Abstract
Microtubules are cytoskeleton filaments consisting of αβ-tubulin heterodimers. They switch between phases of growth and shrinkage. The underlying mechanism of this property, called dynamic instability, is not fully understood. Here, we identified a designed ankyrin repeat protein (DARPin) that interferes with microtubule assembly in a unique manner. The X-ray structure of its complex with GTP-tubulin shows that it binds to the β-tubulin surface exposed at microtubule (+) ends. The details of the structure provide insight into the role of GTP in microtubule polymerization and the conformational state of tubulin at the very microtubule end. They show in particular that GTP facilitates the tubulin structural switch that accompanies microtubule assembly but does not trigger it in unpolymerized tubulin. Total internal reflection fluorescence microscopy revealed that the DARPin specifically blocks growth at the microtubule (+) end by a selective end-capping mechanism, ultimately favoring microtubule disassembly from that end. DARPins promise to become designable tools for the dissection of microtubule dynamic properties selective for either of their two different ends.
Microtubules are dynamic protein assemblies essential for cell morphogenesis, membrane trafficking, and cell division of eukaryotic cells. In vivo, typically 13 straight, parallel, protofilaments interact laterally to form a microtubule. Each protofilament is a longitudinal head-to-tail assembly of αβ-tubulin heterodimers (tubulins), resulting in structurally polar microtubules, with β-tubulin facing the (+) end and α-tubulin being exposed at the (−) end. In vitro experiments with purified tubulin have demonstrated that microtubules switch stochastically between prolonged periods of assembly and disassembly, a phenomenon called dynamic instability (1). Both (+) and (−) ends display this behavior, but show small quantitative differences. In vivo, microtubule dynamics are regulated by different classes of proteins at the different ends. (−) ends are usually capped and do not grow nor shrink (2, 3) whereas (+) ends are dynamic. Their length excursions are affected by polymerases (4) and depolymerases. In addition, a few proteins, such as the kinesin-4 XKLP1 (5) and the kinesin-8 Kif18a (6), have been suggested to pause growth. Their mechanism is unclear and little is known about microtubule pause induction in general. This paucity is in remarkable contrast to the actin cytoskeleton where the importance of filament capping by a variety of proteins is well established (7, 8).
The complex structural events underlying microtubule dynamic instability are difficult to measure at high resolution. Therefore, open questions remain about this fundamental property. One remaining question concerns the mechanism by which GTP promotes microtubule growth. In recent years, the view has emerged that GTP in the exchangeable nucleotide-binding site of tubulin facilitates straightening during incorporation into the microtubule lattice but does not change the curvature of tubulin in solution (9–11). However, only two tubulin high-resolution structures are known. One is straight, in protofilaments of zinc-induced tubulin sheets stabilized by taxol (12); these are very similar to protofilaments in microtubules (13). The other one is curved, in a complex (T2R) of two tubulin heterodimers with the stathmin-like domain (SLD) of the tubulin-sequestering protein RB3 (14) (RB3-SLD). A high-resolution structure of GTP-tubulin alone is not available. As a consequence, the atomic details of the changes occurring during GTP-tubulin incorporation into the microtubule lattice had to be deduced from the available structures, assuming that the same motions occur in the absence of an SLD. Therefore, additional high-resolution data are needed to provide independent evidence for the structure of soluble GTP-tubulin; ideally these data should be obtained with tubulin alone or in complex with a molecule contacting it at a different surface. A molecule targeting a surface specifically exposed at microtubule ends would be particularly interesting as it might be used as a probe of the structure of tubulin at the very tip of microtubules.
A second open question concerns the mechanisms by which regulatory proteins affect microtubule dynamic instability. Recently, good progress has been made on the action of polymerases and ATP-dependent depolymerases (4, 15, 16). However, little is known about the structural aspects underlying the effects of specific end-binding proteins on microtubule dynamics. It is especially unclear how slowdown or even a complete stop of growth may be achieved. Observing the effect on microtubule assembly of a protein that binds in a well-defined manner to a surface exposed at microtubule (+) ends could provide information on possible mechanisms for the control of microtubule dynamics through a specific interaction at that end.
In the course of a screen of designed ankyrin repeat proteins (DARPins) for tubulin binding, biased to bind remote from the longitudinal interface of α-tubulin (a longitudinal interface is the interface between a tubulin subunit and the neighboring heterodimer in a protofilament), we have identified a series of proteins that inhibit microtubule assembly. One of them (D1) structurally caps the microtubule (+) end. Here, we describe the structure of its complex with tubulin at 2.2 Å resolution and its effect on microtubule assembly. The structure reveals that D1 binds to the longitudinal interface of β-tubulin, very different from tubulin-sequestering proteins belonging to the stathmin-like protein family. The structure demonstrates furthermore that GTP-tubulin is curved in this complex. An engineered tandem construct of the DARPin specifically stops growth at the (+) end at a few hundreds of nanomolar concentrations and, as a consequence, causes microtubule disassembly independent of ATP hydrolysis. This well-characterized capping molecule promises to be an interesting tool for the manipulation of microtubule dynamics in a unique way to understand better the regulation of their (+) end.
Results
D1 DARPin Inhibits Microtubule Assembly.
A DARPin library (17) was screened through ribosome display (18, 19) for tubulin binders. Proteins in this library consist of five ankyrin repeats: three randomized variable repeats and two stabilizing ones at their N- and C-terminal ends. Tubulin was immobilized with its α-subunit longitudinal interface facing the surface of the wells of microtiter plates (Methods). As a result, the screen was biased for binding of DARPins to the exposed tubulin β-subunit. After four rounds of ribosome display, the cDNAs of selected DARPins were subcloned for single-clone ELISA screening on crude cell extracts (20). One of the clones that generated the strongest signal yielded a protein, named here D1, which we expressed and purified.
