Abstract
A coordinated innate and adaptive immune response, orchestrated by antigen presenting cells (APCs), is required for effective clearance of influenza A virus (IAV). Although IAV primarily infects epithelial cells of the upper respiratory tract, APCs are also susceptible. To determine if virus transcription in these cells is required to generate protective innate and adaptive immune responses, we engineered IAV to be selectively attenuated in cells of hematopoietic origin. Incorporation of hematopoietic-specific miR-142 target sites into the nucleoprotein of IAV effectively silenced virus transcription in APCs, but had no significant impact in lung epithelial cells. Here we demonstrate that inhibiting IAV replication in APCs in vivo did not alter clearance, or the generation of IAV-specific CD8 T cells, suggesting that cross-presentation is sufficient for cytotoxic T lymphocyte activation. In contrast, loss of in vivo virus infection, selectively in APCs, resulted in a significant reduction of retinoic acid-inducible gene I-dependent type I IFN (IFN-I). These data implicate the formation of virus replication intermediates in APCs as the predominant trigger of IFN-I in vivo. Taking these data together, this research describes a unique platform to study the host response to IAV and provides insights into the mechanism of antigen presentation and the induction of IFN-I.
Keywords: microRNA, miR-142, small viral RNA
Influenza A virus (IAV) primarily infects epithelial cells of the upper and lower respiratory tract. However, IAV is also capable of infecting cells of the immune system, specifically antigen presenting cells (APCs), such as dendritic cells (DCs) and macrophages (1–6). The body’s defense to IAV infection requires a coordinated response from both the innate and adaptive arms of the immune system to control, and ultimately clear, the infection. This process demands cellular recognition of IAV, which is mediated through pathogen recognition receptors (PRRs) that include the Toll- and retinoic acid-inducible gene (RIG)-I like receptors (TLRs and RLRs, respectively) (7). PRRs recognize various pathogen-associated molecular patterns (PAMPs) that, in the case of IAV, can include RNA containing an exposed 5′triphoshate or other RNA structures foreign to the cell (8). PAMP recognition culminates in the activation of three transcription-factor families and assembly of a multisubunit structure, termed the enhancesome, which is responsible for induction of type I IFN (IFN-I) (9, 10). Following IAV infection, DCs and macrophages, as well as infected epithelial cells, produce IFN-I in an effort to curb early virus replication (11, 12). Although IFN-I induction in IAV-infected epithelial cells has been clearly demonstrated to be mediated by RIG-I, it is still unclear whether TLRs play a more dominant role in the induction of IFN-I from hematopoietic cells (8).
Following IAV infection and the induction of IFN-I, macrophages and DCs migrate from the lungs into the draining lymph node (LN), where further cytokine production orchestrates the development of an adaptive immune response (13). Within the draining LN, DC subsets, primarily CD103+ DCs that migrated from the infected respiratory tract, as well as LN resident CD8α+ DCs, present antigen to CD8 T cells (14–18). These DCs can acquire IAV antigens by at least three distinct mechanisms: (i) through direct infection, (ii) through phagocytosis of necrotic or apoptotic IAV-infected epithelial cells, or (iii) through membrane exchange with infected cells and transfer of peptide-loaded MHC I (1–6, 19–22). Importantly, antigen presentation by CD103+ DCs and CD8α+ DC subsets can be mediated by any one of these three processes (23–25). Because IAV has been found to replicate in both lung epithelial cells and APCs in vitro and in vivo (1–6, 20–22), the mechanism of antigen presentation during in vivo infection remains controversial. In an effort to address the hematopoietic requirements for IFN-I induction and antigen acquisition in response to IAV, we used microRNA (miRNA)-mediated targeting to silence replication selectively in APCs.
Noncoding small RNAs, such as miRNAs, are ∼22 nt regulatory RNA species that mediate repression of mRNA with partially complementary sequences (26). Although miRNA expression is generally broad, subsets of miRNAs are expressed in a cell type- or tissue-specific manner (27). For example, miR-122 expression is primarily restricted to the liver and miR-142 to cells of hematopoietic origin, but miR-93 is ubiquitously expressed in all mammalian tissues (27). The diverse expression profiles of miRNAs provide a unique tool by which virus transcription can be controlled through exploitation of the small RNA silencing potential. Previous work from our group and others have demonstrated that insertion of complementary miRNA target sequences into the genome of a virus transforms that miRNA into an effective short interfering RNA (siRNA) (28–34). For example, incorporating miR-93 target sites into the nucleoprotein (NP) gene of IAV demonstrated that virus replication was inversely proportional to miR-93 levels both in vitro and in vivo (28). These data suggested that this approach could be expanded to use cell-specific miRNAs to determine the contribution of IAV replication to the generation of the innate and adaptive immune responses.
