Abstract
Tyrosine phosphorylation is a hallmark for activation of STAT proteins, but their transcriptional activity also depends on other secondary modifications. Type I IFNs can activate both the ISGF3 (STAT1:STAT2:IRF9) complex and STAT3, but with cell-specific, selective triggering of only the ISGF3 transcriptional program. Following a genome-wide RNAi screen, we identified the SIN3 transcription regulator homolog A (Sin3a) as an important mediator of this STAT3-targeted transcriptional repression. Sin3a directly interacts with STAT3 and promotes its deacetylation. SIN3A silencing results in a prolonged nuclear retention of activated STAT3 and enhances its recruitment to the SOCS3 promoter, concomitant with histone hyperacetylation and enhanced STAT3-dependent transcription. Conversely, Sin3a is required for ISGF3-dependent gene transcription and for an efficient IFN-mediated antiviral protection against influenza A and hepatitis C viruses. The Sin3a complex therefore acts as a context-dependent ISGF3/STAT3 transcriptional switch.
STAT3 was originally identified as an IL-6–activated transcription factor in hepatocytes (1–4) and later reported to be activated by many other stimuli, including cytokines [e.g., leukemia inhibitory factor (LIF), IL-10, IFNs], growth factors (e.g., EGF), and hormones (e.g., insulin). Activated STAT3 stimulates the transcription of several genes involved in cell-cycle progression and the antiapoptotic program (5). Therefore, and for its ability to transform normal fibroblast cells and cause tumors in nude mice, STAT3 has been classified as an oncogene (6). The regulation of STAT3 transcriptional activity strongly depends on its posttranslational modification status. The functional role of phosphorylation on hallmark tyrosine and serine residues is by far best understood (7) and correlates in most cases with functional and transcriptionally active STAT3. Beside phosphorylation, STAT3 activity is tightly regulated by several other posttranslational modifications, including lysine methylation (8, 9) and acetylation (10–12). Although STAT3 methylation negatively regulates its activity, lysine acetylation is associated with a positive regulation of STAT3 activity in general, although its precise impact depends on the acetylated residues. STAT3 acetylation is efficiently reverted by histone deacetylases (HDAC)1, HDAC2, and HDAC3, which associate with STAT3 and contribute to its negative regulation (10).
Type I IFNs induce antiviral and antiproliferative responses through the activation of the ISGF3 (STAT1:STAT2:IRF9) transcriptional complex (13). IFN stimulation also leads to STAT3 phosphorylation (14), which is remarkable, given the opposed roles of ISGF3 and STAT3 in regulating cell survival and proliferation. We have previously shown that, in a cell-specific manner, IFN stimulation can induce STAT3 phosphorylation and DNA binding without triggering transcription (15). HDAC1/2 are responsible for this transcriptional repression, as interfering with their expression or activity restored the transcription of STAT3-target genes (15). On the other hand, HDAC activity is required for transcriptional activation of ISGF3-responsive genes and IFN-induced antiviral immunity (16–18).
Here, we report that the SIN3 transcription regulator homolog A (Sin3a) complex represses STAT3 activity by modifying its acetylation status. Sin3a is instead required for IFN-stimulated gene (ISGs) transcription and an efficient antiviral response. Our results unveil a critical role for the Sin3a complex in balancing STAT functions at the transcriptional level.
Results
Genome-Wide RNAi Screen Identifies the Sin3a Complex as a Repressor of STAT3 Transcriptional Activity.
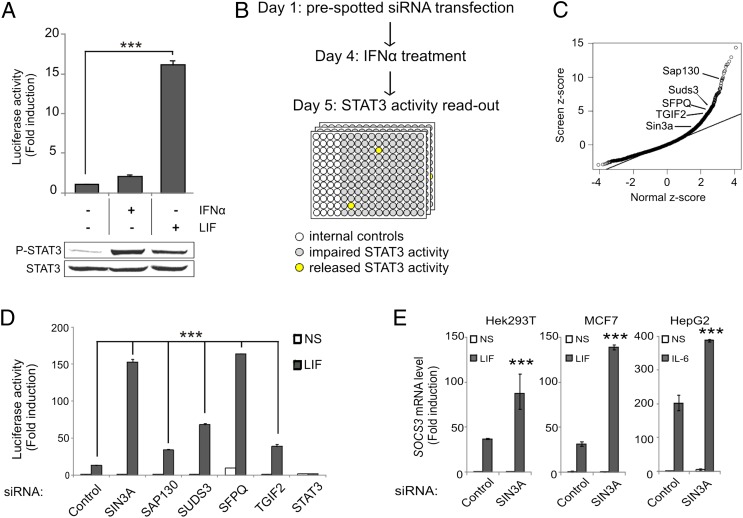
Although type I IFN treatment stimulates STAT3 tyrosine phosphorylation and its binding to STAT3-responsive promoters, the subsequent transcription of canonical STAT3-responsive genes is impaired in certain cell types (15). In line herewith, IFN-α2 stimulation of Hek293T cells failed to activate the transcription of the STAT3-responsive rat pancreatitis-associated protein 1 (rPAP1)-luciferase reporter (Fig. 1A, Upper graph). In contrast, LIF stimulation strongly activated the reporter. Because both cytokines support STAT3 phosphorylation (Fig. 1A, Lower blot), a different regulatory mechanism must account for the impaired STAT3 activity. We performed a genome-wide RNAi screen aimed at identifying putative STAT3 repressors (Fig. 1B). In this setting, the silencing of a candidate repressor would release the STAT3 transcriptional brake and induce rPAP1-luciferase activation. The normalized reporter activation corresponding to each siRNA was expressed by z-scoring (cellHTS2) (19). Strikingly, among the top candidate repressors (Dataset S1), we identified four components of the Sin3a transcriptional repressor complex: SAP130 (z-score: 9.5), SUDS3 (z-score: 6.3), SFPQ (z-score: 5.2), and TGIF2 (z-score: 4.25) (Fig. 1C). SIN3A silencing itself also enhanced the reporter activation. Independent reporter-based experiments confirmed that the Sin3a complex is involved in repressing IFN-α2–activated STAT3 (Fig. S1). Of note, silencing of the Sin3a complex components identified in the screen also led to a robust increase of the LIF-induced rPAP1-luciferase reporter activation (Fig. 1D), as well as the transcription of the endogenous STAT3-responsive SOCS3 gene in both LIF-induced Hek293T and MCF7 cells, and IL-6–stimulated HepG2 cells (Fig. 1E). These results unveil a unique role for the Sin3a complex as a negative regulator of STAT3 transcriptional activity, regardless of the activating stimulus.
