Abstract
In this study we test the hypothesis that mechanically elastic regions in a virus particle (or large biomolecular complex) must coincide with conformationally dynamic regions, because both properties are intrinsically correlated. Hypothesis-derived predictions were subjected to verification by using 19 variants of the minute virus of mice capsid. The structural modifications in these variants reduced, preserved, or restored the conformational dynamism of regions surrounding capsid pores that are involved in molecular translocation events required for virus infectivity. The mechanical elasticity of the modified capsids was analyzed by atomic force microscopy, and the results corroborated every prediction tested: Any mutation (or chemical cross-linking) that impaired a conformational rearrangement of the pore regions increased their mechanical stiffness. On the contrary, any mutation that preserved the dynamics of the pore regions also preserved their elasticity. Moreover, any pseudo-reversion that restored the dynamics of the pore regions (lost through previous mutation) also restored their elasticity. Finally, no correlation was observed between dynamics of the pore regions and mechanical elasticity of other capsid regions. This study (i) corroborates the hypothesis that local mechanical elasticity and conformational dynamics in a viral particle are intrinsically correlated; (ii) proposes that determination by atomic force microscopy of local mechanical elasticity, combined with mutational analysis, may be used to identify and study conformationally dynamic regions in virus particles and large biomolecular complexes; (iii) supports a connection between mechanical properties and biological function in a virus; (iv) shows that viral capsids can be greatly stiffened by protein engineering for nanotechnological applications.
Keywords: nanotechnology, scanning probe microscopy, single-molecule analysis, virus mechanics
Viruses constitute relevant models to study biological nanomachines. A major advance has been the recognition of virus particles as highly dynamic macromolecular assemblies (1–4). During the infection cycle, viral particles may experience biologically relevant structural rearrangements that involve local atomic displacements and reorganization of noncovalent interactions between atomic groups. Limited proteolysis/mass spectrometry and other techniques have been applied to explore virus particle dynamics (5, 6). Unfortunately, detection of subtle, transient dynamic events in a viral particle (or any other large biomolecular complex) and their tracing to specific regions in the particle are still difficult tasks.
Developments in atomic force microscopy (AFM) (7, 8) have recently led to the experimental investigation of the mechanical properties of single virus particles (9–25). The mechanical elasticity of virions and capsids can be probed using the approach pioneered in ref. (9). Individual particles are physically deformed (indented) by application of a controlled force through the tip of an AFM microscope. For small indentations, the particle behaves as an ideal mechanical spring. Using Hooke’s law, the relationship between force and indentation depth is used to calculate the spring constant, ks, which provides a measurement of the local mechanical stiffness of the viral particle, at the region contacted by the tip, along the direction of the applied force. The elastic thin shell model (26) generally provides a good geometrical approximation to a virus capsid and can be used to calculate the Young’s modulus that characterizes the intrinsic stiffness of each region probed (9, 27).
Studies on virus mechanics so far have been undertaken largely by physicists wishing to understand the physical behavior of unique examples of “soft” condensed matter capable of self-assembly into nanoparticles with potential applications in nanotechnology (18, 20, 25). In addition, hypotheses on possible biological roles for some of the elastic features found in different viruses have been put forward (10, 13–15, 19, 24, 25). However, to our knowledge no support for an involvement of the mechanical elasticity of virus particles in biological function had been provided by subjecting any hypothesis to experimental corroboration.
In this study we experimentally challenge the hypothesis that mechanical elasticity and conformational dynamics in a virus particle are inextricably correlated. Not because one is the cause and the other the effect, but because they are two manifestations of the same physical phenomenon: If a region of a viral particle can be varied from its minimal free energy conformation without requiring too much energy, a structural biologist may detect a conformationally dynamic region; whereas a physicist using AFM may detect a mechanically highly elastic region. The phenomenon would be the same, but it would be detected in different ways by probing different properties. If the hypothesis of a phenomenological equivalence between conformational dynamism and mechanical elasticity is correct, regions of higher mechanical elasticity in a virus particle must match conformationally more dynamic regions. Determination by AFM of the ks of different regions in a viral particle could then be used to identify dynamic regions and investigate the role of elasticity/dynamics in biological function.