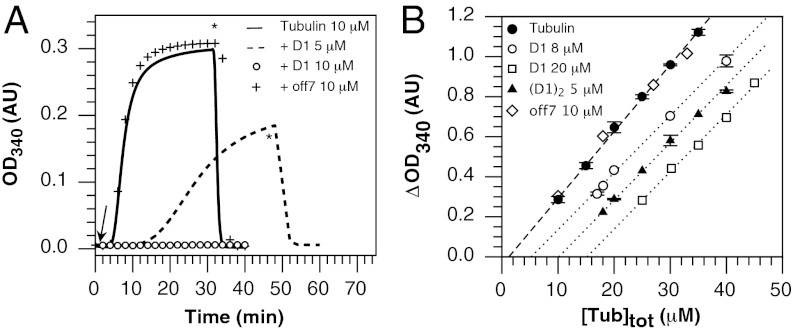
D1 formed a stable 1:1 complex with soluble tubulin as shown by size exclusion chromatography coupled to multi-angle light-scattering analysis (SEC-MALLS) (Fig. S1). By monitoring the time course of the turbidity of a tubulin solution in the presence of different D1 concentrations, we found that D1 inhibited microtubule assembly in a dose-dependent manner (Fig. 1A). This effect is specific as an unrelated DARPin (off7) from the same library as D1, but not selected for tubulin binding (17), had no effect when added at a concentration at which D1 completely blocks microtubule assembly (Fig. 1A). We also monitored the microtubules assembled by measuring the steady-state turbidity of microtubule suspensions at different tubulin concentrations in the absence and presence of D1 or of off7 (Fig. 1B). In contrast to off7, D1 had two effects on the variation of turbidity as a function of tubulin concentration. First, it increased the apparent critical concentration (Cc) of tubulin, i.e., the concentration of tubulin that is not assembled in microtubules. The apparent Cc shifted from 1.2 ± 0.5 μM to 5.2 ± 0.5 μM in the presence of 8 μM D1 and to 15.2 ± 0.8 μM when 20 μM D1 was added, confirming the dose dependence of the effect of D1 on microtubule assembly. Surprisingly, D1 also decreased the slope of the plot significantly. Because turbidity is a function of the amount of microtubular tubulin but also of the distribution of microtubule sizes (21), this result suggests that, in addition to modifying the mass of microtubules assembled, D1 modified their size distribution, possibly by transiently associating with tubulin to growing microtubules.
Fig. 1.
D1 prevents microtubule assembly. (A) In vitro polymerization of 10 μM tubulin alone and in presence of 5 μM or 10 μM D1 and of an unrelated DARPin selected from the same library as D1 (off7, 10 μM), used as a control (20). The arrow indicates the temperature jump from 4 °C to 37 °C and the asterisk indicates the time of the reverse temperature jump. (B) Critical concentration plots of tubulin and of tubulin in presence of 8 μM or 20 μM D1, 10 μM off7, or 5 μM (D1)2. (D1)2 is a D1 tandem repeat; see main text for its complete definition. Error bars represents SDs deduced from at least duplicate experiments.
D1 Binds to Curved GTP-Tubulin at Its β-Subunit Longitudinal Interface.
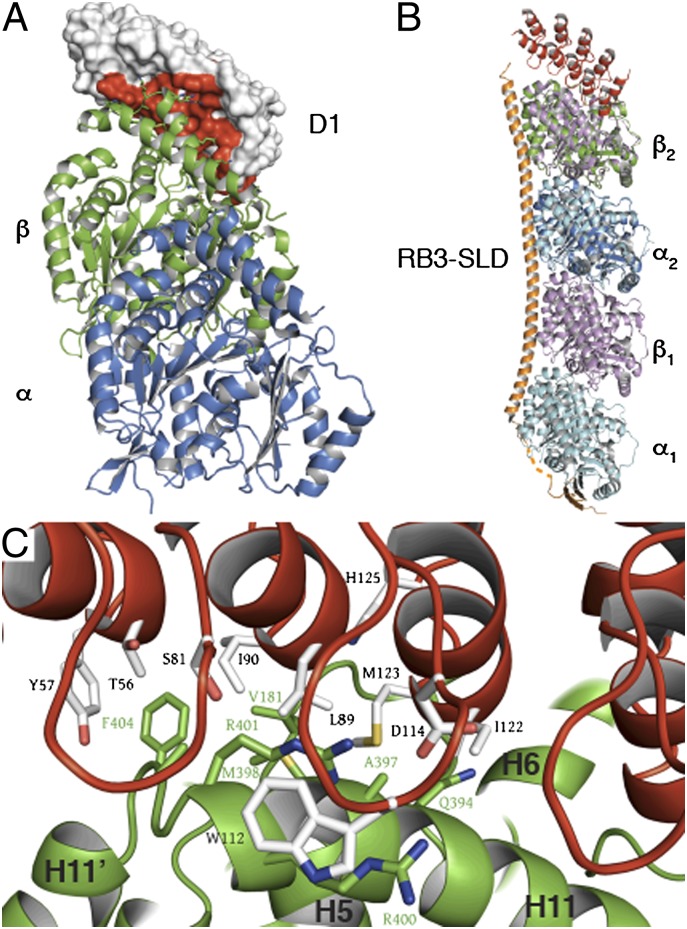
To get further insight into the molecular mechanism by which D1 interferes with microtubule assembly, we determined the structure of the tubulin–D1 complex (Tub-D1) by X-ray crystallography. Tubulin was cleaved by subtilisin (22), which removes C-terminal peptides of both subunits, which are not seen in any of the structures that have been determined (12, 14), and does not change the rest of the tubulin structure in any other respect (10). Removal of these heterogeneous C-terminal peptides has allowed us to obtain crystals that diffracted to 2.2 Å resolution. The structure was solved by molecular replacement and refined to a final R factor of 0.16 (Table S1). Interestingly, D1 contacts only the β-tubulin longitudinal interface (Fig. 2A), in a way that is very different from RB3-SLD in the only other known structure of a soluble tubulin complex (T2R) (10, 14). In T2R, the RB3-SLD structure consists of an N-terminal two-stranded β-sheet, a linker with no regular secondary structure, and a 100-aa long C-terminal α-helix that runs along the two tubulin molecules. The difference between the binding modes of D1 and RB3-SLD is best visualized by superimposing tubulin in Tub-D1 to the heterodimer bound to the C-terminal moiety of the RB3-SLD α-helix in T2R (Fig. 2B). The superposition shows that the two tubulin structures are very similar, with 0.65 Å root mean-squared deviation (862 Cαs superimposed). The angle of the rotation that is required to superimpose α- and β-tubulin is identical in Tub-D1 and T2R and equal to 12°. This result supports the view that the tubulin curvature seen both in T2R and in Tub-D1 is intrinsic to unpolymerized tubulin.