Fig. 1.
A genome-wide RNAi screen identifies the Sin3a complex as negative regulator of STAT3 transcriptional activity. (A) Hek293T cells were transiently transfected with the rPAP1-luciferase reporter. After 24 h, cells were left nonstimulated (NS) or stimulated with LIF or IFN-α2 for 24 h for the luciferase assay or 30 min for phospho-STAT3 detection. Luciferase readout is expressed as a ratio between stimulated and unstimulated values. ***P < 0.001; t test. Total cell extracts were blotted with anti-phospho STAT3 (Tyr705) and anti-STAT3 antibody. (B) Genome-wide RNAi screen strategy. Hek293 cells stably expressing the rPAP1-luciferase reporter were used. siRNAs targeting potential STAT3 repressors will release STAT3 transcriptional brake and activate the rPAP1-luciferase reporter (wells in yellow), but STAT3 activity will be impaired in wells containing unrelated siRNAs (in gray). Wells colored in white were used for the internal controls. The detailed screening procedure is described in the SI Materials and Methods. (C) Most siRNA phenotypes behave as normally distributed data (line). The tail deviating from normal distribution represents siRNA with significant phenotypes (repressor candidate genes). The Sin3a complex components identified in the screen are highlighted. (D) Hek293T cells were transiently transfected with the indicated siRNA and with the rPAP1-luciferase reporter after 24 h. Renilla luciferase siRNA was used as control. After 48 h, cells were left nonstimulated or stimulated with LIF. Luciferase readout is expressed as a ratio over the control-silenced NS condition. ***P < 0.001; one-way ANOVA with Bonferroni test. (E) Hek293T, MCF7, and HepG2 cells were transfected with control (Renilla luciferase) or a SIN3A siRNA. After 72 h, cells were cultured 4 h without FCS, then left nonstimulated or stimulated with LIF or IL-6 for 1 h. Graphs represent mRNA levels relative to NS sample. ***P < 0.001; one-way ANOVA with Bonferroni test. Results are representative of three independent experiments. Error bars indicate SD from triplicates
Sin3a Directly Interacts with STAT3 and Promotes Its Deacetylation.
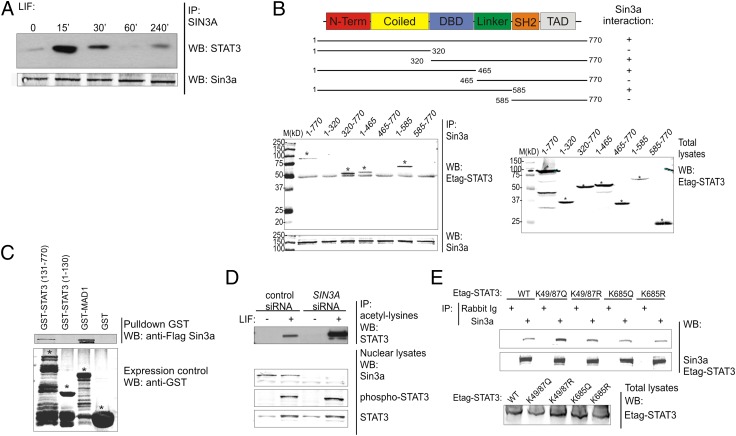
Sin3a associates with several transcription factors promoting their deacetylation and modulating their activity (20). Hence, we asked whether the modulation of STAT3 activity by Sin3a involves a stimulus-dependent physical interaction. Coimmunoprecipitation analysis in Hek293T cells demonstrated that endogenous STAT3 interacts with endogenous Sin3a in a stimulus-dependent manner (Fig. 2A). To identify the functional domain of STAT3 involved in Sin3a binding, Etag-STAT3 deletion mutants were generated and overexpressed in Hek293T along with GFP-Sin3a. Only the constructs containing the STAT3 DNA-binding domain (DBD) coprecipitated with GFP-Sin3a, revealing the importance of the STAT3-DBD for this interaction (Fig. 2B). To verify if Sin3a directly interacts with STAT3, a GST pull-down assay was performed. In vitro produced Flag-Sin3a directly interacted with the GST-tagged C-terminal part of STAT3 (amino acids 131–770) containing the DBD (Fig. 2C), in concordance with the previous experiment. Next, we asked whether Sin3a modified the posttranslational profile of STAT3. Because posttranslational modifications alter the protein isoelectric point (pI), we compared the pI distribution of STAT3 isoforms in cells transfected with control or SIN3A-specific siRNA, using the NanoPro technology (21, 22). SIN3A silencing resulted in a clear pI-shift of STAT3 isoforms toward more acidic values, suggesting increased STAT3 acetylation (Fig. S2A). Confirming this hypothesis, we observed a marked STAT3 hyperacetylation in LIF-stimulated SIN3A-silenced cells, but the phosphorylation status was not significantly altered (Fig. 2D). It has been previously reported that HDACs bind to and negatively regulate acetylated STAT3 (10, 23), so we asked whether the Sin3a-STAT3 interaction could be regulated by STAT3 acetylation. We found that Sin3a interacted more strongly with the acetyl-mimicking STAT3K49/87Q mutant compared with STAT3 WT or the other STAT3 variants (Fig. 2E), indicating that binding of Sin3a preferentially occurs with N-terminally acetylated STAT3. Further analysis on single N-terminal lysine STAT3 mutants identified K87 acetylation as the main regulator of the STAT3-Sin3a interaction (Fig. S2B).