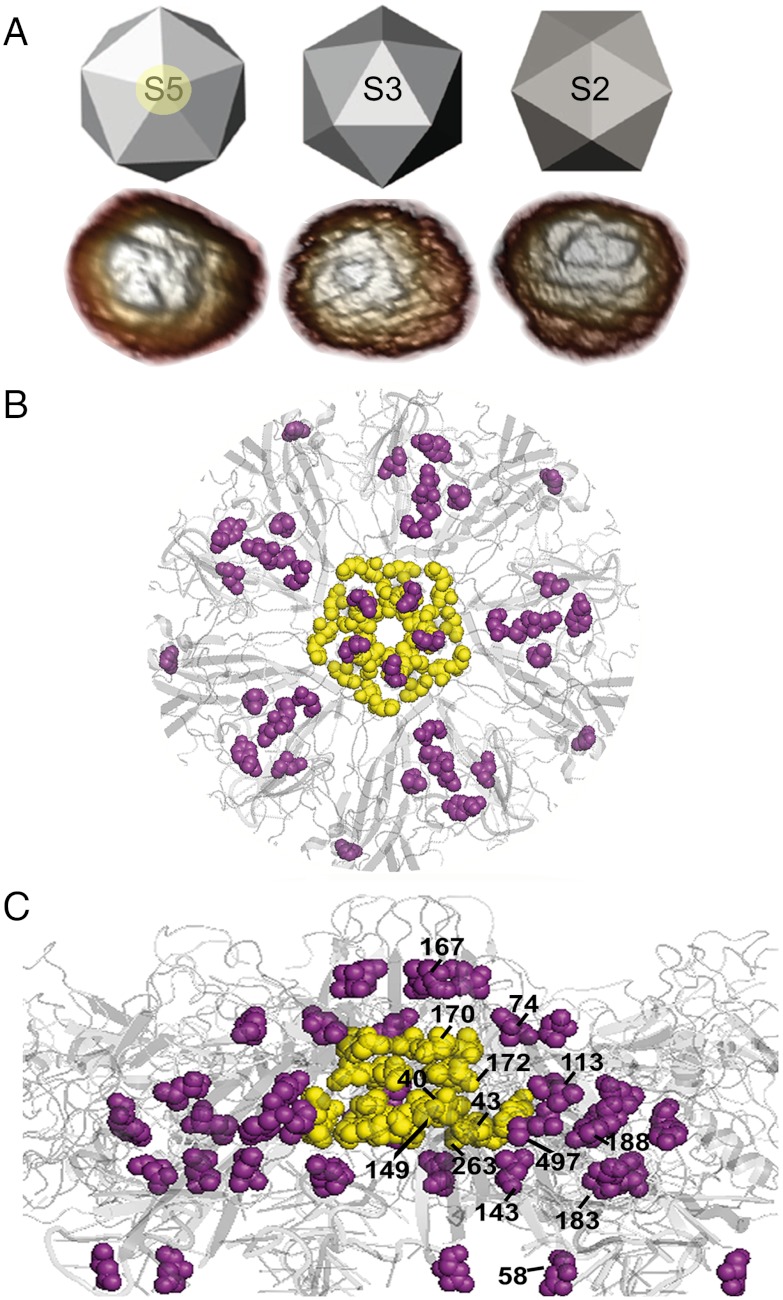
The nonenveloped minute virus of mice (MVM) provides a simple model to corroborate or disprove the above hypothesis. The small, T = 1 icosahedral capsid of MVM is formed by 60 equivalent protein subunits with the same fold (28, 29) (Fig. 1). In the MVM virion, segments of the single-stranded (ss) viral DNA bound to equivalent sites at the capsid inner wall act like molecular buttresses that decrease the mechanical elasticity of most regions in the virus particle (10, 15). However, the regions around the fivefold symmetry (S5) axes in the virion are free from bound DNA segments and remain as elastic as in the empty (DNA-free) capsid. The bound DNA segments also provide the virion with the biological advantage of increased resistance against thermal inactivation (30). Interestingly, the regions in which virion-stabilizing bound DNA segments are absent (the S5 regions) are conformationally highly dynamic.
Fig. 1.
MVM capsid structure and mutations tested. (A) Icosahedral model and AFM images of capsids, viewed along a S5, S3, or S2 symmetry axis. The circle around the labelled S5 axis delimits the region represented as a ribbon model of the crystallographic structure in B (front view) and C (lateral view), with a S5 pore at the center. The residues around the base of the pore or other mutated residues are, respectively, represented as yellow or purple spacefilling models and labeled in C. The program PyMol (DeLano Scientific) and the Protein Data Bank coordinates 1Z1C (29) were used.
In MVM, narrow channels (pores) at the S5 axes are involved in the externalization of molecular signals located in the N-terminal segments (Nt) of capsid proteins VP2 and VP1 (a VP2 subunit with a longer Nt), and in the encapsidation and uncoating of the viral DNA (31–43). These biologically relevant events require a conformationally dynamic structure at the S5 regions to allow the aperture and closing of the pores and other rearrangements. In vivo, externalization of the VP2 Nt, required for nuclear export of the virion and subsequent events in the viral cycle, occurs in response to DNA encapsidation (34, 39). In vitro, the same event can be triggered in DNA-free capsid-like particles by mild heating (32, 35); this event is associated with a conformational transition of the capsid that propagates beyond the pores and produces slight changes in the exposure of tryptophan residues to solvent, which can be detected by fluorimetry (35). Capsid-like particles with amino acid substitutions surrounding the base of the pores still contain the VP2 Nt inside, but do not undergo a heat-induced rearrangement detected by fluorimetry, and show alterations in VP2 Nt externalization (36). Introduction in the virion of those same mutations (36), or others at a nearby position (L172) (37, 38), dramatically reduced infectivity at 37 °C (36, 43), impaired infection at 32 °C, and caused untimely genome uncoating during cell entry (43). In short, mutations of residues at the base of the pores in the MVM capsid impair the biologically relevant conformational dynamics of the S5 (pore) regions. Mutations of residues lining the S5 pores of adeno-associated virus type 2, a related virus, also impair local dynamics (44).
When applied to MVM, the hypothesis that mechanical elasticity and conformational dynamism are intrinsically correlated leads to testable stringent predictions: (i) Any modification in the capsid that impairs the dynamics of the S5 regions (reflected in impairment of the pore-associated conformational transition detected by fluorimetry) should increase the mechanical stiffness of the S5 regions, and vice versa; (ii) any modification that does not impair the pore-associated transition should preserve the mechanical elasticity of the S5 regions, and vice versa; (iii) no correlation should be observed between S5 dynamics and the elasticity of capsid regions different from S5.
Here we have repeatedly tested those predictions in different ways by comparing the conformational and mechanical features of 19 structurally modified MVM capsids with those of the unmodified (WT) capsid. The results provided multiple independent corroborations of the hypothesis under test. This study validates local mechanical elasticity in a virus particle as a fundamental physical manifestation of conformational dynamism at the atomic level, and supports a connection between virus mechanics and biological function.
Results
Topography of Mutant MVM Capsids and Comparison of Mechanical Stiffness.
AFM imaging in physiological buffer revealed for each mutant capsid the major topographic features (spikes at S3, dimples at S2, and cylindric prominences at S5; Fig. 1 and Fig. S1) that served to determine the orientation of individual particles and the region in each particle for which elasticity could be measured (10). No significant alterations in topography were detected for any mutant (Fig. S1).