In this article we demonstrate that targeting IAV NP by the hematopoietic-specific miRNA, miR-142, ablates replication, as defined by sustained RNA-dependent RNA polymerase activity, selectively within immune cells. Generation of this engineered tropism-restricted virus demonstrated that the inability of IAV to replicate within hematopoietic cells did not alter virus clearance or the generation of IAV-specific CD8 T-cell responses. These data suggest that, in vivo, cross-presentation is sufficient to induce functional CD8 T cells. In contrast to antigen acquisition, loss of hematopoietic replication did have consequences on the cytokine response to virus infection. Loss of virus replication in hematopoietic cells resulted in a significant decrease of RIG-I–mediated IFN-I. Taken together, these results demonstrate that virus replication in hematopoietic cells is required for optimal virus sensing but is dispensable for the proper orchestration of the adaptive immune response.
Results
Targeting of IAV by a Hematopoietic Cell-Specific miRNA.
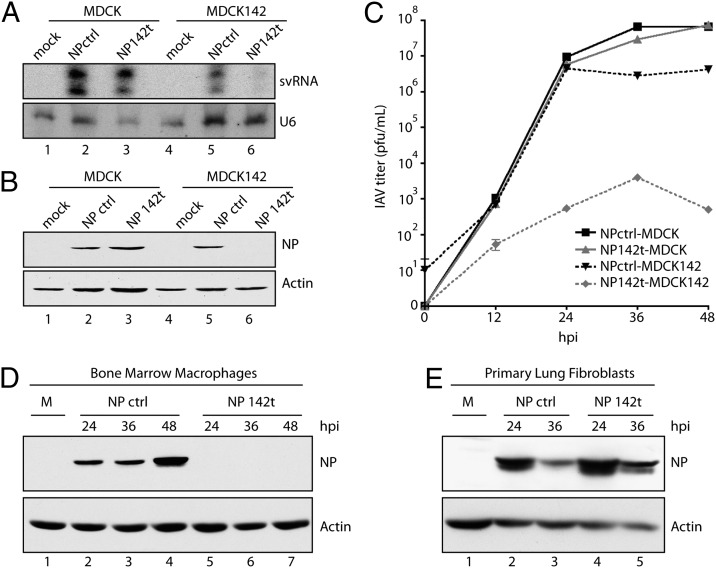
To ensure IAV did not alter endogenous miRNA profiles and, therefore, could be used as a tool to study aspects of the immune response in vivo, human pulmonary epithelial cells were infected and analyzed by deep sequencing. These data demonstrated that the levels of miRNAs were not significantly changed in response to virus infection and confirmed that the utilization of miRNAs to silence infection could provide a useful tool to study the in vivo immune response to IAV (Fig. S1). In an effort to generate a recombinant IAV strain incapable of replicating within cells of hematopoietic origin, we inserted four perfect miR-142 target sites into the NP gene (NP142t). To retain replication competency, we duplicated the packaging sequence of segment five, thereby generating a bona fide 3′ UTR in which to incorporate miRNA targets or length-matched random RNA (NPctrl) (Fig. S2A).
In an effort to monitor the silencing potential of miR-142, we compared virus infection in unmodified Madin Darby canine kidney (MDCK) cells to MDCK cells engineered to express miR-142 (MDCK142) (Fig. S2B). To directly assess virus targeting, we first chose to evaluate the levels of small viral RNA (svRNA), as this IAV-specific product accumulates to greater than 100,000 copies per cell and is required to complete the virus life cycle (35, 36). These data found that, despite the robust signal of svRNA in IAV-infected MDCK cells, MDCK142 selectively suppressed svRNA detection following NP142t infection (Fig. 1A). To confirm the loss of virus replication and measure the degree of primary transcription in the presence of miR-142, we next ascertained the levels of virus cRNA and mRNA, respectively. In accordance with the small RNA profile of IAV, miR-142 expression selectively abolished both viral mRNA and cRNA, suggesting that the incoming virus is targeted rapidly and efficiently (Fig. S2C). To further assess the quantity of viral protein generated in the presence of miR-142, we performed Western blot analysis and found that, as expected, NP levels were undetectable in the context of miRNA-targeting (Fig. 1B). To ensure that the targeted virus did not demonstrate altered fitness in the absence of cognate miRNA, we performed multicycle growth curves in MDCK cells, comparing NP142t to NPctrl. These results demonstrated no discernable difference in virus replication in the absence of miR-142, in stark contrast to the greater than three log-attenuation observed in a heterogeneous population of cells expressing miR-142 (Fig. 1C).