Fig. 2.
The structure of the tubulin–D1 complex. (A) Overall organization. D1 binds to the β-tubulin longitudinal interface. The red surface corresponds to D1 interfacing residues. (B) Superposition of Tub-D1 with T2R (pdb 3RYI). D1 is in red here as well as in C. Tubulin in Tub-D1 (α is in dark blue and β is in green) is superimposed to the α2β2 GDP-bound heterodimer in T2R (α2 is in cyan, as is α1, and β2 is in pink, as is β1); the RB3-SLD is in orange. (C) Residues of the tubulin–D1 interface belong to all five D1 ankyrin repeats and to β-tubulin helices H6, H11, and loop T5 (residues 206–215, 385–397, and 171–181, respectively; for a nomenclature of tubulin secondary structure elements see ref. 45). Residues shown are those that are randomized in the library from which D1 was selected and tubulin residues within 5 Å of them (for a stereoview see Fig. S2).
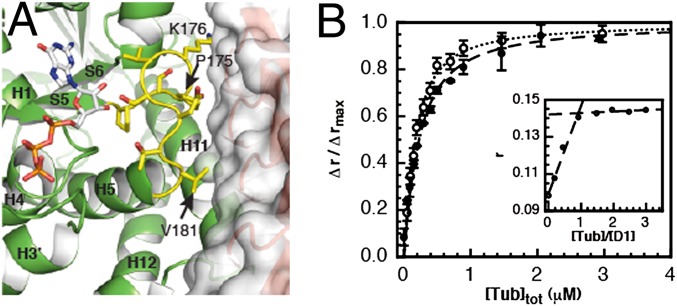
Residues on the tubulin side of the interface belong to three β-tubulin structural elements, helices H11 and H6 and loop T5 (Figs. 2C and 3A). Residues of the interface with D1 in all these structural elements are also involved in longitudinal interdimer interactions between tubulins that are embedded in microtubules (Table S2) (13). The structure therefore predicts that D1 interferes with tubulin longitudinal interactions. This interference provides a mechanistic explanation for the observed microtubule growth inhibition by D1 (Fig. 1). Our Tub-D1 crystals contained GTP bound to β-tubulin (GTP-tubulin) (Fig. S3). The β-tubulin nucleotide state (GDP or GTP) is functionally important, because only GTP-tubulin assembles into microtubules. The comparison of the affinities of D1 for GDP- and GTP-tubulin can provide information on the tubulin structural variations induced by nucleotide exchange, in particular because D1 binds close to the β-tubulin nucleotide-binding site (Fig. 3A). To measure these affinities, we used an assay based on the change of the fluorescence anisotropy of an Oregon 488-labeled D1 upon tubulin binding. Using a 1:1 association model and fitting the variation of the anisotropy with a quadratic equation (Methods) yields the value of the dissociation equilibrium constant from GDP-tubulin (KD = 120 ± 10 nM), which was found to be very similar to that of GTP-tubulin (KD = 155 ± 10 nM) (Fig. 3B). The very similar affinities of D1 for GDP- and GTP-tubulin suggest that the corresponding structures of soluble tubulin are very similar, supporting conclusions drawn from the comparison of these structures, as determined in the context of T2R (10).
Fig. 3.
D1 binds to GDP- and GTP-tubulin with similar affinities. (A) D1 binds close to the β-tubulin nucleotide-binding site and contacts its T5 loop (yellow). T5 is in its “out conformation,” which is fully populated when tubulin is GTP bound (10). T5 residues in contact with D1 are labeled. (B) Variations of fluorescence anisotropy of labeled D1 (120 nM) upon binding to GDP- (open circles) and GTP-tubulin (solid circles). Errors bars represent SDs from duplicate experiments. The lines represent the fit as described in Methods. Inset shows the titration of 1 μM Oregon-labeled D1 by GDP-tubulin, under conditions where the concentration of D1 was much larger than the dissociation equilibrium constant. Under these conditions, the fluorescence anisotropy reached its maximum for a 1:1 tubulin:D1 ratio, consistent with the 1:1 stoichiometry of the complex.
A D1 Tandem Repeat Caps Microtubule (+) Ends and Leads to Disassembly.