The mechanical stiffness of capsids in the same buffer was then determined in indentation experiments using AFM. For each mutant capsid and region analyzed, a high number of measurements (between 41 and 143 indentations of 4 to 20 particles) were carried out. Cantilevers with a kc of 0.1 N/m were used and yielded high quality, reproducible measurements; the average ks values obtained for the reference capsid were indistinguishable from those obtained with softer cantilevers (15). Cantilevers with a higher kc (e.g., 0.3 N/m) led to sample damage. Modifications in capsid preparation had no significant effect in ks, but, even so, the same conditions were used for all variant capsids. As expected from their identical architecture, authentic MVM capsids and capsid-like particles (made only of VP2) showed the same ks (0.59 ± 0.11 N/m and 0.60 ± 0.12 N/m, respectively).
Mutations in the MVM Capsid That Impair the Pore-Associated Conformational Dynamics Increase the Mechanical Stiffness Around the Pores.
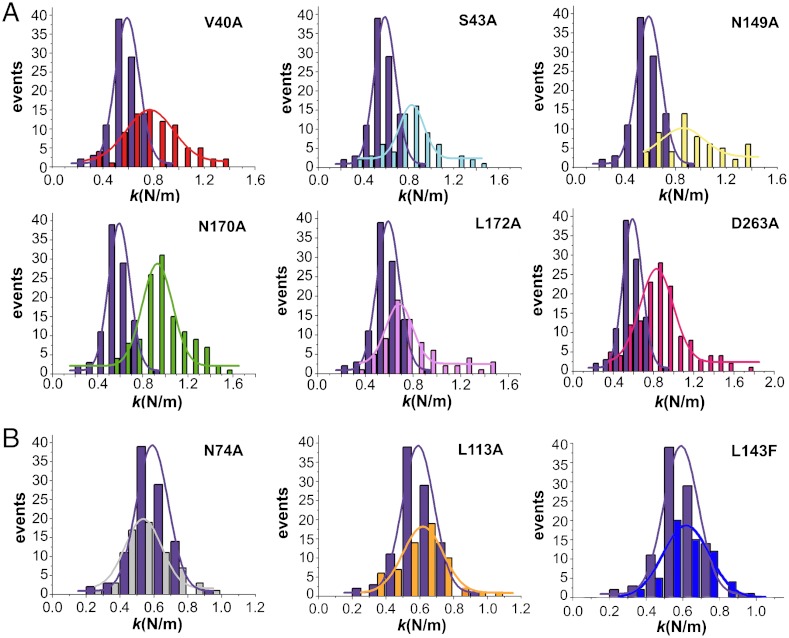
Five mutations (V40A, S43A, N149A, N170A, D263A) at the base of the S5 pores (Fig. 1) had been shown to impair the conformational transition detected by fluorescence and nearly abolish virus infectivity (36; Table 1). We have now constructed capsid-like particles carrying mutation L172A and have shown that it also impairs the pore-associated rearrangement (Fig. 2A and Table 1) as expected from its location at the base of the pores and its negative effect on infectivity (37, 38).
Table 1.
Effect of mutations on virus infectivity, pore-associated conformational rearrangement, and mechanical stiffness at S5 regions in the MVM capsid
| Mutant * | Titer *‡ | Tm (°C) †‡ | ks (N/m)§ | Fz ¶ | n ¶ | T ∥ |
| WT | 1 | 46.1 ± 0.8 | 0.59 ± 0.11 | 99 | 6 | - |
| V40A | 1 × 10-4 | No | 0.81 ± 0.23 | 80 | 7 | 6 × 10-12 |
| S43A | < 4 × 10-5 | No | 0.83 ± 0.23 | 66 | 6 | 1 × 10-11 |
| N149A | < 8 × 10-7 | No | 0.93 ± 0.24 | 58 | 6 | 2 × 10-15 |
| N170A | < 5 × 10-6 | No | 0.98 ± 0.21 | 123 | 5 | 3 × 10-42 |
| L172A | No | No | 0.79 ± 0.27 | 74 | 7 | 6 × 10-08 |
| D263A | < 8 × 10-6 | No | 0.87 ± 0.27 | 138 | 6 | 2 × 10-21 |
| D58A | 0.14 | 44.2 ± 0.2 | 0.57 ± 0.05 | 44 | 5 | 0.76 |
| N74A | 0.93 | 48.5 ± 1.0 | 0.57 ± 0.15 | 62 | 7 | 0.34 |
| L113A | 2.17 | 44.6 ± 0.3 | 0.60 ± 0.15 | 59 | 5 | 0.66 |
| L113F | 0.88 | 43.3 ± 0.5 | 0.57 ± 0.17 | 66 | 5 | 0.30 |
| L143F | 0.99 | 49.5 ± 0.9 | 0.62 ± 0.13 | 59 | 6 | 0.14 |
| I167A | 0.07 | 45.2 ± 1.0 | 0.53 ± 0.15 | 49 | 7 | 0.01 |
| N183A | 0.07 | 45.1 ± 0.2 | 0.57 ± 0.07 | 44 | 5 | 0.95 |
| Y188F | 1.07 | 48.2 ± 0.5 | 0.63 ± 0.10 | 72 | 5 | 0.04 |
| Y188W | 0.56 | 48.8 ± 2.2 | 0.60 ± 0.18 | 49 | 6 | 0.64 |
| M497L | 1.05 | 48.9 ± 1.2 | 0.59 ± 0.11 | 61 | 5 | 0.78 |
*Mutants are grouped in two series. The first (V40A to D263A) includes those whose virions had drastically reduced infectivity; the second (D58A to M497L), those whose virions had an infectivity comparable to WT. Mutants are listed in numerical order in each series. Titer indicates normalized virus infection titer at 37 °C, relative to WT. Mutation L172A was lethal (37).