Fig. 2.
Sin3a interacts with STAT3 and modifies its acetylation pattern. (A) Hek293T cells were cultured 4 h without FCS and then stimulated with LIF for the indicated time points. Nuclear lysates were prepared, endogenous Sin3a was immunoprecipitated, and coprecipitated endogenous STAT3 was revealed with an anti-STAT3 antibody. Precipitated Sin3a was revealed using an anti-Sin3a antibody. (B) Schematic representation of Etag-STAT3 truncated mutants. DBD, DNA binding domain; SH2, Src Homology 2; TAD, transactivation domain. Hek293T cells were transfected with plasmids coding for GFP-Sin3a and Etag-STAT3 truncated mutants. Sin3a was immunoprecipitated and coprecipitated Etag-STAT3 mutants were revealed with anti-Etag antibody. Total lysates were blotted as loading control. Stars indicate the specific bands. (C) Escherichia coli BL21DE3 cells were transformed with plasmids coding for GST-STAT3 N-terminal (1-130), C-terminal (131-770), GST-MAD1 (positive control), or GST alone (negative control). Cell lysates were incubated with the in vitro transcribed and translated Flag-Sin3a protein. Complexes were precipitated using glutathione-agarose beads and revealed with anti-flag antibody. GST-transformed bacterial lysates were blotted with anti-GST antibody as control of protein production and solubility. Stars indicate the specific bands. (D) Hek293T cells were transfected with control (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were cultured 4 h without FCS and then stimulated with LIF for 30 min. Nuclear extracts were prepared and acetylated lysines were immunoprecipitated. Coprecipitated STAT3 was revealed with anti-STAT3 antibody. Endogenous Sin3a and STAT3 levels in the nuclear lysates were detected using specific antibodies. (E) Hek293T cells were transfected with plasmids coding for GFP-Sin3a and Etag-STAT3 WT, acetyl-mimicking (K49/87Q and K685Q), or acetyl-deficient (K49/87R and K685R) mutants. Sin3a was immunoprecipitated and coprecipitated Etag-STAT3 mutants were revealed with an anti-Etag antibody. Total lysates were blotted as loading control. Results are representative of three independent experiments.
Sin3a Complex Regulates a Subset of STAT3-Responsive Genes.
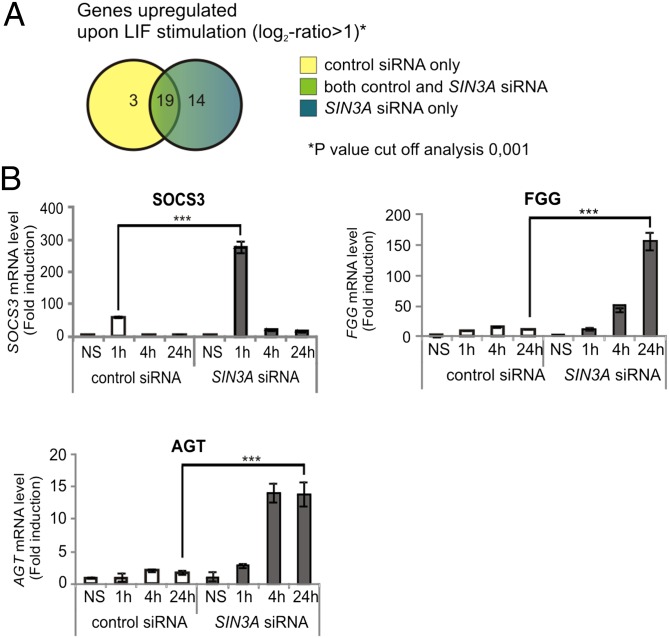
Sin3a is a conserved multifunctional repressor protein complex that regulates gene transcription (20). To gain more insight into the STAT3-responsive genes that are regulated by the Sin3a complex, we performed an Affymetrix microarray-based screen on LIF-treated MCF7 cells that had been transfected with a control siRNA or with SIN3A siRNA. Knockdown of SIN3A and LIF-induced STAT3 phosphorylation were confirmed during sample preparation (Fig. S3). As represented in Fig. 3A, 14 LIF-responsive genes were up-regulated in the near absence of Sin3a. Results were confirmed by independent quantitative RT-PCR experiments, highlighting that the Sin3a complex negatively regulates the transcription of a subset of STAT3-responsive genes. The expression profile of three representative Sin3a-regulated LIF-induced genes (SOCS3, FGG, and AGT) is shown in Fig. 3B. Of note, both early and late genes were subjected to regulation by Sin3a. The expression profile of the remaining LIF-induced Sin3a-regulated genes is summarized in Table 1.