Fig. 1.
Hematopoietic-specific miR-142 inhibits replication of targeted IAV infection. (A) MDCK and MDCK142 cells infected with NPctrl and NP142t IAV at a multiplicity of infection (MOI) of 2 and 16 hours post infection (hpi) analyzed for svRNA (Upper) and U6 (Lower) expression. (B) Cells infected as in A at a MOI of 0.1 and 16 hpi analyzed for NP (Upper) and actin (Lower) protein expression. (C) MDCK and MDCK142 cells infected with NPctrl or NP142t IAV at an MOI of 0.001 and virus titered from supernatants at the indicated time points. (D) BMM infected with NPctrl or NP142t IAV at an MOI of 5 and protein analyzed as in B. (E) Primary lung fibroblasts infected with NPctrl or NP142t IAV at an MOI of 2 and protein analyzed as in B. Data are representative of two to three independent experiments.
Having demonstrated effective targeting of the NP142t virus in cells expressing exogenous miR-142, we next sought to ascertain the silencing potential of an endogenous model. To this end, murine-derived bone marrow macrophages (BMM) were infected with either NPctrl or NP142t IAV and the level of replication was assessed by IAV protein expression (Fig. 1D). Consistent with the in vitro results from Fig. 1A and B and Fig. S2C, NP142t replication was completely inhibited in cells expressing endogenous miR-142. Conversely, primary lung fibroblasts, which do not express miR-142, demonstrated comparable levels of NP between the two recombinant virus strains (Fig. 1E).
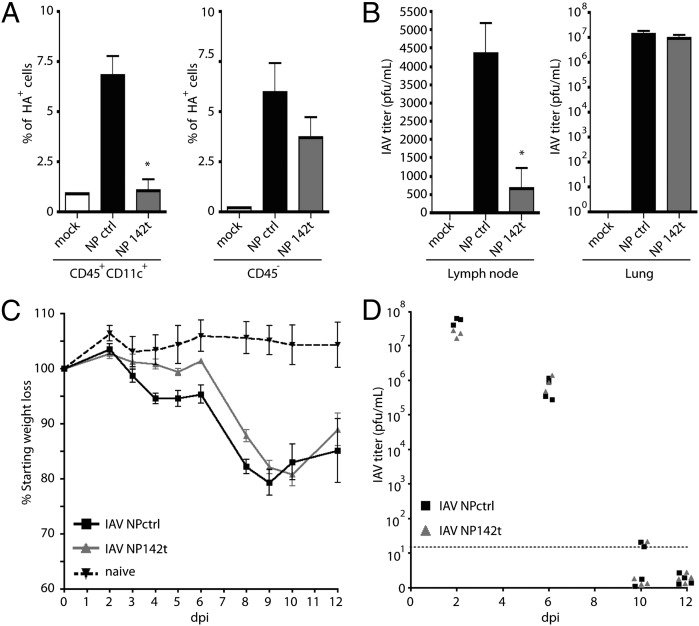
We next determined if miR-142–targeted virus was silenced in vivo. To this end, mice were infected with a high dose (105 pfu) of targeted or untargeted IAV, and CD45+CD11c+ DCs and CD45− nonhematopoietic cells from the lungs were analyzed for surface HA expression, a marker of IAV replication. Although HA surface expression on CD45− cells showed no significant difference in response to control and miRNA-target targeted virus, the DCs revealed a complete loss of NP142t virus replication (Fig. 2A). To further assess the effectiveness of miR-142 silencing of NP142t, we measured the level of virus within the lung draining LN. Because the only cells that migrate to the LN during IAV infection are cells of hematopoietic origin, we reasoned this organ could also be used as an in vivo measure of miR-142 silencing (37). In agreement with in vitro, ex vivo, and DC HA surface expression in vivo results, analysis of the LN also demonstrated a selective block of NP142t virus replication (Fig. 2B), further suggesting that replication in hematopoietic cells is abrogated in vivo.
Fig. 2.
Inhibition of IAV replication in hematopoietic cells in vivo does not alter disease progression or virus titers. (A) Mice were infected intranasally with 105 pfu of NPctrl or NP142t IAV and 2 dpi CD45+CD11c+ (Left) and CD45− cells (Right) from the lungs were analyzed for surface HA expression by FACS; P = 0.0258 and P = 0.3284 for CD45+CD11c+ and CD45− cells, respectively. (B) Mice were infected as in A and titered from lung draining LN (Left) and lungs (Right) 2 dpi. P = 0.0107 and P = 0.282 for LN and lungs, respectively. (C) Mice were infected with NPctrl or NP142t IAV and monitored daily for weight loss. (D) Mice were infected as in C and lungs were removed at indicated time points and virus titers were assessed by plaque assay. Samples not detected were arbitrarily plotted below the limit of detection (LOD). All Data are representative of two independent experiments with three to five mice per group.