†Transition temperature (average ± standard deviation) for the fluorescence-detected conformational transition of the capsid. No: no transition occurred.
‡Titers (except for L113A, L113F, L143F, Y188F, Y188W, M497L) and Tm (except for D58A, N183A) had been previously obtained (36, 45) and are shown to facilitate comparison with the results of mechanical analysis.
§Spring constant ks (average ± standard deviation).
¶Number of events (Fz) and individual viral particles (n) analyzed.
∥Value of T in a Student t-test (see text).
Fig. 2.
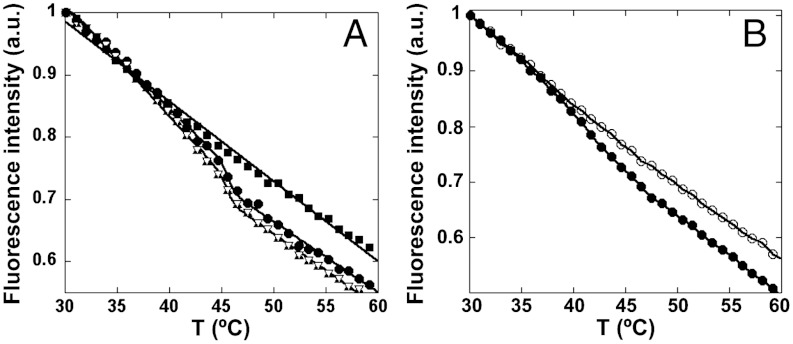
Heat-induced conformational transition in some variant MVM capsid-like particles followed by the variation in tryptophan fluorescence as a function of temperature. (A) Closed circles, WT capsid; squares, mutant L172A; triangles, D58A; inverted triangles, N183A. In the virion, mutation L172A led to a drastically reduced infectivity; mutations D58A and N183A had no severe effect on the infectivity. (B) Closed circles, WT capsid; open circles, cross-linked WT capsid.
According to the first prediction derived from the challenged hypothesis, the impairment of the pore-associated transition caused by any of those six mutations should be paralleled by an increase in the mechanical stiffness of the S5 regions in the capsid. AFM analysis corroborated this prediction for each and every one of those mutants (Fig. 3A and Table 1). The increase in ks ranged from 34% to 66% relative to the WT capsid, and a Student t-test confirmed that these substantial increases in ks are statistically highly significant.
Fig. 3.
Comparison of the mechanical stiffness at the S5 regions between the WT and mutant MVM capsids. Each histogram depicts the number of determinations relative to the ks obtained for a capsid variant. In each graph, the histogram in purple corresponds to the WT capsid, and the one in a different color to a mutant capsid (labeled). (A) Capsids that carry a mutation that impairs both S5 dynamics and virus infectivity relative to the WT (Table 1). (B) Representative examples of capsids that carry a mutation that neither impair S5 dynamics nor virus infectivity relative to the WT. For statistical analysis see Table 1.
Mutations in the MVM Capsid That Have No Effect on the Pore-Associated Conformational Dynamics Preserve Mechanical Elasticity Around the Pores.
It could be argued that the observed increase in S5 stiffness could be not specific of mutations that impair S5 dynamics. Thus, we used eight additional mutants to test the second hypothesis-derived prediction: Any mutation that does not impair the pore-associated dynamics should preserve the elasticity of the S5 regions.
Two mutations (N74A, I167A; Fig. 1) were chosen because they are located in S5, close to the pore wall, but still preserve the pore-associated conformational transition and virus infectivity (36) (Table 1). Six other mutations were chosen because they also preserve the pore-associated transition (45) (Table 1); we have now showed that they have no substantial effect on infectivity either (Table 1). These mutations were designed to increase the size of the cavities in the capsid by reducing the size of the side chain (L113A, Y188F) or reduce the size of the cavity by increasing the size of the side chain (L113F, L143F, Y188W) or modify the shape of those cavities (M497L) (45) (Fig. 1). By analogy with a sponge, we reasoned that these mutations could be prone to modify capsid stiffness. Remarkably, AFM analysis showed that none of those eight mutations had any statistically significant effect on the ks of the S5 regions (Fig. 3B and Table 1), thus corroborating the second prediction.
Two other mutant capsids (D58A, N183A; Fig. 1) provided an opportunity to test this prediction in reverse. These mutations had been previously shown to have no effect on the mechanical elasticity of the S5 regions (15) (Table 1). We have now obtained capsid-like particles containing D58A or N183A and proved by fluorimetry that they do not impair the pore-associated transition either (Fig. 2A and Table 1).
The Correlation Between Dynamics at the Pore Regions and Capsid Elasticity Are not Observed When the Mechanics of Other Capsid Regions Is Probed.
A third prediction of the tested hypothesis was that no correlation should be observed between S5 dynamics and mechanical elasticity of capsid regions other than S5. To test this prediction, we determined the effect of all of the above mutations on the elasticity of the regions around twofold (S2) and threefold (S3) symmetry axes. Many of those mutations led to substantial, statistically validated increases in ks at S2 and/or S3, that ranged from 22% to 139% (Figs. S2 and S3 and Table S1). However, many of these mutations still preserved the pore-associated rearrangement and virus infectivity, thus corroborating the third prediction.
Increasing Mechanical Stiffness at the Pore Region by Chemical Means Impairs the Pore-Associated Conformational Dynamics.
It could still be argued that the increase in S5 stiffness by mutations around the base of the pores could be a coincidental consequence of altering the local structure without being necessarily connected to the impairment of S5 dynamics by the same mutations. Thus, we looked for an alternative method to increase the stiffness of the S5 regions. The aim was to test whether the correlation between S5 stiffness and dynamics occurred irrespective of the type and location of the underlying structural modification.