Fig. 3.
The Sin3a complex negatively regulates the transcription of a subset of STAT3-dependent genes. (A) Venn diagram showing the number of LIF-induced genes in cell transfected with control siRNA (Renilla luciferase, yellow), with SIN3A siRNA (blue), or in both the conditions (green). As cutoff, a log2-ratio > 1 (more than two times up-regulated) was applied to analyze the gene expression profile. (B) MCF7 cells were transfected with control (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were left nonstimulated or stimulated with LIF. Graphs represent mRNA levels relative to NS sample. Results are representative of three independent experiments. Error bars indicate SD from triplicates. ***P < 0.001; one-way ANOVA with Bonferroni test.
Table 1.
Gene-expression analysis identifies several Sin3a-repressed STAT3-dependent genes
| RL siRNA |
SIN3A siRNA |
||||||||||||
| 1 h LIF |
24 h LIF |
1 h LIF |
24 h LIF |
||||||||||
| Gene symbol | Gene ID | Fold-induction | SD+ | SD− | Fold-induction | SD+ | SD− | Fold-induction | SD+ | SD− | Fold-induction | SD+ | SD− |
| SOCS3 | 9021 | 38.19 | 0.5 | 0.5 | 7.14 | 1.4 | 1.2 | 70.77** | 11.7 | 10.0 | 6.99 | 0.8 | 0.7 |
| FGG | 2266 | 8.03 | 0.4 | 0.4 | 11.35 | 0.4 | 0.4 | 12.95 | 1.8 | 1.6 | 155.42*** | 14.7 | 13.4 |
| AGT | 183 | 1.01 | 0.7 | 0.4 | 1.74 | 0.4 | 0.3 | 2.90 | 0.3 | 0.3 | 13.69*** | 1.9 | 1.7 |
| GADD45G | 10912 | 2.56 | 0.2 | 0.2 | 1.12 | 0.8 | 0.5 | 12.25*** | 1.7 | 1.5 | 7.19*** | 0.6 | 0.5 |
| CEBPD | 1052 | 3.59 | 0.2 | 0.2 | 2.12 | 0.1 | 0.1 | 5.92** | 0.4 | 0.4 | 3.33 | 0.8 | 0.6 |
| KLF10 | 7071 | 6.21 | 1.3 | 1.1 | 1.61 | 0.4 | 0.3 | 6.34 | 1.3 | 1.1 | 4.04 | 0.3 | 0.3 |
| CD14 | 929 | 1.95 | 0.2 | 0.1 | 0.64 | 0 | 0 | 8.28*** | 0.2 | 0.6 | 3.26*** | 0.7 | 0.6 |
| SOX9 | 6662 | 2.36 | 0.2 | 0.1 | 1.13 | 0.1 | 0.1 | 5.50*** | 0.5 | 0.5 | 4.79*** | 0.4 | 0.3 |
| RND1 | 27289 | 2.75 | 0.7 | 0.6 | 0.82 | 0.1 | 0.1 | 5.13** | 0.5 | 0.5 | 0.96 | 0.1 | 0.1 |
| STAT3 | 6774 | 0.76 | 0 | 0 | 2.38 | 0 | 0 | 1.50*** | 0 | 0 | 5.12*** | 0.3 | 03 |
| ZFP36 | 7538 | 8.06 | 0.3 | 0.3 | 1.56 | 0.1 | 0.1 | 8.37 | 0.7 | 0.6 | 3.283* | 0.9 | 0.7 |
MCF7 cells were transfected with control (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were left nonstimulated or stimulated with LIF. Fold-inductions represent mRNA levels relative to the NS sample. Results are representative of three independent experiments. SD from triplicates. ***P < 0.001, **P < 0.005, *P < 0.01; one-way ANOVA with Bonferroni test and are highlighted in bold.
Sin3a Complex Regulates STAT3 Nucleocytoplasmic Distribution and Its Recruitment at the SOCS3 Promoter.