Because the affinity of D1 for tubulin is only moderate, one would expect a clearer effect on microtubule assembly with a tighter-binding protein. To generate a tighter-binding DARPin construct, contacting a similar tubulin surface, we made a tandem-repeat DARPin that might cooperatively bind two tubulin heterodimers at the end of two neighboring protofilaments. This binding seemed possible because when tubulin in Tub-D1 is superimposed on the (+) end tubulins of an atomic model of the microtubule (23), the resulting copies of D1 do not interfere with each other (Fig. S4). Because the distance from the C-terminal end of one copy to the N-terminal end of the D1 copy on a neighboring protofilament is 32 Å, we constructed a tandem repeat of D1 separated by three repeats of the G4S motif (24), a sequence expected to be long enough to allow simultaneous binding of two D1 copies to two laterally interacting tubulins. This construct, termed (D1)2, has indeed a stronger effect on microtubule assembly than D1, as shown by a larger shift of the tubulin Cc (Fig. 1B).
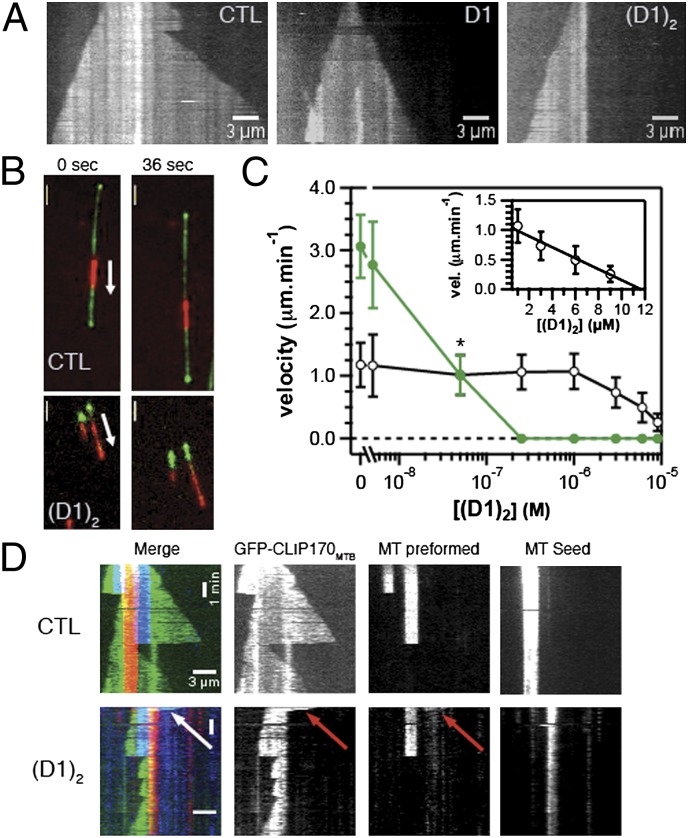
The Tub-D1 structure predicts that the effects of D1 [or (D1)2] at the two ends of a microtubule are different. D1-bound tubulin is not expected to incorporate at the (−) end of a microtubule, because its β-tubulin longitudinal interface is blocked. Therefore, in the presence of D1 as for a typical sequestering protein, the growth rate at the (−) end should solely be a function of the concentration of tubulin not complexed to D1. By contrast, at the (+) end, the Tub-D1 complex is still expected to be added, provided the microtubule end is free, because its α-tubulin longitudinal interface is unchanged compared with tubulin. Further addition of tubulins to the protofilament should be prevented as long as a D1 molecule is bound to the (+) end. To be able to distinguish the effects DARPins have on the two different microtubule ends, we observed microtubule dynamics at the single-microtubule level, using total internal reflection fluorescence (TIRF) microscopy. Dynamic microtubules were grown from stabilized microtubule seeds that were immobilized on chemically functionalized surfaces (25). Kymographs (time–space plots), generated from time-lapse movies in the presence of 20 μM Alexa568-tubulin, demonstrate that in comparison with microtubules growing in the absence of DARPin (Fig. 4A, Left), 0.25 μM D1 decreased the growth rate selectively at one microtubule end (Fig. 4A, Middle). The effect of the same concentration of (D1)2 was more dramatic as growth at one end was blocked completely (Fig. 4A, Right).
Fig. 4.
(D1)2 efficiently inhibits microtubule plus-end growth. (A) Kymographs of Alexa568-labeled microtubules growing from surface-immobilized microtubule seeds in the absence or presence of 0.25 μM D1 or (D1)2 as indicated, observed by TIRF microscopy; total duration displayed is 5 min. The concentration of Alexa568-labeled tubulin (labeling ratio: 6.5%) was 20 μM. (B) TIRF microscopy images of dynamic microtubules being transported by surface-immobilized (−) end-directed kinesin-14 (XCTK2). Microtubules polymerize from Alexa568-labeled GMPCPP microtubule seeds (red) in the presence of 20 μM tubulin and 70 nM Mal3-GFP (green) and either no (Upper) or 0.5 μM (D1)2 (Lower). (Scale bar: 3 μm.) (C) Growth velocity of microtubule (+) (green symbols) and (−) (black symbols) ends in the presence of 20 μM tubulin as a function of increasing (D1)2 concentrations. The data point for 50 nM (D1)2 (marked with an *) is an average of (+)- and (−) end growth velocity, because the two ends could not be distinguished due to similar growth velocities in this condition. Error bars are SD, each experimental value being the average of at least 20 microtubules from three or more independent experiments. (Inset) (−) end growth velocity as a function of (D1)2 concentrations on a linear scale with a linear fit (black line). Note that the (D1)2 concentration required to depolymerize microtubules in these experiments cannot be directly compared with those in turbidity measurements, because these are different types of experiments that were performed under different conditions (Methods). (D) Kymographs show the effect of 0.5 μM (D1)2 on microtubules prepolymerized in the presence of 15 μM Cy5-labeled tubulin (blue) from immobilized Alexa568-labeled stabilized microtubule seeds (red). Imaging started ca. 30 s after replacing labeled with unlabeled tubulin and addition of 65 nM of a GFP-labeled CLIPMTB (green) (Methods) (Lower). The control without (D1)2 is also presented (Upper). Merged kymographs (Left) and the individual one-channel kymographs are shown as indicated. The arrow shows a (+) end that has not depolymerized at the time imaging started. Total time displayed is 500 s.