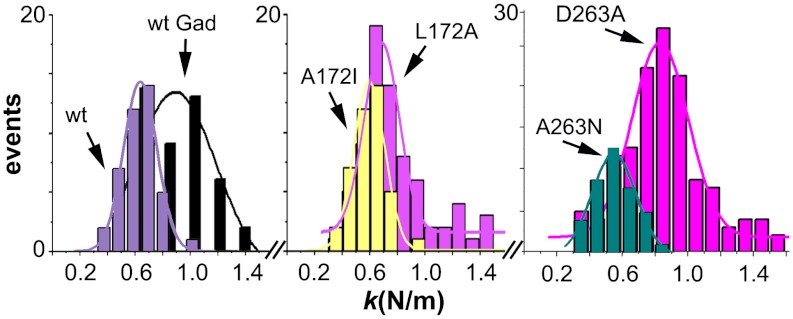
We chose to modify the WT capsid by cross-linking lysine residues with glutaradehyde. Inspection of the capsid structure indicated that no cross-linkable lysines are found at the base of the pores. AFM analysis showed that the ks of S5 regions in cross-linked capsid-like particles was, as intended, significantly higher than in intact capsid-like particles (Fig. 4 and Table 2). Then, the pore-associated conformational transition was probed by fluorimetry of the same particles. As predicted, the increased S5 stiffness obtained by cross-linking was paralleled by the disappearance of the transition (Fig. 2B and Table 2).
Fig. 4.
Comparison of the mechanical stiffness at S5 regions between the WT and variant MVM capsids. Each histogram depicts the number of determinations relative to the ks obtained for a capsid variant. Left: Comparison between unmodified capsid (WT) (that undergoes the conformational transition) and the cross-linked WT capsid (that lost the conformational transition). Center: comparison between mutant capsids L172A (that lost the conformational transition) and A172I pseudo-revertant (that recovered the conformational transition). Right: Comparison between D263A (that lost the conformational transition) and A263N pseudo-revertant (that recovered the conformational transition). For statistical analysis see Table 2.
Table 2.
Effect of cross-linking (wtGad) and pseudo-reversions A172I and A263N on pore-associated rearrangement and mechanical stiffness at S5 regions in the MVM capsid
| Capsid | Tm (°C) * | ks (N/m) † | Fz ‡ | n ‡ | T§ |
| WT | 46.1 ± 0.8 | 0.60 ± 0.12 | 41 | 8 | — |
| wtGad | No | 0.87 ± 0.24 | 46 | 4 | 3 × 10-9 |
| A172I | 45.2 ± 3.4 | 0.63 ± 0.10 | 56 | 5 | 0.02 |
| A263N | 44.9 ± 1.6 | 0.56 ± 0.12 | 40 | 6 | 0.18 |
*Tm, transition temperature (average ± standard deviation) for the fluorescence-detected conformational transition. No: no transition occurred.
†Spring constant ks (average ± standard deviation).
‡Number of events (Fz) and individual viral particles (n) analyzed.
§Value of T in a Student t-test (see text).
The Impaired Pore-Associated Conformational Dynamics and Increased Mechanical Stiffness of S5 Regions Can Be Rescued Together by Pseudo-Reversion.
Finally, we designed a particularly stringent test. Mutations at the base of the pores (including S43A, N170A, L172A, D263A) reduced both conformational dynamics and mechanical elasticity of S5 regions. We aimed now at testing whether the local conformational dynamics and mechanical elasticity of the WT capsid could be restored together by pseudo-reversion at some of those positions. We chose pseudo-reversions A43T, A170L, A172I, and A263N for testing.
Two of those four pseudo-reversions (A172I, A263N) restored as intended the pore-associated conformational transition (Table 2). According to the challenged hypothesis, restoration of S5 dynamics should be paralleled by restoration of S5 elasticity. AFM analysis showed, as predicted, that the ks of the S5 region in A172I and A263N was lower than in L172A and D263A and indistinguishable from that of the WT capsid, and these results were statistically validated (Fig. 4 and Table 2).
Discussion
Some regions in a virus particle may require less energy than others to change the spatial position of the atomic groups in them away from the minimum free energy conformation. The energy may be provided in bulk biochemical experiments by heat or other physicochemical means, and such regions could be described in atomic terms as conformationally more dynamic. The same energy may be provided in single-molecule AFM experiments by the application of mechanical force, and such regions could be described in terms of fundamental mechanics as more elastic. This hypothesis of a phenomenological equivalence between conformational dynamism and mechanical elasticity should not, however, be regarded as obvious. For example, in indentation experiments energy is directionally applied, and mechanical deformation is also directional.
We tested the above hypothesis by comparing predicted and observed effects of many different modifications in the MVM capsid on the conformational dynamics and mechanical elasticity of the S5 (pore) regions. The results corroborated every hypothesis-derived-prediction: (i) Every one of six alanine mutations at the base of each pore and chemical cross-linking of lysines located elsewhere both prevented the conformational rearrangement and increased the mechanical stiffness of the S5 regions; (ii) every one of 10 mutations at other positions in the capsid, including some at S5, close to the pores, preserved the conformational rearrangement and the mechanical elasticity of the S5 regions; (iii) the pseudo-reversion of two mutations at the base of the pores restored both the conformational rearrangement and mechanical elasticity of the S5 region in the WT capsid but in a different genotypic context (in other words, in each of these cases, one of two different mutations of the same residue reduced both S5 dynamics and elasticity, while the other mutation preserved both of them); and (iv) the inextricable correlation between the pore-associated dynamics and mechanical elasticity held only for the S5 (pore) regions and not for other capsid regions.