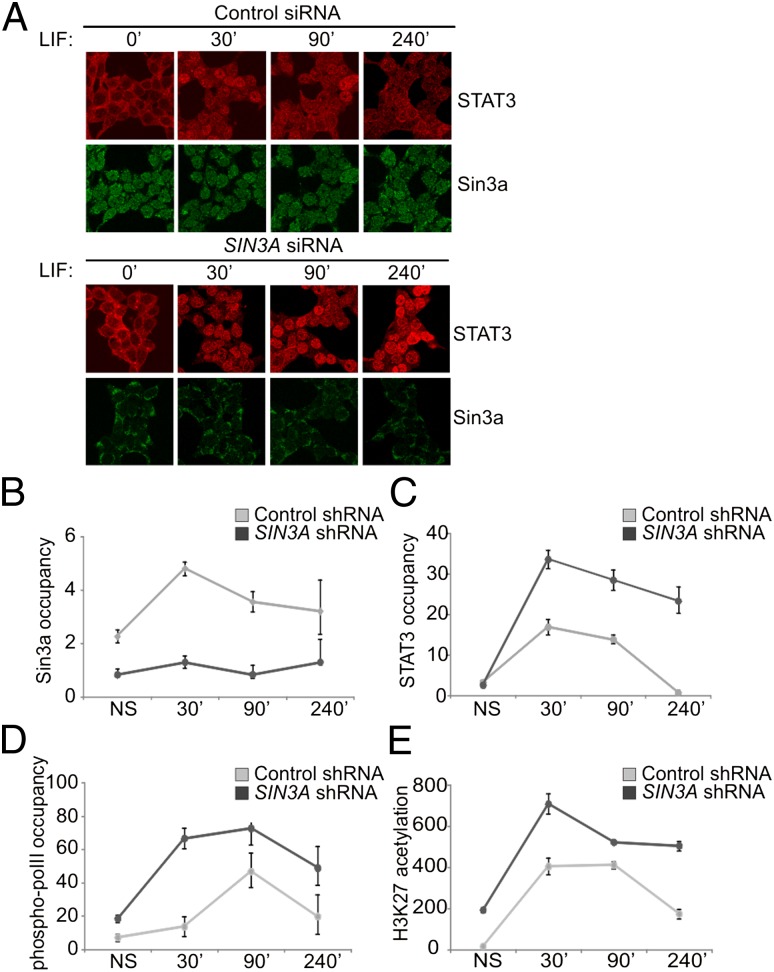
STAT3 activity depends on its nuclear translocation and DNA binding. Interestingly, we found that the LIF-induced nuclear residence time of STAT3 was prolonged in cells transfected with SIN3A-targeting siRNA (Fig. 4A). Thus, we examined whether Sin3a could influence STAT3 binding to responsive promoters. The SOCS3 promoter was used as a representative model for a STAT3-responsive region because its transcription was found to be significantly up-regulated in SIN3A knocked-down cells (Fig. 3B). ChIP assays were performed on Hek293T cells stably transfected with control or SIN3A shRNAmir (Fig. S4). We found that LIF treatment induced the recruitment of Sin3a at the SOCS3 promoter (Fig. 4B), although a basal amount of Sin3a was already present in absence of stimulation. LIF-induced STAT3 recruitment was significantly increased upon SIN3A knockdown (Fig. 4C). The basal levels of phospho-RNA pol II present at the promoter in the uninduced state increased upon LIF stimulation (Fig. 4D) and were further enhanced upon SIN3A silencing. Finally, we observed an enrichment of histone 3 K27 (H3K27) acetylation in SIN3A-silenced cells (Fig. 4E), indicating a more accessible chromatin status. In conclusion, we found that the Sin3a complex regulates SOCS3 transcription, modulating the recruitment of STAT3 and phospho-pol II at the responsive regions of its promoter and influencing the acetylation of associated histones. Silencing of Sin3a complex components leads to an enhanced promoter recruitment of STAT3 and phospho-RNA pol II, histone hyperacetylation, and concomitant enhanced transcription.
Fig. 4.
The Sin3a complex negatively regulates STAT3 nuclear distribution and binding to the SOCS3 gene promoter. (A) Hek293T cells were transfected with control (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were cultured 4 h without FCS and then stimulated with LIF for the indicated times. Cells were fixed and stained and the localization of STAT3 and Sin3a was assessed by confocal analysis. Immunofluorescence of representative cell fields is shown. (Magnification: 63×.) (B–E) Hek293T cells were stably transfected with a scrambled or a SIN3A-targeting shRNAmir. Transfection and silencing efficiency are reported in Fig. S3. Transfected cells were cultured 4 h without FCS and then stimulated with LIF for the indicated time points. ChIP assays were performed to examine the occupancy of Sin3a (B), STAT3 (C), phospho-pol II (D), and the acetylation status of histone 3 (H3) K27 acetylation (E) on the SOCS3 promoters. Immunoprecipitated DNA was quantified by quantitative PCR. Graphs represent occupancy levels relative to NS sample (fold-induction). Results are representative of three independent experiments. Error bars indicate SD from duplicates.
Sin3a Is Necessary for the Transcription of Several ISGs and for Maximal Type I IFN-Induced Resistance to Influenza A and Hepatitis C Viruses.
