjik V. Dynamic map of protein interactions in the Escherichia coli chemotaxis pathway. Mol Syst Biol. 2009;5:238. doi: 10.1038/msb.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopian L, Elisha Y, Nussbaum-Shochat A, Amster-Choder O. Spatial and temporal organization of the E. coli PTS components. EMBO J. 2010;29:3630–3645. doi: 10.1038/emboj.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sourjik V, Vaknin A, Shimizu TS, Berg HC. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 2007;423:365–391. doi: 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- 19.Neumann S, Hansen CH, Wingreen NS, Sourjik V. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 2010;29:3484–3495. doi: 10.1038/emboj.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mesibov R, Ordal GW, Adler J. The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J Gen Physiol. 1973;62:203–223. doi: 10.1085/jgp.62.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu Y, Shimizu TS, Berg HC. Modeling the chemotactic response of Escherichia coli to time-varying stimuli. Proc Natl Acad Sci USA. 2008;105:14855–14860. doi: 10.1073/pnas.0807569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Endres RG, et al. Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Syst Biol. 2008;4:211. doi: 10.1038/msb.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oleksiuk O, et al. Thermal robustness of signaling in bacterial chemotaxis. Cell. 2011;145:312–321. doi: 10.1016/j.cell.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwano M, Taylor BL. Novel sensory adaptation mechanism in bacterial chemotaxis to oxygen and phosphotransferase substrates. Proc Natl Acad Sci USA. 1982;79:11–15. doi: 10.1073/pnas.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greer-Phillips SE, Alexandre G, Taylor BL, Zhulin IB. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology. 2003;149:2661–2667. doi: 10.1099/mic.0.26304-0. [DOI] [PubMed] [Google Scholar]
- 27.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 28.Vladimirov N, Sourjik V. Chemotaxis: How bacteria use memory. Biol Chem. 2009;390:1097–1104. doi: 10.1515/BC.2009.130. [DOI] [PubMed] [Google Scholar]
- 29.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka S, Lerner SA, Lin EC. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967;93:642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.