It should be emphasized at this point that the biunivocal correspondence between local dynamics and elasticity in this model capsid was not the product of finding a posteriori an unforeseen correlation between sets of previously obtained results. Instead, the perfect correlation found had been actually predicted by a hypothesis that we repeatedly subjected to experimental corroboration or disproval. Uncoupling of conformational dynamics and mechanical elasticity for just one single capsid variant out of the 19 tested could have falsified the hypothesis. In fact, the hypothesis was corroborated each time it was challenged, using different approaches. Other explanations to the results obtained could be constructed, but they would require a number of assumptions—e.g., by invoking several unlikely, unwarranted coincidences. The proposed phenomenological identity between conformational dynamics and mechanical elasticity provides the simplest explanation we could find that is consistent with all of the available observations and was corroborated by every one of the experimental results obtained.
The lateral resolution of the indentations at S5 is not enough to determine the mechanical elasticity of the pore walls. What is being measured is the overall elasticity of a wider region around the pore, but this region is involved in the pore-associated dynamics (35, 36). It has been suggested that mutations at the base of the pores may alter local dynamics by keeping the pores effectively frozen in an open configuration; this could facilitate the observed untimely egress of VP1 Nt and release of DNA in virions carrying these mutations (43). The observed S5 stiffening by such mutations (this study) and the fact that they still allow VP2 Nt externalization from capsid-like particles but in a less cooperative process (36) are consistent with that possibility. However, in the absence of crystal structures for the mutant capsids, the structural bases of impaired S5 dynamics and stiffening effects may be only guessed. Small-to-large substitutions that could fill capsid cavities all led to increased stiffness at S2 and S3 regions nearby, as expected. Remarkably, some (not all) large-to-small substitutions surrounding cavities and alanine mutations at the base of the pores also increased local or global capsid stifness. Virtual mutagenesis suggests that removal of a hydroxyl (S43A), an amide (N149A, N170A) or a carboxylate (D263A) at the base of the pores could remove some local interactions and/or steric effects, and the exposed methyl group could minimize its exposure to solvent by local repacking. V40 and L172 interact with each other, and truncation of either apolar side chain may reorganize the pore wall. In each case, the local repacking could lead to increased resistance to mechanical deformation, as observed.
The next step may be to test whether the dynamics-elasticity relationship may be generalized to other capsid regions, viral particles, and nonviral assemblies. Indeed, some preliminary evidence suggests that such an intrinsic relationship can be tentatively invoked to explain the observed effects of the capsid-bound DNA segments in MVM (15, 30). The reduced resistance of the virion to thermal inactivation by mutations that disrupt capsid–DNA interactions appears to be due to the facilitation of a heat-induced nonproductive conformational rearrangement of the particle (30). The mechanical effects of two of those mutations were tested, and both were found to increase the elasticity of S2 and S3 regions in the virion (15). Although further studies are needed in this case, it is tempting to suggest that mechanical elasticity of the S2-S3 regions may be correlated with the virus-inactivating dynamics based on those regions.
From a biological point of view, this study provides strong experimental support for the suggested (15) adaptive role of the DNA-mediated, anisotropic distribution of mechanical elasticity in the MVM virion. MVM may have evolved DNA-binding sites in its capsid because the DNA-mediated structural rigidification of most capsid regions (seen by AFM as an increase in ks at S2 and S3) increases virion thermostability (by impairing a virion-inactivating conformational rearrangement). In contrast, the regions around the capsid pores may have been kept free of bound DNA by negative selection, because a rigidification of these regions (seen by AFM as an increase in ks at S5) impairs the S5 dynamics required for infectivity. To sum up, the anisotropic mechanical stiffness determined by AFM reflects the unequal distribution of structural elasticity/conformational dynamism in the MVM particle and may have arisen as a biological adaptation to prevent virion inactivation without impairing infection.
From a methodological point of view, this study proposes that determination by AFM of the ks of different regions in a viral particle or biomolecular assembly may be used to identify conformationally dynamic regions. Those regions with relatively low ks could be inferred to have a higher conformational dynamism and propensity for structural rearrangements. Mutations could be introduced at these regions, and variations in stiffness determined by AFM (indicating variations in dynamics) could be associated to changes introduced by those same mutations in different properties and functions of the viral particles and their roles in biological function.
From a nanotechnological point of view, this study shows that viral capsids can be greatly stiffened by protein engineering, leading to the mechanically strongest (more rigid) capsids known to date; their calculated Young’s moduli (up to 2.8 Gpa) (10) are about twofold higher than those of the stiffer virus capsids analyzed yet (25). Stiff viral nanoparticles may be useful for applications where structural rearrangements in a virus-based nanomaterial or nanodevice may be undesirable (25).
Materials and Methods
Recombinant Plasmids and Mutagenesis.
Plasmid pSVtk-VP1/2 provided by J. M. Almendral from our institution (46) or bacmid BM-VP2 (35) were, respectively, used for the production of capsids made of VP1 and VP2 or capsid-like particles made of VP2 only. Plasmid pTRp, originally provided by P. Tattersall, Yale University (47), and modified (46), was used for production of virions. Site-directed mutagenesis was carried out using the QuikChange kit (Stratagene). To obtain variant VP1/VP2 capsids or VP2-only capsids, mutations were, respectively, introduced in pSVtk-VP1/2 or in pFB1-VP2 and transferred to BM-VP2. The presence of the mutations was confirmed by sequencing.
Expression and Purification of Empty Capsids and Virions of MVM.
For production of VP2-only capsids, H-5 cells were transfected with BM-VP2, and the capsids were purified (32). For production of VP1/VP2 capsids or virions, NB324K cells were, respectively, transfected with pSVtk-VP1/2 or pTRp, and the capsids were purified (10, 33). Preparations were dialyzed against PBS. Purity and integrity were assessed by electrophoresis, electron microscopy, and/or AFM. For cross-linking of capsid-like particles, these were incubated in PBS with 0.25% glutaraldehyde during 30 min at room temperature and dialyzed.