To identify the blocked end unambiguously, we used a gliding assay in which dynamic microtubules were transported by surface-immobilized molecular motors. Growing (+) and (−) ends were labeled with Mal3-GFP (26). Whereas transported microtubules growing from Alexa568-labeled GMPCPP-tubulin seeds in the presence of Mal3-GFP had both ends strongly labeled (Fig. 4B, Upper), in the additional presence of 0.5 μM (D1)2, only one end was labeled. On surfaces coated with (−) end-directed Xenopus kinesin-14 (XCTK2) (27), this growing end was the lagging end, i.e., the (−) end (Fig. 4B, Lower). This effect was robustly observed at concentrations of 250 nM (D1)2 and higher (Fig. 4C) and demonstrates that (D1)2 blocked microtubule growth selectively at the (+) end. The growth of microtubule (−) ends decreased measurably only at considerably higher concentrations of (D1)2 (1 μM and higher). (−) end growth decreased linearly with the (D1)2 concentration and stopped at a ratio slightly smaller than one (D1)2 per two tubulin dimers as indicated by a linear fit (Fig. 4C, Inset). This result suggests that (D1)2 slows (−) end growth by sequestering free tubulin and that growth at the (−) end stops when the concentration of unbound tubulin is equal to the Cc at that end. D1 also shows a clear dose-dependent inhibitory effect on microtubule growth but it is clearly weaker than that of (D1)2, as expected (Fig. S5).
To test the effect of (D1)2 on the (+) end of a dynamic microtubule, we prepolymerized microtubules in the presence of Cy5-labeled tubulin and GTP from Alexa568-labeled GMPCPP seeds and then replaced labeled by unlabeled tubulin, adding at the same time 0.5 μM (D1)2. The entire length of microtubules was visualized by the addition of a microtubule-binding protein (GFP-CLIPMTB, a GFP-labeled CLIP-170 fragment, previously also called H2) (28). Time-lapse imaging was started ∼30 s after the solution exchange. The stability of (D1)2-capped microtubule (+) ends was assayed in two different ways. First, kymographs of GFP-CLIPMTB–labeled microtubules showed that (D1)2-capped prepolymerized microtubule (+) ends remained stable only for a few tens of seconds before they depolymerized back to the microtubule seed (Fig. 4D, Lower). The (+) end of the seed was unable to regrow (Fig. 4D, Lower), as expected at this concentration of (D1)2 (Fig. 4C). Because DARPin-induced depolymerization affected only the part of the microtubule not stabilized by GMPCPP, (D1)2 differs from ATP-dependent depolymerases that also disassemble stabilized microtubules (16, 29, 30). Minus ends displayed normal dynamic instability behavior as is the case of both microtubule (+) and (−) ends in the absence of (D1)2 (Fig. 4D, Upper). Second, to analyze the stability of capped microtubule (+) ends at a population level, we quantified the number of prepolymerized (+) sections at different time points after solution exchange. In the presence of (D1)2 the number of these sections decreased rapidly, leading to an ∼10-fold reduction after 2 min in comparison with the control without (D1)2 (Fig. S6). Taken together, these experiments demonstrate that inhibition of microtubule (+) end growth by (D1)2 has a destabilizing effect on microtubules.
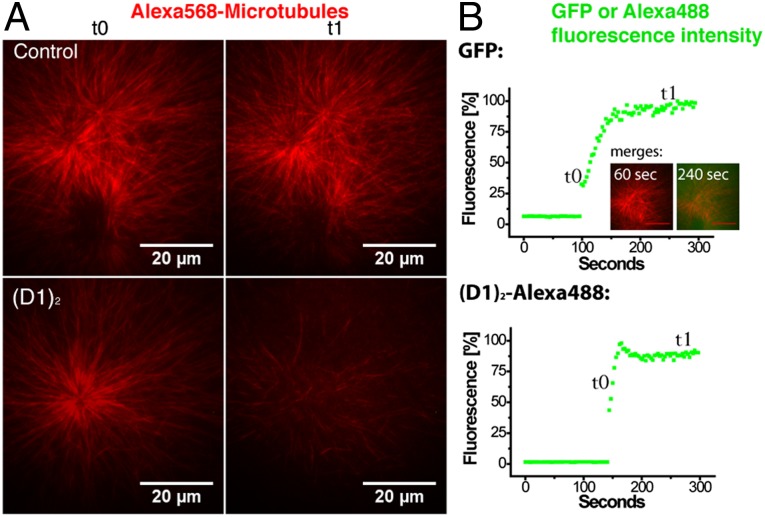
To test whether the destabilizing effect of (D1)2 can also be observed in the presence of other microtubule-binding proteins, we performed experiments in Xenopus egg extract, an established system for the study of in vivo-like microtubule dynamics (31). Addition of (D1)2 to microtubule asters led to a dramatic reduction of the microtubule density in these asters (Fig. 5). Therefore, this microtubule (+) end capping DARPin can interact with microtubule ends also in the presence of natural levels of other microtubule-binding proteins and inhibit their growth, which finally leads to depolymerization.
Fig. 5.