Targeting of NP in Hematopoietic Cells Does Not Alter Virus Pathogenesis.
To ascertain whether the altered tropism of NP142t virus impacted pathogenesis, mice were infected with NPctrl or NP142t viruses to assess morbidity and mortality. We hypothesized that virus replication in APCs may be required for virus sensing by PRRs or direct antigen presentation, either of which could lead to decreased virus clearance and increased pathogenesis. However, intranasal administration of either NPctrl or NP142t viruses did not demonstrate any significant alteration in disease progression, both demonstrating a ∼20% loss of weight between 8–10 d postinfection (dpi) before recovering without any incidence of mortality (Fig. 2C). Similarly, the kinetics of virus replication and clearance from the lungs was nearly identical between targeted and untargeted IAV infection, with maximal titers observed ∼2 dpi and dropping below the limit of detection by 10–12 dpi (Fig. 2D). Importantly, sequencing confirmed that this lack of phenotype was not a result of emergence of an NP142t escape mutant (Fig. S3). Taken together, these results suggest NP142t virus replication is not impaired in nonhematopoietic cells and that, despite its altered tropism, there is no significant change in the morbidity or mortality of the infection.
Normal IAV-Specific CD8 T-Cell Response in the Absence of DC Infection.
Given the importance of hematopoietic cells in coordinating the adaptive immune response, we decided to ascertain if there was any measurable consequence to blocking IAV replication in this cellular compartment. As previously noted, DCs can acquire antigen and induce pathogen-specific CD8 T cells through at least two distinct mechanisms: direct infection or exogenous acquisition from IAV-infected cells (1–6, 19, 20–22, 38). Given the complete loss of targeted virus replication in the presence of miR-142 (Fig. 1), we hypothesized that APC would be unable to directly present antigen to naive CD8 T cells. To test this theory, JAWS II APCs, which express miR-142 (Fig. 3A), were infected with either NPctrl or NP142 and incubated with NP366-specific CD8 T-cell hybridomas. Consistent with our hypothesis and our previous results, there was a significant decrease in the level of direct antigen presentation during targeted IAV infection, with activation levels similar to hybridomas incubated with naive APCs or APCs treated with inactivated virus (Fig. 3B).
Fig. 3.
Loss of virus amplification within hematopoietic cells does not alter generation of the antigen-specific CD8 T-cell response. (A) Northern blot analysis for miR-142 (Upper) and U6 (Lower) of RNA from JAWS II, MDCK, and MDCK142. (B) JAWS II cells infected with NPctrl, NP142t, or UV-inactivated IAV at an MOI of 10 (Right) or pulsed with 1 μM peptide (Left), and 24 h later cells were irradiated and incubated with NP-specific CD8 T-cell hybridomas for 18 h and total number of β-gal+ cells enumerated per well. Data are representative of three independent experiments with three to four samples per group. P = 0.003 for NPctrl vs. NP142t. Mice were infected with IAV NPctrl or IAV NP142t and 9 dpi lung cells analyzed by FACS. (C) Representative FACS plots for CD3+CD8+ cells analyzed for NP366 and PA224 tetramer expression. (D) Frequency (Left) and total number (Right) of total IAV-specific NP366 and PA224 tetramer+ CD8 T cells. (E) Representative plots for CD3+CD8+ cells analyzed for intracellular IFN-γ expression following peptide restimulation. (F) Frequency (Left) and total number (Right) of total IFN-γ+ CD8 T cells after IAV NP366 and PA224 peptide restimulation. Data are representative of four independent experiments for tetramer analysis and two independent experiments for peptide restimulation with three to four mice per group.