Thermodynamic Analysis of Conformational Changes of Viral Particles by Spectrofluorimetry.
Changes in conformation were analyzed by following the variation in intrinsic tryptophan fluorescence at moderate temperatures when subjected to thermal gradients, as described (35). The capsids dissociated only at very high temperatures (75 °C).
Virus Infectivity Assays.
Virions were titrated in duplicate in plaque-formation assays that were reproduced and averaged.
Atomic Force Microscopy of Empty Capsids.
AFM experiments were carried out essentially as described (10, 15). The AFM (Nanotec Electrónica) was operated in jumping mode (48, 49) in liquid. RC800PSA cantilevers (Olympus) with a nominal kc of 0.1 N/m were used. The actual kc of each cantilever used was determined (50). For MVM imaging and mechanical analysis, purified empty capsids adsorbed on glass and kept in PBS were used. The maximum normal force during imaging was about 100 pN. AFM images were processed by using WSxM software (51). Capsid stiffness was determined as described (10). To keep within the range of a linear elastic response and avoid particle damage, only force-vs.-distance (Fz) measurements that involved indentations between 0.5 nm and 2.0 nm were considered. The local stiffness (ks) of the particle was obtained by assuming that the particle and cantilever behave as two ideal springs in series (9–11, 14, 15, 21, 22).
Statistical Analysis.
The ks values followed a normal distribution described by a Gaussian fitting, as corroborated using normality tests, and as observed for other virus particles. The statistical significance of ks differences was evaluated using OriginPro8 (OriginLab). To statistically validate that two ks distributions are different, the two population two-tailed Student t-test was used, with an alpha level = 0.05. Equal variance was not assumed, providing a more stringent test.
Molecular Graphics and Structural Analyses.
The refined Protein Data Bank coordinates of the MVMi virion (1Z1C) and MVMp empty capsid (1Z14) (29) were used.
Supplementary Material
ACKNOWLEDGMENTS.
We thank E. Domingo for critical reading of the original manuscript; P. Tattersall, J. M. Almendral and J. Reguera for providing plasmids; and A. Rodríguez-Huete, M. A. Fuertes, and S. Márquez for technical assistance. M.C. was a fellow from Comunidad de Madrid (CM); R.P. was a recipient of a Juan de la Cierva contract from Ministerio de Ciencia e Innovación (MICINN). This work was supported by Grants BIO2009-10092 from MICINN to M.G.M., S-2009/MAT/1467 from CM to M.G.M. and J.G.-H., MAT2008-02533 and PIB2010US-00233 to P.J.P., Consolider CSD2010-00024 (MICINN) to J.G.-H., and an institutional grant from Fundación Ramón Areces. M.G.M. is an associate member of the Centre for Biocomputation and Physics of the Complex Systems, Zaragoza, Spain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207437109/-/DCSupplemental.
References
- 1.Chiu W, Burnett RM, Garcea RL, editors. Structural Biology of Viruses. New York: Oxford Univ Press; 1997. [Google Scholar]
- 2.Agbandje-McKenna M, McKenna R, editors. Structural Virology. Cambridge, UK: RSC Publishing; 2011. [Google Scholar]
- 3.Johnson JE. Virus particle dynamics. Adv Protein Chem. 2003;64:197–218. doi: 10.1016/s0065-3233(03)01005-2. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JE. Virus particle maturation: Insights into elegantly programmed nanomachines. Curr Opin Struct Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bothner B, et al. Crystallographically identical virus capsids display different properties in solution. Nat Struct Biol. 1999;6:114–116. doi: 10.1038/5799. [DOI] [PubMed] [Google Scholar]
- 6.Bothner B, Hilmer JK. In: Structural Biology of Viruses. Agbandje-McKenna M, McKenna R, editors. New York: Oxford Univ Press; 2011. pp. 41–61. [Google Scholar]
- 7.Hörber JKH, Miles MJ. Scanning probe evolution in biology. Science. 2003;302:1002–1005. doi: 10.1126/science.1067410. [DOI] [PubMed] [Google Scholar]
- 8.Neuman KC, Nagy A. Single molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanovska IL, et al. Bacteriophage capsids: Tough nanoshells with complex elastic properties. Proc Natl Acad Sci USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco C, et al. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci USA. 2006;103:13706–13711. doi: 10.1073/pnas.0601881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michel JP, et al. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kol N, et al. Mechanical properties of murine leukemia virus particles: Effect of maturation. Biophys J. 2006;91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kol N, et al. A stiffness switch in human immunodeficiency virus. Biophys J. 2007;92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanovska I, Wuite G, Jönsson B, Evilevitch A. Internal DNA pressure modifies stability of WT phage. Proc Natl Acad Sci USA. 2007;104:9603–9608. doi: 10.1073/pnas.0703166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrasco C, Castellanos M, de Pablo PJ, Mateu MG. Manipulation of the mechanical properties of a virus by protein engineering. Proc Natl Acad Sci USA. 2008;105:4150–4155. doi: 10.1073/pnas.0708017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arkhipov A, Roos WH, Wuite GJL, Schulten K. Elucidating the mechanism behind irreversible deformation of viral capsids. Biophys J. 2009;97:2061–2069. doi: 10.1016/j.bpj.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roos WH, et al. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc Natl Acad Sci USA. 2009;106:9673–9678. doi: 10.1073/pnas.0901514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roos WH, Wuite GJL. Nanoindentation studies reveal material properties of viruses. Adv Mater. 2009;21:1187–1192. [Google Scholar]
- 19.Cuellar JL, Meinhoevel F, Hoehne M, Donath E. Size and mechanical stability of norovirus capsids depend on pH: A nanoindentation study. J Gen Virol. 2010;91:2449–2456. doi: 10.1099/vir.0.021212-0. [DOI] [PubMed] [Google Scholar]
- 20.Roos WH, Bruinsma R, Wuite GJL. Physical virology. Nat Phys. 2010;6:733–743. [Google Scholar]