(D1)2 promotes MT depolymerization in Xenopus egg extract. (A) Asters in Xenopus egg extract containing Alexa568-tubulin were adsorbed to glass coverslips and observed by TIRF microscopy. (B) After addition of GFP (Upper) or Alexa488-(D1)2 (Lower) to an estimated final concentration of 0.75 μM, the diffusion into the evanescent field was monitored by measuring the GFP or Alexa488 fluorescence intensity. Alexa568-microtubule asters that are still present when the newly added protein just starts to diffuse into the evanescent field (t0 in A or B), largely disappear 150 s later selectively when Alexa488-(D1)2 was added (t1 in A and B). Inserted images in B show merged dual-color images to visualize how GFP (green) diffuses into the region with microtubule asters (red).
Discussion
In this study, we produced a molecule known to selectively bind to the longitudinal interface of β-tubulin. This unique experimental tool has allowed us to address open questions regarding the mechanism of microtubule polymerization and dynamic instability. Four main conclusions can be drawn from this work.
The first one relates to the reason for only GTP-tubulin, as opposed to GDP-tubulin, assembling into microtubules. This property has long been thought to be due to a difference of their overall shapes, GDP-tubulin being curved whereas GTP-tubulin would be straighter and prestructured for lateral interactions (32). More recently, GTP-tubulin and GDP-tubulin in T2R have been found to be similarly curved (10) but a possibility remained that the curvature was influenced by RB3-SLD. Here we find that the curvatures of GTP-tubulin bound to D1 and of GDP-tubulin bound to RB3-SLD are virtually identical. The D1-binding site does not overlap with that of RB3-SLD, is exclusively located on β-tubulin, and is distant by more than 40 Å from the α-β interface where the main differences between curved and straight tubulin are located. Taken together with the similar affinities of D1 for GDP- and GTP-tubulin and with a range of other biochemical evidence (11, 33), our structural data further support the notion that GTP-tubulin in solution is curved similarly to GDP-tubulin and straightens only as it incorporates in microtubules. This result strongly suggests that GTP facilitates the tubulin structural switch that accompanies microtubule assembly but does not trigger it (9–11) and narrows the stage at which the tubulin structural switch occurs down to the initial steps of assembly.
The second conclusion concerns the structure of tubulin at microtubule plus tips, an issue that is not easily addressed at the current resolution of electron microscopy data on microtubules. The interface of β-tubulin with D1 comprises the H6 helix. This helix rotates upon going from curved tubulin to straight protofilaments (14) and is immediately upstream of the long helix that undergoes a piston movement at the tubulin structural switch. The observation that D1 [and (D1)2] adds to the microtubule (+) end (tubulin-bound or on its own) taken together with the very similar structures of β-tubulin when the heterodimer is bound to D1 (this work) or to RB3-SLD (10) means that the longitudinal interface of β-tubulin at the end of a microtubule is very similar to that of β-tubulin in solution. This similarity also suggests that, at the end of a microtubule, the tubulin structural switch has not taken place yet but occurs later on during early stages of microtubule assembly, most likely as lateral interactions between neighboring protofilaments get established.
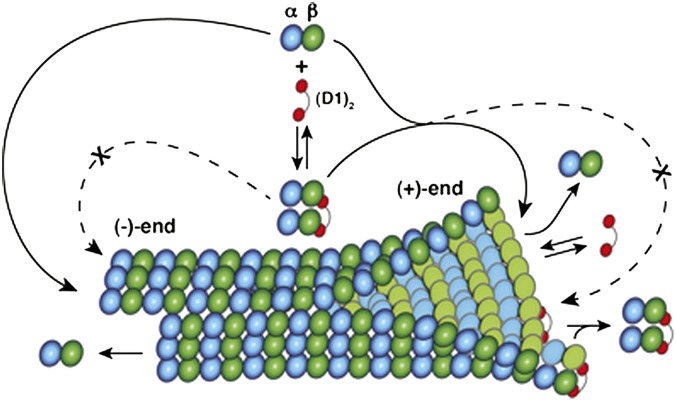
The third conclusion concerns the effects of (D1)2 on microtubule dynamics. (D1)2 inhibits growth at both microtubule ends by strikingly different mechanisms that are readily explained on the basis of the Tub-D1 structure (Fig. 6). As it binds to the β-tubulin longitudinal interface, (D1)2 prevents association at the (−) end of the tubulins it is bound to in solution, by a sequestering effect in which each D1 moiety prevents the assembly of one heterodimer. This effect is similar to that previously described for stathmin. However, different from stathmin, (D1)2 acts as a sequestering protein only at the (−) end. By contrast, at the (+) end (D1)2 acts as a capping protein. There it does not prevent the association of the tubulin it is bound to, because it does not block its α-subunit longitudinal interface. Instead, it blocks the association of many other incoming tubulins (either in complex with DARPins or not), as long as the (D1)2 caps the protofilament (+) end. This result explains why (D1)2 interferes with microtubule growth more drastically at (+) than at (−) ends. It is interesting that, in the presence of 20 μM tubulin, at concentrations as low as 250 nM (D1)2, i.e., at only a 1:80 (D1)2:tubulin ratio, growth is completely blocked at the (+) end. There could be two reasons for that: it prevents association to the protofilaments it is bound to, by capping the β-tubulin longitudinal interface, as described above; and it may also cause the instability of the other protofilaments, because some lateral interactions, those with protofilaments that are capped and do not grow, cannot be established.
Fig. 6.
The mechanism of action of (D1)2. (D1)2 (red) prevents assembly at the (−) end of the one tubulin heterodimer (α, blue; β, green) it is bound to. (D1)2-bound tubulins associate at the (+) end but then block addition of all tubulin heterodimers to a capped protofilament.