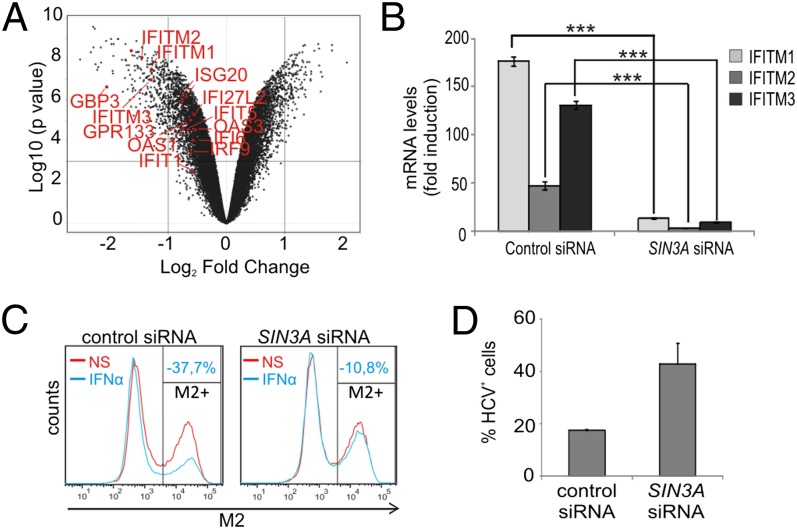
Although HDAC1 and HDAC2 negatively modulate STAT3 signaling (10, 12, 24), their activity is required for IFN-triggered ISGs transcription and antiviral response (16–18). Because the activity of the Sin3a complex depends on its association with HDAC1/2, we asked whether SIN3A silencing would affect the transcription of ISGs involved in the antiviral defense. Interestingly, the basal transcription of several ISGs was found to be down-regulated by SIN3A silencing in the microarray experiment (Fig. 5A). We therefore asked whether SIN3A silencing would also affect the IFN-induced ISG transcription. As summarized in Table 2, expression of all tested ISGs was strongly impaired in SIN3A-silenced cells. Similar results were obtained using a different and nonoverlapping SIN3A-specific siRNA (Fig. S5A). Of note, SIN3A silencing strongly impaired transcription of all three IFN-inducible transmembrane (IFITM) proteins (Fig. 5B). IFITM proteins play a key role in the antiviral defense against influenza A virus and hepatitis C virus (HCV) infection (25, 26). To verify if Sin3a is involved in the IFN-triggered antiviral resistance, control or SIN3A-silenced Hek293T cells were exposed to IFN-α2 before influenza A virus (A/PR8/34, H1N1 subtype) infection. Fig. 5C shows that IFN-α2 treatment potently reduced the infection rate in cells transfected with a control siRNA (−37.7%), but only weakly in SIN3A-silenced cells (−10.8%). SIN3A silencing also increased infectability in untreated and IFN-α2–treated MCF7 cells (Fig. S5B). The key role played by Sin3a in the IFN-induced antiviral response was also observed in HCV-infected Huh7.5 hepatoma cells (Fig. 5D). As for the influenza A virus infection, the antiviral protection triggered by IFN-α was reduced in SIN3A-silenced cells, resulting in an enhanced infection rate compared with control cells. In summary, our data show that the Sin3a complex is required for basal and IFN-α–dependent transcription of several ISGs and, as a consequence, for efficient IFN-α–induced protection against viral infection.
Fig. 5.
The Sin3a complex is required for IFN-stimulated ISGs transcription and efficient antiviral response against influenza A and hepatitis C viruses. (A) Volcano plot representing the differential gene expression profiles (expressed as log2 fold-change) of SIN3A-silenced versus control-silenced MCF7 cells. Down-regulated ISGF3-responsive genes are highlighted. (B) Hek293T cells were transfected with a control siRNA (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were stimulated with IFN-α2 for 24 h and the mRNA levels of the IFITMs were evaluated. Error bars indicate SD from triplicates. ***P < 0.001; one-way ANOVA with Bonferroni test. (C) Hek293T cells were transfected with a control siRNA (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were left nonstimulated or stimulated with IFN-α2 for 24 h before 14-h exposure to influenza A/PR8/34 virus. Infection efficiency was assessed by immunostaining of the viral protein M2 followed by flow cytometric analysis. (D) Huh7.5 cells were transfected with a control siRNA (Renilla luciferase) or SIN3A siRNA. After 72 h, cells were stimulated with IFN-α2 (1 ng/mL) for 24 h before 48-h exposure to HCV. The efficiency of the infection was evaluated by immunostaining of the viral protein NS5A. Results are representative of at least two independent experiments.
Table 2.