Given the lack of direct presentation by APCs infected with targeted IAV, we next sought to determine if presentation through exogenous antigen presentation pathways was capable of generating CD8 T-cell responses in vivo. To quantify this response, mice were infected with NPctrl or NP142t IAV and IAV-specific CD8 T cells were determined by tetramer analysis. NPctrl and NP142t IAV demonstrated no significant difference in the number or frequency of IAV-specific CD8 T cells at 9 dpi in C57BL/6 mice (Fig. 3 C and D) and 10 dpi in BALB/c mice (Fig. S4). To confirm that IAV replication within APCs is not required for CD8 T-cell functionality, the level of intracellular IFN-γ production after peptide restimulation was also assessed. Interestingly, there was a trend of decreased IFN-γ production during targeted IAV infection, but the differences were not great enough to be statistically significant (Fig. 3 E and F). Despite small changes in the cytokine profile, comparable levels of clearance mediated by CD8 T cells (39, 40) indicates that cross-presentation or cross-dressed DCs are sufficient to generate protective CD8 T-cell responses.
IAV Amplification Within Hematopoietic Cells Is Critical for Virus Sensing.
Cells of hematopoietic origin are vital for virus sensing and the orchestration of the IFN-I–induced antiviral state. Because IAV is sensitive to relatively small amounts of IFN-I (41), we decided to determine the scope of the IFN-I–mediated response in NPctrl and NP142t virus infections, despite their comparable level of pathogenesis (Fig. 2D). To determine whether virus replication in hematopoietic cells impacted IFN-I induction, primary BMMs and lung fibroblasts were infected ex vivo with either NPctrl or NP142t viruses. Determining mRNA induction of IFN-β and the IFN-stimulated gene, IRF-7, demonstrated robust induction in both primary cell cultures in response to NPctrl virus infection (Fig. 4 A and B). In contrast, induction of IFN-β and IRF-7 in response to NP142t infection was inversely correlated to miR-142 expression, suggesting virus replication was required for induction of the cell’s autonomous antiviral defenses (Fig. 4 A and B). To extrapolate on these ex vivo results, mice were infected with either NPctrl or NP142t IAV to ascertain the mRNA levels of IFN-β and IRF-7 within the lungs. Interestingly, despite being composed of predominantly nonhematopoietic cells, infection with NP142t resulted in a significant decrease of pulmonary mRNA levels for IFN-β and IRF-7 compared with untargeted virus (Fig. 4 C and D). Given that levels of IFN-β and IRF-7 were similar in nontargeting cells ex vivo (Fig. 4B), these data suggest that IAV replication within hematopoietic cells, despite being a nonproductive site of infection (42), is critical for the induction of the host innate response to virus.
Fig. 4.
Defective induction of IFN-stimulated genes in response to miR-142 targeted IAV. (A) BMMs infected with NPctrl or NP142t IAV at an MOI of 5 and analyzed for IFN-β and IRF-7 mRNA at 24 hpi. P = 0.03 for IFN-β and P = 0.0039 for IRF-7. (B) Primary lung fibroblasts infected with NPctrl or NP142t IAV at an MOI of 2 analyzed for IFN-β and IRF-7 mRNA at 24 hpi by quantitative RT-PCR. Data from A and B are representative of three independent experiments with three to four samples per group. Mice were infected intranasally with NPctrl or NP142t IAV and whole lung analyzed on 48 and 96 hpi for IFN-β (C) and IRF-7 (D) mRNA expression. P = 0.0262 and P = 0.183 for IFN-β at 24 and 48 hpi, respectively. P = 0.06 and P = 0.0024 for IRF-7 at 24 and 48 h, respectively. Data are representative of two independent experiments with four mice per group. (E) BMMs from RIG-I+/+ or RIG-I−/− mice infected with NPctrl or NP142t IAV at an MOI of 5 and analyzed for IFN-β mRNA levels at 24 hpi. P = 0.0045. (F) Mouse embryonic fibroblasts from RIG-I+/+ or RIG-I−/− infected with NPctrl or NP142t IAV at an MOI of 1 and analyzed for IFNβ mRNA levels at 24 hpi by quantitative RT-PCR. Data from E and F are representative of two independent experiments with three to four samples per group.
To extend this data further, we next sought to determine the PRR responsible for the in vivo induction of the innate IFN-I response. Although hematopoietic cells express both TLRs and RLRs, the requirement for virus replication implicated intracellular PAMP detection mediated through RIG-I. To test this hypothesis, we infected primary BMMs and fibroblasts derived from RIG-I−/− or RIG-I+/+ littermate controls with either NPctrl or NP142t. As previously observed, NP142t-mediated IFN-β induction was inversely proportional to miR-142 levels, suggesting that replication was required for the induction of IFN-I (Fig. 4 E and F). In addition, these data demonstrated a complete loss of IFN-β induction in the absences of RIG-I, regardless of the virus model used (Fig. 4 E and F). Importantly, lack of IFN-β induction was not a result of an inherent defect in the ability to produce IFN-I, as transfection of dsRNA, an MDA5 substrate, resulted in high levels of IFN-β message (Fig. S5). Taken together, these data suggest the predominant source of IFN-I during an in vivo infection derives from a RIG-I–dependent, hematopoietic cellular source.