Finally, the capping of microtubules by (D1)2 differs from capping of actin filaments in which barbed ends are efficiently stabilized. Some of the actin barbed-end capping proteins, such as the one named the “capping protein” (34), are heterodimers with a large interface in which the two monomers are rigidly held together and simultaneously and tightly bind to the two actin molecules at the tip of a filament. Uncapping requires either that they dissociate from the filament tip or the simultaneous dissociation of the two actin molecules at the very end of the filament; both are very rare events. By contrast, the relatively long and flexible linker between its two moieties does not allow (D1)2 to make full use of the cooperative binding of two neighboring tubulin molecules to the microtubule tip, likely resulting in the lack of a stabilizing effect of (D1)2 at the microtubule (+) end. Alternatively, the specificities of the two polymers (35) may explain the different behaviors of (D1)2 and actin-capping proteins. The properties of (D1)2 also differ from those of recently characterized proteins that modify the dynamics of (+) ends, although (D1)2 shares characteristics with some of them. (D1)2, the same as yeast Kip3 (16, 29), depolymerizes microtubules exclusively from their (+) end but, different from this kinesin, it is not active on stabilized microtubules. The same as Kif18a (6, 36) and Xklp1 (37), two kinesins that induce pauses, it prevents microtubule growth at the (+) end; different from them, it does not prevent disassembly of dynamic microtubules from that end but, instead, favors it. This is an indirect effect of (D1)2. Its direct effect is to slow growth as it caps protofilaments. The consequence of slowed growth of a dynamic microtubule is that catastrophes become more frequent (38). This consequence ultimately favors disassembly of dynamic microtubules by (D1)2.
DARPins offer possibilities to investigate microtubule assembly in addition to those we described here. It should be possible to mature the affinity of DARPins (39) for tubulin (several orders of magnitude of the Kd may readily be gained). Using higher-affinity derivatives of the DARPins we have in hand to hold tubulin for ribosome display selection, it should be possible to identify new DARPins that bind to other tubulin surfaces, such as the α-tubulin longitudinal interface. Such DARPins would be useful tools to dissect the regulation of microtubule dynamics, both mechanistically and structurally, by probing the effect on microtubule assembly of interactions with various tubulin interfaces.
Methods
Ribosome Display Selection and Binder Identification.
The N3C DARPin library (40) was used to select tubulin binders, using four rounds of ribosome display (18, 19). In these experiments, the target (tubulin) was covalently coupled via a disulphide bond to a biotinylated stathmin-derived peptide (41) and immobilized via neutravidin or streptavidin coated on microtiter plates. Details of the selection are described in SI Methods. Selected DARPins were cloned into pDST67 (18, 20). Clones producing tubulin binders were identified by ELISA on crude cell extracts (20). Details are described in SI Methods.
Protein Purification, Labeling, and Crystallization.
Protein purification, labeling, and complex preparation for crystallization are described in SI Methods. Crystal seeds were obtained after 12 h in 14% (wt/vol) PEG 6000, 0.05 M KCl, 10 mM MgCl2, 0.2 M LiCl. Immediately afterward, microseeding in a drop made with a protein stock at 16–18 mg⋅mL−1 in 10% (wt/vol) PEG 6000, 0.05 M KCl, 10 mM MgCl2, 0.2 M LiCl, 10 mM Hepes, pH 7, yielded crystals within 12 h.
Diffraction Data Collection and Structure Determination.
Data collection and processing are described in SI Methods. Checking data quality with the Xtriage module of Phenix (42) indicated crystal twinning. The structure (space group P21) was solved by molecular replacement, using the AutoMR wizard within Phenix. Starting models were αβ-tubulin [Protein Data Bank (PDB) ID 3RYC] (10) and a N3C DARPin (PDB ID 2XEE) (43) with randomized positions mutated to alanine. The structure was refined in Phenix, alternating twin refinement with manual model building in Coot (44). Final refinement statistics are reported in Table S1. Some regions had no visible electron density: residues 38–46 and 280–284 in α-tubulin and residues 55–61 in β-tubulin as well as residues 1–12 in D1. Coordinates have been deposited in the Protein Data Bank (PDB ID 4DRX). For procedures used to produce figures and to analyze the structure, see SI Methods.
Fluorescence Anisotropy.
Dissociation constant determination was performed by titrating D1-D160C-Oregon Green 488 with increasing amounts of tubulin and monitoring steady-state fluorescence anisotropy as described in SI Methods.
TIRF Microscopy.
Microtubule growth velocity was measured as described (26) and detailed in SI Methods.
Microtubule gliding assay.
Dynamic microtubules polymerizing from Alexa568-labeled GMPCPP-stabilized microtubule seeds in the presence of 20 μM unlabeled tubulin and 70 nM Mal3-GFP in gliding buffer were observed as they were transported by surface-immobilized (−) end-directed XCTK2 motors either in the absence or in the presence of 500 nM unlabeled (D1)2 (SI Methods).
Depolymerization protection assay.
Microtubules were grown from surface-immobilized Alexa568-labeled GMPCPP-stabilized microtubule seeds in the presence of 15 μM Cy5 (Lumiprobe)-labeled tubulin. After 90 s the solution was exchanged and growth continued in 15 μM unlabeled tubulin and 65 nM of GFP-labeled CLIPMTB either in the absence or in the presence of 0.5 μM (D1)2 in standard TIRF buffer (SI Methods). As a measure of microtubule plus-end stability, we quantified how many Cy5-labeled microtubule plus segments [extending from the (+) end of the GMPCPP seed] were still present 2 min after introducing the (D1)2-containing solution (or the control solution). Plus and minus segments were discriminated by their growth velocity.
Microtubule Growth Inhibition in Xenopus Egg Extract.