Down-regulated ISGF3-responsive genes in SIN3A-silenced Hek293T cells
| Control siRNA |
Sin3a siRNA |
||||||
| Gene symbol | Gene ID | Fold-induction | SD+ | SD− | Fold-induction | SD+ | SD− |
| ISG54 | 3433 | 2,574.36 | 40.21 | 39.59 | 303.38*** | 51.54 | 44.06 |
| IFIT1 | 3434 | 265.03 | 12.61 | 12.04 | 62.03*** | 1.27 | 1.24 |
| 2’5′OAS | 4938 | 188.71 | 8.72 | 8.34 | 34.42*** | 1.94 | 1.84 |
| IFITM1 | 8519 | 176.68 | 4.73 | 4.60 | 13.22*** | 0.99 | 0.92 |
| 6–16 | 2537 | 163.14 | 7.76 | 7.41 | 5.94*** | 0.39 | 0.37 |
| IFITM3 | 10410 | 130.69 | 4.17 | 4.04 | 9.32*** | 0.46 | 0.44 |
| IFITM2 | 10581 | 46.69 | 4.48 | 4.09 | 3.19*** | 0.21 | 0.20 |
| IFI27 | 3429 | 46.53 | 0.73 | 0.72 | 2.41*** | 0.44 | 0.37 |
| ISG15 | 9636 | 32.00 | 4.38 | 3.85 | 5.43*** | 0.25 | 0.24 |
| ISG20 | 3669 | 4.07 | 0.36 | 0.33 | 1.97* | 1.44 | 0.83 |
Hek293T cells were transfected with a control (Renilla luciferase) siRNA or SIN3A siRNA. Seventy-two hours after transfection, cells were stimulated with IFN-α2 for 24 h and ISGs transcription was evaluated. Fold-inductions represent mRNA levels relative to the NS sample and are highlighted in bold. ***P < 0.001, *P < 0.01; one-way ANOVA with Bonferroni test.
Discussion
STAT signaling is tightly regulated and imbalances between STAT1 and STAT3 activity can lead to pathologies including immune disorders, cardiomyopathies, and cancer (27). STAT1 and STAT3 reciprocally regulate each other’s expression and activities, thus contributing to maintain the specificity of cytokine signaling (28–30). HDAC1 and HDAC2 exert opposite effects on STAT1- and STAT3-dependent transcription: they negatively regulate STAT3 signaling (23), and they are required for ISGs activation (16–18). Interfering with the activity of those HDACs shifts the ISGF3-dependent IFN response toward a STAT3-dependent response (15).
Type I IFN-induced STAT3 phosphorylation was widely reported (14) yet, in some cell types, IFN-activated STAT3 fails to trigger the transcription of canonical STAT3-responsive genes (15). The transcriptional brake on STAT3 observed upon IFN stimulation allowed us to perform a genome-wide RNAi screen aimed at the identification of novel STAT3 repressors. The Sin3a complex was identified as a repressor of STAT3 activity independently from the STAT3-activating stimulus. Our study provides a rationale for the previous identification of STAT-specific cis-regulatory elements in a subset of Sin3a-regulated genes (31). Although interplay between STAT3 and Sin3a has been suggested for the c-MYC (32) and for glial fibrillary acidic protein (GFAP) (33) promoters, direct proof, and detailed insight into the precise modalities of this interaction have so far been lacking. We show that Sin3a directly interacts with STAT3, promoting its deacetylation, and nuclear exclusion. The stimulus-dependency of the Sin3a-STAT3 interaction and the preferential binding of Sin3a to the acetyl-mimicking STAT3K49/87Q mutant suggest that Sin3a docks to and inhibits N-terminal acetylated STAT3. Consistently, the transcription of the AGT gene, one of the strongest up-regulated genes identified in our microarray analysis in SIN3A-silenced MCF7 cells, was reported to be enhanced by STAT3 acetylation on K49 and K87 and repressed by HDAC1 overexpression (11).
ChIP assays revealed that Sin3a and STAT3 are recruited to the SOCS3 promoter within the same time-frame after LIF stimulation, indicating that these proteins may be recruited at the consensus sequences as a complex. In addition, SIN3A silencing correlates with enhanced recruitment of STAT3 and phospho-RNA pol II to the SOCS3 promoter, which is likely a direct consequence of the STAT3 hyperacetylation status (24). Although the H3K27 hyperacetylation observed in SIN3A-silenced cells may be a direct consequence of the reduced HDAC activity, it was previously described that N-terminal STAT3 acetylation leads to increased recruitment of p300 at the responsive sequences (24), which in turn trigger histone hyperacetylation. Interestingly, among the LIF-induced Sin3a-repressed genes we identified, three (FGG, AGT, and CD14) are known acute-phase response mediators, revealing a possible role for the Sin3a complex in the regulation of this process. However, SIN3A silencing had barely no effect on the induction of these APR genes in IL-6 or LIF-stimulated Huh7 hepatoma cells (Table S1), indicating that the regulation exerted by Sin3a on STAT3 transcriptional activity may depend on the cellular context. Cellular specificity in the Sin3a-dependent modulation of transcription was recently highlighted for the transcription factors Myc and E2F, the target genes of which are derepressed in SIN3A-deficient mouse embryonic fibroblast cells (31), but are repressed in SIN3A-deficient immature myeloid cells (34), compared with WT cells. Such specificity may depend on the complex network of associated transcriptional corepressors and coactivators, essential to guarantee a diverse and cell-type specific response to a unique stimulus.
Although most of the research on the Sin3a complex has focused on its role as a corepressor, increasing evidence suggests that Sin3a may also activate gene transcription (20). Here, we show that Sin3a is required for the transcription of several ISGF3-responsive genes and, in particular, for the basal and IFN-induced transcription of the IFITM proteins, which are crucial mediators of the IFN-induced antiviral response to several pathogens, including influenza A virus (25) and HCV (26). Consistent with these reports, we observed a reduced IFN-mediated protection against these viruses in SIN3A-silenced cells.