Discussion
Understanding the molecular basis for virus pathogenesis consistently relies on manipulation of the host. Although these model systems have provided an invaluable understanding of many immunological processes, the systems are limited by their reliance on loss-of-function data. In an effort to improve upon these methodologies, we chose to manipulate the pathogen rather than the host by exploiting the cell-specific expression of a miRNA. Here we use this technology to address fundamental questions concerning the cellular contribution of cytokine production and antigen presentation during IAV infection.
As IAV has been demonstrated to infect both lung epithelial cells and APCs (1–6, 20–22), the in vivo contributions of these two cellular compartments in cytokine signaling and antigen presentation have remained unclear. Although APCs are known to be critical for both of these processes, virus detection and antigen acquisition can occur through cross-talk between infected epithelial cells or as a result of direct infection. To discern between these two possibilities, we used an endogenous hematopoietic-specific miRNA, miR-142, to restrict IAV tropism in immune cells while not altering fitness in airway epithelial cells. Using this molecular tool, we demonstrate that although IAV replication within immune cells is dispensable for the generation of adaptive immune responses, it is required for optimal virus sensing and the induction of IFN-I and the antiviral state.
Cross-presentation of virus antigens likely evolved as a strategy to prime CD8 T cells from pathogens that replicate poorly in APCs, or to generate CD8-mediated antitumor responses (43, 44). Given that IAV can replicate in APCs, we sought to determine the primary route of antigen presentation to CD8 T cells in vivo. Interestingly, utilization of an IAV strain, incapable of APC replication, and therefore direct antigen presentation, demonstrated no defect in the generation of IAV-specific CD8 T cells or in virus clearance. These data suggest that in vivo, presentation of exogenous antigen through cross-priming is sufficient to generate protective cytotoxic T lymphocyte (CTL) responses. Furthermore, these findings are consistent with previous results demonstrating that, in the absence of cross-presentation, virus-specific CD8 T cells are impaired (45, 46). Additionally it has been demonstrated CD8 T cells generated through cross-presentation are functionally equivalent to those primed by direct presentation (47). Taking these data together, this work explains the lack of phenotype with regards to NP142t pathogenesis and suggests that the overwhelming development of the adaptive response is not mediated by direct presentation.
In contrast to a lack of adaptive immune defects, loss of IAV replication in APCs did result in aberrant innate signaling. The cell’s autonomous response to virus infection is initiated by PRRs, as a result of either intracellular or extracellular PAMP detection, mediated by RIG-I/Mda5 or the TLRs, respectively (48). Although epithelial cells primarily signal as a result of intracellular detection, APCs—with their ability to phagocytose and high basal expression of TLRs—can produce cytokines through either pathway (49). In agreement with past studies demonstrating that myeloid differentiation primary response gene 88 signaling is not required to control early virus infection (50, 51), our results indicate that virus replication and activation of RIG-I is the primary source of virus-induced signaling regardless of the cellular source. It is perplexing that inhibition of virus replication within immune cells leads to such drastic defects in overall IFN-I production, given that IAV replication predominates in lung epithelia, where virus fitness is not altered. This finding may reflect a bias in the ability of IAV to subvert the innate sensing and production of IFN-I through NS1 at the site of replication (52). In this regard, the elevated expression of RLRs and antiviral signaling components in APCs may better equip these cells for the recognition of invading pathogens and provide the necessary redundancy to ensure induction of IFN-I (53, 54).
Materials and Methods
Virus Design, Rescue, and Quantification.
Further information regarding cloning, rescue, and amplification of viruses can be found in SI Materials and Methods.
Small RNA Northern Blot Analysis.
Small RNA Northern blots and probe labeling were performed as previously described (36). Probes used include: anti-svRNA 5′-AAAAANNNCCTTGTTTCTACT-3′, anti–miR-142: 5′- TCCATAAAGTAGGAAACACTACA-3′, and anti-U6: 5′-GCCATGCTAATCTTCTCTGTATC -3′.
Protein and RNA Analysis.
Protein was analyzed by standard Western blot and RNA assessed by quantitative PCR. Further details regarding antibodies, reagents primers and conditions used can be found in SI Materials and Methods.
Mice and Virus Infection.
BALB/c and C57BL/6 mice were purchased from Taconic. RIG-I+/+ and RIG-I−/− mice were kind gifts from Michael Gale, Jr. (University of Washington, Seattle, WA). Mice were anesthetized with isofluorane and infected intranasally with 40 pfu, unless otherwise indicated. All experiments involving animals were performed in accordance with the Mount Sinai School of Medicine Institution of Animal Care and Use Committee.