Microtubules pregrown from sperm nuclei in Xenopus egg extract were supplemented with either Alexa488-(D1)2 or GFP as a negative control and imaged by TIRF microscopy. For details see SI Methods.
Supplementary Material
Acknowledgments
We thank Ingrid Mignot, Iris Lueke, and Caroline Fauquant-Pecqueur for their help in the experimental work; Nicholas Cade for maintenance of the TIRF microscope; Mr. D. Mauchand and Mrs J. Massonneau (Unité Commune d'Expérimentation Animale, Institut National de la Recherche Agronomique) for providing us with the material from which tubulin was purified; and Hiro Mahbubani from the London Research Institute Bio Resources Unit for providing Xenopus egg extract. Diffraction data were collected at the Soleil synchrotron (Saint-Aubin, France) and at the European Synchrotron Research Facility (Grenoble, France). We thank the machine and Proxima 1 beam line groups for making these experiments possible. The research leading to these results was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de Recherches (ANR-09-BLAN-0071), the Fondation pour la Recherche Médicale (DEQ20081213979), and Cancer Research United Kingdom and received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under Grant 227764 (Protein Production Platform).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4DRX).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204129109/-/DCSupplemental.
References
- 1.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin SS, Vale RD. Patronin regulates the microtubule network by protecting microtubule minus ends. Cell. 2010;143:263–274. doi: 10.1016/j.cell.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by γ-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bringmann H, et al. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 6.Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 7.Silacci P, et al. Gelsolin superfamily proteins: Key regulators of cellular functions. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wear MA, Cooper JA. Capping protein: New insights into mechanism and regulation. Trends Biochem Sci. 2004;29:418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Buey RM, Díaz JF, Andreu JM. The nucleotide switch of tubulin and microtubule assembly: A polymerization-driven structural change. Biochemistry. 2006;45:5933–5938. doi: 10.1021/bi060334m. [DOI] [PubMed] [Google Scholar]
- 10.Nawrotek A, Knossow M, Gigant B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J Mol Biol. 2011;412:35–42. doi: 10.1016/j.jmb.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 11.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: Structural studies of αβ- and γ-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Löwe J, Li H, Downing KH, Nogales E. Refined structure of αβ-tubulin at 3.5 Å resolution. J Mol Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- 13.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 14.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 15.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 16.Varga V, et al. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 17.Binz HK, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 18.Dreier B, Plückthun A. Rapid selection of high-affinity binders using ribosome display. Methods Mol Biol. 2012;805:261–286. doi: 10.1007/978-1-61779-379-0_15. [DOI] [PubMed] [Google Scholar]
- 19.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steiner D, Forrer P, Plückthun A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J Mol Biol. 2008;382:1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 21.Hall D, Minton AP. Turbidity as a probe of tubulin polymerization kinetics: A theoretical and experimental re-examination. Anal Biochem. 2005;345:198–213. doi: 10.1016/j.ab.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Knipling L, Hwang J, Wolff J. Preparation and properties of pure tubulin S. Cell Motil Cytoskeleton. 1999;43:63–71. doi: 10.1002/(SICI)1097-0169(1999)43:1<63::AID-CM7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 23.Fourniol FJ, et al. Template-free 13-protofilament microtubule-MAP assembly visualized at 8 Å resolution. J Cell Biol. 2010;191:463–470. doi: 10.1083/jcb.201007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreier B, et al. Her2-specific multivalent adapters confer designed tropism to adenovirus for gene targeting. J Mol Biol. 2011;405:410–426. doi: 10.1016/j.jmb.2010.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bieling P, Telley IA, Hentrich C, Piehler J, Surrey T. Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 2010;95:555–580. doi: 10.1016/S0091-679X(10)95028-0. [DOI] [PubMed] [Google Scholar]
- 26.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 27.Walczak CE, Verma S, Mitchison TJ. XCTK2: A kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel J, et al. Purification and analysis of authentic CLIP-170 and recombinant fragments. J Biol Chem. 1999;274:25883–25891. doi: 10.1074/jbc.274.36.25883. [DOI] [PubMed] [Google Scholar]
- 29.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 30.Hunter AW, et al. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budde PP, Desai A, Heald R. Analysis of microtubule polymerization in vitro and during the cell cycle in Xenopus egg extracts. Methods. 2006;38:29–34. doi: 10.1016/j.ymeth.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 32.Wang HW, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honnappa S, Cutting B, Jahnke W, Seelig J, Steinmetz MO. Thermodynamics of the Op18/stathmin-tubulin interaction. J Biol Chem. 2003;278:38926–38934. doi: 10.1074/jbc.M305546200. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita A, Maeda K, Maéda Y. Crystal structure of CapZ: Structural basis for actin filament barbed end capping. EMBO J. 2003;22:1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver LN, et al. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 38.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: Rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zahnd C, Sarkar CA, Plückthun A. Computational analysis of off-rate selection experiments to optimize affinity maturation by directed evolution. Protein Eng Des Sel. 2010;23:175–184. doi: 10.1093/protein/gzp087. [DOI] [PubMed] [Google Scholar]
- 40.Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol. 2003;332:489–503. doi: 10.1016/s0022-2836(03)00896-9. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, et al. Kif2C minimal functional domain has unusual nucleotide binding properties that are adapted to microtubule depolymerization. J Biol Chem. 2012;287:15143–15153. doi: 10.1074/jbc.M111.317859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kramer MA, Wetzel SK, Plückthun A, Mittl PR, Grütter MG. Structural determinants for improved stability of designed ankyrin repeat proteins with a redesigned C-capping module. J Mol Biol. 2010;404:381–391. doi: 10.1016/j.jmb.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.