- 21.Evilevitch A, et al. Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J Mol Biol. 2011;405:18–23. doi: 10.1016/j.jmb.2010.10.039. [DOI] [PubMed] [Google Scholar]
- 22.Carrasco C, et al. Built-in mechanical stress in viral shells. Biophys J. 2011;100:1100–1108. doi: 10.1016/j.bpj.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanovska IL, Miranda R, Carrascosa JL, Wuite GJL, Schmidt CF. Discrete fracture patterns of virus shells reveal mechanical building blocks. Proc Natl Acad Sci USA. 2011;108:12611–12616. doi: 10.1073/pnas.1105586108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roos WH, et al. Mechanics of bacteriophage maturation. Proc Natl Acad Sci USA. 2012;109:2342–2347. doi: 10.1073/pnas.1109590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateu MG. Mechanical properties of viruses analyzed by atomic force microscopy: A virological perspective. Virus Res. 2012 doi: 10.1016/j.virusres.2012.06.008. Available at http://dx.doi.org/10.1016/j.virusres.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Landau LD, Lifshitz EM. Theory of Elasticity, Course of Theoretical Physics. 3rd Ed. Vol. 7. Boston: Butterworth-Heinemann; 1986. [Google Scholar]
- 27.Schaap IA, Carrasco C, de Pablo PJ, MacKintosh FC, Schmidt CF. Elastic response, buckling, and instability of microtubules under radial indentation. Biophys J. 2006;91:1521–1531. doi: 10.1529/biophysj.105.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- 29.Kontou M, et al. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J Virol. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reguera J, et al. Functional relevance of amino acid residues involved in interactions with ordered nucleic acid in a spherical virus. J Biol Chem. 2005;280:17969–17977. doi: 10.1074/jbc.M500867200. [DOI] [PubMed] [Google Scholar]
- 31.Cotmore SF, D’Abramo AM, Ticknor CM, Tattersall P. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology. 1999;254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- 32.Hernando E, et al. Biochemical and physical characterization of parvovirus minute virus of mice virus-like particles. Virology. 2000;267:299–309. doi: 10.1006/viro.1999.0123. [DOI] [PubMed] [Google Scholar]
- 33.Lombardo E, Ramírez JC, García J, Almendral JM. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J Virol. 2002;76:7049–7059. doi: 10.1128/JVI.76.14.7049-7059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maroto B, Valle N, Saffrich R, Almendral JM. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J Virol. 2004;78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carreira A, Menéndez M, Reguera J, Almendral JM, Mateu MG. In vitro disassembly of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J Biol Chem. 2004;279:6517–6525. doi: 10.1074/jbc.M307662200. [DOI] [PubMed] [Google Scholar]
- 36.Reguera J, Carreira A, Riolobos L, Almendral JM, Mateu MG. Role of interfacial amino acid residues in assembly, stability and conformation of a spherical virus capsid. Proc Natl Acad Sci USA. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farr GA, Tattersall P. A conserved leucine that constricts the pore through the capsid fivefold cylinder plays a central role in parvoviral infection. Virology. 2004;323:243–256. doi: 10.1016/j.virol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Farr GA, Cotmore SF, Tattersall P. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N-terminus and the genome of minute virus of mice. J Virol. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valle N, Riolobos L, Almendral JM. Synthesis, post-translational modification and trafficking of the parvovirus structural polypeptides. In: Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. London: Edward Arnold; 2006. pp. 291–304. [Google Scholar]
- 40.Cotmore S, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv Virus Res. 2007;70:183–232. doi: 10.1016/S0065-3527(07)70005-2. [DOI] [PubMed] [Google Scholar]
- 41.Cotmore SF, Hafenstein S, Tattersall P. Depletion of virion-associated divalent cations induces parvovirus minute virus of mice to eject its genome in a 3′-to-5′direction from an otherwise intact particle. J Virol. 2010;84:1945–1956. doi: 10.1128/JVI.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plevka P, et al. Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a fivefold axis. J Virol. 2011;85:4822–4827. doi: 10.1128/JVI.02598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotmore S, Tattersall P. Mutations at the base of the icosahedral five-fold cylinders of minute virus of mice induce 3′-to-5′ genome uncoating and critically impair entry functions. J Virol. 2012;86:69–80. doi: 10.1128/JVI.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleker S, Sonntag F, Kleinshmidt JA. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol. 2005;79:2528–2540. doi: 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carreira A, Mateu MG. Structural tolerance versus functional intolerance to mutation of hydrophobic core residues surrounding cavities in a parvovirus capsid. J Mol Biol. 2006;360:1081–1093. doi: 10.1016/j.jmb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 46.Ramírez JC, Santarén JF, Almendral JM. Transcriptional inhibition of the parvovirus minute virus of mice by constitutive expression of an antisense RNA targeted against the NS-1 transactivator protein. Virology. 1995;206:57–68. doi: 10.1016/s0042-6822(95)80019-0. [DOI] [PubMed] [Google Scholar]
- 47.Gardiner EM, Tattersall P. Mapping of the fibrotropic and lymphotropic host range determinants of the parvovirus minute virus of mice. J Virol. 1988;62:2605–2613. doi: 10.1128/jvi.62.8.2605-2613.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreno-Herrero F, et al. Scanning force microscopy jumping and tapping modes in liquids. Appl Phys Lett. 2002;81:2620–2622. [Google Scholar]
- 49.Ortega-Esteban A, et al. Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid. Ultramicroscopy. 2012;114:56–61. doi: 10.1016/j.ultramic.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 50.Sader JE, Chon JWM, Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev Sci Instrum. 1999;70:3967–3969. [Google Scholar]
- 51.Horcas I, et al. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum. 2007;78:013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.