MHC Class I Tetramers.
IAV tetramers NP366 (H-2Db/ASNENMETM) and PA224 (H-2Db/SSLENFRAYV) were obtained from National Institute of Allergy and Infectious Disease MHC Tetramer Core Facility (Atlanta, GA).
Flow Cytometry and Antigen Presentation Assay.
Single-cell suspensions from the lungs were stained with monoclonal antibodies or tetramers, fixed and run on a BD FACS Calibur, and analyzed using FlowJo software (TreeStar). Further information regarding antibody clones, peptide restimulation, and staining conditions, as well as antigen presentation assays, can be found in SI Materials and Methods.
Generation of BMMs, Primary Lung Fibroblasts, and MDCK142 Cells.
Details regarding the culture of BMMs, primary lung fibroblasts, and generation of miR-142 expressing cell line can be found in SI Materials and Methods.
Statistical Analysis.
Statistical analysis between datasets was performed using a one-tailed Student’s t test. Differences were considered to be statistically significant at P values at or below 0.05.
Supplementary Material
Acknowledgments
We thank Drs. Michael Gale, Jr., David Woodland, and Peter Palese for providing us with reagents. This work is supported in part by National Institutes of Health Grants A1080624, A1093571, U19AI083025, and U01AI095611 from the National Institute of Allergy and Infectious Diseases (NIAID) Mucosal Immunity Study Team Program; the Center for Research on Influenza Pathogenesis, a NIAID-funded Centers of Excellence for Influenza Research and Surveillance Center for Research on Influenza Pathogenesis Contract HHSN266200700010C; New York University-Mount Sinai School of Medicine Mechanisms of Virus–Host Interactions National Institutes of Health T32 Training Grant AI007647-09 (to R.A.L.); and the Pew Charitable Trust and the Burroughs Wellcome Fund (B.R.t.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206039109/-/DCSupplemental.
References
- 1.Manicassamy B, et al. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci USA. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VanoOsten Anderson R, McGill J, Legge KL. Quantification of the frequency and multiplicity of infection of respiratory- and lymph node-resident dendritic cells during influenza virus infection. PLoS ONE. 2010;5:e12902. doi: 10.1371/journal.pone.0012902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao X, Kim TS, Braciale TJ. Differential response of respiratory dendritic cell subsets to influenza virus infection. J Virol. 2008;82:4908–4919. doi: 10.1128/JVI.02367-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hargadon KM, et al. MHC class II expression and hemagglutinin subtype influence the infectivity of type A influenza virus for respiratory dendritic cells. J Virol. 2011;85:11955–11963. doi: 10.1128/JVI.05830-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moltedo B, Li W, Yount JS, Moran TM. Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 2011;7:e1002345. doi: 10.1371/journal.ppat.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smed-Sörensen A, et al. Influenza A virus infection of human primary dendritic cells impairs their ability to cross-present antigen to CD8 T cells. PLoS Pathog. 2012;8:e1002572. doi: 10.1371/journal.ppat.1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 9.tenOever BR, et al. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 10.Maniatis T, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 11.Donelan NR, Basler CF, García-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Sesma A, et al. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGill J, Heusel JW, Legge KL. Innate immune control and regulation of influenza virus infections. J Leukoc Biol. 2009;86:803–812. doi: 10.1189/jlb.0509368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballesteros-Tato A, León B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat Immunol. 2010;11:216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belz GT, Bedoui S, Kupresanin F, Carbone FR, Heath WR. Minimal activation of memory CD8+ T cell by tissue-derived dendritic cells favors the stimulation of naive CD8+ T cells. Nat Immunol. 2007;8:1060–1066. doi: 10.1038/ni1505. [DOI] [PubMed] [Google Scholar]
- 16.GeurtsvanKessel CH, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TS, Braciale TJ. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS ONE. 2009;4:e4204. doi: 10.1371/journal.pone.0004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belz GT, et al. Distinct migrating and nonmigrating dendritic cell populations are involved in MHC class I-restricted antigen presentation after lung infection with virus. Proc Natl Acad Sci USA. 2004;101:8670–8675. doi: 10.1073/pnas.0402644101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakim LM, Bevan MJ. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature. 2011;471:629–632. doi: 10.1038/nature09863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton-Easton A, Eichelberger M. Virus-specific antigen presentation by different subsets of cells from lung and mediastinal lymph node tissues of influenza virus-infected mice. J Virol. 1995;69:6359–6366. doi: 10.1128/jvi.69.10.6359-6366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert ML, Sauter B, Bhardwaj N. Dendritic cells acquire antigen from apoptotic cells and induce class I-restricted CTLs. Nature. 1998;392:86–89. doi: 10.1038/32183. [DOI] [PubMed] [Google Scholar]