In conclusion, the research described here clearly implicates the Sin3a complex as a cell-specific repressor of STAT3 activity and a crucial player in the innate immune defense against viral infections. Exerting opposite regulation on STAT3 and ISGF3-dependent transcription, Sin3a represents a unique control level determining cytokine-specific responses in different tissues or cells.
Materials and Methods
Detailed protocols are provided in SI Materials and Methods. Hek293T, MCF7, and HepG2 cells were purchased from ATCC, HCV-permissive HuH7.5-RFP-NLS-IPS cells were kindly provided by Charles M. Rice (The Rockefeller University, New York, NY). The list of primer pairs used in this work is in Table S2. The Dharmacon siGenome SMARTpool library was used for the RNAi screen. Microarray datasets generated in this study are available in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/), accession no. GSE35696.
Supplementary Material
Acknowledgments
We thank Prof. Savvas Savvides for his advices on GST-STAT3 constructs expression and Pieter Bogaert for expert assistance with flow cytometry (Methusalem Grant BOF09/01M00709, Ghent University); Angelika Bongers for her help during the RNAi screen; Dominiek Catteeuw for his help in the cloning procedures and Sven Degroeve for his support with the microarray data analysis interpretation; and Dr. Andrea Tu and Dr. David Voehringer from ProteinSimple for analyzing the samples using the NanoPro technology. The anti-NS5A monoclonal antibody 9E10 is a gift from Charles M. Rice, The Rockefeller University, New York; the hepatitis C virus JC1 clone was provided by T. Pietschmann, TWINCORE, 730625 Hannover. Work in the M.B. laboratory was supported by the Helmholtz Alliance for Systems Biology and the German Research Foundation. This work was supported by grants from Ghent University (Concerted Action Grant 01G01712), the Belgian government (Interuniversity Attraction Poles Projects P6/28 and P6/36), the Research Foundation-Flanders (Project G.0521.12N), the ReceptEur FP6 Marie-Curie program, and the Group-ID Multidisciplinary Research Partnerships of Ghent University. K.D.B. is a postdoctoral researcher at the Fonds Wetenschappelijk Onderzoek–Vlaanderen; and J.V. is a predoctoral student at the Fonds Wetenschappelijk Onderzoek-Vlaanderen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.H. is a guest editor invited by the Editorial Board.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35696).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206458109/-/DCSupplemental.
References
- 1.Akira S, et al. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 2.Raz R, Durbin JE, Levy DE. Acute phase response factor and additional members of the interferon-stimulated gene factor 3 family integrate diverse signals from cytokines, interferons, and growth factors. J Biol Chem. 1994;269:24391–24395. [PubMed] [Google Scholar]
- 3.Zhong Z, Wen Z, Darnell JE., Jr Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 4.Lütticken C, et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–1142. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Wen Z, Zhong Z, Darnell JE., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark GR, Wang Y, Lu T. Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res. 2011;21:375–380. doi: 10.1038/cr.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 11.Ray S, Boldogh I, Brasier AR. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology. 2005;129:1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Cherukuri P, Luo J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem. 2005;280:11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 13.Schindler C, Levy DE, Decker T. JAK-STAT signaling: From interferons to cytokines. J Biol Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 14.van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Icardi L, et al. Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2. FASEB J. 2012;26:240–249. doi: 10.1096/fj.11-191122. [DOI] [PubMed] [Google Scholar]
- 16.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HM, et al. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J Biol Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 19.Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silverstein RA, Ekwall K. Sin3: A flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 21.O’Neill RA, et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc Natl Acad Sci USA. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan AC, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray S, Lee C, Hou T, Boldogh I, Brasier AR. Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids Res. 2008;36:4510–4520. doi: 10.1093/nar/gkn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou T, Ray S, Lee C, Brasier AR. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem. 2008;283:30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raychoudhuri A, et al. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regis G, Pensa S, Boselli D, Novelli F, Poli V. Ups and downs: The STAT1:STAT3 seesaw of Interferon and gp130 receptor signalling. Semin Cell Dev Biol. 2008;19:351–359. doi: 10.1016/j.semcdb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Qing Y, Stark GR. Alternative activation of STAT1 and STAT3 in response to interferon-gamma. J Biol Chem. 2004;279:41679–41685. doi: 10.1074/jbc.M406413200. [DOI] [PubMed] [Google Scholar]
- 29.Regis G, et al. IL-6, but not IFN-gamma, triggers apoptosis and inhibits in vivo growth of human malignant T cells on STAT3 silencing. Leukemia. 2009;23:2102–2108. doi: 10.1038/leu.2009.139. [DOI] [PubMed] [Google Scholar]
- 30.Costa-Pereira AP, et al. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci USA. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dannenberg JH, et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres L, et al. In vivo GSH depletion induces c-myc expression by modulation of chromatin protein complexes. Free Radic Biol Med. 2009;46:1534–1542. doi: 10.1016/j.freeradbiomed.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 33.Cheng PY, et al. Interplay between SIN3A and STAT3 mediates chromatin conformational changes and GFAP expression during cellular differentiation. PLoS ONE. 2011;6:e22018. doi: 10.1371/journal.pone.0022018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonel P, Demmers J, Tan DW, Watt F, Hendrich BD. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev Biol. 2012;363:62–73. doi: 10.1016/j.ydbio.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.