- 22.Sauter B, et al. Consequences of cell death: Exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J Exp Med. 2000;191:423–434. doi: 10.1084/jem.191.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedoui S, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10:488–495. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 24.Schnorrer P, et al. The dominant role of CD8+ dendritic cells in cross-presentation is not dictated by antigen capture. Proc Natl Acad Sci USA. 2006;103:10729–10734. doi: 10.1073/pnas.0601956103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez JT, et al. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 29.Pham AM, Langlois RA, tenOever BR. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012;8:e1002465. doi: 10.1371/journal.ppat.1002465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 31.Kelly EJ, Hadac EM, Cullen BR, Russell SJ. MicroRNA antagonism of the picornaviral life cycle: Alternative mechanisms of interference. PLoS Pathog. 2010;6:e1000820. doi: 10.1371/journal.ppat.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J Virol. 2010;84:1550–1562. doi: 10.1128/JVI.01788-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heiss BL, Maximova OA, Pletnev AG. Insertion of microRNA targets into the flavivirus genome alters its highly neurovirulent phenotype. J Virol. 2011;85:1464–1472. doi: 10.1128/JVI.02091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heiss BL, Maximova OA, Thach DC, Speicher JM, Pletnev AG. MicroRNA targeting of neurotropic flavivirus: Effective control of virus escape and reversion to neurovirulent phenotype. J Virol. 2012;86:5647–5659. doi: 10.1128/JVI.07125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. MBio. 2010;1 doi: 10.1128/mBio.00204-10. e00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez JT, et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci USA. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legge KL, Braciale TJ. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18:265–277. doi: 10.1016/s1074-7613(03)00023-2. [DOI] [PubMed] [Google Scholar]
- 38.Yewdell JW, Dolan BP. Immunology: Cross-dressers turn on T cells. Nature. 2011;471:581–582. doi: 10.1038/471581a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topham DJ, Tripp RA, Doherty PC. CD8+ T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol. 1997;159:5197–5200. [PubMed] [Google Scholar]
- 40.Brincks EL, Katewa A, Kucaba TA, Griffith TS, Legge KL. CD8 T cells utilize TRAIL to control influenza virus infection. J Immunol. 2008;181:4918–4925. doi: 10.4049/jimmunol.181.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronni T, Sareneva T, Pirhonen J, Julkunen I. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol. 1995;154:2764–2774. [PubMed] [Google Scholar]
- 42.Bender A, et al. The distinctive features of influenza virus infection of dendritic cells. Immunobiology. 1998;198:552–567. doi: 10.1016/S0171-2985(98)80078-8. [DOI] [PubMed] [Google Scholar]
- 43.Kurts C, Robinson BW, Knolle PA. Cross-priming in health and disease. Nat Rev Immunol. 2010;10:403–414. doi: 10.1038/nri2780. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Aymeric L, Locher C, Kroemer G, Zitvogel L. The dendritic cell-tumor cross-talk in cancer. Curr Opin Immunol. 2011;23:146–152. doi: 10.1016/j.coi.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Wilson NS, et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat Immunol. 2006;7:165–172. doi: 10.1038/ni1300. [DOI] [PubMed] [Google Scholar]
- 46.Singh R, Cresswell P. Defective cross-presentation of viral antigens in GILT-free mice. Science. 2010;328:1394–1398. doi: 10.1126/science.1189176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W, et al. Cross-priming of CD8+ T cells by viral and tumor antigens is a robust phenomenon. Eur J Immunol. 2004;34:194–199. doi: 10.1002/eji.200324257. [DOI] [PubMed] [Google Scholar]
- 48.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 49.Kohlmeier JE, Woodland DL. Immunity to respiratory viruses. Annu Rev Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 50.Seo SU, et al. MyD88 signaling is indispensable for primary influenza A virus infection but dispensable for secondary infection. J Virol. 2010;84:12713–12722. doi: 10.1128/JVI.01675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koyama S, et al. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179:4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 52.Geiss GK, et al. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc Natl Acad Sci USA. 2002;99:10736–10741. doi: 10.1073/pnas.112338099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luber CA, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32:279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Schmid S, Mordstein M, Kochs G, García-Sastre A, tenOever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.